Abstract
In the decade since their discovery, the two major breast cancer susceptibility genes BRCA1 and BRCA2, have been shown conclusively to be involved in a significant fraction of families segregating breast and ovarian cancer. However, it has become equally clear that a large proportion of families segregating breast cancer alone are not caused by mutations in BRCA1 or BRCA2. Unfortunately, despite intensive effort, the identification of additional breast cancer predisposition genes has so far been unsuccessful, presumably because of genetic heterogeneity, low penetrance, or recessive/polygenic mechanisms. These non-BRCA1/2 breast cancer families (termed BRCAx families) comprise a histopathologically heterogeneous group, further supporting their origin from multiple genetic events. Accordingly, the identification of a method to successfully subdivide BRCAx families into recognizable groups could be of considerable value to further genetic analysis. We have previously shown that global gene expression analysis can identify unique and distinct expression profiles in breast tumors from BRCA1 and BRCA2 mutation carriers. Here we show that gene expression profiling can discover novel classes among BRCAx tumors, and differentiate them from BRCA1 and BRCA2 tumors. Moreover, microarray-based comparative genomic hybridization (CGH) to cDNA arrays revealed specific somatic genetic alterations within the BRCAx subgroups. These findings illustrate that, when gene expression-based classifications are used, BRCAx families can be grouped into homogeneous subsets, thereby potentially increasing the power of conventional genetic analysis.
Although germ-line mutations in BRCA1 and BRCA2 account for most familial breast-ovarian cancer cases, these mutations can only explain a small proportion of familial site-specific breast cancer susceptibility (1). Although a small proportion of non-BRCA1/2 hereditary breast cancers evolve in individuals with rare multicancer syndromes, very little is known about the genetic basis of non-BRCA1/BRCA2 breast cancer (BRCAx) families. Genetic linkage analysis of BRCAx families has been performed and identified several chromosomal regions potentially harboring a breast cancer susceptibility gene, including 8p12-p22 (2), 13q21 (3), and 2q31-q33 (P. Huusko, personal communication). However, these loci have either subsequently been excluded as major predisposing loci on a global perspective (4, 5), or remain to be confirmed, emphasizing genetic heterogeneity and population-specific effects within BRCAx kindreds (6). Other difficulties of linkage detection are a high rate of sporadic cases, alternate modes of inheritance, and influence of low penetrance or modifying factors. We have previously shown that the genotypes of hereditary breast cancers are reflected in different gene expression profiles (7), and this has also been illustrated in the study by van't Veer et al. (8). These findings indicate that the separation of the heterogeneous group of BRCAx breast cancers into subgroups based on gene expression profiling may be possible, and may facilitate the future search for breast cancer predisposing genes in more homogeneous groups of families. Histopathological studies have revealed that BRCAx tumors are cytologically heterogeneous, but are generally of lower grade, with less nuclear pleomorphism and lower mitotic activity as compared with BRCA1 and BRCA2 tumors and unselected controls (9). Moreover, unlike the preponderance of negative estrogen (ER) and progesterone (PgR) receptors within BRCA1 tumors (10), and positive hormone receptors in BRCA2 tumors, BRCAx tumors display variable levels of these receptors (11). Thus, it is likely that the different tumor phenotypes reflect the multiple genetic origins of BRCAx cancers.
Materials and Methods
Breast Cancer Specimens.
Tumors were obtained from pathology departments within the southern Sweden health care region. The grossly dissected tumors were snap frozen in liquid nitrogen within 30–60 min of surgical excision and stored at −80°C until further processed. Touch imprints were made to confirm the presence of neoplastic cells in each tumor specimen. Patients diagnosed with breast cancer and with a family disease history compatible with a dominant mode of inheritance were referred to the Oncogenetic Clinic at the Department of Oncology, Lund University Hospital (Lund, Sweden) for genetic counseling or visited a research clinic for familial breast cancer. The patients provided blood samples, and mutation analysis for BRCA1 and BRCA2 was performed as described (12). Individuals with no mutations identified in these genes (non-BRCA1/2) are referred to as BRCAx. Eight families were included in the present study, and two or three tumors from each family were analyzed, with the exception of Lund 5, from which only one tumor was available for analysis. Tumors were from different individuals, except families Lund 111 (bilateral breast cancer, of which only one was included in the expression analysis) and Lund 502 (primary breast cancer and metastasis). The pathological review (Table 1) was performed by a single pathologist (G.C.). These studies were approved by the Research Ethical Committee of the Medical Faculty of Lund University (Lund, Sweden) and the Institutional Review Board of the National Human Genome Research Institute of the National Institutes of Health.
Table 1.
Characteristics of BRCAx breast cancer samples
| Group* | Family | Sample | Age† | Histological type | Histological grade (score)‡ | Ploidy§ | SPF¶ | ER‖ | PgR‖ |
|---|---|---|---|---|---|---|---|---|---|
| A | L5** | 9275‡‡ | 57 | Ductal | 2 (6) | D | 4.6 | +++ | +++ |
| L16 | 7676‡‡ | 60 | Ductal | 1 (5) | NA | NA | +++ | +++ | |
| 9981 | 79 | Metaplastic | 3 (8) | ND | 4.1 | − | − | ||
| 10785 | 75 | Ductal | 3 (9) | ND | 10 | − | − | ||
| L99 | 6874‡‡ | 54 | DCIS | x | D | 5.5 | + | ++ | |
| 7650 | 45 | Ductal | 1 (5) | NA | NA | − | + | ||
| 12096 | 57 | Lobular | 2 (7) | ND | 3.4 | ++ | +++ | ||
| B | L101 | 10463 | 50 | Ductal | 3 (8) | ND | 20 | − | − |
| 12237 | 78 | Medullary | 3 (9) | ND | 23 | − | − | ||
| L111 | 12143‡‡†† | 43 | Ductal | 2 (7) | ND | 11 | ++ | ++ | |
| 15564‡‡ | 47 | Ductal | 2 (7) | ND | 5.1 | ++ | ++ | ||
| L414 | 15401 | 57 | Mucinous, ductal | 3 (8) | ND | 4.2 | ++ | − | |
| 15478‡‡ | 58 | DCIS, ductal | 2 (7) | D | 3.8 | +++ | ++ | ||
| L502 | 8984 | 36 | Ductal | 3 (8) | D | 4.9 | ++ | − | |
| 11954‡‡ | 39 | Metastasis | x | D | 5.1 | − | − | ||
| L505 | 12314 | 36 | Ductal | 2 (7) | D | 7 | ++ | +++ | |
| 14316‡‡ | 62 | Ductal | 3 (9) | ND | 22 | ++ | ++ |
Group as found by our gene expression-based class discovery analysis.
Age at surgery of analyzed tumor sample.
Histological grade was based on the aggregate score for three variables (mitotic counts, nuclear pleomorphism, and tubular differentiation) and was as follows: grade 1 indicated a well-differentiated tumor (3–5 points); grade 2 indicated a moderately differentiated tumor (6 or 7 points); and grade 3 indicated a poorly differentiated tumor (8 or 9 points).
D, diploid; ND, nondiploid.
Percent of cells in S-phase. NA, not available.
ER and PgR status, respectively: −, negative; +, weakly positive; ++, positive; +++, strongly positive.
This family was previously found to be linked to 13q21 (3).
Included in CGH analysis.
Not included in the expression analysis.
Gene Expression Analysis and Class Discovery.
cDNA microarrays were constructed as described (13), and contained ≈6,500 sequence verified cDNA clones obtained under a CRADA with ResGen (Huntsville, AL). Gene names are listed according to UniGene (available at www.ncbi.nlm.nih.gov/UniGene). The 6,500 clones represent ≈4,700 unique known genes and 1,700 ESTs.
RNA was extracted from the frozen tumors after homogenization in TRIzol Reagent (Invitrogen) followed by RNeasy Maxi (Qiagen, Valencia, CA) according to the manufacturers' recommendations. The breast cancer cell line BT-474 (American Type Culture Collection, Manassas, VA) was harvested at subconfluency and used as a reference. Microarrays were hybridized and scanned, and image analysis was performed as described (13, 14). Hierarchical clustering and multidimensional scaling analysis was performed as described (7, 15, 16).
We filtered clones by requiring that a clone should have an average spot quality (17) >0.5 as well as tumor and reference intensities >20 across all experiments. The number of clones that passed this filter was 4,795. Each slide was then normalized, such that the log intensity ratios were mean-centered. The expression data for these 4,795 well measured clones are available in Table 3, which is published as supporting information on the PNAS web site, www.pnas.org. A common approach to finding a difference in gene expression patterns between known classes of samples is to show that there is an overabundance of genes that separate the classes, as compared with what would be expected by chance (7, 16, 18, 19). Subgroupings in classes of gene expression can thus be discovered by seeking partitions of samples with an overabundance of differentially expressed genes (20). We used the method by Ben-Dor et al. (20) modified as follows. For a given partition of samples into two classes (with nA and nB samples), a discriminative weight was calculated for each gene by using the signal-to-noise statistic (18). Genes were ranked according to this weight. To test the statistical significance of the weights, sample labels were randomly permuted and the weight for each gene was computed again. This random permutation of sample labels was performed 5,000 times to generate a weight distribution that could be expected for two classes with nA and nB samples under the null assumption of random gene expression. The weight values for the two actual classes were then assigned Ps based on the weight distribution from the random permutations. Candidate partitions of the data were scored with the number of statistically significant (P < 0.001) weights, i.e., the number of genes significantly different in expression between samples in the two classes. We used a simulated annealing (21) scheme, in which each step consists of changing the class of a randomly selected sample, to find the highest scoring partition of samples into two classes. This approach can easily be extended to accommodate more than two classes.
Copy Number Analysis Using cDNA Microarrays.
cDNA microarrays used for CGH analysis were constructed at Agilent Technologies (Palo Alto, CA), and contained 11,367 cDNA clones (representing ≈8,700 unique known genes and 2,000 ESTs) from the previously mentioned collection. DNA was extracted from breast tumors after the RNA extraction with TRIzol according to the manufacturer's recommendations. The DNA was further purified by several rounds of phenol-chloroform extractions before use. Random-prime labeling of DNA was performed according to the original protocol (22), using normal placental DNA as a reference. Scanning and image analysis was performed as for gene expression analysis.
We filtered clones by requiring that a clone should have an intensity larger than 100 fluorescence units in the reference channel across all experiments; 8,057 clones passed this filter. Each slide was then normalized, such that the log copy number ratios were mean-centered. By using the signal-to-noise statistic (18), genes were ranked based on differences in copy number between the BRCAx subgroups, and a weighted list of genes was generated. A permutation test was used to estimate the probability (α) that a gene got a larger weight for a random labeling of the samples (23), as compared with the BRCAx subclasses. The signal-to-noise statistic is designed to find genes consistently amplified within a group (as indicated by a small α). We evaluated whether there was a significant overabundance of highly ranked genes in a given cytoband by using a null hypergeometric model (24) (see Supporting Text, which is published as supporting information on the PNAS web site). This method has recently been applied to identify regulatory elements (25, 26).
Chromosomal Mapping.
To localize the chromosomal positions of the clones on our arrays, we aligned clone sequences with the draft human genome sequence (27), as assembled in the August 6, 2001, freeze of the Genome Browser at UCSC (ref. 28 and http://genome.ucsc.edu), using blat (ref. 29 and http://genome.ucsc.edu). We only aligned clones assigned to a Unigene cluster (build 142). If the cluster contained an mRNA sequence in the RefSeq dataset (ref. 30 and http://www.ncbi.nlm.nih.gov/LocusLink/refseq.html), that sequence was used. Requiring at least 94% sequence identity, we aligned 7,216 of the 8,057 genes to the genome sequence.
Results and Discussion
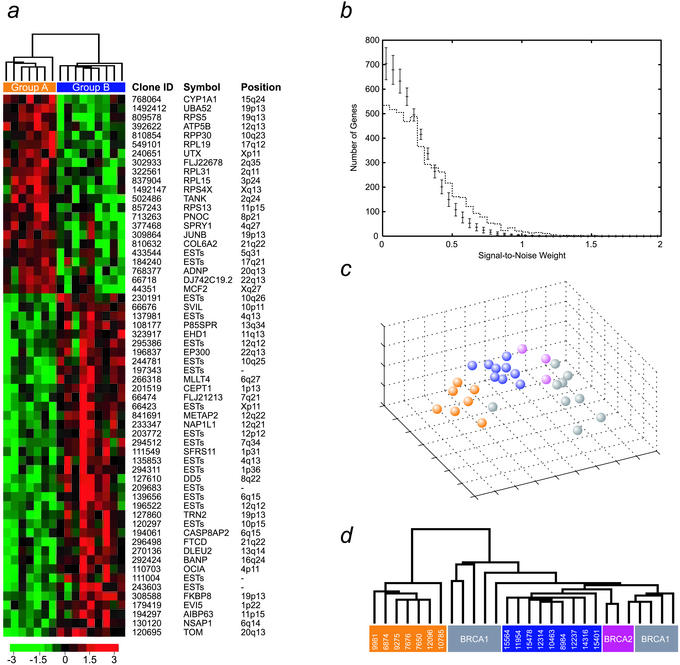
It has been anticipated for some time that BRCAx breast cancers constitute a heterogeneous group, most likely reflecting multiple subclasses. Kainu et al. (3) approached this heterogeneity by using chromosomal CGH and linkage analysis. In this study, we used the complementary strategy of global gene expression profiling followed by various mathematical methods to subclassify BRCAx families into genetically more homogeneous subgroups. Sixteen tumors from eight BRCAx families were analyzed by using cDNA microarrays (Table 1). Based on the gene expression profiles, our class discovery method (20) identified two distinct groups of tumors, comprising seven and nine samples, respectively. This classification was supported by 60 statistically significant (P < 0.001) genes (Fig. 1a), whereas only five genes were expected by chance (see Materials and Methods and Fig. 1b). Furthermore, all of the families in which multiple family members were examined remained intact. Given this data set, the probability that the families would remain intact in a random partitioning into two groups is only 1/256. Further studies are needed to pinpoint whether the partitioning reflects on a hereditary predisposition or is a manifestation of similarities in tumor progression within each group. Nevertheless, the family grouping supports an underlying hereditary cause for the partitioning. It is reasonable to expect the non-BRCA1/BRCA2 tumors to comprise more than two distinct groups. However, given the small sample set, we did not attempt to further subdivide the samples into additional classes. Because we expect, for small sample sets, each partition to by random chance be supported by a large number of discriminatory genes, further partitioning of <10 samples will not result in robust classes using this class discovery approach.
Figure 1.
Gene expression-based class discovery of BRCAx breast cancers. (a) Based on 16 BRCAx tumors, the most significant separation into two classes (see Materials and Methods) resulted in classes with seven (group A, yellow) and nine (group B, blue) samples, respectively. Group A consists of families L5, L16, and L99, and group B of families L101, L111, L414, L502, and L505 (see Table 1). Sixty statistically significant genes (P < 0.001), which were found to separate the groups, are listed. Expression levels for each gene are normalized across the samples such that the mean is 0 and the variance is 1. Expression levels greater than the mean are pseudocolored red, and those below are pseudocolored green. The scale indicates SDs above or below the mean. (b) The number of genes separating BRCAx cancers into two subgroups (dotted line) is plotted as a function of the signal-to-noise weight. The bars (1 SD) show the number of genes expected by chance. There is a clear overabundance of genes separating the BRCAx subgroups. (c and d) Based on the 60 genes that best separated BRCAx tumors into two groups, multidimensional scaling analysis and hierarchical clustering of the 16 samples together with BRCA1 (gray) and BRCA2 (purple) tumors is shown. The BRCAx subgroups were separated from one another as well as from the BRCA1 and BRCA2 tumors, reflecting the difference between BRCAx and BRCA1/2 tumors.
The most sensitive sample in our partitioning was 11954, which in the second best partitioning was the only affected sample. In addition, it was the only sample sensitive to changing the level of significance to a less stringent selection of discriminatory genes. This finding may be explained by the fact that this sample was a brain metastasis from one of the breast cancers (8984). Nevertheless, the global gene expression profile of the brain metastasis was highly correlated with its primary tumor. Moreover, similar results were obtained when a rank-based gene discriminatory score was used, indicating that the partitioning is relatively insensitive to noise. Based on the 60 genes found to separate the BRCAx tumors into two groups, we generated a hierarchical dendrogram (31) illustrating the differences between the groups (Fig. 1a). Interestingly, many of the genes with increased expression in group A as compared with group B are ribosomal, possibly indicating different capacities for protein biosynthesis between these groups. A study of gene expression profiles in ovarian cancer found that a group of well differentiated tumors that clustered together with normal ovarian epithelial samples showed overexpression of a large number of ribosomal genes (32). Furthermore, up-regulation of ribosomal genes correlated with down-regulation of a cluster of proliferative genes, further supporting a less aggressive phenotype in these ovarian cancers. In our data, the two groups were not significantly different from each other with respect to clinical and histopathological factors (Table 1). However, although not statistically significant, individuals in group A tended to have a later age at onset of disease and the tumors had a low percentage of cells in S-phase. This may indicate a less aggressive appearance of these tumors, in agreement with the findings for ovarian tumors reported by Welsh et al. (32). Moreover, groups A and B manifested significantly different expression of the CYP1A1 gene, which encodes a phase I cytochrome P450 enzyme known to play a central role in the metabolism of a wide range of compounds, including steroids. About 10% of Caucasians have a highly inducible form of the enzyme that is associated with an increased risk of lung cancer in smokers (33), but which may also play a role in hormonal carcinogenesis. It would be of interest to investigate whether CYP1A1 overexpression and its susceptible genotype constitute a high risk for breast cancer development. In addition, many of the genes with low expression in group A are ESTs, and need to be further characterized.
To further exclude the possibility that our discriminatory genes were related to unidentified BRCA1 or BRCA2 mutations, we included a number of tumors from known BRCA1 and BRCA2 mutation carriers in our analysis. We performed multidimensional scaling analysis (15) and hierarchical clustering with the 60 genes that best separated the BRCAx tumors into two groups, and found that neither BRCA1 nor BRCA2 tumors were mixed with the BRCAx samples (Fig. 1 c and d), supporting their underlying difference. Additionally, the proximity to the BRCA2 gene of the recently proposed novel breast cancer locus on 13q21 (3), has lead some investigators to believe that the families linked to this region are in fact BRCA2 families without identified mutations (5). One of the families included in our study was previously found to be linked to the 13q21 locus (Table 1); however, this sample was completely separated from the BRCA2 tumors. The generally lower grade of BRCAx breast tumors makes them histologically more similar to the moderate grade BRCA2 tumors than the high grade BRCA1 tumors. Histopathological reexamination of the BRCAx tumors revealed that, as expected, they were heterogeneous with respect to histological and clinical parameters (Table 1). However, of interest, in this study the percentage of high grade tumors, particularly with high mitotic counts, was found to be greater than has been previously described for this group of tumors (9). Numerous studies have shown that the most significant impact on gene expression profiles of sporadic breast cancers is from ER status and ER correlated genes (8, 34, 35). On the other hand, we have previously shown that BRCA1 and BRCA2 breast tumors can be correctly identified based on genes that are unable to separate ER-positive from ER-negative sporadic cases (7). Interestingly, the two groups identified here show variable hormone receptor levels. Therefore, the addition of sporadic samples in the class discovery would likely have a confounding effect on the analysis.
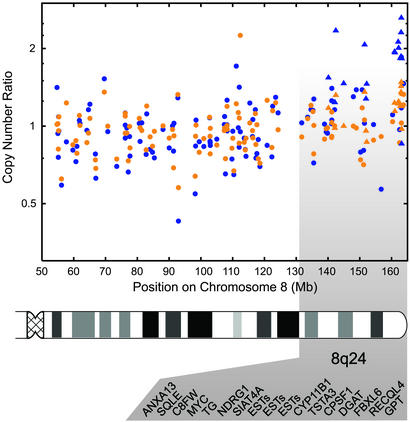
Analyses of genomic aberrations have revealed several chromosomal regions that are common to most breast cancers, but also those that are unique to BRCA1, BRCA2, and BRCAx tumors (3, 36, 37). It should be noted that the regions of genomic aberrations characteristic for BRCA1 or BRCA2 tumors do not harbor the predisposing gene(s). This implies that the specific nature of the predisposing gene influences the subsequent somatic genetic tumor progression pathway, or alternatively, that the various hereditary tumors stem from cells of different lineage. We therefore determined the genomic content of eight individual tumors included in our study using microarray-based CGH analysis (Table 1). The limited number of samples available for CGH analysis precluded the possibility of discovering subclasses using only the CGH data. Instead, to further confirm the molecular differences between the subgroups, we identified 262 cDNA clones that displayed significant differences (α < 0.02) in copy number ratio between the groups (see Table 4, which is published as supporting information on the PNAS web site). These differences were not dominated by any individual sample, but were consistently found within the subgroups (see Table 4). The locations of these genes suggest the presence of common regions of alterations within the BRCAx subgroups (Table 2), confirming their molecular differences. In particular, a large number of clones were located on 8q24, and this region was significantly amplified (P < 10−11) in group B as compared with group A (Fig. 2). Three of the tumors have previously been analyzed by using conventional CGH, and the findings for chromosome 8 confirm our array-based results (data not shown). According to Kainu et al. (3), 8q23–24 amplification is seen in hereditary as well as unselected breast cancers. However, only approximately one-third of BRCAx cancers displayed 8q amplification in that study, whereas over half of BRCA1, BRCA2, and sporadic breast cancers showed gain of this chromosomal region (3, 36). This observation may be explained by the absence of this aberration in group A identified in our classification. Our finding that group A is a distinct and distant branch on the expression-based dendrogram (Fig. 1d) further supports this possibility. Moreover, it has been suggested that c-MYC is the target of this amplification, and 8q24 amplification was recently shown to be associated with poor survival in breast cancer (38). However, even though c-MYC had a significantly higher copy number ratio in this group (α = 0), it did not differ in expression between the groups. Furthermore, c-MYC was on average amplified to a lesser extent (1.5-fold) than other clones (2-fold) located closer to the 8q telomere. Interestingly, a novel gene in this region (PRL-3) associated with metastasis of colorectal cancer has recently been identified as a candidate target for 8q amplification (39). Unfortunately, PRL-3 was not present on our arrays. It has recently been shown that although copy number has a pervasive effect on gene expression, only ≈10% of the variation in gene expression in breast cancer can be attributed to copy number alterations (40, 41). Analysis of larger sample sets using microarrays that are denser in the regions of copy number change should identify candidate target genes for the observed amplifications. Taken together, these findings suggest that fine mapping with cDNA microarray-based CGH analysis can be used to delineate chromosomal aberrations at a high resolution, thereby enabling the identification of more specific amplicon boundaries. It remains to be seen whether group A completely lacks 8q amplification, or displays it more infrequently than other groups of breast cancer. However, the paucity of this chromosomal aberration further supports the less aggressive behavior of the tumors in this group.
Table 2.
Regions of genomic alterations differing between BRCAx subgroups
Chromosome position as aligned to the draft of the human genome sequence (see Materials and Methods).
P value as determined by using a method based on the hypergeometric function (see Supporting Text).
Figure 2.
cDNA microarray-based CGH analysis of chromosome 8q in BRCAx subgroups. Average copy number ratios for group A (yellow) and group B (blue) are shown for clones on chromosome 8q. The position of the clones on chromosome 8 is shown in Mbs from the 8p telomere, as given by alignment to the draft human genome sequence (see Materials and Methods). Seventeen clones on 8q24 that were found to be different (α < 0.05) in copy number ratio between groups A and B are listed in positional order below and are highlighted (triangle). All but one (RECQL4) had higher ratios in group B than in group A.
In summary, we have used cDNA microarrays to identify two classes of familial BRCAx breast cancers that differ in their expression of a large number of genes. Tumors from individual patients/families remained clustered within the groups, and did not mix with tumors from BRCA1 and BRCA2 mutation carriers. Additionally, we show that the identified subclasses were associated with differences in gene amplification patterns. The results clearly require additional experiments using a larger sample set from diverse populations to pinpoint and validate the optimal subclasses and their molecular characteristics. In particular, whether high expression of ribosomal genes is correlated with less aggressive familial breast cancer warrants further investigation. We expect that the groups identified may split into subgroups as additional samples are analyzed. Nevertheless, given the heterogeneity of BRCAx tumors, and the previous difficulty in subsetting this group in a meaningful way, our data suggest that using large-scale gene expression based class discovery, followed by conventional positional linkage/candidate gene analysis may be an effective approach to finally identify novel breast cancer predisposition genes.
Supplementary Material
Acknowledgments
We thank the South Sweden Breast Cancer Group for supplying tumors, B. Baldetorp for flow cytometric analyses, and M. Fernö for coordinating the pathological review. This study was supported by a grant from the Swedish Cancer Society. M.R. was supported by a postdoctoral fellowship from the Swedish Research Council.
Abbreviations
- BRCAx, non-BRCA1/BRCA2 breast cancer gene(s)
ER, estrogen receptor
- PgR
progesterone receptor
Footnotes
To whom correspondence should be addressed at: Translational Genomics Research Institute, 400 North Fifth Street, Suite 1600, Phoenix, AZ 85004. E-mail: jtrent@tgen.org.
References
- 1.Ford D, Easton D F, Stratton M, Narod S, Goldgar D, Devilee P, Bishop D T, Weber B, Lenoir G, Chang-Claude J, et al. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seitz S, Rohde K, Bender E, Nothnagel A, Kolble K, Schlag P M, Scherneck S. Oncogene. 1997;14:741–743. doi: 10.1038/sj.onc.1200881. [DOI] [PubMed] [Google Scholar]
- 3.Kainu T, Juo S H, Desper R, Schaffer A A, Gillanders E, Rozenblum E, Freas-Lutz D, Weaver D, Stephan D, Bailey-Wilson J, et al. Proc Natl Acad Sci USA. 2000;97:9603–9608. doi: 10.1073/pnas.97.17.9603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rahman N, Teare M D, Seal S, Renard H, Mangion J, Cour C, Thompson D, Shugart Y, Eccles D, Devilee P, et al. Oncogene. 2000;19:4170–4173. doi: 10.1038/sj.onc.1203735. [DOI] [PubMed] [Google Scholar]
- 5.Thompson D, Szabo C I, Mangion J, Oldenburg R A, Odefrey F, Seal S, Barfoot R, Kroeze-Jansema K, Teare D, Rahman N, et al. Proc Natl Acad Sci USA. 2002;99:827–831. doi: 10.1073/pnas.012584499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nathanson K L, Weber B L. Hum Mol Genet. 2001;10:715–720. doi: 10.1093/hmg/10.7.715. [DOI] [PubMed] [Google Scholar]
- 7.Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi O P, et al. N Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
- 8.van't Veer L J, Dai H, van de Vijver M J, He Y D, Hart A A, Mao M, Peterse H L, van der Kooy K, Marton M J, Witteveen A T, et al. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 9.Lakhani S R, Gusterson B A, Jacquemier J, Sloane J P, Anderson T J, van de Vijver M J, Venter D, Freeman A, Antoniou A, McGuffog L, et al. Clin Cancer Res. 2000;6:782–789. [PubMed] [Google Scholar]
- 10.Johannsson O T, Idvall I, Anderson C, Borg Å, Barkardottir R B, Egilsson V, Olsson H. Eur J Cancer. 1997;33:362–371. doi: 10.1016/s0959-8049(97)89007-7. [DOI] [PubMed] [Google Scholar]
- 11.Loman N, Johannsson O, Bendahl P O, Borg Å, Fernö M, Olsson H. Cancer. 1998;83:310–319. [PubMed] [Google Scholar]
- 12.Hakansson S, Johannsson O, Johansson U, Sellberg G, Loman N, Gerdes A M, Holmberg E, Dahl N, Pandis N, Kristoffersson U, et al. Am J Hum Genet. 1997;60:1068–1078. [PMC free article] [PubMed] [Google Scholar]
- 13.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y, Dougherty E R, Bittner M L. J Biomed Optics. 1997;2:364–374. doi: 10.1117/12.281504. [DOI] [PubMed] [Google Scholar]
- 15.Khan J, Simon R, Bittner M, Chen Y, Leighton S B, Pohida T, Smith P D, Jiang Y, Gooden G C, Trent J M, et al. Cancer Res. 1998;58:5009–5013. [PubMed] [Google Scholar]
- 16.Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, Radmacher M, Simon R, Yakhini Z, Ben-Dor A, et al. Nature. 2000;406:536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Kamat V, Dougherty E R, Bittner M L, Meltzer P S, Trent J M. Bioinformatics. 2002;18:1207–1215. doi: 10.1093/bioinformatics/18.9.1207. [DOI] [PubMed] [Google Scholar]
- 18.Golub T R, Slonim D K, Tamayo P, Huard C, Gaasenbeek M, Mesirov J P, Coller H, Loh M L, Downing J R, Caligiuri M A, et al. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Dor A, Bruhn L, Friedman N, Nachman I, Schummer M, Yakhini Z. J Comput Biol. 2000;7:559–583. doi: 10.1089/106652700750050943. [DOI] [PubMed] [Google Scholar]
- 20.Ben-Dor A, Friedman N, Yakhini Z. In: Fifth Annual International Conference on Computational Molecular Biology (RECOMB) Lengauer T, Sankoff D, Istrail S, Pevzner P, Waterman M, editors. Montreal: ACM Press; 2001. pp. 31–38. [Google Scholar]
- 21.Kirkpatrick S, Gelatt C D, Vecchi M P. Science. 1983;220:671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 22.Pollack J R, Perou C M, Alizadeh A A, Eisen M B, Pergamenschikov A, Williams C F, Jeffrey S S, Botstein D, Brown P O. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 23.Allander S V, Nupponen N N, Ringner M, Hostetter G, Maher G W, Goldberger N, Chen Y, Carpten J, Elkahloun A G, Meltzer P S. Cancer Res. 2001;61:8624–8628. [PubMed] [Google Scholar]
- 24.DeGroot M. Probability and Statistics. New York: Addison–Wesley; 1986. [Google Scholar]
- 25.Hughes J D, Estep P W, Tavazoie S, Church G M. J Mol Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- 26.Barash Y, Bejerano G, Friedman N. In: First International Workshop on Algorithms in Bioinformatics. Gascuel O, Moret B M E, editors. Berlin: Springer; 2001. pp. 278–293. [Google Scholar]
- 27.Lander E S, Linton L M, Birren B, Nusbaum C, Zody M C, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 28.Kent W J, Sugnet C W, Furey T S, Roskin K M, Pringle T H, Zahler A M, Haussler D. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent W J. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruitt K D, Maglott D R. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Welsh J B, Zarrinkar P P, Sapinoso L M, Kern S G, Behling C A, Monk B J, Lockhart D J, Burger R A, Hampton G M. Proc Natl Acad Sci USA. 2001;98:1176–1181. doi: 10.1073/pnas.98.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McLemore T L, Adelberg S, Liu M C, McMahon N A, Yu S J, Hubbard W C, Czerwinski M, Wood T G, Storeng R, Lubet R A, et al. J Natl Cancer Inst. 1990;82:1333–1339. doi: 10.1093/jnci/82.16.1333. [DOI] [PubMed] [Google Scholar]
- 34.Perou C M, Sorlie T, Eisen M B, van de Rijn M, Jeffrey S S, Rees C A, Pollack J R, Ross D T, Johnsen H, Akslen L A, et al. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 35.Gruvberger S, Ringnér M, Chen Y, Panavally S, Saal L H, Borg Å, Fernö M, Peterson C, Meltzer P S. Cancer Res. 2001;61:5979–5984. [PubMed] [Google Scholar]
- 36.Tirkkonen M, Johannsson O, Agnarsson B A, Olsson H, Ingvarsson S, Karhu R, Tanner M, Isola J, Barkardottir R B, Borg Å, et al. Cancer Res. 1997;57:1222–1227. [PubMed] [Google Scholar]
- 37.Monni O, Bärlund M, Mousses S, Kononen J, Sauter G, Heiskanen M, Paavola P, Avela K, Chen Y, Bittner M L, et al. Proc Natl Acad Sci USA. 2001;98:5711–5716. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jain A N, Chin K, Borresen-Dale A L, Erikstein B K, Eynstein Lonning P, Kaaresen R, Gray J W. Proc Natl Acad Sci USA. 2001;98:7952–7957. doi: 10.1073/pnas.151241198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha S, Bardelli A, Buckhaults P, Velculescu V E, Rago C, Croix B S, Romans K E, Choti M A, Lengauer C, Kinzler K W, et al. Science. 2001;294:1343–1346. doi: 10.1126/science.1065817. [DOI] [PubMed] [Google Scholar]
- 40.Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringnér M, Sauter G, Monni O, Elkahloun A, et al. Cancer Res. 2002;62:6240–6245. [PubMed] [Google Scholar]
- 41.Pollack J R, Sorlie T, Perou C M, Rees C A, Jeffrey S S, Lonning P E, Tibshirani R, Botstein D, Borresen-Dale A L, Brown P O. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.