Abstract
Phenotypic variation that cannot be explained by genetic or environmental heterogeneity has intrigued geneticists for decades. The molecular basis of this phenomenon, however, is largely a mystery. Axin-fused (AxinFu), first identified in 1937, is a classic example of a mammalian allele displaying extremely variable expression states. Here we demonstrate that the presence or absence of its characteristic phenotype, a kinked tail, correlates with differential DNA methylation at a retrotransposon within AxinFu and identify mutant transcripts arising adjacent to the retrotransposon LTR that are likely to be causative of the phenotype. Furthermore, the epigenetic state at AxinFu can be inherited transgenerationally after both maternal and paternal transmission. This is in contrast to epigenetic inheritance at the murine agouti-viable yellow (Avy) allele, which occurs through the female only. Unlike the egg, the sperm contributes very little (if any) cytoplasm to the zygote, and therefore paternal inheritance at AxinFu argues against the possibility that the effects are due to cytoplasmic or metabolic influences. Consistent with the idea of transgenerational inheritance of epigenetic marks, we find that the methylation state of AxinFu in mature sperm reflects the methylation state of the allele in the somatic tissue of the animal, suggesting that it does not undergo epigenetic reprogramming during gametogenesis. Finally, we show that epigenetic inheritance is influenced by strain background. These findings enable us to propose a model for transgenerational epigenetic inheritance in mammals.
It is generally assumed that a phenotype is determined by the interaction of a specific genotype and a specific environment, but there are a number of examples where variable expressivity and incomplete penetrance cannot be explained by genetic or environmental heterogeneity. One of the earliest documented examples is the axin-fused (AxinFu) allele, first identified in 1937 (1). Axin regulates embryonic axis formation in vertebrates by inhibiting the Wnt signaling pathway (2). AxinFu is a dominant gain-of-function allele that has a 5.1-kb intracisternal-A particle (IAP) retrotransposon (subtype IΔ1) inserted in an antisense orientation (relative to the axin locus) in intron 6 (3). The characteristic AxinFu phenotype is kinks in the tail (Fig. 1A) caused by axial duplications during embryogenesis (1, 2). The phenotype is variably expressed among AxinFu individuals, and in some mice the tails appear completely normal; i.e., the mutant phenotype is silent.
Figure 1.
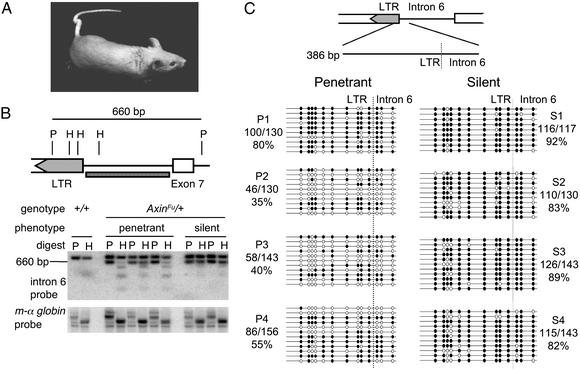
The tail-kink phenotype correlates with the methylation state at the AxinFu allele. (A) An AxinFu/+ mouse of the 129P4/RrRk strain with a kinked tail. (B Upper) The intron 6 probe (checkered box) and the PstI (P) and HhaI (H) sites in the region of the IAP insertion within intron 6 of AxinFu. (B Lower) Tail DNA was digested with PstI alone or in combination with HhaI (sensitive to CpG methylation) and then transferred and hybridized with the intron 6 probe. Digestion with PstI alone produces a 917-bp fragment at the wild-type axin allele and a 660-bp fragment at the AxinFu. In penetrant AxinFu/+ mice this band is digested by HhaI, indicating hypomethylation. In silent AxinFu/+ mice the 660-bp band is not digested by HhaI, indicating hypermethylation. The data shown are representative of seven penetrant and eight silent mice. The membrane was rehybridized with a probe to the mouse α-globin gene to ensure equal digestion of all samples. (C) Methylation profiles of 13 CpG dinucleotides at the IAP–intron 6 junction of the AxinFu allele. The methylation state of each CpG was obtained by sequencing PCR clones from bisulfite-treated genomic DNA. Open and filled circles represent unmethylated and methylated CpGs, respectively. The numbers in the parentheses represent the proportion of methylated CpG sites relative to all CpG sites for which sequence was obtained. Each line represents the sequence of one clone, and each block of clones represents the data from one mouse. The data shown are from four penetrant (P) and four silent (S) mice but are representative of those obtained for six penetrant and five silent mice.
The variable expressivity of AxinFu is reminiscent of that observed for the Aiapy, Ahvy, and Avy alleles of the agouti locus, all of which contain IAP (subtype IΔ1) insertions upstream of the agouti gene (4–6). The coats of isogenic mice carrying these alleles vary from wild-type agouti to completely yellow, with a spectrum of intermediate mottled coats (4–7). The variable expressivity correlates with differential DNA methylation at a cryptic promoter within the long terminal repeat (LTR) of the IAP, which can override the endogenous agouti promoters (4, 5, 8). Hypomethylation is associated with ectopic agouti expression and consequently a completely yellow coat, whereas hypermethylation correlates with normal agouti expression, resulting in an agouti coat. Furthermore, at the Avy allele the epigenetic state can be inherited transgenerationally after maternal transmission, resulting in the inheritance of phenotype, i.e., the range of coat colors of the offspring correlates with the coat color of the dam (8).
Avy is the only endogenous mammalian allele known to display transgenerational epigenetic inheritance, leading many to question the veracity of this phenomenon. We suspected that AxinFu might provide even stronger support for the idea that variable expressivity and inheritance of phenotype in mammals can have an epigenetic basis. Here we demonstrate that the AxinFu phenotype correlates with differential DNA methylation at the associated IAP LTR. We also identify mutant transcripts that are likely to be causative of the phenotype. Most importantly, we demonstrate inheritance of parental phenotype after transmission of the allele through both sexes in an inbred strain. Finally, analysis of the methylation state of the AxinFu and Avy alleles in mature sperm, in conjunction with the finding that epigenetic inheritance at these alleles is influenced by strain background, allows us to propose a model for transgenerational epigenetic inheritance in mammals.
Materials and Methods
Classification of Phenotypes.
Inbred 129P4/RrRk AxinFu/+ mice (The Jackson Laboratory) were classified according to their tail phenotypes as strongly penetrant (several tail kinks with an angle of more than ≈30° to the main tail axis), mildly penetrant (one to several small tail kinks with an angle of less than ≈30° to the main tail axis), or silent (no visible kink). Inbred C57BL/6J Avy/a mice (Oak Ridge National Laboratory, Oak Ridge, TN) were classified according to their coat-color phenotypes as described (8).
Genotyping.
Mice were genotyped for the AxinFu and wild-type axin alleles by multiplex PCR using primers P23 (5′-cg-gagctattccgaggaacg-3′), G245 (5′-gaccagagcccaagaaaaaccc-3′), and IAP forward (5′-gcgcatcactccctgattg-3′), a kind gift from Anatoly Ruvinsky (University of New England, New South Wales, Australia). The thermal parameters were 94°C/60 s followed by 30 cycles of 94°C/30 s, 58°C/10 s, 72°C/80 s, and one final cycle of 72°C/5 min. Genotyping for the Avy allele was performed by multiplex PCR using the IAP forward primer (see above), Agouti3′ (5′-tggccaggaaagaaggaaac-3′), and Agouti5′ (5′-catggctacagcatcctgac-3′). The thermal parameters consisted of 94°C/60 s followed by 32 cycles of 94°C/20 s, 58°C/10 s, 72°C/45 s, and one final cycle of 72°C/5 min.
Statistics.
P is the value from the χ2 distribution for the statistic and the appropriate degrees of freedom.
Methylation-Sensitive Restriction Digests.
Adult tail and kidney DNA were extracted as described (8). DNA was digested overnight with PstI either alone or with HhaI. The fragments were separated on a 2% agarose gel and analyzed by Southern transfer. The membrane was hybridized to a 300-bp radiolabeled fragment from a 750-bp PCR clone of the AxinFu allele (intron 6 probe). After exposure, the membrane was stripped and hybridized with a 2.1-kb HinfI–SacI fragment containing the mouse α-globin gene to control for equivalent digestion of samples.
Bisulfite Sequencing.
Bisulfite sequencing of the AxinFu allele was performed as follows: 10 μg of genomic DNA from kidney was digested with BamHI and treated with sodium bisulfite as described (9). The bisulfite-treated DNA was resuspended in 50 μl of deionized water, 2 μl of which was used in the primary PCR to amplify a 500-bp fragment of the LTR and intron 6 region of AxinFu. The primers used were the LTR primer (5′-ggagtaagagtgtaagaagtaagagagagag-3′) and int6L (5′-ccttcaacccactataaaaactaaaacc-3′). The PCR protocol involved five cycles of 94°C/1 min, 59°C/1 min, and 72°C/1 min followed by 30 cycles of 94°C/30 s, 59°C/30 s, and 72°C/30 s. The PCR contained BSA. One microliter of the products from the first PCR then were used in a seminested PCR that amplified a 386-bp fragment. The PCR conditions were 35 cycles of 94°C/30 s, 59°C/30 s, and 72°C/30 s. The primer set was LTR primer (see above) and int6 (5′-cctcctcatccaatcataacaaaacc-3′). The 386-bp PCR fragment was subcloned into the p-GEM-T Easy vector (Promega) and sequenced.
Mature sperm were isolated from the epididymis of adult males (each contained 106 to 107 spermatocytes). Sperm samples were found to contain >95% spermatocytes as determined by light microscopy. Epididymal sperm DNA was digested with BamHI, ethanol-precipitated, and resuspended in 10 μl of Milli-Q. The DNA was the embedded in agarose (2%) and subjected to bisulfite treatment as described above. The bisulfite-treated DNA was resuspended in 10 μl of deionized water and heated at 80°C/1 min before use in the PCR.
Bisulfite sequencing of the Avy allele in mature sperm was performed essentially as for the AxinFu allele with modifications. In the primary PCR, the primers used were the mbbis upstream primer (5′-cggaattcgaaaagagagtaagaagtaagagagagag-3′) and mbbis downstream (5′-gctctagaaaaatttaacacataccttctaaaaccccc-3′). The PCR protocol involved 94°C/2 min followed by 35 cycles of 94°C/30 s, 58°C/30 s, and 72°C/30 s. One microliter of the products from the first PCR then were used in a seminested PCR that amplified a 290-bp fragment. The PCR conditions were 35 cycles of 94°C/30 s, 62°C/30 s, and 72°C/30 s. The primer set was mbbis upstream (see above) and mbbis internal (5′-actccctcttctaaaactacaaaaactc-3′). The 290-bp PCR fragment was subcloned into p-GEM-T Easy vector and sequenced.
PCR primers and conditions were tested for unbiased amplification of methylated and unmethylated sequences by comparison to Southern blots. Sequenced clones were included in the analysis only if they contained no more than four non-CpG located cytosines per clone that were unconverted by the bisulfite treatment, i.e., a conversion efficiency of at least 94%. Only one AxinFu clone of sperm DNA (a penetrant sample), three AxinFu clones of tail DNA (one penetrant and two silent), and two Avy clones (one yellow and one agouti) were rejected on this basis.
5′ RNA Ligase-Mediated Rapid Amplification of cDNA Ends (RLM RACE).
Total RNA was prepared from kidney with TRI reagent (Sigma). RLM-RACE was performed with an Ambion (Austin, TX) FirstChoice RLM-RACE kit. The gene-specific reverse outer (5′-cactgcatgatcttctggttct-3′) and reverse inner (5′-gattcagccttcttggtgtttc-3′) primers used in the PCR were designed to anneal to exon 7. PCR products were subcloned into p-GEM-T Easy and sequenced.
Northern Transfer.
The intron 6 probe (described above) was added to ExpressHyb hybridization solution (CLONTECH) and incubated overnight at 68°C with Hybond XL (Amersham Pharmacia) blots prepared with adult kidney poly(A)+ mRNA. After exposure, the membrane was stripped and reprobed with a mouse GAPDH probe.
In Vitro Transcription/Translation.
The construct pFu1.1 contains a 1,109-bp insert subcloned into the ApaI site of p-GEM-T Easy, downstream of the T7 promoter. The insert was generated by RT-PCR from total RNA with primers M4fwd (5′-ctagggccccgtggtcctgtggtgacttaat-3′) and stoprev (5′-ctagggccccagtgctcagtccaccttttc-3′), both of which contain ApaI sites incorporated into their 5′ ends. The insert encompasses a region starting in intron 6, 7 bp downstream of the transcriptional start site identified by RLM RACE, and ends in exon 10 at the wild-type axin stop codon. An ATG-to-CTC mutation was carried out on pFu1.1 by using the QuikChange site-directed mutagenesis kit (Stratagene), with the plusmut (5′-ccagagtgacctcctctctcttgttctcccagtg-3′) and minusmut (5′-cactgggagaacaagagagaggaggtcactctgg-3′) primers. The TnT T7-Coupled Reticulocyte Lysate system (Promega) and [35S]methionine (Amersham Pharmacia) was then used to examine translation from pFu1.1. In vitro-translated proteins were resolved by SDS/PAGE on a 12% acrylamide Tris-glycine gel.
Results
The AxinFu Tail-Kink Phenotype Correlates with DNA Methylation at the LTR.
Previous studies demonstrating variable expressivity of AxinFu were performed in mixed genetic backgrounds (1, 10). In our study, we sought to exclude the effects of any genetic differences at modifier loci by investigating the AxinFu allele in the inbred 129P4/RrRk strain. We observed a wide spectrum of tail deformities among these isogenic AxinFu/+ mice, including mice with completely normal tails. Methylation-sensitive restriction-enzyme digests of tail DNA from seven penetrant and eight silent mice (examples are shown in Fig. 1B) and bisulfite sequencing of DNA from six penetrant and five silent mice (examples are shown in Fig. 1C) revealed that the LTR/intron 6 region is heavily methylated in the somatic tissue of silent AxinFu/+ mice, whereas in penetrant AxinFu/+ mice this region is relatively hypomethylated but with some degree of interclone and intermouse variation. We did not find any particular CpG site that was consistently unmethylated in penetrant mice or consistently methylated in silent mice. Methylation-sensitive restriction digests were also performed with kidney DNA, and the results were similar (data not shown). Overall, our findings are consistent with AxinFu being an epigenetically sensitive allele at which the establishment of an epigenetic mark is stochastic, to a degree, and that the epigenetic state correlates with the tail phenotype.
Aberrant Transcripts Associated with the IAP LTR in AxinFu Are Absent in Silent Mice.
If the kinky-tail phenotype is due to aberrant transcription arising from the IAP LTR in AxinFu, analogous to what is observed at the Avy allele, then any such aberrant transcription should be absent in silent AxinFu mice. Although it is known that in penetrant mice the AxinFu allele produces a wild-type transcript and several mutant transcripts (3), differences in transcriptional activity arising from the AxinFu allele between penetrant and silent mice have not been studied previously.
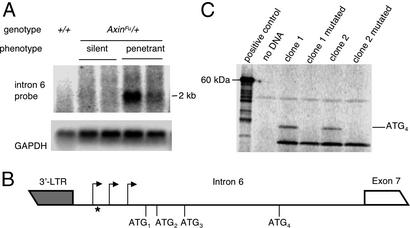
Poly(A)+ Northern analysis, using an intron 6-specific probe, revealed a broad band at ≈2 kb (which may be made up of more than one transcript) in penetrant mice only (Fig. 2A). The size of the mutant band is consistent with that expected of transcripts that initiate within the LTR/intron 6 region and contain all the remaining exons (exons 7–10). This band was more prominent in strongly penetrant mice than mildly penetrant mice (data not shown) but, importantly, was never observed in silent or wild-type mice. RLM RACE produced PCR products in samples from penetrant mice only (data not shown), and sequencing of these products revealed several mutant transcripts that initiate from various sites within intron 6, just downstream of the LTR (Fig. 2B). The ability of an IAP LTR to induce transcriptional initiation at multiple downstream start sites has been noted (11).
Figure 2.
Mutant transcripts expressed in penetrant mice. (A) Northern blot of adult kidney poly(A)+ RNA from wild-type (+/+) silent and penetrant AxinFu/+ adult mice. The blot was probed with the intron 6 probe described in Fig. 1. The GAPDH control of the same blot is also shown. The data presented are representative of that obtained for three wild-type, four penetrant, and four silent mice. (B) RLM-RACE (data not shown) revealed several mutant transcripts (≈2 kb) in penetrant mice only that initiate within intron 6 (arrows), +33, +64, and +97 bp downstream of the LTR at the 3′ end of the IAP insertion. ATG1 and ATG3 are out-of-frame with the axin-coding sequence. Translation initiated from ATG2 would result in a 39-aa peptide only. ATG4 is in-frame and could be used to generate a truncated form of the wild-type Axin protein that lacks the amino-terminal domain encoded by exons 1–6. (C) An in vitro transcription/translation assay was performed by using a construct containing a 1,109-bp cDNA insert derived from the AxinFu allele. The insert encompasses a region starting in intron 6 (marked with * in B), 40 bp downstream of LTR, and ending in exon 10 at the wild-type axin stop codon. The in vitro-translated protein that results from initiation of translation at ATG4 is indicated. The experiment was repeated after site-directed mutagenesis of the ATG4 site to CTC. Transcription/translation of the luciferase gene was used as a positive control.
Sequencing of the mutant transcripts identified by RLM RACE revealed that in all of them the first three start codons are either out-of-frame or would generate only a 39-aa peptide (Fig. 2B). However, the fourth start codon is in-frame and potentially could generate a mutant protein that contains exons 7–10. We performed in vitro eukaryotic transcription/translation experiments using a clone that contained all four potential start codons in intron 6, followed by exons 7–10 (Fig. 2C). We found that translation can be initiated from the fourth start codon despite the presence of the three preceding start codons. Site-directed mutagenesis of the fourth ATG site to CTC abolished production of this peptide (Fig. 2C).
Previous studies (12) and our own analysis (data not shown) have demonstrated that the mutant tail phenotype is not caused by a reduction of wild-type axin transcription, and antisense transcripts arising from the LTR at the 5′ end of the IAP insertion have not been detected either by us (data not shown) or others (3).
Our transcriptional analyses suggest that mutant transcripts that arise within intron 6 of the AxinFu allele are associated with the presence of the LTR, because aberrant transcription was not observed in wild-type mice; hypermethylation at the LTR correlates with abrogation of aberrant transcription, because mutant transcripts were not observed in silent AxinFu/+ mice. Presumably, differential epigenetic modifications, such as but not necessarily just DNA methylation, influence the activity of a cryptic promoter/enhancer within the LTR. Amino-terminal-deficient axin transcripts, similar to the ones associated with AxinFu, have been found to generate axial duplications during early development in Xenopus (2). Ventral injection into Xenopus embryos of mutant axin mRNA that contains a deletion of the regulator of the G protein signaling (RGS) domain (located in the amino-terminal end) induces axial duplications (2) similar to those observed in AxinFu mice. Therefore, the mutant transcripts arising adjacent to the IAP LTR, in penetrant mice only, are likely to be causative of the kinky-tail phenotype.
Transgenerational Inheritance of Epigenetic States at the AxinFu Allele.
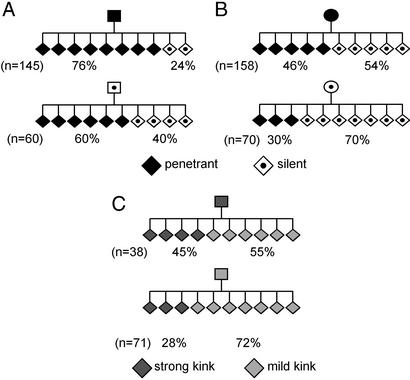
It has been observed that the phenotype of the AxinFu parent influences the range of tail phenotypes of the offspring (1, 10). However, because those studies were performed in mixed backgrounds, the possibility that the inheritance of phenotype was due to the inheritance of unlinked genetic modifiers in some mice could not be ruled out. To investigate the inheritance of AxinFu phenotypes in an inbred strain we set up matings between congenic AxinFu/+ and +/+ mice of the 129P4Rr/Rk strain. We found that penetrant sires produced 76% penetrant and 24% silent offspring, whereas silent sires produced 60% penetrant and 40% silent offspring (Fig. 3A). Likewise, penetrant dams produced 46% penetrant and 54% silent offspring, whereas silent dams produced 30% penetrant and 70% silent offspring (Fig. 3B). In all cases, the differences are significant (P < 0.05). Therefore, AxinFu transgenerational epigenetic inheritance occurs after both maternal and paternal transmission. We also observed that the severity of the phenotype of a penetrant sire is inherited; strongly penetrant sires gave rise to more strongly penetrant offspring than mildly penetrant sires (Fig. 3C). This further strengthens the case for inheritance of phenotype at AxinFu. A similar trend is also observed after female transmission (data not shown).
Figure 3.
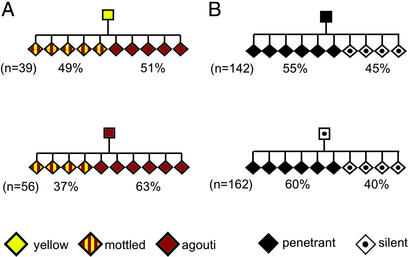
Inheritance of parental phenotype. AxinFu/+ 129P4/RrRk mice of the indicated phenotypes were crossed with congenic +/+ mice, and the percentage of offspring of each phenotype was scored. The number of total AxinFu/+ progeny of each cross is indicated (n); +/+ offspring have been omitted from the pedigrees. (A) The proportions of offspring phenotypes arising from penetrant and silent sires differ significantly (P < 0.05). The data were collated from 18 different matings. (B) The proportions of offspring phenotypes arising from penetrant and silent dams differ significantly (P < 0.05). The data were collated from 29 different matings. (C) The data presented in A were reanalyzed according to the severity of the penetrant phenotype. The severity of the phenotype of the sire influences the severity of the phenotypes in the penetrant offspring (P < 0.05). Silent AxinFu/+ and wild-type mice have been omitted from the pedigree.
Others have suggested that inheritance of the Avy phenotype is due to differences in maternal metabolism between phenotypically different dams (7). In our previous study on the Avy allele in an inbred strain (8), embryo-transfer experiments argued for the inheritance of phenotype being due to the transgenerational inheritance of an epigenetic state and not differences in maternal metabolism. The finding of epigenetic inheritance at AxinFu after both maternal and paternal transmission makes it even more unlikely that inheritance of phenotype is due to metabolic or cytoplasmic differences among fertilized eggs of an inbred strain, because the sperm contributes very little if any cytoplasm to the egg.
It has been argued that epigenetic inheritance in mammals could be due to the inheritance of unlinked genetic modifiers present in some mice that influence, in this case, the epigenetic status of IAPs. Because our studies were performed in inbred strains, these putative modifiers, if present at all, would be rare and therefore could only realistically exert their effects if they were dominant alleles. We carried out reciprocal crosses between heterozygous penetrant and silent AxinFu mice in the 129P4/RrRk strain with heterozygous yellow, mottled, or agouti Avy mice in the C57BL/6J strain (12 different types of matings) and analyzed the tail and coat-color phenotypes of the compound heterozygous F1 offspring, i.e., F1 offspring that have inherited both the AxinFu and Avy alleles. If the variable expressivity of these alleles is due to differences in unlinked genetic modifiers among the mice, then we would expect a compound heterozygous F1 mouse that has a silent tail phenotype to carry modifiers that increase methylation at IAPs and consequently should also display an agouti coat-color phenotype. The compound heterozygous F1 offspring (n = 203) displayed all possible combinations of coat-color and tail phenotypes e.g., mice were yellow with kinked tails, yellow with straight tails, agouti with straight tails, etc., without any evidence for the tail phenotype correlating with the coat color (data not shown). This argues that the epigenetic states at the AxinFu and Avy alleles are independent of each other, and therefore it is unlikely that modifiers, of the type described above, could explain the patterns observed in the breeding studies with the AxinFu or Avy alleles.
The high frequency with which AxinFu parents produce offspring of different phenotypes argues against the idea that the different tail phenotypes are due to genetic mutation at the AxinFu allele. Moreover, we sequenced the region of the LTR that was analyzed for differential methylation by using DNA samples from six penetrant and six silent mice. Twelve clones were sequenced for each mouse, and only two base differences, of the 144 clones sequenced (each clone represents 362 bp of sequence), were found (data not shown). This is consistent with the hypothesis that the different tail phenotypes are due to epigenetic differences rather than genetic differences at the AxinFu allele.
Analysis of the Methylation State of the AxinFu and Avy Alleles in Mature Sperm.
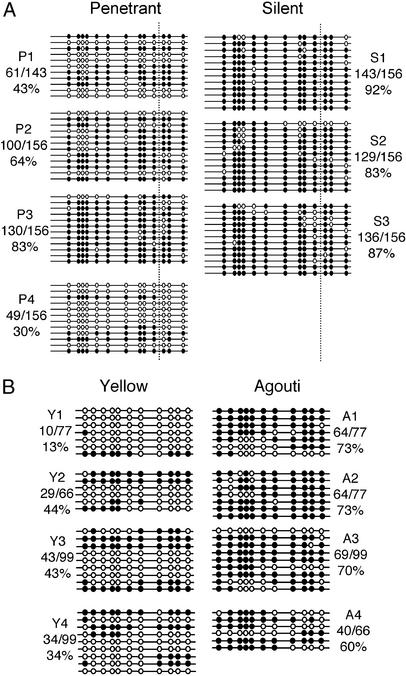
Transgenerational epigenetic inheritance necessitates that epigenetic differences at the allele are maintained in the mature gametes. We investigated the methylation state of AxinFu in the mature sperm of individual silent and penetrant AxinFu/+ males by bisulfite sequencing (Fig. 4A). The AxinFu allele in the sperm of silent males (three mice) was always heavily methylated (83–92% of CpG sites analyzed were methylated), and samples from penetrant mice (four mice) were relatively hypomethylated but with considerable interclone and intermouse variation (30–83% of CpG sites analyzed were methylated). Methylation-sensitive restriction digests of sperm DNA (from another four penetrant and four silent mice, data not shown) were consistent with the bisulfite sequencing results. The fact that the allele is heavily methylated in the sperm of silent mice and relatively hypomethylated in the sperm of penetrant mice is consistent with penetrant sires producing more penetrant offspring relative to silent sires.
Figure 4.
The methylation state of AxinFu and Avy in mature sperm reflects the methylation state of the alleles in the somatic tissue of the animal. (A) DNA from mature sperm of penetrant (P) and silent (S) 8-week-old AxinFu/+ males was subjected to bisulfite sequencing. One line represents the sequence from one clone and the methylation profile of the LTR/intron 6 region (refer to Fig. 1D) in one mature sperm. Each block of lines represents the methylation data of sperm DNA obtained from one adult male mouse. (B) DNA from mature sperm of yellow (Y) and agouti (A) 8-week-old Avy/a males was subjected to bisulfite sequencing. All the CpG sites shown for the Avy allele are contained within the LTR.
We also analyzed the methylation state of the Avy allele (in the C57BL/6J strain) in mature sperm (Fig. 4B). The Avy allele was heavily methylated in the sperm of agouti Avy males and relatively hypomethylated in the sperm of yellow Avy males. This finding was particularly interesting, because transgenerational epigenetic inheritance of Avy is not observed after paternal transmission in the C57BL/6J strain (8). Therefore, the methylation state of the AxinFu and Avy alleles in germ cells seems to be a reflection of the methylation state of the allele in the somatic tissue of that animal.
Transgenerational Epigenetic Inheritance Is Influenced by Strain Background.
It is interesting that Avy (in the C57BL/6J strain) displays epigenetic inheritance after maternal transmission only (8), but AxinFu (in the 129P4/RrRk strain) displays epigenetic inheritance after both maternal and paternal transmission. This is surprising in light of the observation that the methylation state of both alleles in mature sperm reflects the methylation state of the allele in the somatic tissue of that animal. We were uncertain whether the lack of inheritance of Avy after paternal transmission was due to some intrinsic property of the allele itself or a strain-specific effect. In the F1 generation (obtained by crossing AxinFu/+ 129P4/RrRk mice with Avy/a C57BL/6 mice as described above) we observed inheritance of the coat-color phenotype after paternal transmission of the Avy allele (i.e., a cross between Avy/a C57BL/6J sires and AxinFu/+ 129P4/RrRk dams) (Fig. 5A), but there was no inheritance of phenotype after paternal transmission of the AxinFu allele (i.e., a cross between AxinFu/+ 129P4/RrRk sires and Avy/a C57BL/6J dams) (Fig. 5B). These findings suggest that C57BL/6J-fertilized eggs completely erase epigenetic marks on paternally inherited alleles, because there is no epigenetic inheritance after paternal transmission of Avy in the C57BL/6J strain or AxinFu in F1 mice. On the other hand, 129P4/RrRk-fertilized eggs do not erase epigenetic marks on paternally inherited alleles completely, because epigenetic inheritance does occur after paternal transmission of AxinFu in the 129P4/RrRk strain and Avy in F1 mice.
Figure 5.
Transgenerational epigenetic inheritance is influenced by strain background. (A) Avy/a C57BL/6J males of the indicated phenotypes (eight yellow sires and six agouti sires) were mated to 129P4/RrRk females (129P4/RrRk mice are Aw/Aw at the agouti locus; Avy is dominant over Aw), and the offspring's coat color was recorded. The number of total Avy/Aw progeny of each cross is indicated (n); mice that did not carry the Avy allele have been omitted from the pedigree. The phenotype of the sire influences the range of phenotypes in the offspring (P < 0.05). (B) AxinFu/+ 129P4/RrRk sires of the indicated phenotypes (five penetrant sires and five silent sires) were mated to C57BL/6J dams (+/+ at the axin locus), and the F1 offspring's tail phenotype was recorded. The number of total AxinFu/+ progeny from each cross is indicated (n); +/+ mice have been omitted from the pedigree. The proportions of phenotypes arising from penetrant and silent sires do not differ significantly (P = 0.48).
A Model for Transgenerational Epigenetic Inheritance.
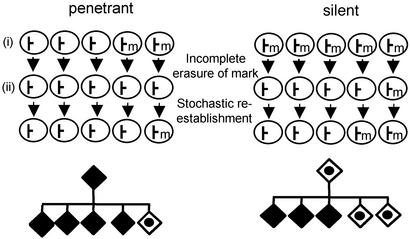
Assuming that methylation is, or is a reflection of, the primary epigenetic mark, we now can propose a model for transgenerational epigenetic inheritance (Fig. 6). Epigenetic marks at alleles such as AxinFu and Avy seem to be resistant to demethylation during gametogenesis, and therefore the epigenetic state of the allele in the gamete correlates, to a considerable extent, with the phenotype of the individual. Then postfertilization, the marks are not cleared completely, and after stochastic reestablishment there would be some memory of the epigenetic state that existed in the gametes of the parent. Indeed, it has been reported recently that IAPs are relatively resistant to demethylation during gametogenesis and preimplantation development (13). However, that study was performed in one particular strain, and our data suggest that murine strains differ in their ability to reprogram epigenetic marks at IAPs postfertilization. It remains unclear whether the C57BL/6J- and 129P4/RrRk-fertilized eggs differ in their capacity to erase epigenetic marks at IAPs associated with the AxinFu and Avy alleles exclusively, or whether the differences would affect other IAPs in the genome.
Figure 6.
A model for transgenerational epigenetic inheritance. We can consider a hypothetical allele that is marked at a single site by methylation (m). The silent phenotype corresponds to a methylated state, and the penetrant phenotype corresponds to the nonmethylated state. During gametogenesis (i), the marks escape demethylation, and therefore the overall epigenetic profile of the allele in mature sperm (five gametes shown for each male) correlates with the epigenetic state of the allele in somatic tissue of that animal. After fertilization (ii), the inability of the cell to erase marks at initially unmethylated alleles is inconsequential, but some alleles that were methylated initially will not be cleared completely. Ultimately, this will result in some memory of the epigenetic state of the parent's allele. This transgenerational epigenetic inheritance will not occur if the epigenetic marks are cleared completely postfertilization (not indicated in the figure), e.g., paternal copies of AxinFu and Avy in the C57BL/6J-fertilized egg.
Discussion
The work described here demonstrates that the variable expressivity of the kinky-tail phenotype of AxinFu correlates with differential epigenetic states of the LTR at the 3′ end of the IAP insertion. We have identified mutant transcripts starting within intron 6, just downstream of the IAP insertion, which are expressed in penetrant mice only. It is likely that these transcripts code for a truncated form of Axin that has a dominant gain-of-function effect during early development, resulting in tail kinks. The inference from these observations is that hypermethylation within the LTR at the 3′ end of the IAP insertion correlates with cryptic promoter inactivity and therefore a lack of mutant transcripts, resulting in a wild-type phenotype, in this case a normal-tailed mouse. More importantly, we also observed transgenerational epigenetic inheritance at AxinFu after both paternal and maternal transmission. Consistent with this phenomenon was the finding that the methylation state of the AxinFu allele in mature sperm reflects that of the allele in the somatic tissue of the animal. We also observed that the methylation state of the Avy allele in mature sperm reflects that of the allele in the somatic tissue, which was surprising because epigenetic inheritance at this allele does not occur after paternal transmission in the C57BL/6J strain. However, our F1 breeding studies showed that epigenetic inheritance at the AxinFu and Avy alleles is influenced by the strain background: The 129P4/RrRK-fertilized egg allows inheritance of epigenetic marks on paternal alleles, whereas the C57BL/6-fertilized egg does not.
Consistent with previous studies (1, 10), we found that AxinFu displays parent-of-origin effects: The penetrance of the kinky-tail phenotype is greater after paternal compared with maternal transmission (Fig. 2). Avy also displays parent-of-origin effects (7, 8). These effects probably arise because the resistance of IAPs to epigenetic reprogramming differs between the male and female germ line and also between maternal and paternal genomes postfertilization (13). Our results are consistent with this idea. Indeed, it is known already that the maternal and paternal genomes are treated differently with regard to DNA methylation immediately postfertilization (14, 15).
Inheritance of phenotype has been demonstrated previously in yeast (16), plants (17), and Drosophila (18) and after transgenesis (19, 20) and nuclear transplantation (21) in mice. Our work on the AxinFu allele presented here, in combination with our previous findings at Avy, demonstrates that transgenerational epigenetic inheritance can occur at endogenous mammalian alleles. It is difficult to assess the contribution epigenetics makes to phenotypic states in humans because of our extreme genetic heterogeneity. However, conventional genetics cannot explain, for example, the high degree of phenotypic discordance sometimes observed between monozygotic twins (22). Recently there has been a report of increased susceptibility to diabetes due to epigenetic influences (23), and it has been shown also that the methylation state at a few random sites in the human genome varied between individuals, and these states showed some degree of inheritance to the next generation (24). Finally, it is worth noting that the draft human genome sequence revealed 8% of our DNA to be retroviral in origin (25). It is conceivable that inheritance of variable epigenetic states at such sequences contributes to the phenotype of an individual.
Acknowledgments
We thank Anatoly Ruvinsky (University of New England, New South Wales, Australia) for helpful discussions. V.K.R. was supported by an international postgraduate award. M.E.C. and H.D.M. were supported by Australian postgraduate awards. This work was supported by a grant from the National Health and Medical Research Council of Australia (to E.W.).
Abbreviations
- AxinFu
axin-fused
- IAP
intracisternal-A particle
- Avy
agouti-viable yellow
- RLM RACE
5′ RNA ligase-mediated RACE
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Reed S C. Genetics. 1937;22:1–13. doi: 10.1093/genetics/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek T J, Perry W L, Tilghman S M, Costantini F. Cell. 1997;90:181–192. doi: 10.1016/s0092-8674(00)80324-4. [DOI] [PubMed] [Google Scholar]
- 3.Vasicek T J, Zeng L I, Zhang T, Costantini F, Tilghman S M. Genetics. 1997;147:777–786. doi: 10.1093/genetics/147.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Argeson A C, Nelson K K, Siracusa L D. Genetics. 1996;142:557–567. doi: 10.1093/genetics/142.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michaud E J, van Vugt M J, Bultman S J, Sweet H O, Davisson M T, Woychik R P. Genes Dev. 1994;8:1463–1472. doi: 10.1101/gad.8.12.1463. [DOI] [PubMed] [Google Scholar]
- 6.Duhl D M, Vrieling H, Miller K A, Wolff G L, Barsh G S. Nat Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- 7.Wolff G L. Genetics. 1978;88:529–539. doi: 10.1093/genetics/88.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgan H D, Sutherland H G E, Martin D I K, Whitelaw E. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 9.Clark S J, Harrison J, Paul C L, Frommer M. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belyaev D K, Ruvinsky A O, Borodin P M. J Hered. 1981;72:107–112. doi: 10.1093/oxfordjournals.jhered.a109436. [DOI] [PubMed] [Google Scholar]
- 11.Christy R J, Huang R C C. Mol Cell Biol. 1988;8:1093–1102. doi: 10.1128/mcb.8.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry W L, III, Vasicek T J, Lee J J, Rossi J M, Zeng L, Zhang T, Tilghman S M, Costantini F. Genetics. 1995;141:321–332. doi: 10.1093/genetics/141.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lane, N., Dean, W., Erhardt, E., Hajkova, P., Surani, A., Walter, J. & Reik, W. (2003) Genesis, in press. [DOI] [PubMed]
- 14.Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 15.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 16.Grewal S I S, Klar A J S. Cell. 1996;86:95–101. doi: 10.1016/s0092-8674(00)80080-x. [DOI] [PubMed] [Google Scholar]
- 17.Brink R. Genetics. 1956;41:872–889. doi: 10.1093/genetics/41.6.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavalli G, Paro R. Cell. 1998;93:505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 19.Allen N D, Norris M L, Surani M A. Cell. 1990;61:853–861. doi: 10.1016/0092-8674(90)90195-k. [DOI] [PubMed] [Google Scholar]
- 20.Hadchouel M, Farza H, Simon D, Tiollais P, Pourcel C. Nature. 1987;329:454–456. doi: 10.1038/329454a0. [DOI] [PubMed] [Google Scholar]
- 21.Roemer I, Reik W, Dean W, Klose J. Curr Biol. 1997;7:277–280. doi: 10.1016/s0960-9822(06)00124-2. [DOI] [PubMed] [Google Scholar]
- 22.Machin G A. Am J Med Genet. 1996;61:216–228. doi: 10.1002/(SICI)1096-8628(19960122)61:3<216::AID-AJMG5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 23.Bennett S T, Wilson A J, Esposito L, Bouzekri N, Undlien D E, Cucca F, Nistico L, Buzzetti R, Bosi E, Pociot F, et al. Nat Genet. 1997;17:350–355. doi: 10.1038/ng1197-350. [DOI] [PubMed] [Google Scholar]
- 24.Silva A J, White R. Cell. 1988;54:145–152. doi: 10.1016/0092-8674(88)90546-6. [DOI] [PubMed] [Google Scholar]
- 25.International Human Genome Sequencing Consortium. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]