Abstract
Ends-in and ends-out refer to the two arrangements of donor DNA that can be used for gene targeting. Both have been used for targeted mutagenesis, but require donors of differing design. Ends-out targeting is more frequently used in mice and yeast because it gives a straightforward route to replace or delete a target locus. Although ends-in targeting has been successful in Drosophila, an attempt at ends-out targeting failed. To test whether ends-out targeting could be used in Drosophila, we applied two strategies for ends-out gene replacement at the endogenous yellow (y) locus in Drosophila. First, a mutant allele was rescued by replacement with an 8-kb y+ DNA fragment at a rate of ≈1/800 gametes. Second, a wild-type gene was disrupted by the insertion of a marker gene in exon 1 at a rate of ≈1/380 gametes. The I-SceI endonuclease component alone is not sufficient for targeting: the FLP recombinase is also needed to generate the extrachromosomal donor. When both components are used we find that ends-out targeting can be approximately as efficient as ends-in targeting, and is likely to be generally useful for Drosophila gene targeting.
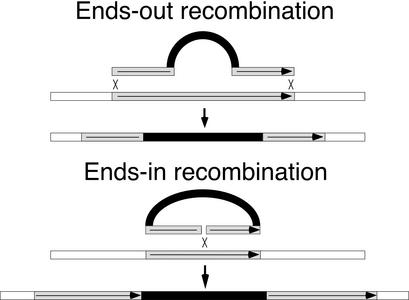
Gene targeting is the modification of an endogenous gene sequence by recombination between an introduced DNA fragment and the homologous target gene. Over the past 25 years, gene targeting has been widely used in model eukaryotes, first in yeast and then in mice (1, 2), but the difficulty of introducing a linear DNA molecule into germ-line cells hindered its development for Drosophila. Recently, a method to generate such a linear fragment in vivo was reported, accompanied by a demonstration of ends-in or insertional gene targeting (3). This occurs when a DNA double-strand break (DSB) is made in a donor DNA fragment within a stretch of DNA that is homologous to the target locus, and results in the insertion of the donor to generate a duplication of the targeted region. An alternative arrangement, where DSBs are provided at each end of a homologous segment, is termed ends-out targeting, and causes a segment of chromosome to be replaced with an introduced segment (Fig. 1). In mouse and in yeast some studies show that ends-out targeting is less efficient than ends-in (4–6), whereas others indicate that the two types can be equally efficient (7, 8). Doubts about the efficacy of ends-out targeting in Drosophila have been raised because of a previous failure (9). We undertook this work to determine whether ends-out targeting could be usefully implemented in Drosophila.
Figure 1.
Two general forms of gene targeting. Donor DNA molecules are diagramed above their targets, along with the expected products of recombination. The light gray region is the target-homologous DNA; the black region is the positive marker gene.
Materials and Methods
Ends-Out Donor Constructs.
The yellow rescue construct.
Two pairs of oligos, 5′-GTACATTACCCTGTTATCCCTA-3′, 5′-GTACTAGGGATAACAGGGTAAT-3′ and 5′-TAGGGATAACAGGGTAATTGCA-3′, 5′-ATTACCCTGTTATCCCTATGCA-3′, were annealed and cloned into the Acc65I and PstI sites of pw8 (10), to insert two I-SceI recognition sites. The primers 5′-GAGAAAGGATCCAAGCATGCTGCGACGTGAACAGTGAGCTGTA-3′ and 5′-GTTAGAGGATCCCCGCATGCAGCTCGTTACAGTCCGGTGCGTTTTTGGT-3′ were used to add SphI termini to the FLP recombination target (FRT) by PCR. Primers 5′-GTCATAGAATTCACGCACTATGCCGTTCTTCTCATG-3′ and 5′-GAGCATGAATTCGTTTGTGGAAGCGGTATTCGCAA-3′ were used to add EcoRI termini to the FRT by PCR. As template, a plasmid carrying a single copy of the FRT from the yeast 2μ plasmid, with ≈300 bp of 2μ-flanking DNA on each side, was used. The amplified FRTs were cut with SphI and EcoRI respectively and ligated into the SphI and EcoRI sites of the polylinker in the modified pw8. Clones were chosen in which the two FRTs were in the same direction, generating the vector pw30, a P element vector carrying a polylinker flanked by I-SceI sites, and then by FRTs, adjacent to a white+ (w+) gene. pw30 was cut with XhoI, and an 8 kb SalI y+ genomic fragment from pSIG (11) was cloned into that site.
The yellow disruption construct.
Two oligos, 5′-CTCGAGGGTACCGCGGCCGCGCATGCCTGCA-3′, 5′-GGCATGCGCGGCCGCGGTACCCTCGAGTGCA-3′, were annealed and cloned into the PstI site of Carnegie 4 (12). By DNA sequencing, a plasmid was chosen with the new sites inserted as (HindIII)XhoI-Acc65I-NotI-SphI(PstI-SalI), where sites in parentheses were already present in Carnegie 4. Subsequently, two pairs of oligos, 5′-TAGGGATAACAGGGTAAT-3′, 5′-ATTACCCTGTTATCCCTA-3′ and 5′-GTACATTACCCTGTTATCCCTA-3′, 5′-GTACTAGGGATAACAGGGTAAT-3′, were annealed and cloned into the SmaI and Acc65I sites of the polylinker, respectively, generating two I-SceI recognition sites. The PCR was used to add EcoRI termini to the FRT, as above. After digestion with EcoRI, it was ligated into the EcoRI site of the polylinker. Then, the w+ gene of pw6 (10) was removed as a PstI–SphI fragment and cloned into these sites of the polylinker. An XhoI-flanked FRT was produced by using the primers 5′-AGCACTCGAGTGCGACGTGAACAGTGAGCTGTA-3′ and 5′-TACCCTCGAGAGCTCGTTACAGTCCGGTGCGTTTTTGGT-3′ to amplify the FRT. After digestion with XhoI, it was ligated into the XhoI site of the polylinker. A clone whose two FRTs lay in the same direction was chosen, generating the vector pw35, a P element vector that carries a w+ gene flanked by I-SceI recognition sites, and FRTs outside of those. Unique sites of NotI and SphI upstream and BamHI downstream of w+ are available for cloning in pw35. Primers 5′-AGCAGCGGCCGCCGAAAGGGGGATGTGCTGCAAG-3′ and 5′-TACCGCATGCGCACTTAGCTCTAAGCTGACAATC-3′ were used in the PCR with pS/G as template, to add NotI and SphI termini to a 3.05-kb y DNA fragment (from −3,043 to −2 bp upstream of the start codon), which was then cloned into those sites of pw35. Next, the oligos 5′-GATCCACGTACGAGGCGCGCC-3′ and 5′-GATCGGCGCGCCTCGTACGTG-3′ were annealed and cloned into the BamHI site of the polylinker, and selected the plasmid whose BamHI site was next to the XbaI site of the polylinker, generating BamHI-BsiWI-AscI sites in the polylinker. Primers 5′-TACCCGTACGCGTCTTGGGCTGCTTACAAACTTC-3′ and 5′-AGCAGGCGCGCCTATGTTGTGTGGAATTGTGAGCGG-3′ were used to add BsiWI and AscI termini to a 4.77-kb y DNA fragment (including part of the first exon and the intron and second exon) by PCR. This y fragment was cloned into the corresponding sites downstream of w+.
The yellow rescue construct without FRTs.
Two pairs of oligos, 5′-AATTTAGGGATAACAGGGTAAT-3′, 5′-AATTATTACCCTGTTATCCCTA-3′, and 5′-TAGGGATAACAGGGTAATTGCA-3′, 5′-ATTACCCTGTTATCCCTATGCA-3′, were annealed and cloned into the EcoRI and PstI sites of pw8, respectively, destroying the two restriction sites and creating two I-SceI recognition sites. Then, the 8-kb SalI y+ fragment was cloned into the XhoI site of this vector.
Genetics and Heat Shocks.
Crosses for targeting were carried out in standard 25-mm-diameter vials, with three to six females per vial and a corresponding number of the appropriate males. Heat shocks were performed in a circulating water bath as described (13). All constructs were transformed into the germ line of Drosophila melanogaster by using standard methods (14).
Southern Blotting.
For verification of targeting, DNA was prepared from males carrying the targeted allele. For analysis of I-SceI cutting in vivo, genomic DNA was prepared from larvae either 0, 2, 4, 8, or 16 h after heat shock, and from larvae that had not been heat- shocked. The genomic DNAs were digested as indicated, separated by agarose gel electrophoresis, and transferred to nylon membranes. The membranes were probed with a Dig-labeled 8 kb SalI y DNA fragment from pS/G, and hybridization was detected by chemiluminescence using the DIG system (Roche).
Results
Mutation Rescue.
The two enzyme constructs used in this work are the same as those used for ends-in targeting: heat-inducible FLP recombinase (13) and I-SceI endonuclease (3) transgenes (70FLP and 70I-SceI). We also constructed donor transgenes that carry sequence from the locus to be targeted.
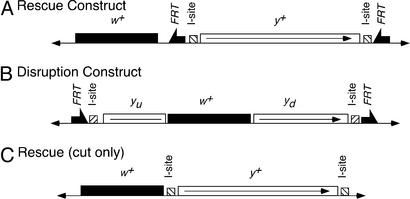
We first made and transformed a y+ P element donor construct to rescue y1, which has a mutation in its first codon. This construct carries a y+ gene flanked by I-SceI recognition sequences and FRTs, adjacent to a w+ gene that lies outside the FRTs (Fig. 2A). For targeting, FLP and I-SceI expression was induced to excise y+ from the chromosome and to generate the DSBs that stimulate HR. We carried out test crosses as diagramed in Fig. 3 and screened for y+ w progeny to recover events that converted the endogenous y1 allele to y+. In other words, we screened for y+ to move from its original location, next to the w+ gene on an autosome, to the X chromosome, and then segregate from w+ in meiosis. In this experiment y+ was efficiently excised from the donor chromosome, with almost all white+ progeny showing loss of y+ (33,668 of 33,790 = 99.6% in 772 vials counted). A total of 43 independent targeting events that converted y1 into y+ were recovered at an average rate of approximately one independent event per 16 vials screened (unweighted; Table 1). Each vial produced ≈52 white-eyed progeny, translating to 1 targeting event in 832 gametes. Four independent nontargeted y+ w events were also recovered. In these, y+ did not map to the X, and they were not examined further. They may have included insertion of y+ at other locations, or loss of w+ from the donor construct after an I-SceI cut, without excision of y+. Targeted recombination events outnumbered nontargeted events by better than 10:1 and were confirmed by Southern blotting (Fig. 4A).
Figure 2.
Constructs for ends-out targeting. Locations of y+ and w+ genes are indicated, along with the FRTs and the I-SceI recognition sequences (I-site). yu and yd indicate the upstream and downstream portions of y. The small arrowheads at the left and right ends of each construct indicate the P element inverted repeat termini.
Figure 3.
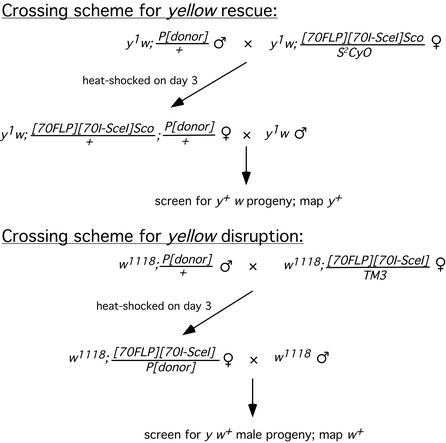
Crossing schemes. Typical crosses used for the rescue and disruption experiments are shown. For the y rescue experiments, females with eye pigment were selected for the second cross, ensuring that they all carried the donor. For the disruption crosses, in some cases the donor element was hemizygous in the males, and so only half of the females used in the second cross carried the donor. The targeting frequency was adjusted accordingly in Table 1. In other cases we selected females exhibiting some degree of pigmentation in the eye, ensuring that they carried the donor; or the males used in the first cross carried the donor heterozygous with a dominantly marked balancer chromosome, thus allowing the selection of females that carried the donor for the second cross.
Table 1.
Recovery of targeting events
| FLP I-SceI | Donor* | n | T | NT | Approximate frequency/vial |
|---|---|---|---|---|---|
| Rescue construct (Fig. 2A) | |||||
| on 2 | 1A (3) | 652 | 22 | 1 | 1/30 |
| on 2 | 2A (2) | 147 | 10 | 2 | 1/15 |
| on 3 | 2A (2) | 135 | 11 | 1 | 1/12 |
| Disruption construct (Fig. 2B) | |||||
| on 3 | 6C† (3) | 147 | 48 | 0 | 1/3 |
| on 3 | 5C† (3) | 79 | 29 | 0 | 1/3 |
| on 2 | 5C (3) | 99 | 14 | 0 | 1/7 |
| on 2 | 4A (3) | 90 | 8 | 0 | 1/11 |
| on 3 | 4A (3) | 84 | 7 | 0 | 1/12 |
| Rescue (Fig. 2C) | |||||
| on 2 | 43 (3) | 131 | 0 | 0 | – |
| on 3 | 43 (3) | 116 | 0 | 0 | – |
| Disruption construct (Fig. 2B, but using I-SceI expression only) | |||||
| on 3 | 6C (3) | 112 | 0 | 0 | – |
| on 3 | 5C (3) | 120 | 0 | 0 | – |
n, number of vials scored for targeting; T, number of vials with targeting events, which is taken to be the minimum estimate of independent targeting events; NT, number of vials with nontargeted events.
The chromosome bearing the donor is indicated in parentheses.
Only half of the tested females carried the donor. The number of vials tested has been multiplied by 0.5 to correct. Each vial contained four to seven pairs of heat-shocked females and males.
Figure 4.
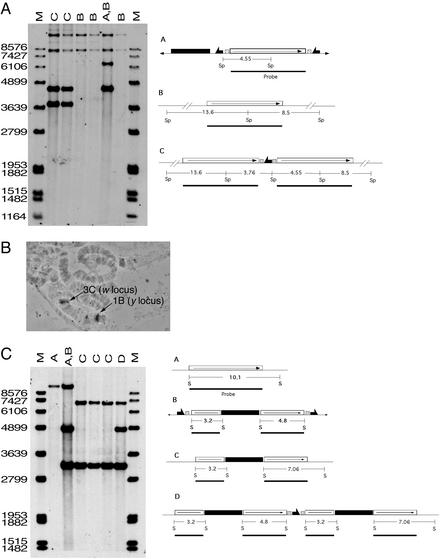
Verification of targeting. (A) Genomic Southern blotting to verify targeted y rescue. Letters above each lane indicate the genotype examined and refer to the expected structures of the donor construct, the y target gene (which is unchanged after the expected single-copy rescue event), and a tandem rescue event indicated to the right. The region used as a probe is indicated as a solid line below each structure, along with the expected sizes of the fragments produced by SphI (Sp) digestion. The left and right lanes (M) carry molecular weight markers with sizes indicated at the left. The first four experimental lanes are targeted alleles; the next two are y w flies carrying the donor, and y w alone. (B) Cytological verification of targeted disruption. The w gene was used to probe polytene chromosomes of a larva carrying a targeted y allele. The two sites of hybridization, and their cytological designations, are indicated. (C) Genomic Southern blotting to verify targeted y disruption. The expected structures of the target locus, the donor construct, a targeted allele, and a tandem-insertion targeted allele are indicated to the right, along with the expected sizes of bands produced by SalI (S) digestion. The first two experimental lanes represent genomic DNA from y+ w1118 flies and from y+ w1118 flies carrying the donor construct. The remainder are examples of targeting.
The majority of targeting events resulted from a straightforward replacement of y1 with y+, but ≈20% (9 of 43) had two copies of the y gene at the target locus. Integration of donor dimers has been seen previously with ends-in targeting in yeast and flies (3, 15, 16), and ends-out targeting in mouse ES cells (4, 17, 18). Concatemer formation between multiple copies of the donor appears to be the cause. The flies used in this experiment carried a single copy of the donor P element (verified by Southern blotting). However, in G2 of the cell cycle, two copies will be present on the replicated chromatids, providing the opportunity for two donors to dimerize by FLP-mediated or homologous recombination, or by nonhomologous end-joining, and undergo targeting.
Gene Disruption.
To extend this method to generate mutant alleles of target genes we constructed and transformed a donor element carrying the y gene disrupted by the insertion of a w+ gene (Fig. 2B). Approximately 50 bp of y coding sequence, including the start codon, were also eliminated in the construction. Targeting with this construct is expected to generate a mutant y allele and carry w+ into the locus.
We screened for disruption of the endogenous y+ gene in a y+ w background by using donors on chromosome 3, and looking for y w+ progeny by test-crossing (Fig. 3). As in the previous experiments, excision of the donor was very efficient (>99%), as judged by loss of w+. We recovered 106 independent targeting events at a rate of ≈1 in 5 vials (unweighted average; Table 1). Each vial produced ≈76 male offspring, or 1 targeting event in 380 gametes. In every case the w+ gene mapped to the X chromosome as expected for targeting at y. Four lines were examined by chromosomal in situ hybridization, and in all four the w gene sequence was detected at its normal location of 3C on the X chromosome, and also at 1B, the normal location of y, confirming that recombination had integrated w+ at the target locus (Fig. 4B). Southern blotting of 89 cases confirmed that all resulted from targeted homologous recombination at y (Fig. 4C). As in the previous experiments, some duplicated alleles were recovered (2 of 89).
In these crosses we screened for disruption of y+ and integration of w+ simultaneously, and recovered only targeted events. But it should be nearly as easy to recover targeting events by screening only for mobilization of a w+ marker. In three previous sets of studies using a variety of target genes (3, 16, 19), and additionally with the y rescue experiments reported above, we have seen that donor integration in females occurs mostly at the target locus. In general, we expect that the majority of events detected by w+ mobilization will be inserted at the target locus.
Does Targeting Require FLP?
Because I-SceI cuts with high efficiency (3), we reasoned that a y+ gene could be readily liberated from the chromosome by two I-SceI cuts, and then act as the donor to convert the endogenous y1 to y+. We built a construct similar to the first rescue construct, but without FRTs (Fig. 2C). A donor insertion on chromosome 3 was used to detect yellow rescue by testcrossing as in the previous experiment (Fig. 3). We observed no targeting events in 247 vials (Table 1). Surprisingly, y w+ flies were very rare among the w+ offspring (10 of 1,154 from 22 vials counted = 0.9% of all w+ progeny), although we expected them to be quite frequent as a result of excision and loss of y+. One possible explanation supposes that repair of the cut chromosomal ends is very inefficient, and although targeting may have occurred, those cells died because of a failure to fix the chromosomal DSB at the donor site. Alternatively, repair of a DSB generated by I-SceI may be extremely efficient so that it is rare for both I-SceI sites to be cut at the same time, which is necessary to generate the extrachromosomal donor.
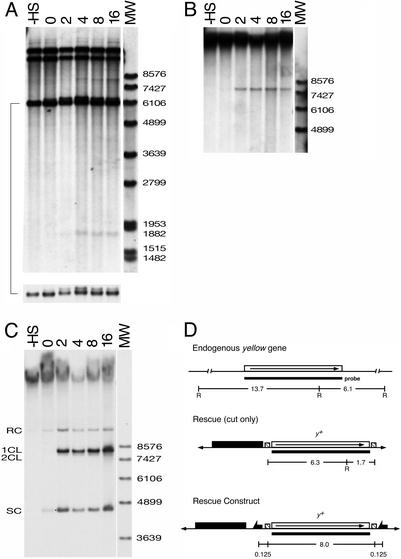
To distinguish these possibilities we carried out a physical analysis of cutting at the I-SceI sites. Second and third instar larvae were heat shocked at 38°C for 1 h, and genomic DNA was prepared from samples of these larvae at various times after the heat shock. We digested the genomic DNA with EcoRI (which cuts once within y), and after electrophoretic separation and Southern blotting, probed with the y gene clone to assess the frequency of DSBs within the donor element by scanning of the exposed film (Fig. 5A). A small fraction of the donor (≈5%) was observed as an extrachromosomal 8-kb band; it must have been circular to generate this full-length band after EcoRI digestion and it is unlikely that it would participate in targeting. It was probably produced by double-cutting followed by repair that joined the two ends. We also estimated the approximate proportion of donor with a DSB at each site. A cut at the left site will generate a 6.3-kb fragment; a cut at the right site will generate a 1.7-kb fragment. At the peak of cutting, about 4 h after heat shock, ≈30% of the donor exhibited a DSB at the left site and ≈5% at the right site. This predicts that only 1–2% of donor elements have two DSBs at the same time. To confirm this we carried out blotting and hybridization with undigested genomic DNA from the same experiment (Fig. 5B). Only ≈7% of the donor is found in the extrachromosomal linear form 8 h after heat shock. It is not the case that the extrachromosomal donor was produced and degraded because we saw no significant reduction in the quantity of donor hybridizing material during the course of the experiment. Thus, the extrachromosomal linear donor element was infrequently produced by I-SceI cutting alone, and at its peak only 2–7% of the donor element existed in this form.
Figure 5.
Efficiency of donor generation. Larvae carrying donor constructs and 70I-SceI (A and B) or 70FLP and 70I-SceI (C) were heat shocked at 38°C for 1 h. Genomic DNA was prepared at various times after heat shock (indicated in hours above the lanes). This experiment was carried out with a mixture of male and female larvae that were heterozygous for the donor element: 60% of the total yellow signal derives from the endogenous yellow gene; the remaining 40% comes from the donor. (A) Assay of DSBs at the I-SceI sites in the cut-only rescue construct (Fig. 2C). Genomic DNA was digested with EcoRI. The 13.7-kb band (the largest band on the gel) and the 6.1-kb band are produced by the endogenous y gene. The band at ≈12 kb is one of the chromosomal donor bands generated by EcoRI digestion (the other is not visible on this gel). The 6.3-/6.1-kb doublet is shown again at a reduced exposure at the bottom. (B) Genomic DNA from larvae carrying 70I-SceI and the cut-only construct, blotted without restriction digestion. The film was overexposed to visualize the 8.0-kb linear donor freed by I-SceI digestion. (C) Genomic DNA from larvae carrying 70FLP and 70I-SceI and the y rescue construct with FRTs (Fig. 2A), blotted without restriction digestion. RC, relaxed circle; 1CL, single-cut linear; 2CL, double-cut linear; SC, supercoiled circle. (D) The structures of the endogenous y gene and the two donor constructs. All blots were probed with the 8-kb y probe indicated by the solid bar beneath the y gene. In the rescue construct, a single FRT adds ≈250 bp. R, EcoRI; -HS, not heat-shocked; MW, molecular weight markers.
If the failure to obtain targeting with the cut-only construct resulted only because this particular donor is inefficiently cut relative to constructs with successful targeting, it seems reasonable to expect that targeting could succeed by using I-SceI, and not FLP, with the y+ disruption construct (Fig. 2B). Accordingly, we tested donors 5C and 6C of the disruption construct for targeting by using I-SceI expression alone. No targeting events were obtained in 232 vials (Table 1), though we would have expected >60 if targeting occurred as efficiently as it did when FLP was also used with these same donors. It is most probable that targeting fails when using I-SceI alone because the extrachromosomal donor is not produced efficiently without FLP.
To test this more directly, we examined the generation of extrachromosomal donor after expression of FLP and I-SceI in larvae carrying the first yellow rescue construct (Fig. 2A). By Southern blotting of genomic DNA prepared without further digestion, it is seen that the extrachromosomal form of the donor appears rapidly: within 2 h the majority of donor is found out of the chromosome (Fig. 5C and other results not shown) in four bands. These probably represent relaxed circles, single and double cut linear forms, and supercoiled circles. At all time points, from 2 h after heat shock and beyond, more than 50% of the extrachromosomal donor is found in the linear form. The extrachromosomal donor molecules, including the linear forms, are generated much more efficiently by FLP and I-SceI than by I-SceI alone. The most likely explanation for failed targeting with I-SceI expression alone is that I-SceI-generated breaks in the chromosome are rapidly and efficiently repaired, and that there is only a small chance of having both sites cut simultaneously to free the donor.
Our results do not rule out the possibility that targeting can occur by I-SceI cutting only, as there was a small fraction of freed donor detected. But if it can occur, it is clearly a much less efficient process than when FLP is used to excise the donor from its chromosomal site.
Discussion
These results demonstrate that ends-out gene targeting can be used efficiently for gene knockouts in D. melanogaster. In this work we used the same stretch of y-homologous DNA that we had previously used for ends-in targeting of y, and the efficiency was not vastly different from that previously obtained. In females, ends-in targeting of yellow occurred at a rate of ≈1 independent event in four to five vials, or ≈1 in 500 gametes (3). In this work we also observed targeting at a rate of ≈1 in 5 vials, or ≈1 in 380 gametes with the ends-out disruption construct, and at a rate of ≈1 in 16 vials, or ≈1 in 800 gametes, for the ends-out rescue construct. These sets of experiments were not controlled in such a way so as to allow a rigorous comparison of frequencies with the previous work, but they do clearly demonstrate that ends-out gene targeting occurs at a usable frequency in Drosophila. Because we and others (16, 20) have shown that ends-in targeting can be used to modify loci throughout the genome, we expect that ends-out targeting will be equally useful.
In a previous attempt at ends-out targeting, Bellaiche et al. (9) failed to recover gene targeting events when screening for ends-out disruption of w+. A consideration of that failure may provide useful insight into constraints on the use of gene targeting in Drosophila. In our estimation, the most significant difference is that Bellaiche and colleagues chose to drive FLP and I-SceI expression with the β2-tubulin promoter, a male germ line-specific promoter that drives transcription in primary spermatocytes (21). Our previous experiments have all produced more efficient targeting in females than in males (3, 16, 19), and this may be partly responsible. When we did recover targeting events from males, those events must have occurred in mitotic, not meiotic cells, because the heat shock promoter that we used is limited in its activity to the earliest stages of spermatogenesis (22, 23). In contrast, β2-tubulin-promoted expression of FLP appears to be predominantly postmeiotic (24), and postmeiotic spermatids may be a very unfavorable environment for targeting.
Bellaiche and coworkers also had a large nonhomologous stretch of DNA at one end of the donor DNA, whereas we did not. But this does not pose a significant impediment to targeting efficiency in yeast or in mouse ES cells (refs. 1 and 8, and K. Thomas, personal communication). Thus we favor the idea that expression of the recombinase and endonuclease in late spermatogenesis was mainly responsible for their failure to obtain targeting.
In this work the phenotype of the target gene was known, but in most cases it will not be. Targeting can then be detected in testcrosses by the mobilization of a marker gene that is inserted within the target-homologous sequences (as with the construct of Fig. 2B). This also has the virtue that the mutant allele that is produced is distinguished by insertion of a marker gene (Fig. 1A). This can be highly desirable for tracking the mutant allele in crosses, but may cause undesired side-effects on the regulation of the target locus or its neighbors. One solution to this problem has been to apply site-specific recombination to remove the marker gene from the chromosome (25–28). The Cre-lox recombinase system may be used at this step (29), and we have constructed a Drosophila targeting vector carrying loxP sites for this purpose (unpublished data).
The use of ends-out targeting, also referred to as replacement or substitution type targeting, provides significant new capabilities for Drosophila genome modification. It gives a very direct route to the generation of a mutant allele, in a single step the target gene can be disrupted by insertion of a marker gene within its coding region. The cloning steps are relatively simple because it is not necessary to engineer point mutations within the coding sequence of the gene, as is typically the case with ends-in targeting (although not always: see refs. 1 and 19). Constructing donors to make deletions should also be straightforward: segments of DNA that flank, but do not include, the target gene are placed to the left and right of the marker gene in the donor construct (1, 2, 30, 31). Homologous recombination then replaces the target gene with the marker gene.
One of the reasons that ends-out targeting is preferred in mouse ES cells is that it allows the application of positive–negative selection to enrich for targeted recombinants (32). In addition to the positively selectable marker gene within the target-homologous region (w+ in our scheme) a negatively selectable marker gene is located outside the target-homologous DNA. Random integration tends to incorporate both genes, but homologous recombination excludes the negative marker. Applying both selections simultaneously greatly enriches for the targeted events. Because, in our experience, most donor insertion events in Drosophila are targeted, this is not needed. However a positive-negative screening method could be used to facilitate recovery of targeting events in Drosophila. One marker gene, inserted in the targeting DNA, could be used to track mobilization of that segment, whereas a second marker gene, located outside the FRTs, would be left behind after FLP-mediated excision and would mark the donor chromosome. Segregation of these two markers would provide an easy screen to recover events in which donor DNA has transposed. Most of these would be legitimate targeting events. This is similar to the method we used in the yellow rescue experiment, where we screened for mobilization of y+, detected by its segregation from w+.
There are yet cases where ends-in targeting may be preferred. Ends-in procedures can be carried out so that all exogenous DNA is eliminated from the altered locus. After a tandem duplication is generated by the initial targeting event (Fig. 1A), typically accompanied by the introduction of an engineered mutation, a site-specific DSB is generated between the duplicated regions. Repair of the DSB often occurs by a mechanism that reduces the duplication to a single copy of the target gene and eliminates the intervening marker gene. Some of these reduction events carry the introduced mutation. Introducing specific missense changes without alteration of the surrounding DNA sequence is easily achieved with this allelic substitution procedure (16). Such precise changes may be useful for examining the function of proteins with specific amino acid substitutions, or for knocking out a specific member of a family of alternatively spliced transcripts without disturbing normal transcriptional regulation. There are also examples in Drosophila where one gene lies within the intron of another gene. In such cases it may be difficult to employ ends-out targeting to generate a null allele of one gene without impinging on the regulation of the second gene.
In sum, the choice between an ends-in or ends-out targeting protocol must be made on a case-by-case basis, with the investigator taking into account variables such as the local genetic structure, the desired type of mutant allele, the crosses used to analyze those mutants, and the time required for different types of targeting. This demonstration of ends-out gene targeting in flies provides Drosophila investigators with that choice.
Acknowledgments
We thank Yikang Rong, Simon Titen, Mary Golic, and Keith Maggert for help and advice during this work. This work was supported by National Institutes of Health Grants GM60700 and GM65604 and by the Stowers Institute for Medical Research.
Abbreviations
- DSB
double-strand break
- FRT
FLP recombination target
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rothstein R. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 2.Muller U. Mech Dev. 1999;82:3–21. doi: 10.1016/s0925-4773(99)00021-0. [DOI] [PubMed] [Google Scholar]
- 3.Rong Y S, Golic K G. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 4.Hasty P, Rivera-Pérez J, Chang C, Bradley A. Mol Cell Biol. 1991;11:4509–4517. doi: 10.1128/mcb.11.9.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hastings P J, McGill C, Shafer B, Strathern J N. Genetics. 1993;135:973–980. doi: 10.1093/genetics/135.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung W-Y, Malkova A, Haber J E. Genetics. 1997;94:6851–6856. doi: 10.1073/pnas.94.13.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas K R, Capecchi M R. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 8.Deng C, Capecchi M R. Mol Cell Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellaiche Y, Mogila V, Perrimon N. Genetics. 1999;152:1037–1044. doi: 10.1093/genetics/152.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klemenz R, Weber U, Gehring W J. Nucleic Acids Res. 1987;15:3947–3959. doi: 10.1093/nar/15.10.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geyer P K, Corces V G. Genes Dev. 1987;1:996–1004. doi: 10.1101/gad.1.9.996. [DOI] [PubMed] [Google Scholar]
- 12.Rubin G M, Spradling A C. Nucleic Acids Res. 1983;11:6341–6351. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golic K G, Lindquist S. Cell. 1989;59:499–509. doi: 10.1016/0092-8674(89)90033-0. [DOI] [PubMed] [Google Scholar]
- 14.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 15.Orr-Weaver T, Szostak J W, Rothstein R J. Proc Natl Acad Sci USA. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rong Y S, Titen S W, Xie H B, Golic M M, Bastiani M, Bandyopahdyay P, Olivera B M, Brodsky M, Rubin G M, Golic K G. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scwartzberg P L, Robertson E, Goff S P. Proc Natl Acad Sci USA. 1990;87:3210–3214. doi: 10.1073/pnas.87.8.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas K R, Deng C, Capecchi M R. Mol Cell Biol. 1992;12:2919–2923. doi: 10.1128/mcb.12.7.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rong Y S, Golic K G. Genetics. 2001;157:1307–1312. doi: 10.1093/genetics/157.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seum C, Pauli D, Delattre M, Jaquet Y, Spierer A, Spierer P. Genetics. 2002;161:1125–1136. doi: 10.1093/genetics/161.3.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michiels F, Gasch A, Kaltschmidt B, Renkawitz-Pohl R. EMBO J. 1989;8:1559–1565. doi: 10.1002/j.1460-2075.1989.tb03540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonner J J, Parks C, Parker-Thornburg J, Mortin M A, Pelham H R B. Cell. 1984;37:979–991. doi: 10.1016/0092-8674(84)90432-x. [DOI] [PubMed] [Google Scholar]
- 23.Golic M M, Golic K G. Genetics. 1996;143:385–400. doi: 10.1093/genetics/143.1.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golic M M, Rong Y S, Petersen R B, Lindquist S L, Golic K G. Nucleic Acids Res. 1997;25:3665–3671. doi: 10.1093/nar/25.18.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dale E C, Ow D W. Proc Natl Acad Sci USA. 1991;88:10558–10562. doi: 10.1073/pnas.88.23.10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu H, Marth J D, Orban P C, Mossman H, Rajewsky K. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 27.Meyers E N, Lewandowski M, Martin G R. Nat Genet. 1998;18:136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 28.Bunting M, Bernstein K E, Greer J M, Capecchi M R, Thomas K R. Genes Dev. 1999;13:1524–1528. doi: 10.1101/gad.13.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siegal M L, Hartl D L. Genetics. 1996;144:715–726. doi: 10.1093/genetics/144.2.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Surosky R, Tye B-K. Proc Natl Acad Sci USA. 1985;82:2106–2110. doi: 10.1073/pnas.82.7.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matzuk M M, Finegold M J, Su J G, Hsueh A J, Bradley A. Nature. 1992;360:313–319. doi: 10.1038/360313a0. [DOI] [PubMed] [Google Scholar]
- 32.Mansour S L, Thomas K R, Capecchi M R. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]