Abstract
Granzyme B (GzmB) is a serine protease involved in many pathologies, including viral infections, autoimmunity, transplant rejection, and antitumor immunity. To measure the extent of genetic variation in GzmB, we screened the GzmB gene for polymorphisms and defined a frequently represented triple-mutated GzmB allele. In this variant, three amino acids of the mature protein Q48P88Y245 are mutated to R48A88H245. In CD8+ cytotoxic T lymphocytes, GzmB was expressed at similar levels in QPY homozygous, QPY/RAH heterozygous, and RAH homozygous individuals, demonstrating that RAH GzmB is a stable protein. Active RAH GzmB expressed in glioblastoma cell lines displayed proteolytic activity, but in contrast to QPY GzmB, it did not accumulate in the nucleus and was unable to induce Bid cleavage, cytochrome c release, or apoptosis. Molecular modeling showed that the three amino acid substitutions clustered near the C-terminal α-helix of the protein, indicating that this region of the protein may be involved in the intracellular targeting of GzmB. The triple-mutated GzmB allele that we describe appears to be incapable of inducing apoptosis in tumor cell lines, and its presence could, therefore, influence both the prognosis of cancer patients and the success rates of antitumor cellular immunotherapy.
Granzyme B (GzmB) is a protease of the chymotrypsin family expressed by differentiated cytotoxic T lymphocytes (CTL; ref. 1), as well as certain other cell types, such as activated keratinocytes (2). In cytotoxic cells, GzmB is a component of cytolytic granules (3). It is released upon target cell recognition, then specifically endocytosed by target cells via the cation independent mannose 6-phosphate receptor (4). In the presence of perforin (5), which is also released from cytolytic granules (6), GzmB escapes from the endolysosomal compartment and gains access to a number of key proteins involved in the execution of the apoptotic program.
GzmB has a substrate specificity unique among serine proteases. It cleaves after aspartate residues, and experiments combining synthetic peptide and phage libraries have defined the extended specificity of GzmB as IEPD/xG (7). This hexapeptide is remarkably similar to the optimal substrate for caspase-8, an initiator caspase at the apex of the apoptotic signaling cascade activated by the ligation of the Fas death receptor, and this similarity explains the proapoptotic properties of GzmB. In particular, cleavage of Bid by GzmB activates the mitochondrial pathway of programmed cell death (8, 9), cleavage of caspase-3 directly generates cytoplasmic caspase activity (10), and caspase-activated DNase (CAD) is released by GzmB cleavage of the inhibitor protein, ICAD, leading to nuclear DNA fragmentation (11, 12).
Like other cytotoxic granule contents, GzmB is involved in several pathologies including viral infections (13), graft rejection, graft-vs.-host disease (14–17), and rheumatoid arthritis (18, 19). GzmB is also likely to play an important role in antitumor immune responses. The perforin/GzmB pathway efficiently induces tumor cell death in vitro and is also able to induce apoptosis in multiple-drug-resistant and death-receptor-resistant cell lines (20, 21). However, no congenital human diseases have been linked to the disruption of the GzmB gene, and GzmB knockout mice have no developmental or hematological abnormalities (22). Therefore, we hypothesized that, in contrast to mutations in the perforin gene (23), coding polymorphisms in the GzmB gene might be relatively well tolerated in humans and, if present, could influence the killing of tumor cells.
To investigate this hypothesis, we screened the GzmB gene for single-nucleotide polymorphisms (SNPs) and thereby identified a common GzmB allele coding for a triple-mutated protein, the structure of which was investigated by molecular modeling. The expression of GzmB and the cytolytic activity in CTL from individuals homozygous and heterozygous for the variant allele was investigated. The enzymatic activity and proapoptotic functions of variant GzmB were tested in transfected glioblastoma lines, which revealed striking functional differences between the two alleles.
Materials and Methods
Patient Samples.
HIV+ patient DNA samples were obtained from the French Immunoco cohort, and control DNA samples were from healthy blood donors. DNA samples from West African and Vietnamese individuals were from HIV seropositive and seronegative donors. All DNA samples were obtained from individuals enrolled in clinical studies approved by the relevant local or national ethical review board. DNA was extracted from peripheral blood mononuclear cell (PBMC) by using QIAamp mini extraction kits (Qiagen, Valencia, CA).
Resected tumor tissue was obtained from adult glioblastoma patients undergoing surgery at the Department of Neurosurgery, Laennec Hospital (Nantes, France). Primary glioblastoma lines were derived from tumor samples as described (24).
PCR and Mutation Detection.
A 4.0-kb PCR product encompassing exons 1–5 of the GzmB gene was amplified using the ELT PCR kit (Roche Molecular Biochemicals). This PCR product was then used as a template for amplification of 200– to 450-bp segments encompassing each exon of the gene. PCR products were dehybridized/rehybridized, and heteroduplex formation was detected by denaturing HPLC (dHPLC) by using a Transgenomic Wave instrument. Mutations were identified by sequencing of PCR products from heterozygous individuals. Linkage disequilibrium was confirmed by sequencing of long PCR products cloned into the TA cloning vector (Invitrogen).
Individuals heterozygous for SNPs in exon 2 (48Q = >R) and exon 3 (88P = >A) and exon 5 (245Y = >H) of the gene were typed by dHPLC. Individuals homozygous for mutations in these exons were identified by hybridizing PCR products with a standard wild-type PCR product and then typed by dHPLC. The C3057A SNP in intron 3 was detected by NgoM IV restriction fragment-length polymorphism (RFLP).
Flow Cytometry.
PBMCs were thawed and then stained for surface CD8 and intracellular GzmB either immediately or after 4 days culture in RPMI medium 1640 containing 10% (vol/vol) FCS (Invitrogen), 1% (vol/vol) MEM nonessential amino acids (Invitrogen), 1 mM sodium pyruvate (Invitrogen), 2 mM l-glutamine (Invitrogen), 100 units/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B, and 1 μg/ml PHA-P (Sigma). Cells were surface-labeled with anti-CD8 APC (PharMingen) and anti-CD56 PE-Cy5 (Beckman Coulter), washed, and then permeabilized by using the Fix and Perm kit (Caltag, South San Francisco, CA). During permeabilization, cells were incubated with 10 μg/ml anti-GzmB PE (clone GB12, Caltag) or 10 μg/ml IgG1 PE isotype control. Cells were analyzed on a FACSCalibur (Becton Dickinson) instrument, and GzmB staining was quantified by using Quantibrite-PE fluorescent beads (PharMingen).
Cytotoxicity Assays.
HIV-specific CTL lines were generated from PBMC of HIV-seropositive patients enrolled in the long-term asymptomatic (ALT) cohort as described (25). Briefly, PHA-stimulated PBMC were irradiated and then cocultured with unstimulated PBMC at a ratio of 1:5. Exogenous IL-2 was added every 3 days, and CTL activity was tested in a 51Cr release assay after 15–20 days of culture. Autologous Epstein–Barr virus (EBV)-immortalized B-lymphoma lines infected with different recombinant vaccinia viruses (Transgene, Strasbourg, France) were used as targets. CTL responses against HIV antigens were considered positive if they exceeded the mean of wild-type vaccinia-specific lysis by 3 SDs and by 10%.
Expression of Active Recombinant QPY and RAH GzmB in Glioblastoma Lines.
Total RNA was extracted from the PBMC of a heterozygous individual, full-length GzmB cDNA was amplified by using Pfu polymerase, and then ligated into the TA cloning vector (Invitrogen). Clones coding for QPY and RAH GzmB were selected, and the inserts subcloned into pAlter-Max (Promega). The six nucleotides coding for the two amino acid precursor peptide were deleted by site-directed mutagenesis to produce the constructs pAlt-GzmBWtΔGE and pAlt-GzmBMutΔGE.
These plasmids were transfected into primary glioblastoma lines which had been cultured for 7 to 15 passages in RPMI medium 1640 containing 10% (vol/vol) FCS, 2 mM l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and G-5 supplement (Invitrogen). A total of 5 × 105 cells were electroporated at 200 V and 250 μF with 5 μg of plasmid, then cultured for 32 h before analysis. GzmB expression was visualized by confocal microscopy using the 2C5 monoclonal (Chemicon), followed by Alexa 568 coupled antimouse IgG (Molecular Probes). Nuclei were counterstained with TOPRO1 (Molecular Probes), and images were acquired on a Leica DM IRBE confocal microscope.
Western Blotting.
Crude cell extracts were prepared by Nonidet P-40 lysis followed by centrifugation for 15 min at 12,000 × g and recovery of supernatants. Mitochondria were prepared as described (26).
Proteins were separated on 15% polyacrylamide gels and then transferred onto a poly(vinylidene difluoride) membrane with a semidry transfer system (Bio-Rad). Membranes were blocked in PBS 0.05% Tween 20 containing 10% (vol/vol) skim milk and then incubated with the following mouse monoclonal antibodies: anti-human GzmB (clone 2C5, Santa Cruz Biotechnology); anti-actin (Chemicon); anti-cytochrome c (R & D Systems); anti-cytochrome oxidase subunit IV (Molecular Probes), anti-ICAD/DFF-45 (Santa Cruz Biotechnology), anti-caspase-3 (PharMingen), anti-caspase-8 (PharMingen) or a polyclonal rabbit antibody against human Bid (kindly provided by J. C. Martinou, University of Geneva, Geneva). Bands were visualized by chemiluminescence (Amersham Pharmacia) after incubation with the appropriate peroxidase-conjugated secondary antibodies.
Cell Death and GzmB Activity Assays.
Cell death was quantified by using the Cytotox 96 NonRadioactive Cytotoxicity Assay kit (Promega). Caspase 3 (DEVDase) activity in Nonidet P-40 lysates was measured by using the Caspace Assay kit (Promega) containing the fluorescent substrate DEVD-7-amino-4-methyl coumarin (AMC). GzmB activity in cell lysates was measured by monitoring cleavage of IETD-AMC. In some experiments, caspase activity was blocked by the addition of 1 mM iodoacetamide, 50 μM zVAD-fmk (Bachem), or 100 μM CHO-IETD (Bachem). Experiments were carried out in triplicate in a 96-well format. Plates were incubated at 37°C, and fluorescence emission was measured at 15-min intervals for 90 min by using a Fluorolite 1000 fluorimeter (Dynatech). Specific activity was calculated from the gradients of the curves obtained, which remained linear during the assay period, and expressed as fluorescence units per hour per microgram of protein.
Molecular Modeling.
The coordinates of the triple mutant were constructed by using the SMD procedure (27), starting from the structure of the wild type, determined by x-ray crystallography at a resolution of 3.1 Å (PDB ID code: 1fq3). Figures were prepared by using PROTEIN EXPLORER software (www.umass.edu/microbio/chime/explorer/).
Results
Identification of a GzmB Allele Encoding Three Amino Acid Substitutions.
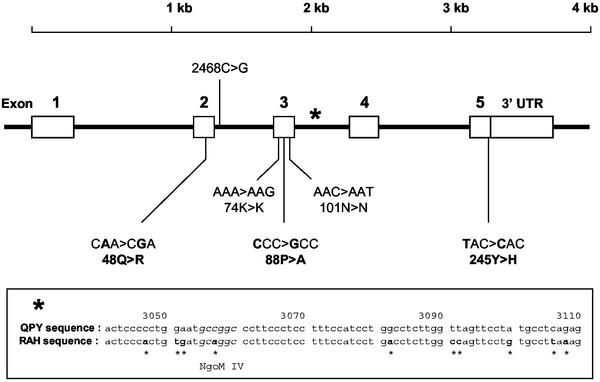
To identify genetic polymorphisms in GzmB, we screened DNA extracted from 48 individuals enrolled in the IMMUNOCO cohort for mutations in the coding sequences of the GzmB gene by PCR and dHPLC of amplification products, followed by sequencing to identify mutations. Three coding mutations were detected: in exon 2, an A→G substitution resulted in the mutation of glutamine (CAA) 48 (numbering with reference to the bovine chymotrypsinogen A sequence) to arginine (CGA); in exon 3, a C→G substitution changed proline 88 (CCC) to alanine (GCC); and in exon 5, a T→C substitution altered tyrosine 245 (TAC), the last amino acid in the protein, to histidine (CAC). Two noncoding SNPs in exon 3, a SNP in intron 2, and multiple substitutions in intron 3 were also detected (Fig. 1). One of the substitutions in intron 3 removed an NgoM IV restriction site, which facilitated typing of this mutation by RFLP.
Figure 1.
SNPs in the coding region of the GzmB gene. Nucleotide positions are numbered according to GenBank accession no. M28879. Amino acid positions are numbered according to the bovine chymotrypsinogen A sequence. Nucleotide positions of the SNPs located in exons are as follows: 2364A>G (48Q>R); 2893A>G (74K>K); 2933C>G (88P>A); 2974C>T (101N>N); 4243T>C (245Y>H). (Inset) *, The presence of multiple substitutions in intron 3.
Two types of data indicated that all of these mutations were in strong linkage disequilibrium. Firstly, individuals heterozygous for the Tyr245→His (Y245H) substitution were almost always heterozygous for the Q48R and P88A mutations, and H245 homozygotes were also R48 and A88 homozygotes. Secondly, cloning and sequencing of long PCR products spanning exons 1–5 of the GzmB gene from an individual heterozygous at all three positions allowed us to confirm that one chromosome coded for Q48 P88 Y245, whereas the other coded for R48 A88 H245. These three coding SNPs, therefore, define an allele of the GzmB gene in which three amino acids of the mature protein Q48P88Y245 are mutated to R48A88H245.
To investigate the geographic distribution of GzmB alleles, Caucasian, West African, and Vietnamese donors were genotyped for the Q48R, P88A, and Y245H SNPs, as well as the noncoding SNPs in exon 3 and introns 2 and 3. In a group of 90 Caucasians, the QPY allele was found at a frequency of 71.1%, and the RAH allele was found at a frequency of 25.5%. Other alleles were found at frequencies of less than 2% (Table 1). However, DNA from West African individuals presented more diverse haplotypes. The QPY allele was still the most frequent (53.6%), followed by RAH (27.3%), but the RAY allele was also frequently represented (9.1%), as was RPY (7.3%). In Vietnamese donors, QPY (65.7%) was again the dominant allele, followed by RAH (29.6%) and RPY (2.8%). Other alleles were found at frequencies of less than 1%. The global distribution of the RAH allele suggests that it is extremely ancient, probably originating before the expansion of modern humans from the African continent 50,000–100,000 years ago.
Table 1.
Global distribution of GzmB alleles
| Allele | Allelic frequency by geographical location
|
||
|---|---|---|---|
| France | Vietnam | West Africa | |
| QPY | 0,711 | 0,657 | 0,536 |
| QPH | 0,011 | 0,009 | 0,018 |
| QAH | 0,006 | 0,009 | 0,009 |
| RPY | ND | 0,028 | 0,073 |
| RAY | 0,017 | ND | 0,091 |
| RAH | 0,251 | 0,296 | 0,273 |
Individuals from France (n = 95, 190 chromosomes), Vietnam (n = 54, 108 chromosomes), and West Africa (n = 61, 122 chromosomes) were genotyped for the 48Q→R, 88P→A, and 245Y→H mutations by dHPLC. Individual genotypes were assembled into allele combinations by maximum parsimony. For example, an individual with the genotype 48Q/Q, 88P/A, 245Y/H was judged to carry one wild-type QPY allele and one rare QPH allele, rather than two rare alleles (QPH and QAR).
Expression of GzmB and Lytic Activity in CTL from Individuals with Different GzmB Genotypes.
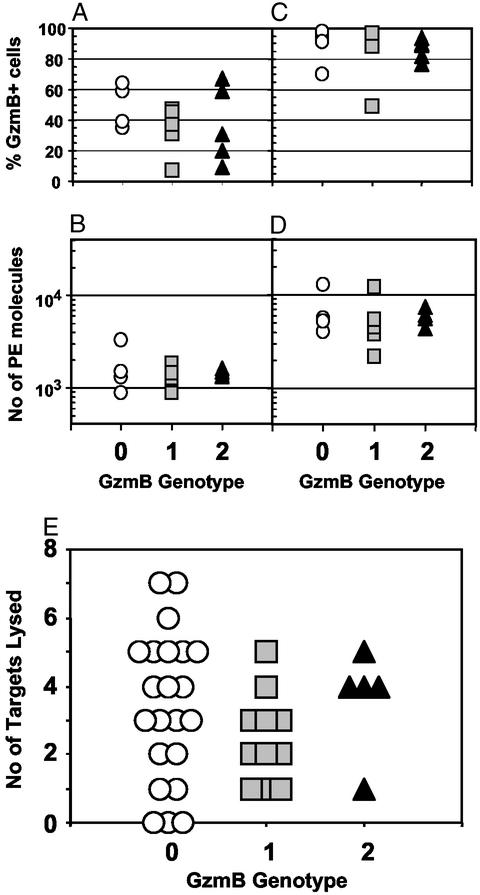
GzmB is strongly expressed by differentiated CTL. To test whether GzmB genotype affected protein expression, GzmB in PBMC from HIV+ patients enrolled in the ALT cohort was measured by flow cytometry. These individuals had not received any antiretroviral treatment at the time the PBMC sample was taken, and it was expected that relatively large numbers of differentiated CD8+ CTL would be present in these samples because of the ongoing anti-HIV immune response. Cells were stained for GzmB and CD8 directly after thawing and after a 4-day culture in the presence of 1 μg/ml PHA-P to investigate GzmB expression in cells stimulated in vivo and in vitro. After thawing, the percentage of CD8+ lymphocytes expressing GzmB was highly variable (median = 37%; range = 7–64%), with no correlation between genotype and the percentage of GzmB+ CD8+ cells (Fig. 2A). The amount of GzmB expressed was more stable (median number of PE molecules bound = 1,365; range = 885–3,286) and showed no relationship to genotype (Fig. 2B). After culture in the presence of PHA, GzmB expression was up-regulated, with both the percentage of GzmB+ cells (median = 91%; range = 49–98%) and the amount of GzmB expressed per cell (median = 5,385; range = 2,186–12,868) significantly increased (Fig. 2 C and D). Once again, no difference was observed between genotypes.
Figure 2.
Expression of GzmB and CTL activity by GzmB genotype. PBMC from untreated long-term asymptomatic HIV+ patients enrolled in the ALT cohort were thawed, and GzmB expression in CD8+ lymphocytes was measured by intracellular fluorescence-activated cell sorter staining either directly (A and B) or after 4 days culture in the presence of 1 μg/ml PHA-P (C and D). Results are shown as the percentage of CD8+ lymphocytes staining positive for GzmB (A and C), and as the number of PE molecules bound per cell in the GzmB-positive population (B and D). Each symbol represents one patient. Anti-HIV CTL activity in cell lines derived from PBMC of HIV+ patients enrolled in the ALT cohort was tested against autologous EBV-immortalized B-lymphoma lines infected with recombinant vaccinia viruses expressing HIV gag, pol, env, nef, rev, tat, or vif proteins. For each HIV protein, the results of a 51Cr release assay were noted as positive or negative, and the number of positive results were scored for each patient (E). Each symbol represents one patient. GzmB genotypes are as follows: 0, QPY/QPY; 1, QPY/RAH; 2, RAH/RAH.
The HIV-specific lytic activity of CTL lines from 38 patients enrolled in the ALT cohort (22 QPY/QPY, 11 QPY/RAH, 5 RAH/RAH) was also tested. Seven 51Cr release assays were performed for each individual by using autologous EBV-immortalized B-lymphoma lines expressing HIV gag, pol, env, nef, rev, tat, or vif proteins as targets. There was a tendency toward less lysis in CTL from QPY/RAH heterozygous individuals, but this was not confirmed in patients homozygous for the RAH allele (Fig. 2E). CTL expressing RAH GzmB were, therefore, able to kill target cells.
Proteolytic and Apoptosis-Inducing Activity of QPY and RAH GzmB Proteins.
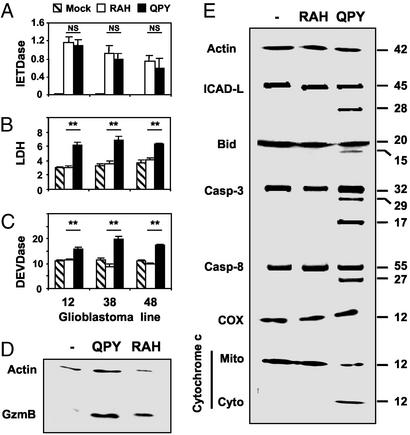
To study the functions of the RAH allele in more detail, we subcloned full-length QPY and RAH GzmB cDNAs from a heterozygous donor. The two amino acid precursor peptide was deleted by site-directed mutagenesis so that active GzmB could be produced in the absence of dipeptidyl peptidase I. These constructions were used to transfect primary glioblastoma cell lines which showed no endogenous expression of GzmB, but did express Bax and Bak and were, therefore, susceptible to GzmB-induced apoptosis. Similar levels of GzmB were expressed by cells transfected with RAH and QPY GzmB (Fig. 3D), and both RAH and QPY proteins displayed proteolytic activity, shown by cleavage of the fluorescent substrate, IETD-AMC (Fig. 3A). This activity was observed in the presence of iodoacetamide, an inhibitor of all cysteine proteases, and the broad spectrum caspase inhibitors zVAD and CHO-IETD (data not shown), indicating that caspases were not responsible for the cleavage of IETD-AMC. However, despite similar expression levels and enzymatic activity, apoptosis induction differed greatly between RAH and QPY GzmB. As expected, QPY GzmB induced apoptosis, characterized by the appearance of DEVDase activity in cell lysates and the release of LDH from dying cells (Fig. 3 B and C). In contrast, RAH GzmB did not induce DEVDase activity or cell lysis above that observed in control transfected cells. These results were observed in three independent cell lines derived from different patients. GzmB is known to induce apoptosis in target cells by the cleavage of Bid, which leads to the release of cytochrome c into the cytosol; however, cells expressing active RAH GzmB displayed neither of these hallmarks of mitochondrial apoptosis (Fig. 3E). Other GzmB substrates relevant for the induction of apoptosis, caspase3, caspase8, and ICAD-L, were also cleaved in cells transfected with QPY GzmB but remained intact in cells transfected with RAH GzmB (Fig. 3E).
Figure 3.
Proteolytic activity and apoptosis induction by QPY and RAH GzmB. Three independently derived glioblastoma lines (numbered 12, 38, and 48) were transfected with plasmids coding for QPY or RAH GzmB, or mock- transfected with pCDNA3, as indicated. (A) IETDase activity measured in crude lysates of transfected cells in the presence of 1 mM iodoacetamide. (B) DEVDase activity in lysates of transfected cells. (C) LDH activity in supernatants of transfected cells. Enzyme activities are expressed as fluorescence units per hour per microgram of protein. NS, no significant difference in enzymatic activity. **, P < 0.005 (Student's unpaired t test). (D) Western blot detection of GzmB in cells transfected with QPY and RAH GzmB, but not in mock-transfected cells. Bands in this blot were quantified three times. Actin/GzmB ratios were 0.87 ± 0.04 and 0.83 ± 0.04 for QPY and RAH GzmB, respectively. (E) Cleavage of ICAD-L, Bid, caspase-3, and caspase-8 and release of cytochrome c from mitochondria were detected only in cells transfected with QPY GzmB.
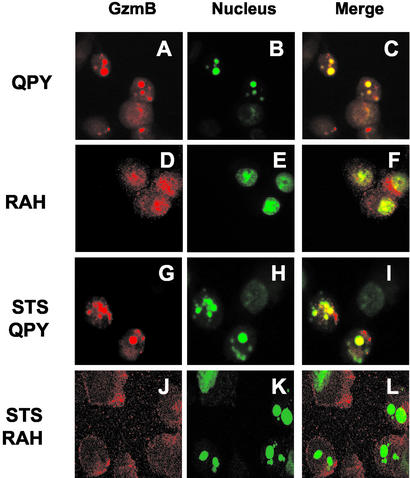
To investigate why RAH GzmB was unable to induce apoptosis despite retaining proteolytic activity in vitro, we examined the intracellular distribution of RAH and QPY GzmB in transfected cells by confocal microscopy. Both QPY and RAH GzmB were observed in the cytoplasm of transfected cells with viable morphology; however, cells with condensed nuclei typical of apoptosis were only found after transfection with QPY GzmB, and in these cells, GzmB was localized exclusively in the nucleus (Fig. 4 A–C). RAH GzmB was also detected in the nucleus of transfected cells (Fig. 4 D–F) but was principally located in the cytoplasm. To test whether the difference in subcellular distribution of GzmB was a consequence or a cause of apoptosis, cell death was induced in transfected cells by treatment with 1 μM staurosporine. Apoptotic nuclear condensation was seen in cells transfected with both QPY and RAH GzmB; however, strong nuclear localization of GzmB was only observed in cells transfected with QPY GzmB. RAH GzmB was, therefore, unable to accumulate in the nucleus, even in apoptotic cells.
Figure 4.
Subcellular localization of QPY and RAH GzmB. Primary glioblastoma cells were transfected with QPY (A–C and G–I) or RAH (D–F and J–L) GzmB, cultured for 14–18 h in the presence (G–L) or absence (A–F) of 1 μM staurosporine, then stained for GzmB (A, D, G, and J) and nuclear DNA (B, E, H, and K). Merged images are shown in C, F, I, and L.
Comparison of QPY and RAH GzmB Structures.
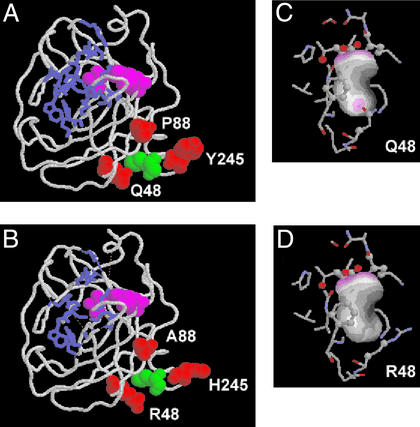
The published structure of wild-type (QPY) human GzmB shows that the three amino acids substituted in the RAH allele are clustered together close to the C-terminal α-helix (Fig. 5A). None of the three positions contribute to the active site or the substrate-binding pocket, which explains why RAH GzmB showed normal enzymatic activity. In addition, the positioning of the side chains of the triple mutant, calculated by using the SMD procedure (27), did not reveal any steric incompatibility of the new side chains with the conformation of the wild type. However, the side chain of Q48 is involved in a hydrogen bond with M242, and this interaction, which could tether the C-terminal α-helix to the surface of the protein, appears to be disrupted by the R48 mutation (Fig. 5 C and D).
Figure 5.
Structure of QPY and RAH GzmB. (A and B) Representations of the complete QPY (A) and RAH (B) GzmB molecules. The catalytic triad H57, D102, S195 is shown in magenta, and side chains contributing to the substrate-binding pocket (30) are shown in a blue stick representation. The three amino acids that differ between the two proteins (Q/R48, P/A88, and Y/H245, as indicated) are shown in red, and M242 is shown in green. The three mutated amino acids do not affect the structure of the active site or the substrate binding cleft, and are clustered together toward the C-terminal α-helix. (C and D) Contact surface of M242 in QPY (C) and RAH (D) GzmB. Regions of M242 involved in hydrogen bonds are indicated by purple patches. The hydrogen bond formed between M242 and Q48 is not present in RAH GzmB.
Discussion
We describe a GzmB allele incorporating three amino acid substitutions which codes for a stable protein that retains enzymatic activity in vitro but is unable to induce apoptosis in vivo. This triple-mutated allele is found in European, African, and Asian populations at an allelic frequency of 25–30%. In contrast to the worldwide distribution of the RAH allele, the intermediate RPY and RAY alleles were found almost exclusively in a West African population. This finding is consistent with an ancient African origin of the GzmB RAH allele in which successive mutations transformed the QPY sequence first to RPY, then to RAY, and finally to RAH. However, it is also possible that the RAH allele is the ancestral form, and that the more widely distributed QPY allele arose more recently, in which case the H245Y mutation would have been the first to arise, followed by A88P, then R48Q. Interestingly, arginine at position 48 is conserved in both rat and mouse GzmB as well as the closely related protease cathepsin G, which also contains alanine at position 88. The QPY allele, therefore, shows more sequence divergence from closely related proteins than the RAH allele and, hence, may have arisen more recently.
Equivalent levels of GzmB expression were found in CD8+ CTL from QPY/QPY homozygous, QPY/RAH heterozygous, and RAH/RAH homozygous patients. The RAH allele is, therefore, actively transcribed, and its precursor RNA is correctly processed, despite the extensive substitutions in intron 3 of the RAH allele. Normal expression of the RAH allele also shows that the coding mutations in the RAH allele are not in linkage disequilibrium with polymorphisms in the promoter region of the gene that affect GzmB expression, and that RAH GzmB is a stable protein which accumulates to similar levels in activated CTL as QPY GzmB.
RAH GzmB was unable to induce apoptosis in primary glioblastoma cell lines established from patient tumors, at least at the early time points (up to 24 h after transfection) we considered. However, because GzmB is able to provoke caspase-independent cell death in some circumstances (28, 29), RAH GzmB may have induced cell death at later time points in the absence of morphological or biochemical evidence of apoptosis. Nevertheless, RAH GzmB did differ functionally from QPY GzmB and, in particular, was unable to cleave biologically relevant substrates, such as Bid, in vivo, despite retaining proteolytic activity against peptide substrates. Two explanations can be proposed to resolve this paradox. Firstly, it is possible that RAH GzmB is unable to cleave protein substrates, even though it can cleave a short peptide. The Q48R, P88A, and Y245H mutations do not affect the catalytic site nor the extended substrate-binding pocket of the enzyme, including the residues that make up the P′ binding site (30), so it is unlikely that proteolysis is directly compromised in RAH GzmB. However, the three mutations comprising RAH GzmB may alter a site on the surface of GzmB involved in binding its protein substrates, or conceivably an as-yet-unidentified molecule which facilitates GzmB interactions with proteins such as Bid. Alterations to this site, which appears to include the C-terminal α-helix of the protein where the three mutations are clustered together, would, therefore, inhibit cleavage of important protein substrates without altering the active site of the enzyme.
On the other hand, RAH GzmB may retain proteolytic activity against important protein substrates, such as Bid, but may be physically sequestered from them in transfected cells. The subcellular localization of GzmB is known to be important for its proapoptotic function. In the absence of perforin, for example, exogenous GzmB is taken up by target cells and retained in an endocytic compartment (5) without inducing apoptosis. The role of perforin seems to be to target GzmB to cellular compartments where its substrate proteins are found: the cytosol and, in particular, the nucleus (31, 32). QPY GzmB colocalized with apoptotic nuclei in transfected cells, whereas RAH GzmB did not accumulate in the nucleus, even when apoptosis was induced by staurosporine. However, it is unclear whether RAH GzmB failed to accumulate in the nucleus because of a defect in nuclear transport per se, or whether the principal barrier to RAH GzmB transport occurred at an earlier stage. In particular, the absence of Bid cleavage in cells transfected with RAH GzmB could imply that RAH GzmB did not gain access to the cytoplasm and, hence, may have been retained in an undefined intracellular compartment.
Although our results are consistent with the hypothesis that the intracellular targeting of RAH GzmB is altered, further experiments with recombinant QPY and RAH GzmB proteins will be required to determine whether RAH GzmB can cleave protein substrates and whether the intracellular trafficking of QPY and RAH GzmB differs in the presence of perforin. If RAH GzmB can efficiently induce apoptosis in the presence of perforin, this may explain why we did not observe differences in the lytic activity of HIV-specific CTL derived from RAH/RAH homozygous patients compared with QPY/QPY homozygous patients. On the other hand, GzmB is only one weapon in the cytotoxic cell's arsenal, so one would not expect RAH/RAH homozygous CTL to be totally inactive. Indeed, CTL from GzmB knockout mice can efficiently kill target cells (22), and our results are consistent with those from knockout mouse studies.
The discovery of a relatively common allele of GzmB incapable of inducing apoptosis has relevance for all pathologies in which GzmB is involved and, in particular, for cancer, because we tested the function of RAH GzmB in primary cancer cell lines. Moreover, because the GzmB/perforin pathway is able to induce apoptosis in cells that have become resistant to death-receptors or anticancer drugs, the absence of effective GzmB function in a subset of patients may be clinically relevant. In particular, cellular therapies based on the induction of antitumor CTL are currently being tested in phase I/II trials involving patients bearing multiresistant tumors in whom conventional therapies are ineffective. The success of such cellular immunotherapies depends on the efficacy of the cytotoxic machinery expressed by the patient's CTL. The triple-mutated GzmB allele that we describe seems to be inactive in inducing apoptosis in tumor cell lines, and its presence could, therefore, influence both the prognosis of cancer patients and the success rates of antitumor cellular immunotherapy.
Acknowledgments
We thank Philippe Pellet for technical assistance with subcloning and sequencing, and Olivia Bonduelle for help generating and testing CTL lines. D.McI. was the recipient of a Moniteur d'Etudes Biologiques postdoctoral fellowship financed by the Agence Nationale de Recherche Contre le SIDA (ANRS). P.-F.C. was supported by a fellowship from the Ligue Contre le Cancer du Doubs-Montbéliard. This work was supported by grants from the ANRS and the Ligue Contre le Cancer Grand-Ouest.
Abbreviations
- GzmB
granzyme B
- CTL
cytotoxic T lymphocyte
- CAD
caspase-activated DNase
- SNP
single-nucleotide polymorphism
- PBMC
peripheral blood mononuclear cell
- ALT
long-term asymptomatic
References
- 1.Peitsch M C, Tschopp J. Methods Enzymol. 1994;244:80–87. doi: 10.1016/0076-6879(94)44007-7. [DOI] [PubMed] [Google Scholar]
- 2.Berthou C, Michel L, Soulie A, Jean-Louis F, Flageul B, Dubertret L, Sigaux F, Zhang Y, Sasportes M. J Immunol. 1997;159:5293–5300. [PubMed] [Google Scholar]
- 3.Masson D, Tschopp J. Cell. 1987;49:679–685. doi: 10.1016/0092-8674(87)90544-7. [DOI] [PubMed] [Google Scholar]
- 4.Motyka B, Korbutt G, Pinkoski M J, Heibein J A, Caputo A, Hobman M, Barry M, Shostak I, Sawchuk T, Holmes C F, et al. Cell. 2000;103:491–500. doi: 10.1016/s0092-8674(00)00140-9. [DOI] [PubMed] [Google Scholar]
- 5.Pinkoski M J, Hobman M, Heibein J A, Tomaselli K, Li F, Seth P, Froelich C J, Bleackley R C. Blood. 1998;92:1044–1054. [PubMed] [Google Scholar]
- 6.Browne K A, Blink E, Sutton V R, Froelich C J, Jans D A, Trapani J A. Mol Cell Biol. 1999;19:8604–8615. doi: 10.1128/mcb.19.12.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris J L, Peterson E P, Hudig D, Thornberry N A, Craik C S. J Biol Chem. 1998;273:27364–27373. doi: 10.1074/jbc.273.42.27364. [DOI] [PubMed] [Google Scholar]
- 8.Heibein J A, Goping I S, Barry M, Pinkoski M J, Shore G C, Green D R, Bleackley R C. J Exp Med. 2000;192:1391–1402. doi: 10.1084/jem.192.10.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutton V R, Davis J E, Cancilla M, Johnstone R W, Ruefli A A, Sedelies K, Browne K A, Trapani J A. J Exp Med. 2000;192:1403–1414. doi: 10.1084/jem.192.10.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darmon A J, Nicholson D W, Bleackley R C. Nature. 1995;377:446–448. doi: 10.1038/377446a0. [DOI] [PubMed] [Google Scholar]
- 11.Thomas D A, Du C, Xu M, Wang X, Ley T J. Immunity. 2000;12:621–632. doi: 10.1016/s1074-7613(00)80213-7. [DOI] [PubMed] [Google Scholar]
- 12.Sharif-Askari E, Alam A, Rheaume E, Beresford P J, Scotto C, Sharma K, Lee D, DeWolf W E, Nuttall M E, Lieberman J, Sekaly R P. EMBO J. 2001;20:3101–3113. doi: 10.1093/emboj/20.12.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mullbacher A, Waring P, Tha Hla R, Tran T, Chin S, Stehle T, Museteanu C, Simon M M. Proc Natl Acad Sci USA. 1999;96:13950–13955. doi: 10.1073/pnas.96.24.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pascoe M D, Marshall S E, Welsh K I, Fulton L M, Hughes D A. Transplantation. 2000;69:2547–2553. doi: 10.1097/00007890-200006270-00013. [DOI] [PubMed] [Google Scholar]
- 15.Li B, Hartono C, Ding R, Sharma V K, Ramaswamy R, Qian B, Serur D, Mouradian J, Schwartz J E, Suthanthiran M. N Engl J Med. 2001;344:947–954. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- 16.Graubert T A, DiPersio J F, Russell J H, Ley T J. J Clin Invest. 1997;100:904–911. doi: 10.1172/JCI119606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higaki Y, Yamada O, Okamura T, Mizoguchi H, Kawashima M. Dermatology. 2001;202:94–98. doi: 10.1159/000051606. [DOI] [PubMed] [Google Scholar]
- 18.Tak P P, Kummer J A, Hack C E, Daha M R, Smeets T J, Erkelens G W, Meinders A E, Kluin P M, Breedveld F C. Arthritis Rheum. 1994;37:1735–1743. doi: 10.1002/art.1780371205. [DOI] [PubMed] [Google Scholar]
- 19.Ronday H K, van der Laan W H, Tak P P, de Roos J A, Bank R A, TeKoppele J M, Froelich C J, Hack C E, Hogendoorn P C, Breedveld F C, Verheijen J H. Rheumatology (Oxford) 2001;40:55–61. doi: 10.1093/rheumatology/40.1.55. [DOI] [PubMed] [Google Scholar]
- 20.Johnstone R W, Cretney E, Smyth M J, Shtil A A, Turner J G, Dalton W S, Yu H. Blood. 1999;93:1075–1085. [PubMed] [Google Scholar]
- 21.Shtil A A, Turner J G, Durfee J, Dalton W S, Yu H. Blood. 1999;93:1831–1837. [PubMed] [Google Scholar]
- 22.Heusel J W, Wesselschmidt R L, Shresta S, Russell J H, Ley T J. Cell. 1994;76:977–987. doi: 10.1016/0092-8674(94)90376-x. [DOI] [PubMed] [Google Scholar]
- 23.Stepp S E, Dufourcq-Lagelouse R, Le Deist F, Bhawan S, Certain S, Mathew P A, Henter J I, Bennett M, Fischer A, de Saint Basile G, Kumar V. Science. 1999;286:1957–1959. doi: 10.1126/science.286.5446.1957. [DOI] [PubMed] [Google Scholar]
- 24.Cartron P F, Oliver L, Martin S, Moreau C, LeCabellec M T, Jezequel P, Meflah K, Vallette F M. Hum Mol Genet. 2002;11:675–687. doi: 10.1093/hmg/11.6.675. [DOI] [PubMed] [Google Scholar]
- 25.Nixon D F, Townsend A R, Elvin J G, Rizza C R, Gallwey J, McMichael A J. Nature. 1988;336:484–487. doi: 10.1038/336484a0. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Liu X, Bhalla K, Kim C N, Ibrado A M, Cai J, Peng T I, Jones D P, Wang X. Science. 1997;275:1129–1132. doi: 10.1126/science.275.5303.1129. [DOI] [PubMed] [Google Scholar]
- 27.Tuffery P, Etchebest C, Hazout S, Lavery R, Kawashima M. J Biomol Struct Dyn. 1991;8:1267–1289. doi: 10.1080/07391102.1991.10507882. [DOI] [PubMed] [Google Scholar]
- 28.Sarin A, Williams M S, Alexander-Miller M A, Berzofsky J A, Zacharchuk C M, Henkart P A. Immunity. 1997;6:209–215. doi: 10.1016/s1074-7613(00)80427-6. [DOI] [PubMed] [Google Scholar]
- 29.Trapani J A, Jans D A, Jans P J, Smyth M J, Browne K A, Sutton V R. J Biol Chem. 1998;273:27934–27938. doi: 10.1074/jbc.273.43.27934. [DOI] [PubMed] [Google Scholar]
- 30.Rotonda J, Garcia-Calvo M, Bull H G, Geissler W M, McKeever B M, Willoughby C A, Thornberry N A, Becker J W. Chem Biol. 2001;8:357–368. doi: 10.1016/s1074-5521(01)00018-7. [DOI] [PubMed] [Google Scholar]
- 31.Trapani J A, Jans P, Smyth M J, Froelich C J, Williams E A, Sutton V R, Jans D A. Cell Death Differ. 1998;5:488–496. doi: 10.1038/sj.cdd.4400373. [DOI] [PubMed] [Google Scholar]
- 32.Pinkoski M J, Heibein J A, Barry M, Bleackley R C. Cell Death Differ. 2000;7:17–24. doi: 10.1038/sj.cdd.4400604. [DOI] [PubMed] [Google Scholar]