Abstract
Narnavirus 23S RNA is a persistent positive-stranded RNA virus found in yeast Saccharomyces cerevisiae. The viral genome (2.9 kb) only encodes its RNA-dependent RNA polymerase, p104. Here we report the generation of 23S RNA virus, with high frequency, from a vector containing the entire viral cDNA sequence. When the conserved GDD (Gly-Asp-Asp) motif of RNA-dependent RNA polymerase was modified, the vector failed to generate the virus, indicating that an active p104 is essential for replication. Successful launching required transcripts having the proper viral 3′ terminus generated in vivo. This was accomplished through in vivo processing of the primary transcripts by the hepatitis delta virus antigenomic ribozyme directly fused to the 3′ terminus of the 23S RNA genome. Although the primary transcripts also contained extra nucleotides at their 5′ ends derived from the vector, the launched virus possessed the authentic 5′ terminus of the viral genome without these extra nucleotides. Modifications of the genome sequence at the 5′ and 3′ termini abolished viral generation, indicating that the viral genome has cis-acting signals for replication at both termini. The great ease to generate the virus will facilitate the identification of these cis-acting signals. Furthermore, the virus, once generated, can be transmitted to daughter cells indefinitely without the vector or any selection, which makes the 23S RNA virus-launching system particularly useful for investigating the basis for RNA virus persistence.
The availability of versatile genetics and well developed molecular biology techniques makes the yeast Saccharomyces cerevisiae one of the most powerful model organisms among eukaryotes. In the field of RNA virology, this organism has provided valuable information on the mechanism of replication of its endogenous L-A double-stranded RNA (dsRNA) virus and on the complex virus–host interactions (1). Recent developments (2–4) include launching RNA viruses of animal and plant origin from expression vectors in yeast as a surrogate host. These have begun to provide rich information on virus–host interactions that is difficult or impossible to obtain from their native hosts. The yeast cells possess two types of endogenous RNA viruses: dsRNA virus totivirus (type species L-A) and positive (+)-strand RNA virus Narnavirus. Unfortunately, however, to date no launching or RNA-infection systems have been available to generate these viruses in yeast. Among fungal RNA viruses, to our knowledge such systems are only available in the dsRNA hypovirus of the chestnut blight fungus Cryphonectria parasitica, the infection of which causes virulence attenuation of the fungus. By using reverse genetics, the molecular basis of hypovirulence has been pursued (reviewed in ref. 5).
Most laboratory strains of S. cerevisiae harbor 20S RNA (ScNV-20S), and fewer strains carry 23S RNA (ScNV-23S). Both belong to the genus Narnavirus of the Narnaviridae family (6). The family also includes the genus Mitovirus, the members of which reside in fungal mitochondria (7, 8). 20S and 23S RNA viruses have a number of characteristics in common. Their RNA genomes are small (2,891 and 2,514 nt for 23S and 20S RNA, respectively) and share five nucleotide sequences at their 5′ ends (5′-GGGGC… ) and also at their 3′ ends (… GCCCC-OH) (9). Each genome encodes only a single protein: a 104-kDa protein (p104) for 23S RNA and a 91-kDa protein (p91) for 20S RNA. Both proteins contain four amino acid motifs well conserved among RNA-dependent RNA polymerases (RDRPs) (10, 11). Because the viral genomes do not encode coat proteins, the RNA genomes are not encapsidated into viral particles. Instead they form ribonucleoprotein complexes with their cognate RDRPs in a 1:1 stoichiometry and reside in the host cytoplasm (12). Both viruses are compatible in the same host and can be maintained stably without excluding each other. They are transmitted vertically to daughter cells during mitosis or horizontally during mating. Additionally, typical of fungal viruses, they do not kill the host nor render phenotypic changes to the host, which makes their genetic manipulation difficult or infeasible. However, their interactions with the host can be glimpsed by the following observations: the copy numbers of 20S and 23S RNAs can be increased greatly by transferring the host cells to nitrogen starvation conditions (13, 14) and are also elevated by host mutations such as ski2 (15). Nitrogen starvation is commonly used to induce sporulation in diploid cells, and increase in viral load may contribute to the efficient distribution of these viruses to meiotic descendents. The SKI2 gene was identified originally as a suppressor of a satellite RNA of the L-A virus (16). It has been proposed that mutations in this gene increase the copy number of dsRNA and single-stranded RNA viral genomes either by decreasing a 3′-5′ exoribonuclease activity (17, 18) or by increasing the translational efficiencies of the viral RNAs that lack poly(A) tails (19–21).
In the present study we established a launching system to generate 23S RNA virus in vivo from a yeast expression vector. The launched virus can be transmitted to daughter cells indefinitely without a vector or any selection and can be induced under nitrogen-starvation conditions like the native 23S RNA virus. Our data indicate that the 5′ and 3′ termini of the viral genome contain cis-acting signals essential for replication. This launching system will pave the way to introduce a selectable marker into the viral genome and will allow us to investigate not only the virus–host interactions but also the mechanism of 23S RNA virus persistence with the aid of powerful yeast genetics.
Materials and Methods
Strains and Media.
Yeast strains (obtained from J. C. Ribas, Instituto de Microbiologia Bioquimica CSIC, Salamanca, Spain) were L-A-o derivatives of strain 2928 (a ura3 trp1 his3, 20S RNA, 23S RNA-o) (22) and 2927 (a ura3 trp1 his3 ski2-2, 20S RNA, 23S RNA-o). Cells were grown in either rich YPAD (1% yeast extract/2% peptone/0.04% adenine/2% glucose) or tryptophan-omitted synthetic (H-Trp) medium (23). Nitrogen starvation was performed as described (14).
Northern Hybridization.
Cells were broken with glass beads (12), and RNA was extracted from cell lysates in the presence of 0.3% SDS once with phenol and twice with phenol/chloroform and precipitated with ethanol. RNA was separated in a native agarose gel, blotted onto a neutral nylon membrane (Hybond-N, Amersham Pharmacia), and hybridized with a 32P-labeled 23S RNA positive or negative strand-specific probe (24). Because negative strands are less prevalent in the cells compared with positive strands, the autoradiograms with the latter probe were exposed 5–10 times longer. We analyzed 10–20 independent transformants in each experiment. All the mutations introduced into the launching plasmid described in this article produced clear-cut results. For the sake of simplicity, only a representative of each experiment is presented in the figures. Especially in the case of the G-8 plasmid, we detected 23S RNA virus in all 20 colonies analyzed, although the launching efficiency was much less compared with the control plasmid as described in the text.
Plasmids.
Launching plasmid pRE637 was constructed as follows: The complete cDNA sequence of 23S RNA (2,891 bp), the internal two SmaI sites of which had been modified (C966U and C2067U), was inserted at the Klenow-treated EcoRI site of pI2 (22) downstream of the PGK1 promoter. The plasmid carries TRP1 as a selective marker. Then a unique SmaI site was created at the 3′ end of the 23S RNA sequence (pRE620). A chemically synthesized 82-bp hepatitis delta virus antigenomic ribozyme (25) was first subcloned between the SmaI and EcoRI sites of the pBluescript KS(+) vector, and then an SmaI–KpnI fragment containing the ribozyme sequence was inserted between the SmaI and KpnI sites of pRE620, thus resulting in pRE637. 23S RNA positive and negative strand-specific probes were made by T7 and T3 run-off transcription from pRE473 predigested with appropriate restriction enzymes. pRE473 contains an SmaI fragment of 23S RNA cDNA (nucleotides 966–2,070) in the SmaI site of pBluescript KS(+) vector. In vitro mutagenesis was done as described in ref. 26. All the mutations introduced were confirmed by DNA sequencing.
Primer Extension.
Primer extension analysis was performed as described (9).
Results
Launching Plasmid.
We constructed a yeast expression plasmid (pRE637) to generate 23S RNA virus in vivo by inserting the entire 23S RNA cDNA sequence (2,891 bp) downstream of the constitutive PGK1 promoter of pI2 (ref. 22; Fig. 1A). Viral (+) strands could be transcribed from the promoter. The major transcription start site (27) is located at −33 relative to the viral genome. An 82-nt hepatitis delta virus antigenomic ribozyme sequence (25) was fused directly to the 3′ end of the 23S RNA sequence. Thus, intramolecular cleavage by the ribozyme was expected to create transcripts having the precise 3′-end terminus of the viral genome in vivo. The cDNA sequence of 23S RNA had been tagged by silent mutations at the two SmaI sites (C966U and C2067U, numbering from the 5′ end of the viral genome). These modifications do not change the amino acid sequence of p104, nor do they affect the computer-predicted secondary structure of the RNA.
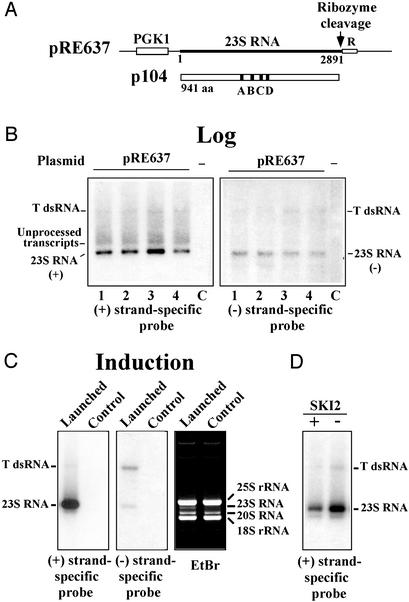
Figure 1.
Diagram of 23S RNA launching plasmid (A) and launched RNA detected by Northern blots (B–D). (A) pRE637 contains the complete 23S RNA cDNA sequence (bold line) downstream of the PGK1 promoter (PGK1). The antigenomic hepatitis delta virus ribozyme (R), its cleavage site (vertical arrow), p104, and its conserved RDRP motifs (A–D) are shown. The viral genome contains 6 and 59 nt in the 5′ and 3′ untranslated regions, respectively. (B–D) Detection of launched viral RNA in growing (B) or induced (C) cells or in a ski2 strain (D) by Northern blots hybridized with 23S RNA positive or negative strand-specific probes. In B, four independent transformants (1–4) and recipient cells without plasmid (C) are shown. In C, ethidium-bromide staining of the gel is also shown. The positions of 23S RNA, its double-stranded form (T dsRNA), as well as yeast rRNAs and 20S RNA are indicated. For clarity the autoradiogram in D was exposed five times shorter than the one in C with the same probe.
Generation of 23S RNA Virus.
Yeast cells negative for 23S RNA virus were transformed with the launching plasmid pRE637. Twenty independent colonies were grown in H-Trp liquid medium selective for the plasmid. RNA was extracted from growing cells (it is estimated that these cells had been growing 25–30 generations after receiving the plasmid) and analyzed by Northern blot hybridization. A 23S RNA (+) strand-specific probe detected an RNA band with the mobility of 23S RNA viral genome in all the colonies analyzed. The intensity of the band varied slightly among the isolates (Fig. 1B). All these colonies also possessed T dsRNA, the double-stranded form of 23S RNA (11, 28). A specific probe for 23S RNA negative (−) strand also detected the single-strand (−) genome as well as T dsRNA in all these colonies. Recipient cells without the plasmid did not give these signals. These results indicate that 23S RNA (−) strands were replicated from the (+)-strand transcripts produced in vivo and suggest the generation of 23S RNA virus from the vector. We confirmed, as described below, that 23S RNA virus was generated from the plasmid and that the generated virus possessed the same properties known for endogenous virus. Transformants growing 25–30 generations in the presence of the launching plasmid were streaked for single-colony isolation on rich nonselective (YPAD) plates to eliminate the plasmid. When the plasmid-free colonies were analyzed, 20–50% of them retained 23S RNA virus. Once generated, the virus replicates autonomously and can be maintained stably in the yeast cells for more than 100 generations (thus-far examined) in the absence of the plasmid. The generated virus can be induced under nitrogen-starvation conditions, and the viral RNA can be seen directly by ethidium-bromide staining in an agarose gel (Fig. 1C). The amount of induced RNA and (+)/(−)-strand ratio are similar to those found in endogenous 23S RNA virus. We noticed that a ski2 mutation did not affect the launching efficiency. Because the ribozyme would create a precise viral 3′ terminus and the majority of p104 would be translated from the primary transcripts [with CAPs and poly(A) tails], SKI2, according to its two proposed functions, is expected to be irrelevant to the launching process. Once the virus is generated, its copy number in the ski2 strain is several-fold higher than that in the SKI2 strain (Fig. 1D).
Because the viral genome in the plasmid was tagged by silent mutations, we examined the presence of these tags in the launched viral RNA from plasmid-cured cells (Fig. 2). Endogenous or launched viral genome was gel-purified and treated with DNase I. Then a 644-bp cDNA fragment encompassing one of the two SmaI sites was reverse-transcribed and amplified from the RNA. The cDNA derived from the launched viral RNA was resistant to SmaI digestion, whereas the one from the endogenous viral RNA was sensitive to the enzyme, confirming that the launched 23S RNA originated from the plasmid. We also examined the effects of these silent tags on the generation of 23S RNA virus. When either one of the two SmaI sites or both together were restored by in vitro site-directed mutagenesis, all the resulting plasmids had efficiencies of virus generation similar to that of the original launching plasmid (data not shown).
Figure 2.
Proof of 23S RNA virus generation from the launching plasmid. (A) Diagram of 23S RNA and one of two SmaI sites analyzed by RT-PCR amplification. Two oligonucleotides used for amplification are shown by the arrows. The RT-PCR products (644 nt) from endogenous virus will produce two fragments (394 and 250 bp) after SmaI digestion, whereas those from launched virus are expected to be resistant to the enzyme. (B and C) RT-PCR products from endogenous and launched 23S RNA viruses before (B) and after (C) SmaI treatment. Ethidium-bromide staining of agarose gels is shown. Lambda HindIII markers and the sizes of digested and undigested PCR products are indicated. In the third lane of C (mixture) the PCR products from endogenous and launched virus are mixed together and then digested with the enzyme. Note that in C the amount of the larger fragment (394 bp) appears less than that of the smaller fragment (250 nt) because of comigration of the bromophenol-blue dye.
p104 Is Essential.
The 23S RNA genome encodes a single protein, p104. p104 has the four sequence motifs well conserved among RDRPs. When one of the motifs (GDD) was changed to EFD in the launching plasmid, the modified plasmid failed to produce 23S RNA virus (Fig. 3). The recipient strain also harbors 20S RNA virus, which belongs to the same genus as 23S RNA. Its RDRP (p91) shares a high degree of amino acid similarity with p104 that extends beyond the RDRP consensus motifs (29). Nevertheless, the failure to launch 23S RNA in this strain with the modified sequence implies that p91 cannot substitute for p104 in replication of 23S RNA virus. This is consistent with our previous observation that each RDRP interacts only with its cognate viral RNA (12). p104 has a stretch of 8 aa in the GDD region identical to those present in p91 except for one mismatch (496-RICGDDLI-503 in p104 and 457-RVCGDDLI-464 in p91). When I497 was replaced with V, the modified plasmid could generate 23S RNA virus (Fig. 3). The launching efficiency and the amount of RNA produced were indistinguishable from those with the unmodified plasmid. Thus 23S RNA virus can tolerate a change between these aliphatic amino acids at a position close to the well conserved GDD motif. Therefore, these results indicate that p104 is essential for replication and suggest the usefulness of the launching system to perform structure-function analysis of the RNA polymerase.
Figure 3.
p104 is essential for launching. (A) Mutations were introduced into the well conserved GDD motif (EFD) or its neighboring amino acid (I497V) of p104. Transformants with launching plasmids having the modified p104 were grown and induced for 23S RNA. RNA was extracted from the induced cells, separated in an agarose gel, and analyzed by hybridization using a 23S RNA positive or negative strand-specific probe. (B) The mutations introduced to the p104 sequence are shown.
Importance of Viral 3′ End.
We have not observed the generation of 23S RNA virus from plasmids previously even though they contained the complete cDNA sequence (perhaps because they lacked the means to generate transcripts ending with the precise viral 3′ terminus). To demonstrate the importance of the 3′ terminus in launching, we used in vitro mutagenesis to construct two variants of the launching plasmid (Fig. 4D). In one variant [ribozyme (−)] the GGG sequence 3′ to the cleavage site was substituted with AAA. This substitution destroys or modifies the substrate-bearing PI helix as well as a G⋅U wobble at the cleavage site in the ribozyme core structure (25, 30, 31). In the second variant (3′ end), three of four Cs clustered at the 3′ end of the viral genome were substituted with three As (Fig. 4D). The last C at the 3′ end was left unmodified, because a nucleotide change at this position may affect cleavage by the ribozyme (31). When these two variant plasmids were introduced into yeast cells, we did not observe 23S RNA virus generation (Fig. 4 A and B).
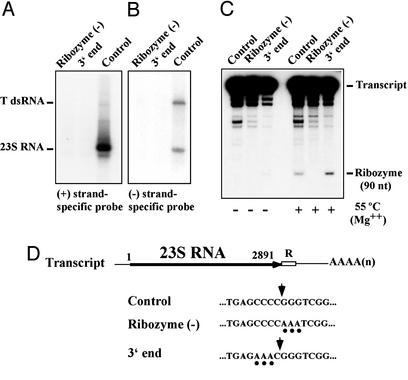
Figure 4.
Generation of the precise viral 3′ terminus is important for launching. (A and B) Several nucleotides in the ribozyme [ribozyme (−)] or at the viral 3′ end of the launching plasmid were modified. RNAs from transformants with the modified or unmodified launching plasmids were analyzed with a positive (A) or negative (B) strand-specific probe for 23S RNA as described in the Fig. 3 legend. (C) A fragment containing the complete sequences of 23S RNA and ribozyme from the modified or unmodified launching plasmid was subcloned into the pBluescript vector downstream of the T7 promoter. Uniformly labeled, run-off T7 transcripts were made in vitro. The transcripts with or without incubation at 55°C in the presence of 5 mM Mg2+ were separated in a 5% acrylamide gel and detected by autoradiography. The 90-nt cleavage products containing the ribozyme sequence are indicated. (D) Diagram of the primary transcript from the launching plasmid and modifications introduced thereon. The modified nucleotides and cleavage site by the ribozyme are indicated with filled circles and vertical arrows, respectively.
To confirm that the ribozymes in these variant RNAs functioned as intended in the context of the 23S RNA sequence, we subcloned the entire 23S RNA and ribozyme sequence from these variant plasmids into the pBluescript KS(+) vector downstream of the T7 RNA polymerase promoter. When run-off T7 transcripts were made in vitro, the 3′-end variant RNA produced a 90-nt cleavage fragment containing the ribozyme sequence, and its cleavage was increased greatly by incubation at 55°C in the presence of Mg2+ (Fig. 4C). The efficiency of cleavage was equivalent to or better than that of the control RNA, which indicates that the ribozyme in the 3′-end variant RNA is functional. On the other hand, the ribozyme (−) variant RNA failed to produce the fragment even at 55°C. Therefore these results indicate that the 3′ terminus of the (+) strands contains a cis-acting signal(s) for viral replication, the activity of which is impaired by the substitution of CCC with AAA in the 3′-end variant RNA. Because the ribozyme (−) variant RNA failed to launch 23S RNA virus, extra nucleotides at the 3′ end may impede the activity of the cis signal(s). Alternatively, the RNA terminus of the (+)-strand genome itself may constitute a part of the cis signal. Finally, because we did not detect (−)-strand single-stranded RNA or T dsRNA in the cells transformed with the 3′-end or ribozyme (−) variant plasmids, their failure to launch 23S RNA virus is most likely due to their inability to produce appropriate template RNAs for the (−)-strand synthesis.
5′-End Signal for Replication.
The major transcription start site in the launching plasmid is located at position −33, relative to the 5′ terminus of 23S RNA genome. To examine the effects of these extra nucleotides and the importance of the viral 5′ end on the generation of 23S RNA virus, we constructed two derivatives of the launching plasmid (Fig. 5A). The 23S RNA genome possesses four Gs at the 5′ end. In one derivative (5′ end) the four Gs were substituted with CAAA. In the second derivative (G-8) a cluster of 8 Gs was inserted at the 5′ end of the viral genome, thus creating a stretch of 12 Gs. Oligo G tracts are thought to form strong secondary structures that inhibit progression of the Xrn1p/Ski1p 5′-3′ exoribonuclease, a major player in the mRNA degradation pathway in S. cerevisiae (17, 32, 33). The cells transformed with the 5′-end plasmid did not produce a band corresponding to the 23S RNA (+)- or (−)-strand genome (Fig. 5 B and C). On the other hand, cells transformed with the G-8 plasmid showed the band corresponding to the 23S RNA (+)-strand genome, although its intensity was much weaker than that in the cells transformed with the control plasmid. The presence of 23S RNA (−) strands and T dsRNA in these cells indicates (−)-strand synthesis, suggesting that the 23S RNA virus was generated from the G-8 plasmid. To confirm this, we cured the G-8 plasmid from the cells. Among plasmid-negative colonies some contained 23S RNA virus, although their occurrence was 5–10 times lower than the launching efficiency of the control plasmid. However, once generated the virus in these G-8 plasmid-cured cells was indistinguishable in terms of quantity and stability from the one generated from the control plasmid.
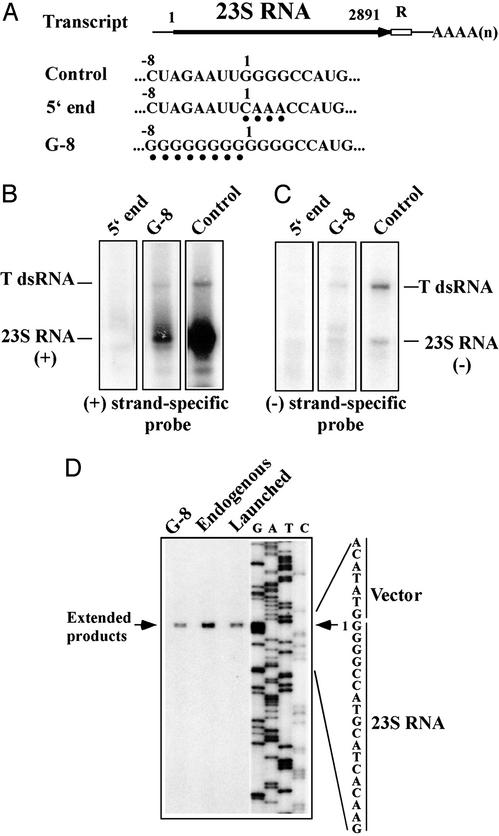
Figure 5.
Launched viruses have the correct 5′ end termini. (A) Diagram of the primary transcripts from the launching plasmid and modifications introduced at the 5′ end of the viral genome. The nucleotides substituted in the 5′ end and the insertion of a poly-G tract in the G-8 plasmid are indicated by filled circles. The first nucleotide at the viral 5′ end is numbered 1. (B and C) Cells were transformed with the modified 5′ end or G-8 or unmodified (control) launching plasmid. RNA was extracted from transformants and analyzed as described in the Fig. 3 legend by using a positive (B) or negative (C) strand-specific probe for 23S RNA. (D) Determination of the 5′ ends of launched viral RNAs by primer extension. The 5′ ends of the viral RNAs generated from the unmodified (Launched) and modified (G-8) launching plasmids were analyzed by reverse transcription using a 5′-labeled primer. The extended products were separated in an 8% sequencing gel and detected by autoradiography. As references, primer-extension products obtained from endogenous 23S RNA virus (Endogenous) and sequence ladders were run in the same gel.
The viral RNAs generated from the control and G-8 plasmids were isolated, and their 5′ ends were compared with that of endogenous 23S RNA virus by primer extension. As shown in Fig. 5D, 23S RNA viruses both generated from the G-8 and control plasmids possessed exactly the same (+)-strand 5′ end as native 23S RNA virus. These results indicate that the extra nucleotides at the 5′ end of the primary transcripts were effectively eliminated from the generated virus during the launching process. The insertion of an oligo G8 stretch at the 5′ terminus of the viral genome reduced the launching efficiency most likely by blocking the progression of a 5′-3′ exonuclease. Alternatively, it could be an effect of the G8 stretch on 5′ end recognition by the RNA polymerase. Although there are four consecutive Gs at the 5′ end of the viral genome, our results indicate that additional Gs at the 5′ end are harmful or unfavorable for replication. Finally, because the 5′ end plasmid failed to produce 23S RNA virus, the four consecutive Gs at the 5′ end seem to be a part of the essential 5′ cis signal(s) for replication.
Discussion
We established a launching system to generate 23S RNA virus from a cDNA vector. Similar to native virus, the virus generated from a cDNA copy can be induced by nitrogen starvation and has an increased copy number of the RNA genome in ski2 strains. Once generated, the virus does not require the launching vector and can be transmitted stably to daughter cells without any selection. The launched virus retained the silent mutations tagged for identification, proving that it was derived from the cDNA.
A potential use of this launching system to investigate RNA polymerase structure and function is demonstrated by two mutations introduced in the p104 amino acid sequence. The substitution of the GDD motif with EFD failed to generate 23S RNA virus. However, a point mutation (I497V) adjacent to the GDD motif did not affect the launching. Crystal structures of RDRPs show remarkable similarity to those of DNA-dependent RNA or DNA polymerases and of reverse transcriptases (34–36). These structures resemble that of a right hand, and the four motifs conserved among RDRPs including the GDD motif are located within a subdomain termed palm. The first aspartic acid residue of the GDD motif coordinates two catalytic Mg2+ ions, which carry out the polymerization reaction. Thus, the inability of the EFD mutant to generate 23S RNA virus is caused by the destruction of the catalytic site of the polymerase. On the other hand, the I497V mutation produced an active polymerase and generated the virus. This was partly expected, because a valine can be found at the same position relative to the GDD motif in p91. A short stretch of amino acids that includes the GDD motif forms a β-hairpin structure, with the GDD motif located at the hairpin loop. Therefore, it suggests that the exchange between the aliphatic amino acids at position 497 did not alter the hairpin structure significantly, thus keeping the overall polymerase structure intact.
We have demonstrated that there are cis-acting signals for replication at the 5′- and 3′-terminal regions of the 23S RNA genome. Although the characterization of these signals and their functions awaits further investigation, available data indicate that their locations at the RNA termini are crucial. Transcripts from the expression vector have extra nonviral 33 nts at their 5′ ends. These nucleotides were eliminated from the viral genome during the launching process probably by cellular 5′-3′ exonucleases. In fact, the insertion of an oligo G8 at the viral 5′ end in the vector, which created a stretch of twelve Gs, decreased the launching efficiency. Primer extension indicated that the virus generated possessed only four Gs at the 5′ end, the authentic viral 5′ terminus, and that all the extra Gs added had been eliminated. This indicates that the addition of extra Gs to the 5′ end or the elimination of the 5′-terminal Gs is harmful for replication. Consistently, the substitution of the 5′-terminal GGGG with CAAA completely abolished the generation of the virus. Therefore, these nucleotides are a part of the 5′-end cis signal(s). Similarly, the viral 3′ terminus has a cis signal(s), because the substitution of the 3′-terminal CCCC-OH with AAAC-OH abolished launching. The transcription termination site for the FLP gene (37) of the 2 μM plasmid is located 0.7 kb downstream of the 23S RNA viral genome in the vector. The failure in launching of the ribozyme (−) plasmid or of plasmids without the ribozyme indicates that the 3′ cis signal needs to be located at or close to the 3′ terminus. We now can create RNAs in vivo having a defined number of extra nucleotides at their 3′ (or 5′) ends and assess their effects on launching. Therefore, this launching system will provide information not only on the cis signals for virus replication but also on the cellular exonucleases involved in the launching process.
The five nucleotide sequences at the 5′ and 3′ termini of the 23S RNA genome are complementary. This may simply reflect that the polymerase machinery would require the same 3′-terminal sequence for synthesis of both the positive and negative strands. Alternatively, the 5′ and 3′ termini may form a panhandle structure that could be the real template for the polymerase machinery. These possibilities can be examined by carefully defining the cis signals and measuring their individual effects on positive- and negative-strand synthesis in vivo.
The persistence of fungal RNA viruses would involve many interactions with the host at different levels. Because they have no extracellular transmission pathway, cytotoxicity and uncontrolled utilization of host machinery that is harmful to the host must be avoided. The virus should be invisible to host surveillance. Furthermore, when the host cells divide, the virus has to replicate and needs to be transmitted to daughter cells. In the daughter cells, the virus has to replicate and adjust its copy number to be ready for another round of cell division. When nutrients become scarce, the virus has to cease replication. Alternatively, in the case of Narnavirus, the virus may need to increase its copy number to ensure efficient transmission to meiotic spores. Therefore, fungal viruses interact extensively with the host cells and monitor their condition constantly. The launching system of 23S RNA virus provides a good opportunity to introduce a selectable marker to the viral RNA. Once introduced, it will enable the utilization of yeast genetics to understand the complex virus–host interactions. Because 23S RNA is a native virus of yeast, we expect that their interactions are genuine, not imposed artificially. Some of these interactions could be observed even in infectious RNA viruses, because infectious viruses also have to obey certain rules as long as they use host machinery for their replication.
Acknowledgments
We thank Drs. D. Martín-Zanca, L. M. Esteban, and N. Swaminathan for thoughtful reading and criticisms of the manuscript. This work was supported by Spanish Ministry of Science and Technology Grants PB98-1121 and BMC2001-1065. T.F. is the recipient of a contract from the Spanish Research Program “Ramón y Cajal.”
Abbreviations
- dsRNA
double-stranded RNA
- RDRP
RNA-dependent RNA polymerase
References
- 1.Wickner R B. In: Fields Virology. Knipe D M, Howley P M, editors. Vol. 1. Williams & Wilkins, Philadelphia: Lippincott; 2001. pp. 629–658. [Google Scholar]
- 2.Price B D, Rueckert R R, Ahlquist P. Proc Natl Acad Sci USA. 1996;93:9465–9470. doi: 10.1073/pnas.93.18.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishikawa M, Diez J, Restrepo-Hartwig M, Ahlquist P. Proc Natl Acad Sci USA. 1997;94:13810–13815. doi: 10.1073/pnas.94.25.13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noueiry A O, Chen J, Ahlquist P. Proc Natl Acad Sci USA. 2000;97:12985–12990. doi: 10.1073/pnas.240460897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawe A L, Nuss D L. Annu Rev Genet. 2001;35:1–29. doi: 10.1146/annurev.genet.35.102401.085929. [DOI] [PubMed] [Google Scholar]
- 6.Wickner R B, Esteban R, Hillman B I. In: Virus Taxonomy: Seventh Report of the International Committee on Taxonomy of Viruses. Van Regenmortel M H V, Fauquet C, Bishop D H L, Carsten E, Estes M, Lemon S M, Maniloff J, Mayo M, McGeoon D, Pringle C, Wickner R B, editors. New York: Academic; 2000. pp. 651–656. [Google Scholar]
- 7.Polashock J J, Hillman B I. Proc Natl Acad Sci USA. 1994;91:8680–8684. doi: 10.1073/pnas.91.18.8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong Y, Dower S L, Cole T E, Brasier C M, Buck K W. Virology. 1999;258:118–127. doi: 10.1006/viro.1999.9691. [DOI] [PubMed] [Google Scholar]
- 9.Rodríguez-Cousiño N, Solórzano A, Fujimura T, Esteban R. J Biol Chem. 1998;273:20363–20371. doi: 10.1074/jbc.273.32.20363. [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez-Cousiño N, Esteban L M, Esteban R. J Biol Chem. 1991;266:12772–12778. [PubMed] [Google Scholar]
- 11.Esteban L M, Rodríguez-Cousiño N, Esteban R. J Biol Chem. 1992;267:10874–10881. [PubMed] [Google Scholar]
- 12.Solórzano A, Rodríguez-Cousiño N, Esteban R, Fujimura T. J Biol Chem. 2000;275:26428–26435. doi: 10.1074/jbc.M002281200. [DOI] [PubMed] [Google Scholar]
- 13.Kadowaki K, Halvorson H O. J Bacteriol. 1971;105:826–830. doi: 10.1128/jb.105.3.826-830.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wejksnora P J, Haber J E. J Bacteriol. 1978;134:246–260. doi: 10.1128/jb.134.1.246-260.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumoto Y, Fishel R, Wickner R B. Proc Natl Acad Sci USA. 1990;87:7628–7632. doi: 10.1073/pnas.87.19.7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toh-e A, Guerry P, Wickner R B. J Bacteriol. 1978;136:1002–1007. doi: 10.1128/jb.136.3.1002-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacobs Anderson J S, Parker R. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown J T, Bai X, Johnson A W. RNA. 2000;6:449–457. doi: 10.1017/s1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Widner W R, Wickner R B. Mol Cell Biol. 1993;13:4331–4341. doi: 10.1128/mcb.13.7.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masison D C, Blanc A, Ribas J C, Carroll K, Sonenberg N, Wickner R B. Mol Cell Biol. 1995;15:2763–2771. doi: 10.1128/mcb.15.5.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Searfoss A M, Wickner R B. Proc Natl Acad Sci USA. 2000;97:9133–9137. doi: 10.1073/pnas.97.16.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wickner R B, Icho T, Fujimura T, Widner W R. J Virol. 1991;65:155–161. doi: 10.1128/jvi.65.1.155-161.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wickner R B. Cell. 1980;21:217–226. doi: 10.1016/0092-8674(80)90129-4. [DOI] [PubMed] [Google Scholar]
- 24.Fujimura T, Esteban R, Esteban L M, Wickner R B. Cell. 1990;62:819–828. doi: 10.1016/0092-8674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- 25.Perrota A T, Been M D. Nature. 1991;350:434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- 26.Esteban R, Fujimura T, Wickner R B. EMBO J. 1989;8:947–954. doi: 10.1002/j.1460-2075.1989.tb03456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derynck R, Singh A, Goeddel D V. Nucleic Acids Res. 1983;11:1819–1837. doi: 10.1093/nar/11.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wesolowski M, Wickner R B. Mol Cell Biol. 1984;4:181–187. doi: 10.1128/mcb.4.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esteban R, Rodríguez-Cousiño N, Esteban L M. Prog Nucleic Acid Res Mol Biol. 1993;46:155–182. doi: 10.1016/s0079-6603(08)61021-1. [DOI] [PubMed] [Google Scholar]
- 30.Ferre-D'Amare A R, Zhou K, Doudna J A. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 31.Perrotta A T, Been M D. Nucleic Acids Res. 1996;24:1314–1321. doi: 10.1093/nar/24.7.1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muhlrad D, Decker C J, Parker R. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 33.Hsu C L, Stevens A. Mol Cell Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale R L, Mathieu M, De Francesco R, Rey F A. Proc Natl Acad Sci USA. 1999;96:13034–13039. doi: 10.1073/pnas.96.23.13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butcher S J, Grimes J M, Makeyev E V, Bamford D H, Stuart D I. Nature. 2001;410:235–240. doi: 10.1038/35065653. [DOI] [PubMed] [Google Scholar]
- 36.Ng K K, Cherney M M, Vazquez A L, Machin A, Alonso J M, Parra F, James M N. J Biol Chem. 2002;277:1381–1387. doi: 10.1074/jbc.M109261200. [DOI] [PubMed] [Google Scholar]
- 37.Hartley J L, Donelson J E. Nature. 1980;286:860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]