Abstract
B7-1 and B7-2 are homologous costimulatory ligands expressed on the surfaces of antigen-presenting cells. Their interactions with CD28/CTLA-4 receptors expressed on T cell surfaces are crucial for the proper regulation of T cell activity. B7-1 and B7-2 display distinct roles in immune regulation, although they are usually considered to have redundant functions. Here, we report the crystal structure of the receptor-binding (Ig V-type) domain of human B7-2 at 2.7-Å resolution. Structures of unliganded and liganded B7-1 and B7-2 suggest a physical–chemical basis for the observed functional similarities and differences between these two costimulatory ligands. Of particular note, whereas the majority of the residues mediating B7-1 dimerization are hydrophobic, the B7-2 dimer observed in the B7-2/CTLA-4 complex displays a very hydrophilic dimer interface. These differences provide a mechanism for preventing the formation of B7-1/B7-2 heterodimers. The divergence at the putative dimer interface is also consistent with the lower tendency of B7-2 to dimerize, as shown by the monomeric state of unliganded B7-2 both in solution and crystalline form, and may result in detailed differences in signaling mechanisms associated with B7-1 and B7-2.
The initiation and regulation of the cell-mediated immune response depend on two distinct signals. The first signal results from the specific interaction between the T cell receptor and its antigenic peptide bound to an MHC molecule. Whether this signal leads to T cell activation or anergy is determined by the concurrent delivery of a second signal provided by costimulatory receptor–ligand interactions (1). CD28 and CTLA-4, which share ≈30% identity, are the most well characterized costimulatory receptors present on the surface of T cells (2, 3). Upon binding its B7-1 and B7-2 ligands, CD28 delivers a costimulatory signal to the responding T cell, enhancing T cell proliferation and cytokine secretion and preventing the induction of T cell anergy (4, 5). In contrast, the engagement of CTLA-4 by these same ligands results in down-regulation of the response that is essential for maintaining T cell homeostasis and self-tolerance (6–8).
B7-1 and B7-2, which share ≈25% sequence identity, are type I transmembrane glycoproteins expressed on the surfaces of antigen-presenting cells (APCs; refs. 9 and 10). Both B7 ligands bind CTLA-4 with 100- to 1,000-fold higher affinity than they do CD28, and B7-1 has ≈10-fold higher affinity for both receptors than does B7-2 (11, 12). Whereas B7-1 and B7-2 mainly have overlapping functions in T cell costimulation, comparative studies have suggested distinct immunoregulatory roles for these two proteins. For instance, although B7-2 is constitutively expressed on dendritic cells and can be rapidly up-regulated upon immune activation, B7-1 is only poorly expressed on resting dendritic cells, and its up-regulation occurs significantly later than that of B7-2. Furthermore, B7-2 expression is predominantly restricted to hematopoietic cells, whereas B7-1 is also expressed in nonhematopoietic cells such as the parenchymal cells of the pancreas, heart, and liver. The different kinetics and expression patterns of B7-1 and B7-2 have led to the hypothesis that B7-2, upon binding CD28, provides the costimulatory signals needed for the activation of naive T cells within draining lymphoid tissues, whereas B7-1 plays a major role in amplifying and regulating T cell activity at peripheral inflammatory sites (3). In addition, previous studies have suggested that whereas B7-2 plays an essential role in IL-4 secretion and T helper 2 responses, B7-1 preferentially induces IL-2 release and T helper 1 differentiation (13, 14). Another functional difference between these costimulatory ligands is that the engagements of B7-1 and B7-2 deliver distinct outside-in transmembrane signals to B cells. Crosslinking of B7-2 with anti-B7-2 mAb promotes the phosphorylation of the cytoplasmic tail of B7-2 and the enhancement of B cell proliferation and production of murine IgG1 and IgG2a or human IgG4 and IgE isotypes (3, 15, 16). In contrast, mAb crosslinking of B7-1 results in blocking both B cell proliferation and production of IgG1 and IgG2a isotypes (16, 17). The distinct signals transmitted to APCs by B7-1 and B7-2 may be due to differences in their cytoplasmic tails and/or to the structure and potential oligomerization state of their extracellular regions, which would control the organization of the associated intracellular signaling complexes.
Both B7-1 and B7-2 are composed of membrane distal Ig V-type and membrane proximal Ig C-type domains in their extracellular regions. The recently reported crystal structures of the B7-1/CTLA-4 complex (18) and the complex between CTLA-4 and the Ig V-type domain of B7-2 (19) have demonstrated that B7-1 and B7-2 bind CTLA-4 in a very similar manner, with the majority of the binding interface formed by the front face of the Ig V-type domain of B7-1/B7-2 and the CDR3 analogous (99MYPPPY104) loop of CTLA-4. However, several studies have shown that whereas the Ig V-type domain of B7-2 binds CD28 and CTLA-4 with the same affinities as those of the full-length extracellular region, both the Ig V-type and C-type domains of B7-1 are required for optimal binding to these receptors (20, 21). The crystal structures of unliganded human B7-1 and the B7-1/CTLA-4 complex have revealed numerous atomic contacts between the Ig V-type and C-type domains of B7-1, which may help stabilize the conformation of its variable domain (18, 22). This indirect effect may explain the requirement of the Ig C-type domain of B7-1 for optimal receptor binding.
Both biochemical and structural studies have shown that B7-1 is a dimer in the crystalline and solution states, albeit with weak affinity (20–50 μM; refs. 18 and 22). In the crystal structure of the B7-2/CTLA-4 complex, B7-2 exhibits a dimer interface reminiscent of that observed in B7-1 (19). The apparent dimeric nature of B7 molecules is critical for the formation of the alternating B7 and CTLA-4 array observed in the complex crystal structures and may play a role in the signaling complexes involving these receptor–ligand pairs in the immunological synapse (18, 19). Here, we report the structure of the unliganded, receptor-binding Ig V-type domain of human B7-2 to 2.7-Å resolution. Interestingly, in contrast to B7-2 in the B7-2/CTLA-4 complex and to both liganded and unliganded B7-1, the present structure does not suggest any significant B7-2 dimer interface and is consistent with our biochemical data showing that B7-2 is monomeric in solution. These observations suggest a lower tendency of B7-2 to dimerize and possibly a different signaling mechanism associated with this molecule. The structures of unbound B7-1 and B7-2, as well as the structures with which these ligands complexed to CTLA-4 are compared, extend our understanding of the structure–function relationships of these molecules in immune modulation.
Materials and Methods
Expression, Refolding, and Purification of the Receptor-Binding Domain of Human B7-2.
The receptor-binding domain (Ig V-like domain) of human B7-2, consisting of residues 1–109, was subcloned into the BamHI and NdeI restriction sites of the bacterial expression vector pET3a (Novagen) and transformed into the Escherichia coli strain BL21(DE3) and the methionine auxotroph B834(DE3) for the production of native and selenomethionyl-substituted proteins, respectively. Protein was expressed as inclusion bodies, and solubilized, refolded B7-2 was purified by anion-exchange chromatography, as described (23). Protein identity and purity were confirmed by mass spectrometry and N-terminal sequencing (data not shown). Gel filtration chromatography showed that this recombinant protein was monomeric in solution at concentrations up to 10 mg/ml (23).
Analytical Ultracentrifugation.
Sedimentation equilibrium experiments were performed at 20°C in an analytical ultracentrifuge using an An-60 Ti rotor and 1.2-cm six-channel charcoal-Epon centerpieces (Beckman Coulter). The receptor-binding domain of human B7-2 was concentrated and exchanged into 10 mM Tris⋅HCl/100 mM NaCl, pH 8.0 by using an Amicon concentrator with a 5-kDa cut-off membrane. Data were collected at protein concentrations of 77.9, 233.8, and 389.6 μM (1, 3, and 5 mg/ml) and rotor speeds of 25,000 and 34,000 rpm. Interference scans were taken after 24 and 26 h; equilibrium was assumed to have been reached if these scans were unchanged. Typically, two scans were taken at 26 h at each rotor speed. Data analysis was performed by using Beckman Coulter XL-A/XL-I V.4 data analysis software with the protein partial specific volume 0.7339 cm3·g−1 and the buffer density 1.0024 cm3·g−1. Partial specific volume was calculated from amino acid residue composition by using the program SEDNTERP V.1.03, and the buffer density was determined by using a Mettler DE40 density meter operated at the experimental temperature. Analysis consisted of the six scans taken of the three different nominal concentrations at each of the two rotor speeds.
Crystallization and Data Collection.
The Ig V-type domain of B7-2 was crystallized in buffer composed of 0.1 M NaAc, 0.1 M Tris⋅HCl, and 16–26% polyethylene glycol (PEG) 4000, pH 8.5 at 4°C, as described (23). For data collection, crystals of selenomethionyl-substituted proteins were flash-cooled at 100 K in crystallization buffer supplemented with 15% glycerol under a stream of nitrogen gas. Three-wavelength multiple wavelength anomalous dispersion data were collected at beamline X9B (National Synchrotron Light Source, Brookhaven National Laboratory, Upton, NY) by using a 2 × 2 charge-coupled device detector (Area Detector Systems Corp., Poway, CA). Data were collected at wavelengths 0.96110, 0.98000, and 0.98019 Å to a 2.7-Å resolution and reduced with the HKL suite (24). Diffraction of the crystals is consistent with orthorhombic space group P21212 (a = 56.69 Å, b = 63.01 Å, c = 58.61 Å, with two molecules per asymmetric unit, 40% solvent content).
Structure Determination and Refinement.
Seven of eight selenium sites were located and refined, and initial phases were calculated to a 3.0-Å resolution with the program SOLVE (25). The electron density map was improved by solvent flattening and twofold noncrystallographic symmetry (NCS) averaging using the program DM (26), allowing most β-strands and some loops to be identified. Two human CD2 variable domain models (PDB code: 1QA9) with all non-Gly residues mutated to Ser were docked into the density map by using a six-dimensional exhaustive phased-translation function (B. Strokopytov and S.C.A., unpublished work).
Native data set to 3.0-Å resolution extracted by SOLVE were used for refinement with CNS (27), including torsion angle dynamics simulated annealing refinement, strict NCS, individual B-factor refinement, and a bulk solvent correction. Of the total reflections, 10% were set aside for calculating Rfree. After the Rfree dropped below 30%, the refinement was extended to 2.7-Å resolution, and the NCS restraints were gradually loosened. The final model contains all residues from the B7-2 Ig V-type domain, including the initiator Met encoded by the bacterial expression vector, with an Rcryst and Rfree of 21.43% and 27.8%, respectively. The model has good geometry, with rms deviations from ideality of 0.008 Å for bond lengths and 1.4° for bond angles, with all other stereochemical parameters comparable with or exceeding those of structures at similar resolution. Data collection and refinement statistics are summarized in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org.
Molecular superimpositions and rms deviation calculations were performed with FIT (G. Lu, http://bioinfo1.mbfys.lu.se/∼guoguang/fit.html). Structure-based sequence alignments were performed with DALI, and the figure was generated with ALSCRIPT (28, 29). Molecular surfaces were generated with GRASP (30). Other figures were prepared with SETOR (31).
Results and Discussion
Overall Structure of the Receptor-Binding Domain of Human B7-2.
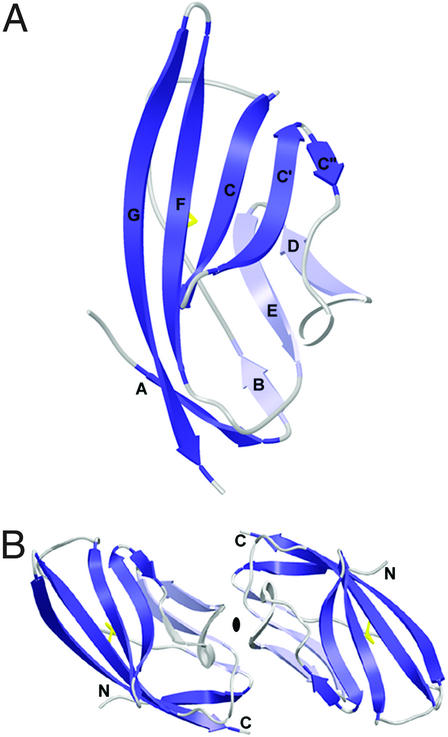
The B7-2 monomer in the present structure exhibits the same chain topology (two-layer β-sandwich) as that of B7-2 bound to CTLA-4 described in the complex crystal structure, with front and back sheets composed of AGFCC′C′′ and BED strands, respectively (Fig. 1A; ref. 19). There are two B7-2 Ig V-type domains related by 2-fold NCS in the asymmetric unit (Fig. 1B) which have very similar structures (as evidenced by the low rms deviation of 0.61 Å between equivalent Cα atoms). The two independent B7-2 Ig V-type domains in the structure of the B7-2/CTLA-4 complex exhibit an rms deviation of 0.32 Å, whereas all pairwise comparisons of the liganded and unliganded B7-2 molecules show rms deviations ranging from 0.70 to 0.88 Å (Fig. 2). The high structural similarity between the liganded and unliganded B7-2 indicates that B7-2 does not undergo a significant conformational change upon binding its receptor. The only pronounced structural difference between liganded and unliganded B7-2 is observed in the CDR3 analogous loop (FG loop) of B7-2, which makes several direct contacts with CTLA-4, as seen in the structure of the B7-2/CTLA-4 complex (19). Compared with that of unliganded B7-2, the CDR3 loop of complexed B7-2 adopts a somewhat “open” conformation (Fig. 2A), which may be required for better accommodation of the 99MYPPPY104 loop of CTLA-4, allowing for optimal interaction. Based on the near sequence invariance of the ligand-binding sites in CD28 and CTLA-4, it is predicted that the B7/CTLA-4 complexes provide a reliable model for the B7/CD28-binding interface (19).
Figure 1.
Structure of the receptor-binding domain of human B7-2. (A) Ribbon diagram of the structure of the B7-2 monomer. Strands from the front sheet (AGFCC′C′′) and the back sheet (BED) are dark and light blue, respectively. The intrachain disulfide is yellow. (B) The two monomers in the asymmetric unit are related by twofold NCS symmetry, with the twofold axis (denoted by the black oval) perpendicular to the plane of the page. N and C termini of the two monomers are labeled with N and C, respectively.
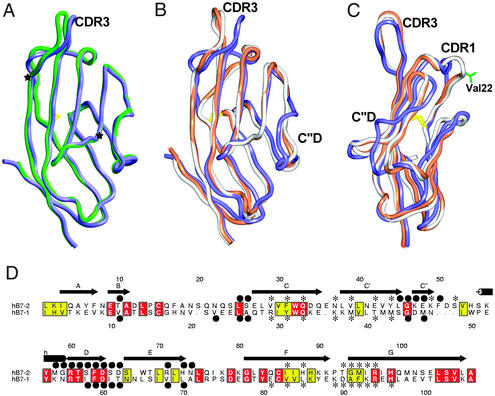
Figure 2.
Comparison of the receptor-binding domains of B7-1 and B7-2. (A) Superimposition of the unliganded B7-2 monomer (dark blue) and B7-2 monomer from the B7-2/CTLA-4 complex (green), demonstrating the high structural similarity between unliganded and liganded B7-2. The two β-bulges at Leu-38 and Arg-97 in B7-2 are highlighted by two black stars. (B and C) Two orthogonal views of a superimposition of B7-2 (dark blue), unbound B7-1 (pink), and B7-1 from the B7-1/CTLA-4 complex (gray). Regions with significant structural difference between B7-1 and B7-2 are labeled. (D) Structure-based sequence alignment of the receptor-binding domain of human B7-2 and B7-1. Residues from B7-2 participating in the dimer interface (●) and CTLA-4 binding (*) are indicated above the sequences, whereas those from B7-1 are indicated below the sequences. The numbering schemes for B7-2 and B7-1 are also shown above and below the sequences, respectively. Conserved (yellow) and identical (red) residues are highlighted.
Three-dimensional structural comparison with DALI (28) revealed that the B7-2 Ig V-type domain is most similar to the membrane distal domains of human B7-1 (PDB code: 1DR9; rms deviation of 2.3 Å for 103 equivalent Cα atoms), CD4 (PDB code: 1CDY; rms deviation of 1.7 Å for 93 equivalent Cα atoms), CD58 (PDB code: 1CCZ-A; rms deviation of 2.0 Å for 90 equivalent Cα atoms), and CD2 (PDB code: 1QA9; rms deviation of 2.0 Å for 95 equivalent Cα atoms). Similar to the membrane distal domains of B7-1 and CD2, the Ig V-type domain of B7-2 contains two β-bulges at Leu-38 (equivalent to Met-38 in B7-1) and Arg-97 (equivalent to Arg-94 in B7-1) immediately C-terminal to the CC′ and FG loops, respectively (Fig. 2A; refs. 22 and 32). These features are not typical of Ig V-type domains, in which the two β-bulges are generally located within the C′ and G strands, resulting in a more extensive twist to the front sheet than that observed for CD2, B7-1, and B7-2.
Comparison of B7-1 and B7-2.
Superimpositions of the receptor-binding domain of B7-2 with the analogous domain of both unliganded and liganded B7-1 show that B7-2 has similar overall structure to that of B7-1 (rms deviations of 2.5 and 2.3 Å for 104 equivalent Cα atoms, respectively), with significant differences in the CDR1, C′′D, and CDR3 loops (Fig. 2 B and C; refs. 18 and 22). The CDR1 segments of B7-1 and B7-2 are both composed of coil and α-helix; however, the CDR1 of B7-1 contains two α-helical turns, whereas that of B7-2 contains only a single turn. The additional helical turn in B7-1 results in the protrusion of several residues; in particular, Val-22 makes multiple contacts with the other monomer in the dimer and serves to stabilize the dimer interface. The C′′D loop of B7-2 contains three additional residues compared with the analogous loop in B7-1. These three residues in B7-2 form a bulge at the edge of the receptor-binding face that has been shown to be involved in receptor binding (19).
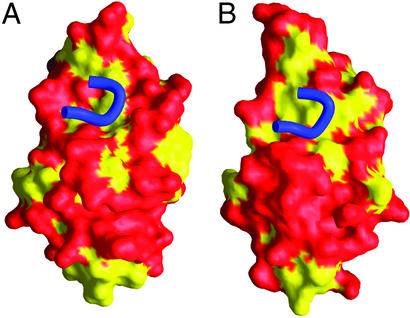
Even though B7-1 and B7-2 bind their CD28/CTLA-4 receptors with only ≈10-fold different affinities, they have low sequence homology in their receptor-binding domains (≈23% identity), with the majority of residues in B7-2 that contribute to receptor binding not conserved in B7-1 (Fig. 2D). Despite the apparent sequence divergence, the front faces of both B7-1 and B7-2 form shallow concave surfaces, which serve to accommodate binding of the 99MYPPPY104 loop in CTLA-4, as observed in the crystal structures of the B7-1/CTLA-4 and B7-2/CTLA-4 complexes (Fig. 3). Among the few conserved residues at the receptor-binding interface, the hydrophobic Phe-31 in B7-2 and Tyr-31 in B7-1 have been shown to be crucial for receptor binding by mutagenesis studies (33). Consistent with this observation, the crystal structures of the B7/CTLA-4 complexes demonstrate that the aromatic rings of Phe-31 and Tyr-31 pack against the side-chain pyrrolidine ring of Pro-102 in CTLA-4 at the core of the interfaces, potentially contributing significant binding energy to these receptor–ligand interactions (18, 19). In addition to the general architectural similarity of the receptor-binding faces of B7-1 and B7-2, physical similarities are also observed. Nonpolar residues (Val-29, Phe-31, Val-39, Ile-86, Met-95, and Ile-96) on the front face of B7-2 form a hydrophobic patch centered around Phe-31, which makes numerous van der Waals interactions with the hydrophobic 99MYPPPY104 loop of CTLA-4, as observed in the structure of the B7-2/CTLA-4 complex (Fig. 3). Similarly, hydrophobic residues Tyr-31, Met-38, Met-43, Val-83, Leu-85, Ala-91, and Phe-92 on the front face of B7-1, most of which occupy positions analogous to those of the residues in B7-2 that form the hydrophobic patch, seem to play the same role in receptor binding as shown by the structure of B7-1/CTLA-4 complex. The hydrophobic patches on the front faces of both B7-1 and B7-2 are surrounded by a ring of hydrophilic residues reminiscent of a classic protein–protein interface (34). Overall physical and chemical similarity of the B7-1 and B7-2 receptor-binding faces, combined with their detailed sequence differences, may provide an explanation for the fact that these ligands interact with the same receptors by using a similar binding mode, but with different affinities.
Figure 3.
Comparison of the receptor-binding interfaces of B7-2 and B7-1. In both B7-2 (A) and B7-1 (B), hydrophobic and hydrophilic residues are yellow and red, respectively. The CDR3 analogous loop (99MYPPPY104) of CTLA-4 is shown in blue in each figure to indicate the binding modes of B7-2/CTLA-4 and B7-1/CTLA-4.
Mutagenesis studies have shown that a conservative substitution of Tyr-100 in CTLA-4 with Phe selectively eliminated binding to B7-2, without affecting B7-1 binding (35). The structure of the B7-2/CTLA-4 complex demonstrates that the hydroxyl group of Tyr-100 forms a hydrogen-bonding network with Glu-42 and Lys-49 in B7-2, which may contribute significantly to the stability of the interaction between B7-2 and CTLA-4. Superimposition of B7-1 and B7-2 structures shows that the equivalent residues are Thr-41 and Asn-48 in B7-1, which have much shorter side chains than their respective counterparts in B7-2 and, as a consequence, cannot form hydrogen bonds with the hydroxyl groups of Tyr-100 in CTLA-4, as demonstrated by the crystal structure of the B7-1/CTLA-4 complex. This observation suggests that the hydroxyl group of Tyr-100 does not contribute to the B7-1/CTLA-4 interaction and explains why the Tyr-100→Phe mutation only affects the binding of CTLA-4 to B7-2. Similar behavior is exhibited by the CD28 Tyr-100→Phe mutant and further supports the proposed similarity between the B7/CTLA-4 and B7/CD28 interfaces (36).
Oligomerization State of the Unliganded Receptor-Binding Domain of B7-2.
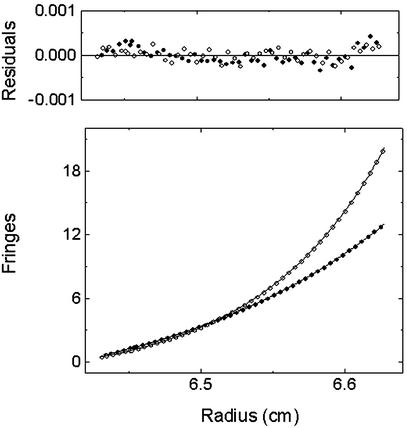
Previous biochemical and structural studies have shown that B7-1 is dimeric both in the crystalline state and in solution with a modest Kd in the 20–50 μM range (18, 22). In contrast, gel filtration chromatography showed that the receptor-binding domain of B7-2 elutes as a single monodispersed peak of ≈13 kDa, indicating that B7-2 exists as a monomeric species in solution (23). This result was confirmed by sedimentation equilibrium experiments (Fig. 4). Fitting the data to a single-component model yielded values within experimental error of the theoretical monomeric molecular mass (12.8 kDa) of the B7-2 receptor-binding domain. Additional data fitting to self-association models (i.e., monomer ↔ dimer, monomer ↔ trimer, and monomer ↔ tetramer) failed to converge. These results demonstrate that the receptor-binding domain of human B7-2 is a stable monomer and does not measurably oligomerize in solution.
Figure 4.
Determination of the oligomerization state of the receptor-binding domain of human B7-2 in solution by sedimentation equilibrium. Black points (●) and white points (○) represent data collected at 25,000 and 34,000 rpm, respectively. For clarity, every sixth point is plotted. Lines represent the best fit to a single-component sedimentation model, yielding an estimate of the apparent molecular mass, 12.6 kDa. The residuals of the corresponding points are randomly distributed, indicating that there is no oligomerization occurring and that the sample consists of only monomers.
In the crystal of the receptor-binding domain of B7-2, two types of dimers form as the consequence of crystal packing. One dimer is composed of the two monomers in the asymmetric unit that are related by twofold NCS (Fig. 1). The other dimer results from crystallographic twofold symmetry. However, based on known B7/CTLA-4-binding mode, neither of these two dimers accommodates receptor binding from an opposing T cell membrane (not shown). In addition, the NCS dimer is a tail-to-tail anti-parallel dimer with the C terminus of each monomer pointing in opposite directions, and it is apparent that two monomers from a single cell cannot form such a dimer (Fig. 1). These observations, in combination with the fact that each of these two dimers has a small dimer interface (only ≈700 and ≈500 Å2 buried solvent-accessible surface area for the NCS and crystallographic dimer, respectively), support the notion that neither of these dimers is physiologically relevant. Given that the dimer interface observed in the B7-1 crystals is formed exclusively by contacts between residues in its Ig V-type domains (18, 22), we believe that it is unlikely that the monomeric state of B7-2 is due to the absence of the Ig C-type domain in our recombinant protein.
Comparison of the Dimer Interfaces of B7-1 and B7-2 in the Complex Structures.
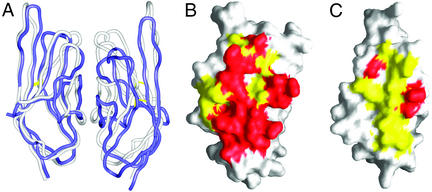
In contrast to unliganded B7-2, the crystal structure of the B7-2/CTLA-4 complex demonstrated that liganded B7-2 forms a dimer that is similar in many aspects to that observed in the B7-1 structures (18, 19, 22). The dimer interfaces of both B7-1 and B7-2 are predominantly formed by their back strands (B, E, and D) and loops and bury similar total solvent-accessible surface areas of 1,220 and 1,405 Å2, respectively (Figs. 1 A and 5). After superimposition of one monomer from each dimer (based on 104 Cα atoms), the remaining monomers show a minor displacement described by an 11° rotation and a 1.7-Å translation (Fig. 5). This displacement reflects the fact that, whereas the B7-1 dimer shows proper twofold symmetry, the two monomers in the B7-2 dimer are not related by perfect twofold rotational symmetry, which may be a consequence of crystal lattice-packing effects. Thus, the B7-1 dimer might serve as a better model for the actual dimerization mode of B7-2. It is worthwhile to point out that simple modeling based on the dimerization modes of both B7-1 and B7-2 suggests that the potential glycosylation of B7-2 does not interfere with its dimerization (not shown). Based on our structural sequence alignment, the majority of residues contributing to the dimer interfaces in B7-1 and B7-2 occupy the same positions in their respective primary sequences (Fig. 2D); however, the chemical properties of the dimer interfaces of B7-1 and B7-2 are very different. In the B7-1 dimer, 11 of 13 residues making contacts in the dimer interface are hydrophobic (Figs. 2 and 5). In contrast, 15 of 20 residues contributing to the B7-2 dimer interface are hydrophilic, including seven charged residues. Consistent with our biochemical data, these observations indicate that B7-2 would be a weak dimer because of the more polar nature of its dimer interface compared with that of B7-1, which is predominantly hydrophobic (37). Importantly, the chemical differences at the respective dimer interfaces provide a potential mechanism to prevent the formation of B7-1 and B7-2 heterodimers, which would have unknown functional consequence given the different roles B7-1 and B7-2 play in the regulation of T cell and B cell activity. In light of the above considerations (i.e., the analytical ultracentrifugation results, and the asymmetric and weak dimer interface), the physiological relevance of the apparent B7-2 dimer remains to be firmly established.
Figure 5.
Comparison of the B7-2 dimer from the B7-2/CTLA-4 complex and the B7-1 dimer. (A) Superimposition showing the conformational similarity between the B7-2 dimer (blue) and B7-1 dimer (gray). The superimposition was performed based on one monomer from each dimer, which resulted in a fair overlay between the two dimers, with a minor displacement of an 11° rotation and a 1.7-Å translation between the remaining two monomers. (B and C) Surface properties of the dimer interfaces of B7-2 and B7-1. Hydrophobic and hydrophilic residues involved in the dimer interfaces are yellow and red, respectively. It is obvious that the B7-2 dimer interface is dominated by hydrophilic residues, whereas that of B7-1 is predominantly formed by hydrophobic residues.
Taken together, biochemical and crystallographic analyses suggest that both B7-1 and B7-2 have a weak tendency to dimerize under some conditions, although the B7-1 dimer interface appears to be considerably stronger than that of B7-2. The free and complexed B7 monomers exhibit high structural similarity, suggesting that receptor binding does not promote or enhance B7 dimerization through local or global conformational changes. However, the dimerization of B7 isoforms may be favored at the T cell/APC interface, because this two-dimensional arrangement of B7 molecules on the cell surface decreases the entropic penalty for oligomerization by constraining its orientation to one more favorable for dimerization, compared with the random distribution of individual B7 molecules in solution. In addition, CD28 and, subsequently, CTLA-4 are enriched in the immunological synapse during T cell activation (38, 39). These receptors at the immunological synapse may bind and recruit B7 molecules, bringing them into proximity, increasing their local concentrations, and further constraining them in a more productive orientation for dimerization. These entropic effects may provide an efficient conformation-independent mechanism for B7 dimerization on the surfaces of APCs.
Several lines of evidence implicate B7-2 as an APC-signaling molecule. It has been shown that crosslinking of B7-2 with anti-B7-2 antibody increases cellular proliferation and IgG4 and IgE secretion by activated B cells (15, 16). Consistent with its potential role in signaling, B7-2 has a long cytoplasmic tail with at least three potential protein kinase C phosphorylation sites, which have been shown to be phosphorylated after B cell activation (3). Thus, the putative receptor-induced dimerization of B7-2 (perhaps as observed in the B7-2/CTLA-4 crystal structure) may not only promote the assembly of stable inhibitory signaling complexes but also provide a mechanism for B7-2 signaling in APCs. A similar mechanism has been proposed for class II MHC, also a weak dimer, which may form a more stable dimer upon binding its cognate T cell receptor, resulting in signal transduction to APCs (40, 41). The oligomerization state of B7-2 will also affect the potential signaling mechanism involved in CTLA-4/CD28 functions, as monomeric B7-2 would only support the formation of a trimeric complex (one dimeric receptor and one B7-2 molecule) or a tetrameric complex (one dimeric receptor and two B7-2 molecules), instead of the long-range ordered arrays of receptor–ligand pairs associated with the dimeric B7-2 molecule (19). Monomeric B7-2 would only function in localizing individual CTLA-4/CD28 dimers to the immunological synapse rather than leading to the formation of the ordered signaling lattice. In contrast to B7-2, considerable crystallographic and solution data suggest a preformed B7-1 dimer on cell surfaces, precluding a receptor-induced dimerization mechanism for signaling. In further contrast to B7-2, several studies have shown that the engagement of B7-1 results in signals to inhibit B cell proliferation and antibody secretion (16, 17). The higher-order clustering of B7-1 by the binding of anti-B7-1 antibody or bivalent CTLA-4/CD28 might be a major mechanism for B7-1 signaling. The differences in their cytoplasmic tails and/or signaling mechanisms may explain the different signals B7-1 and B7-2 deliver to B cells. Of course, these discussions must be considered speculative, given the lack of experimental data concerning the oligomerization state of B7-1 or B7-2 on cell surfaces.
Supplementary Material
Acknowledgments
We thank Dr. S. Porcelli and M. Roden for insightful discussions, Dr. K. Rajashankar for assistance with data collection, and Dr. M. Brenowitz and S. Morris for assistance with analytical ultracentrifugation experiments. This work was supported by National Institutes of Health Grants AI07289 and P30CA13330 and National Cancer Institute Training Grant T32CA09173.
Abbreviations
- APC
antigen-presenting cell
- NCS
noncrystallographic symmetry
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID 1NCN).
References
- 1.Chambers C A, Kuhns M S, Egen J G, Allison J P. Annu Rev Immunol. 2001;19:565–594. doi: 10.1146/annurev.immunol.19.1.565. [DOI] [PubMed] [Google Scholar]
- 2.Brunet J F, Denizot F, Luciani M F, Roux-Dosseto M, Suzan M, Mattei M G, Golstein P. Nature. 1987;328:267–270. doi: 10.1038/328267a0. [DOI] [PubMed] [Google Scholar]
- 3.Lenschow D J, Walunas T L, Bluestone J A. Annu Rev Immunol. 1996;14:233–258. doi: 10.1146/annurev.immunol.14.1.233. [DOI] [PubMed] [Google Scholar]
- 4.Thompson C B, Lindsten T, Ledbetter J A, Kunkel S L, Young H A, Emerson S G, Leiden J M, June C H. Proc Natl Acad Sci USA. 1989;86:1333–1337. doi: 10.1073/pnas.86.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linsley P S, Brady W, Grosmaire L, Aruffo A, Damle N K, Ledbetter J A. J Exp Med. 1991;173:721–730. doi: 10.1084/jem.173.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walunas T L, Lenschow D J, Bakker C Y, Linsley P S, Freeman G J, Green J M, Thompson C B, Bluestone J A. Immunity. 1994;1:405–413. doi: 10.1016/1074-7613(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 7.Waterhouse P, Penninger J M, Timms E, Wakeham A, Shahinian A, Lee K P, Thompson C B, Griesser H, Mak T W. Science. 1995;270:985–988. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 8.Tivol E A, Borriello F, Schweitzer A N, Lynch W P, Bluestone J A, Sharpe A H. Immunity. 1995;3:541–547. doi: 10.1016/1074-7613(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 9.Freeman G J, Freedman A S, Segil J M, Lee G, Whitman J F, Nadler L M. J Immunol. 1989;143:2714–2722. [PubMed] [Google Scholar]
- 10.Freeman G J, Gribben J G, Boussiotis V A, Ng J W, Restivo V A, Jr, Lombard L A, Gray G S, Nadler L M. Science. 1993;262:909–911. doi: 10.1126/science.7694363. [DOI] [PubMed] [Google Scholar]
- 11.Linsley P S, Greene J L, Brady W, Bajorath J, Ledbetter J A, Peach R. Immunity. 1994;1:793–801. doi: 10.1016/s1074-7613(94)80021-9. [DOI] [PubMed] [Google Scholar]
- 12.Greene J L, Leytze G M, Emswiler J, Peach R, Bajorath J, Cosand W, Linsley P S. J Biol Chem. 1996;271:26762–26771. doi: 10.1074/jbc.271.43.26762. [DOI] [PubMed] [Google Scholar]
- 13.Freeman G J, Boussiotis V A, Anumanthan A, Bernstein G M, Ke X Y, Rennert P D, Gray G S, Gribben J G, Nadler L M. Immunity. 1995;2:523–532. doi: 10.1016/1074-7613(95)90032-2. [DOI] [PubMed] [Google Scholar]
- 14.Kuchroo V K, Das M P, Brown J A, Ranger A M, Zamvil S S, Sobel R A, Weiner H L, Nabavi N, Glimcher L H. Cell. 1995;80:707–718. doi: 10.1016/0092-8674(95)90349-6. [DOI] [PubMed] [Google Scholar]
- 15.Jeannin P, Delneste Y, Lecoanet-Henchoz S, Gauchat J F, Ellis J, Bonnefoy J Y. J Biol Chem. 1997;272:15613–15619. doi: 10.1074/jbc.272.25.15613. [DOI] [PubMed] [Google Scholar]
- 16.Suvas S, Singh V, Sahdev S, Vohra H, Agrewala J N. J Biol Chem. 2002;277:7766–7775. doi: 10.1074/jbc.M105902200. [DOI] [PubMed] [Google Scholar]
- 17.Hirokawa M, Kuroki J, Kitabayashi A, Miura A B. Immunol Lett. 1996;50:95–98. doi: 10.1016/0165-2478(96)02526-6. [DOI] [PubMed] [Google Scholar]
- 18.Stamper C C, Zhang Y, Tobin J F, Erbe D V, Ikemizu S, Davis S J, Stahl M L, Seehra J, Somers W S, Mosyak L. Nature. 2001;410:608–611. doi: 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz J C, Zhang X, Fedorov A A, Nathenson S G, Almo S C. Nature. 2001;410:604–608. doi: 10.1038/35069112. [DOI] [PubMed] [Google Scholar]
- 20.Rennert P, Furlong K, Jellis C, Greenfield E, Freeman G J, Ueda Y, Levine B, June C H, Gray G S. Int Immunol. 1997;9:805–813. doi: 10.1093/intimm/9.6.805. [DOI] [PubMed] [Google Scholar]
- 21.Ellis J H, Burden M N, Vinogradov D V, Linge C, Crowe J S. J Immunol. 1996;156:2700–2709. [PubMed] [Google Scholar]
- 22.Ikemizu S, Gilbert R J, Fennelly J A, Collins A V, Harlos K, Jones E Y, Stuart D I, Davis S J. Immunity. 2000;12:51–60. doi: 10.1016/s1074-7613(00)80158-2. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Schwartz J C, Almo S C, Nathenson S G. Protein Expression Purif. 2002;25:105–113. doi: 10.1006/prep.2002.1616. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger T C, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowtan K, Main P. Acta Crystallogr D. 1998;54:487–493. doi: 10.1107/s0907444997011980. [DOI] [PubMed] [Google Scholar]
- 27.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Sander C. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 29.Barton G J. Methods Enzymol. 1990;183:403–428. doi: 10.1016/0076-6879(90)83027-7. [DOI] [PubMed] [Google Scholar]
- 30.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 31.Evans S V. J Mol Graphics. 1993;11:127–134. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- 32.Bodian D L, Jones E Y, Harlos K, Stuart D I, Davis S J. Structure (London) 1994;2:755–766. doi: 10.1016/s0969-2126(94)00076-x. [DOI] [PubMed] [Google Scholar]
- 33.Peach R J, Bajorath J, Naemura J, Leytze G, Greene J, Aruffo A, Linsley P S. J Biol Chem. 1995;270:21181–21187. doi: 10.1074/jbc.270.36.21181. [DOI] [PubMed] [Google Scholar]
- 34.Bogan A A, Thorn K S. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 35.Harris N, Peach R, Naemura J, Linsley P S, Le Gros G, Ronchese F. J Exp Med. 1997;185:177–182. doi: 10.1084/jem.185.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kariv I, Truneh A, Sweet R W. J Immunol. 1996;157:29–38. [PubMed] [Google Scholar]
- 37.Jones S, Thornton J M. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bromley S K, Iaboni A, Davis S J, Whitty A, Green J M, Shaw A S, Weiss A, Dustin M L. Nat Immunol. 2001;2:1159–1166. doi: 10.1038/ni737. [DOI] [PubMed] [Google Scholar]
- 39.Egen J G, Allison J P. Immunity. 2002;16:23–35. doi: 10.1016/s1074-7613(01)00259-x. [DOI] [PubMed] [Google Scholar]
- 40.Brown J H, Jardetzky T S, Gorga J C, Stern L J, Urban R G, Strominger J L, Wiley D C. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- 41.Schafer P H, Malapati S, Hanfelt K K, Pierce S K. J Immunol. 1998;161:2307–2316. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.