Abstract
SWI/SNF complexes in yeast and higher eukaryotes are thought to facilitate gene activation and transcription factor binding by disrupting repressive chromatin structures. Little is known, however, about how these complexes target specific genes for activation. We now have purified a specialized SWI/SNF-related complex (PYR complex) from murine erythroleukemia (MEL) cell nuclear extract that binds pyrimidine-rich elements at the human and murine β-globin loci. PYR complex DNA-binding activity is restricted to definitive hematopoietic cells and is both DNA sequence- and length-dependent. Mass spectrometric identification of purified peptides and antibody supershift assays indicate that PYR complex contains at least four known mammalian SWI/SNF subunits: BAF57, INI1, BAF60a, and BAF170. PYR complex broadly footprints a 250-bp pyrimidine-rich element between the human fetal and adult β-globin genes. A short intergenic deletion that removes this element from a human globin locus cosmid construct results in delayed human fetal-to-adult globin gene switching in transgenic mice. Taken together, the data suggest that PYR complex may act through this intergenic element to facilitate human fetal-to-adult globin gene switching, presumably by opening the locus in the region of the adult genes to permit the binding of β-globin transcriptional activators.
Keywords: chromatin remodeling, gene regulation
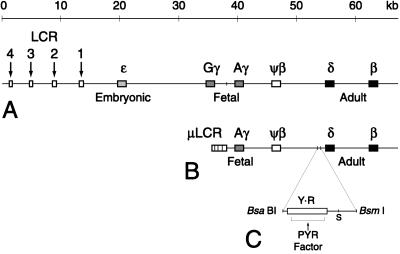
The human β-globin locus is comprised of embryonic (ɛ), fetal (Gγ and Aγ), and adult (δ and β) genes arranged in the developmental order of their expression downstream of a powerful tissue-specific enhancer, the locus control region (LCR, Fig. 1A). Expression of the β-globin-like genes is characterized by two developmental “switches,” first from ɛ- to γ-globin expression in early fetal life, then from γ- to δ- and β-globin expression just after birth (1, 2). Although a number of erythroid- and hematopoietic cell-specific DNA-binding factors have been described that are known to function in the transcriptional activation of the locus, there is no evidence for an individual factor that controls switching (2, 3). GATA-1 and NF-E2, critical factors required for erythroid cell development and differentiation, are present at relatively constant levels throughout blood cell development and appear to function as general activators of globin gene transcription (2, 3). EKLF, a factor that specifically activates the β-globin gene, also is expressed throughout hematopoiesis but is unable to exert its effect on the β-globin gene in embryonic and fetal cells (4–7). This finding suggests that a level of control exists for the locus beyond that of the presence or absence of transcription factors.
Figure 1.
Maps of the human β-globin locus, DNA constructs, and the PYR factor binding site upstream of the δ-globin gene. (A) The human β-globin locus on chromosome 11. The LCR is characterized by four erythroid-specific DNase I hypersensitive sites (arrows) upstream of the embryonic ɛ gene. There are duplicated fetal genes (Gγ and Aγ), a pseudogene (ψβ), and two adult genes (δ and β). The vertical line upstream of Aγ marks a HindIII site. (B) Map of the μLCRAγψβδβ (wt) mini-locus construct used in transgenic mice. It consists of a 2.5-kb cassette containing each of the four LCR DNase I hypersensitive sites (μLCR) linked to a 29-kb HindIII–KpnI restriction fragment containing the Aγ through β genes. Vertical lines upstream of the δ gene delineate the 511-bp BsaBI–BsmI restriction fragment removed from the construct to create μLCRAγψβδβ-Δ (Δ). (C) Expanded view of the deleted BsaBI–BsmI fragment showing the long pyrimidine-rich domain (rectangle Y⋅R) and the region footprinted by PYR factor. The segment from the BsaBI site to a Sau3AI site (S) has been analyzed for transcription factor binding sites by DNase I footprinting.
It has been proposed that transcriptional control at the level of chromatin structure may be an important component of globin gene switching (8, 9). The control of higher-order chromatin structure is a well-established mechanism for the regulation of a number of gene loci and is probably critical in the regulation of sets of genes that require very tight “all-or-none” expression between different tissues (8, 10). Developmental regulation of the Drosophila homeotic genes, which, analogous to the globin genes, are organized in their spatial order of expression, is tightly controlled at the level of chromatin structure. Homeotic gene silencing is maintained by Polycomb group (PcG) proteins, which form closed chromatin complexes that maintain stable, heritable states of transcriptional repression in the locus (11, 12). Homeotic gene activation requires trithorax group (trxG) proteins such as BRAHMA, which act as components of large protein complexes homologous to yeast SWI/SNF to disrupt chromatin-mediated transcriptional repression and facilitate the binding of transcription factors (11, 13). Homologues to PcG and trxG proteins and the yeast SWI/SNF complex recently have been shown to exist in mammalian cells (14).
There is evidence that the mammalian globin genes may be controlled by similar mechanisms (8). In mouse embryonic erythroblast-murine erythroleukemia (MEL) cell hybrids, a heritable state of β-globin silencing similar to Polycomb group-mediated repression has been described (15). In addition, naturally occurring mutations in the ATRX (XH2) gene, which is structurally similar to Drosophila brahma and yeast SNF2, are associated with a form of α-thalassemia in which the structure of the α-globin locus itself is normal (16). Thus, the inherited lack of a trans-activator protein that is very likely a mammalian SWI/SNF complex subunit is associated with loss of α-globin expression.
We previously have reported a DNA-binding activity restricted to adult hematopoietic cells (PYR factor) that recognizes a long pyrimidine-rich sequence between the human fetal and adult β-globin-like genes (Fig. 1C) (17). PYR factor activity is abundant in adult erythroid (MEL), myeloid (HL60), T- and B-lymphoid (EL4, CEM, and Daudi), and megakaryocytic (CMK) cell lines. It is only very faintly present in embryonic-fetal erythroid (K562) cells and is absent in nonhematopoietic cell lines (HeLa, 3T3, and COS). More importantly, there is only very weak PYR factor activity in nuclear extracts from embryonic day 11 (E11) mouse yolk sac (a primitive hematopoietic tissue, expressing primarily mouse embryonic globin), but strong PYR factor activity in E14 mouse fetal liver (a definitive hematopoietic tissue, expressing only adult globin) that is not found in adult liver (17). We now have purified PYR factor from MEL cell nuclear extract and show that it is a large protein complex (PYR complex) that contains at least four known mammalian SWI/SNF complex subunits. PYR complex binds specifically to pyrimidine-rich DNA sequences in a DNA length- and sequence-dependent manner, suggesting that it recognizes both DNA sequence and structure. In addition, deletion of a short intergenic element containing the PYR complex binding site from a human β-globin locus construct delays human γ-to-β globin gene switching in transgenic mice. Taken together, the data suggest that PYR complex acts through this intergenic element to facilitate fetal-to-adult globin gene switching, most likely through an effect on higher-order chromatin structure.
MATERIALS AND METHODS
DNA Binding Studies.
MEL cells (GM00086E) were obtained from the National Institute of General Medical Sciences Cell Repository, Camden, NJ. Nuclear extracts were prepared as described (17, 18). Gel shift assays were performed as described (17, 19) and electophoresed on nondenaturing 4% polyacrylamide gels. For supershift assays, crude nuclear extract or purified fractions were incubated with 1–2 μl of undiluted antibody for 1–2 hr before the addition of DNA probe. Antibodies were obtained commercially (RbAp46/48, GeneTex, San Antonio, TX; HMGI(Y), Santa Cruz Biotechnology), and from Gerald Crabtree (Stanford University, Stanford, CA; BAF57, BAF60a, BAF155, and BAF170) and Ganjam Kalpana (Albert Einstein Medical College, Bronx, NY; INI1 PB3). Supershift assays were electrophoresed on 3.9% polyacrylamide gels. Probes and competitor DNAs (sense strand shown) used are: δ99, CCTCCATCCCTTCCATCCTCTCTCTTCCCCTCTTCCTTCCTTCCTTTCTCCATTTCTTCCTCCTCTTTCCCTCAATCCTTCCTTTTGGATATGCTCATG; δ60, GATCCTCTCTCTTCCCCTCTTCCTTCCTTCCTTTCTCCATTTCTTCCTCCTCTTTCCCTC; δ60ym, GATCCTCTCTCTTCCCCTCTTCCTTCCTTCCTTTCCTTATTTCTTCCTCCTCTTTCCCTC; δ46, TTCCATCCTCTCTCTTCCCCTCTTCCTTCCTTCCTTTCTCCATTTC; δ49, GATCCCTCTTCCTTCCTTCCTTTCTCCATTTCTTCCTCCTCTTTCCCTG; and YY1, ACGTCGCTCCGCGGCCATCTTGGCGGCTGGT.
Purification of PYR Complex.
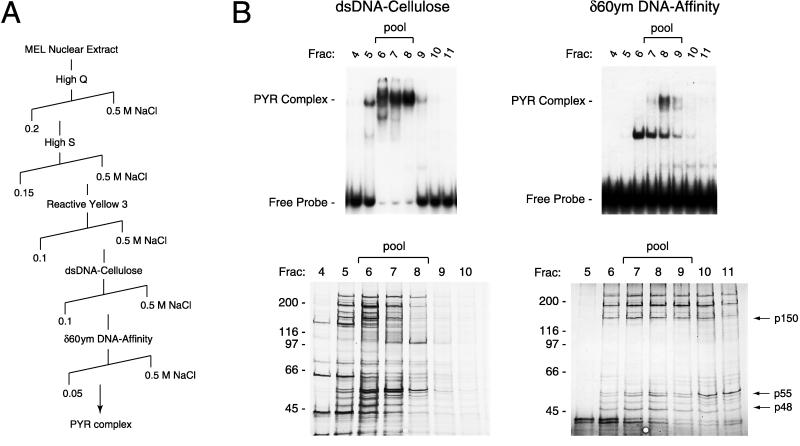
PYR complex was purified from approximately 800 mg of MEL crude nuclear extract in five chromatographic steps (Fig. 2). Column fractions from each step were assayed for PYR complex DNA-binding activity by using gel shift assays with a radiolabeled 60-bp double-stranded oligonucleotide, (δ60ym, see above) derived from the PYR complex binding site that has a mutation in a YY1 recognition sequence also present at that site. Fractions containing peak activity were pooled and concentrated overnight in Centriplus or Centricon centrifugal concentrators (Amicon) to preserve PYR complex activity, which is lost if the samples become too diluted. To construct the DNA affinity column used for the final step, approximately 400 μg of double-stranded δ60ym oligonucleotide was coupled to Sepharose CL-2B as described (20). Peak activity off the affinity column was pooled and precipitated with trichloroacetic acid, electrophoresed on a 4–15% SDS polyacrylamide gel, and electroblotted onto a nitrocellulose membrane.
Figure 2.
Purification of PYR complex from MEL nuclear extract. (A) Purification scheme. PYR complex was purified from 800 mg of MEL crude nuclear extract in five chromatographic steps, using continuous NaCl gradients (diagonal lines) to elute PYR complex activity from each column. (B) Analysis of fractions from the final two purification steps by gel shift assay (Upper) and silver-stained 7.5% SDS/PAGE (Lower). Fractions from the double-stranded DNA (dsDNA) cellulose column containing peak PYR complex activity (fractions 6–8, Left) were pooled and applied to a 1-ml δ60ym DNA-affinity column. High molecular weight PYR complex activity peaks off the affinity column in fraction 8 (Right), with a low molecular weight δ60ym-binding activity, most probably a smaller subunit of PYR complex that still retains DNA-binding activity, eluting in earlier fractions (peak in fraction 6). Bands with approximate molecular masses of 48, 55, and 150 kDa (p48, p55, and p150, arrows) from pooled fractions 7–9 subsequently were identified as SWI/SNF complex subunits INI1 (p48) BAF57 (p55), and BAF155 (p150) by mass spectrometric analysis.
Mass Spectrometric Analysis of Proteolytic Peptides.
Bands were processed for internal sequence analysis as described (21). The tryptic peptide mixture then was partially fractionated on a Poros 50 R2 RP-microtip (22), and resulting peptide pools were analyzed by matrix-assisted laser desorption ionization (MALDI) reflectron time-of-flight (reTOF) MS, using a REFLEX III instrument (Bruker-Franzen, Bremen, Germany). Electrospray ionization (ESI) MS was done on an API 300 triple quadrupole instrument (PE-SCIEX, Thornhill, Canada), modified with an injection adaptable fine ionization source as described (23). Selected mass values from the MALDI–reTOF experiments were taken to search a protein nonredundant database (EBI, Hinxton, UK) using the peptidesearch (24) algorithm. MS/MS spectra from the ESI triple quadrupole analyses were inspected for y" ion series and the resultant information transferred to the sequencetag (25) program and used as a search string. Any protein identification thus obtained was verified by comparing the computer-generated fragment ion series of the predicted tryptic peptide with the experimental MS/MS data.
Analysis of Human β-Globin Locus Constructs in Transgenic Mice.
The 32-kb wild-type (wt) mini-locus (26) is propagated as a cosmid, pHC79-μLCRAγψβδβ (pHC79-wt). The mutant cosmid (pHC79-Δ) was constructed by digesting pHC79-wt with AgeI and SalI, which removes a 1.4-kb fragment from the Aγ-δ intergenic region, and subcloning the AgeI–SalI fragment into an AgeI–SalI-digested pUC-19 plasmid modified to contain an AgeI site between its BamHI and EcoRI sites. The resulting plasmid was digested with BsaBI and BsmI to remove the 511-bp region containing the pyrimidine-rich sequence and PYR complex binding site, blunt-ended with T4 DNA polymerase, and religated. The insert (now 886 bp) then was excised with AgeI and SalI and ligated back into AgeI–SalI-digested pHC79-wt.
Cosmid inserts were excised with NotI and KpnI, purified by sucrose gradient centrifugation, and used to create transgenic mice as described (27). One line of mice carrying wt (wt33) and three lines carrying Δ (Δ23, Δ27, and Δ32) were established. A second line of wt transgenics (wt1) was generously supplied by George Stamatoyannopoulos (University of Washington, Seattle). Transgene copy number for each line was determined by PhosphorImager quantitation of DNA dot blots. The wt1 and Δ27 lines carry three transgene copies, Δ32 and Δ23 one transgene copy, and wt33 50 transgene copies per haploid genome. All lines were mapped by Southern blot (26) and PCR to ensure that the transgenic constructs were intact. wt mice were distinguished from Δ mice by Southern blot of EcoRI-digested genomic DNA using a 1.9-kb PstI–EcoRI upstream δ-globin probe (2.3-kb band in wt mice, 5.0-kb band in Δ mice). The 5′ and 3′ ends of the mini-locus were assayed for by PCR using primer sets for LCR HS4 (GGTGGACTCCAGAGACTCTC and GCCAGTCAATGAGTCTCAGGT, 425-bp PCR product) and for a KpnI repeat downstream of the human β-globin gene (CGGTGGCTCACACCTGCAAT and CGTCACTCACGATGGGAGCT, 365-bp product).
Hemizygous transgenic offspring were sacrificed at time points from E11.5 to 10 days after birth. Total RNA was prepared from hematopoietic tissues (micro RNA kit, Stratagene), and primer extensions were performed as described (28), using a mixture of four end-labeled primers complementary to human β-globin (CCACAGGGCAGTAACGGCAGA), human γ-globin (CCAGCATCTTCCACATTCACC), mouse βh1-globin (ATAGCTGCCTTCTCCTCAGCT), and mouse β major (βmaj) globin (TGATGTCTGTTTCTGGGGTTGTG). Reactions were electrophoresed on 6% polyacrylamide sequencing gels, and signals were quantitated on a Molecular Dynamics PhosphorImager.
RESULTS AND DISCUSSION
PYR Factor Is a SWI/SNF-Related Complex (PYR Complex).
PYR factor was determined to have a native molecular mass of approximately 800 kDa by gel filtration chromatography on Sephacryl S300 and Superose 6 (Amersham Pharmacia; data not shown). UV crosslinking and deoxycholate dissociation experiments indicated that PYR factor is a multisubunit complex (PYR complex) with a 60- to 70-kDa DNA-binding subunit (not shown). To identify its subunits, we purified PYR complex from MEL cell nuclear extract in five chromatographic steps (Fig. 2A): High Q and High S Macro-Prep (Bio-Rad), Reactive Yellow 3 agarose (Sigma), double-stranded calf thymus DNA cellulose (Sigma), and a sequence-specific DNA-affinity column (δ60ym-Sepharose). PYR complex activity was monitored in column fractions by gel shift assay using radiolabeled δ60ym probe.
Analysis of fractions from the final two purification steps is shown in Fig. 2B. Aliquots from each fraction were tested for PYR complex activity by gel shift assay (Upper) and analyzed for purity by silver-stained SDS/PAGE (Lower). Two bands are seen in the gel shift assay of the δ60ym affinity column fractions (Upper Right). The higher-mobility band (peak in fraction 6) that binds the δ60ym column less tightly (elutes at lower salt) represents partially dissociated PYR complex. The lower mobility band, which represents the complete complex, peaks in fraction 8. Fractions containing the complete complex (fractions 7–9) were pooled, trichloroacetic acid-precipitated, electrophoresed on an SDS gel, and blotted onto a nitrocellulose membrane that was used for mass analysis.
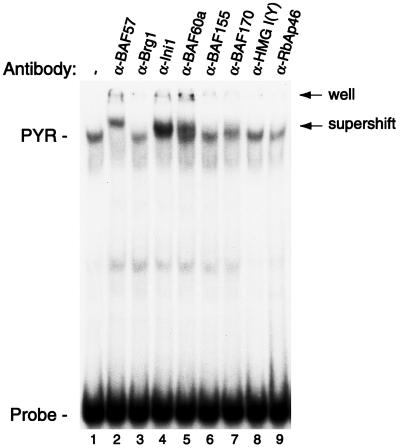
Approximately 26 bands were detected on this membrane for identification. Initial experiments focused on the characterization of three bands with approximate molecular masses of 48, 55, and 150 kDa (Fig. 2B, arrows). Two independent mass spectrometric techniques, peptide mass fingerprinting using matrix-assisted laser desorption ionization–reflectron time-of-flight mass spectrometry and sequencetag database searching using limited amino acid sequence data obtained by electrospray ionization tandem mass spectrometry, were used to identify the 48-, 55-, and 150-kDa proteins as, respectively, INI1, BAF57, and BAF155, all known mammalian SWI/SNF complex subunits (29–31). Also identified in band p55 was RbAp46, a known component of a histone deacetylase complex (32, 33). To determine whether these proteins were components of PYR complex or whether they had simply copurified with it, we tested the ability of antibodies to each of them to interact with PYR complex in gel supershift assays (Fig. 3). Strong supershifts are seen with antibodies to INI1 and BAF57, indicating that these are PYR complex subunits. No clear supershifts are seen when using antibodies to BAF155 or RbAp46, so we cannot confirm their presence in the complex. However, because it is possible that the epitopes recognized by these antibodies may be inaccessible because of the presence of other complex subunits, we also cannot rule out the possibility that PYR complex might contain BAF155 or RbAp46 when using this assay.
Figure 3.
PYR complex contains at least four known mammalian SWI/SNF subunits. Gel supershift assay using MEL nuclear extract and labeled δ60ym DNA as probe. Nuclear extract was preincubated for 1 hr at room temperature with antibodies to known SWI/SNF complex subunits (lanes 2–7) and other nuclear factors, two of which are shown (lanes 8 and 9). Strong supershifts are seen with anti-BAF57, anti-INI1, and anti-BAF60a antibodies, with a weaker supershift using antibody to BAF170. No clear supershift is seen with anti-BRG1, anti-BAF155, anti-HMG I(Y), or anti-RbAp46.
Antibodies to other known mammalian SWI/SNF components also were tested to determine whether PYR complex contains additional SWI/SNF subunits (Fig. 3). We see strong reactivity with anti-BAF60a antibody and a clear but weaker supershift with anti-BAF170. No clear supershift is seen with anti-BRG1. A number of antibodies to proteins that have not been associated with SWI/SNF complexes also have been tested, including antibodies to YY1, GATA-1, NF-E2, and EKLF, and have given negative results to date (data not shown). These experiments indicate that PYR complex contains at least four known SWI/SNF subunits, BAF57, INI1, BAF60a, and BAF170, identifying it as a mammalian SWI/SNF complex.
SWI/SNF in yeast is a very large (2,000 kDa) complex of 11 protein subunits that is thought to facilitate the activation of genes through nucleosome displacement and the disruption of repressive chromatin structures (34, 35). Genes encoding the various subunits of SWI/SNF are highly conserved in eukaryotes, with homologues in Drosophila, avian, and mammalian cells (11, 34, 36, 37). SWI/SNF complexes have been purified from mammalian cells, and like those in yeast, have the ability to disrupt nucleosomes in an ATP-dependent manner (30, 38). It has been shown that mammalian SWI/SNF complexes vary considerably between different cell types, both in the number and the identity of their subunits, and multiple forms have even been shown to exist within individual cells (30, 37). Presumably, this diversity leads to specialization of function. PYR complex is a unique example of a SWI/SNF-related complex that binds DNA in a tissue- and developmental stage-specific manner. We do not as yet know the mechanism of this specificity, but it may be the result of restricted expression of one or more PYR complex subunits in definitive hematopoietic cells.
PYR Complex DNA Binding Depends on Both DNA Length and Sequence.
SWI/SNF-related complexes are not known to bind DNA in a sequence-specific manner. To better define the binding activity of PYR complex, we constructed a series of mutant probes and competitor DNAs derived from a 99-bp binding site, δ99, described previously (17). A series of linker scanning mutants, mutated near the center of δ99 in the region of a strongly footprinted TTCC repeat, retain full binding affinity for PYR complex (data not shown). Truncation mutants of δ99 result in a gradual decrease in affinity for PYR complex as the probe is progressively shortened from either end, indicating that PYR complex binding is DNA length-dependent (not shown). This finding is a characteristic described for yeast SWI/SNF and high mobility group (HMG) proteins, and suggests that PYR complex recognizes a DNA structure (39–41).
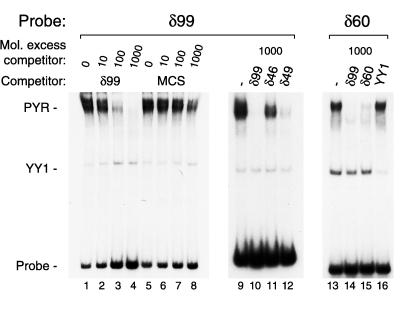
These results also could indicate that PYR complex might simply bind DNA nonspecifically, recognizing any piece of DNA longer than 80 bp. This possibility is not the case, however, because random sequence DNA of similar length to δ99 (from the pBluescript II multicloning site) competes very poorly for the binding of PYR complex in gel shift assays (Fig. 4). Similar results have been obtained with other random DNA sequences (not shown) and indicate that PYR complex specifically recognizes pyrimidine-rich DNA. When double-stranded 46- and 49-bp oligonucleotides derived from δ99 (δ46 and δ49, Fig. 4) are used as competitor, δ49 clearly shows higher affinity for PYR complex than δ46, suggesting that PYR complex prefers some pyrimidine-rich sequences to others. PYR complex similarly binds other pyrimidine-rich sequences in the human and murine β-globin loci: at the exon 1-intron 1 junction of both human γ-globin genes and at a long polypyrimide sequence 2.5 kb upstream of the murine βmaj-globin gene (data not shown).
Figure 4.
PYR complex specifically binds pyrimidine-rich DNA sequences. Lanes 1–8, competition gel shift assay, δ99 probe, and MEL nuclear extract. Preincubation with increasing amounts of unlabeled δ99 (lanes 2–4) or 105-bp pBluescript II multicloning site (MCS, lanes 6–8) shows that PYR complex DNA-binding depends on DNA sequence. Lanes 9–12, competition gel shift assay, δ99 probe, and MEL nuclear extract. Overlapping oligonucleotide competitors (δ46 and δ49) from the center of δ99 have different affinities for PYR complex, with δ49 competing for PYR complex binding more strongly than δ46. Lanes 13–16, a 60-bp probe (δ60) from the center of δ99 binds PYR complex almost as well as δ99. A 100-fold molar excess of unlabeled δ99 competes all PYR complex binding to δ60 probe, whereas a 100-fold molar excess of unlabeled δ60 competes well but not completely.
The binding of PYR complex to specific pyrimidine-rich DNA sequences is a unique example of a sequence requirement for the assembly of a SWI/SNF complex on DNA. This finding relates to a fundamental question about SWI/SNF complexes: the mechanism by which they target specific genes for activation. It has been proposed that they are either recruited by sequence-specific transcription factors (42), bind directly to the RNA polymerase II holoenzyme (43), or have their own intrinsic DNA-binding activity (41). Our results indicate that at least some SWI/SNF-related complexes can target specific DNA sequences on their own by recognizing a combination of DNA sequence and structure.
A Short Intergenic Deletion Including the PYR Complex Binding Site Delays Human Fetal-to-Adult Globin Gene Switching in Transgenic Mice.
The developmental stage-specific pattern of PYR complex DNA-binding activity, the location of its binding sites in the human and murine β-globin loci, and its identity as a SWI/SNF-related complex suggest that PYR complex might function in the regulation of hemoglobin switching. In an initial attempt to define whether such a function exists for PYR complex, we examined the effect of deleting its intergenic binding site, 1 kb upstream of the δ-globin gene, on human fetal-to-adult globin gene switching in transgenic mice. We used a human β-globin mini-locus construct, μLCRAγψβδβ (wt), that contains a condensed version of the LCR (μLCR) linked to a 29-kb HindIII–KpnI fragment containing the Aγ through β genes (Fig. 1B). Because PYR complex broadly footprints the 250-bp pyrimidine-rich sequence upstream of the δ gene, we removed this entire sequence by deleting a 511-bp BsaBI–BsmI fragment from the wt mini-locus (Fig. 1B). This deletion also removes binding sites for two known transcription factors, YY1 and GATA-1, which are within the pyrimidine-rich region of interest (17). The deletion construct, Δ, has greater than 98% identity to wt.
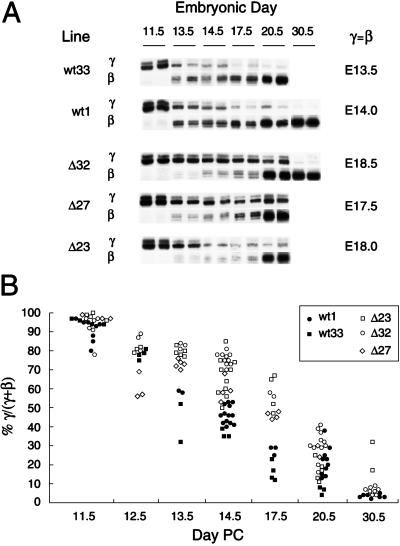
Mice carrying the wt mini-locus previously have been shown to switch from human γ- to β-globin expression during fetal liver erythropoiesis, with the midpoint of the switch, defined as the point of equal γ and β expression, occurring reproducibly at or near E14 (26). Using a primer extension assay, we see a marked delay in the γ-to-β switch in all three Δ lines, with γ = β at E13.5 and E14 for the two wt lines but at E17.5, E18, and E18.5 for the three Δ lines (Fig. 5A). Differences between wt and Δ mice are evident as early as E13.5, peaking at E17.5 and persisting at the time of birth. The ratio of human γ- to β-globin expression at each time point is very consistent for lines carrying the same construct, irrespective of position of integration or copy number, with the delay in the switch reproducibly seen after analyzing more than 100 mice from multiple separate litters (Fig. 5B). The deleted lines eventually complete the switch, although Δ23 mice show a persistently high percentage of γ-globin expression as late as E30.5. We also examined the effect of the deletion on the absolute levels of human γ and β expression, normalized to the level of endogenous mouse β-globin (βh1 plus βmaj), to determine whether the delayed switch is caused by impaired γ gene silencing, impaired β gene activation, or both. There is primarily a delay in β gene activation in Δ23 mice, whereas the delayed switch in Δ27 mice mostly is caused by impaired γ gene silencing, and a nearly equal combination of the two is seen in Δ32 mice (data not shown). Thus, it appears that the site of chromosomal integration of the transgene has an effect on whether the γ or β gene is primarily affected.
Figure 5.
Delayed human γ-to-β globin gene switching in transgenic mice after a 511-bp intergenic deletion including the PYR complex binding site. (A) Primer extension assay of human globin mRNA expression in wt and Δ transgenic mice during development. Two lines of mice carrying the wt construct (wt1 and wt33) and three lines carrying the Δ construct (Δ23, Δ27, and Δ32) were analyzed at several time points during development. Developmental stage is indicated by embryonic day (E), which is the number of days postcoitus (PC). Birth is at E19.5. Bands corresponding to primer extension products of human γ- and β-globin mRNA are shown, with two representative samples for each line at each time point. The midpoint of the fetal-to-adult switch, defined as the embryonic day at which equal amounts of human γ and β message are being expressed (γ = β), is shown for each transgenic line at the right, and is delayed on average by 4 days in the Δ lines compared with the wt lines. (B) Summary of primer extension analysis of more than 100 wt and Δ transgenic offspring from five separate lines. Points represent the ratio of human γ- to β-globin mRNA levels, defined as % γ globin/total human globin [% γ/(γ+β)]. Each point on the graph represents a separate mouse. Values for γ and β globin mRNA levels were determined by PhosphorImager quantitation of primer extension signals. There is a significant difference in % γ/(γ+β) between wt and Δ mice at E13.5 (P < 0.05), E14.5 (P < 0.001), and E17.5 (P < 0.001).
These experiments do not prove that PYR complex per se is mediating this effect, although PYR complex is the only factor known to bind this element with a developmental stage-specific pattern of activity. Much of the deleted element is footprinted by PYR complex, but it also includes binding sites for YY1 and GATA-1 (17), and approximately one-fourth of the deleted DNA has not been mapped by footprinting or gel shift assay and potentially could bind other protein factors. Thus, although the data suggest that PYR complex is mediating the observed effect on switching, we cannot rule out the possibility that factors other than or in addition to PYR complex also act at this site. The transient nature of the observed effect suggests that the deleted element is required to facilitate γ-to-β switching, but is not required for switching to eventually occur. It should be noted, however, that PYR complex binds elsewhere in the locus, at the very least within both γ-globin genes, and this binding may have a profound negative effect on γ-globin expression that is not addressed in the deletion construct we have used. Also, it is known that the distance between the LCR and the human γ and β genes can affect their expression (44, 45). If the deleted element is required to bring the adult genes closer to the LCR, the reduced distance between the LCR and the β gene in our construct compared with the native β-globin locus could minimize the effect of the deletion in our experiments.
In summary, we describe a specialized SWI/SNF-related complex specific to definitive hematopoietic cells that binds pyrimidine-rich elements in the human and murine β-globin loci. This complex is a unique example of tissue-and developmental stage-specific DNA binding by a SWI/SNF-related complex and is a SWI/SNF-like complex described to recognize DNA sequence as well as structure. We show that a short deletion that removes the intergenic PYR complex binding site from a human β-globin mini-locus construct delays human fetal-to-adult globin gene switching in transgenic mice. We propose that PYR complex may function at this site to facilitate human γ-to-β globin switching by disrupting the chromatin structure of the β-globin locus late in erythroid development, permitting transcriptional activators such as EKLF access to the adult genes. It will be of interest to characterize the complete subunit composition of PYR complex, the mechanism of its tissue- and developmental stage-specific DNA-binding activity, its architectural effects on DNA upon binding, and the effect of targeted mutations of genes that encode its subunits on globin gene expression in mice. We also hope to define other PYR complex binding sites, both within the globin loci and near other hematopoietic cell-specific genes, and to determine in more detail the cis elements responsible for the delay in γ-to-β globin gene switching we have observed.
Acknowledgments
We thank Samantha Atkins, Eugene Leung, and Frederic Marrache for their excellent technical assistance; Chengyu Liu and the Columbia transgenic mouse facility for their help with the production of transgenic mice; Lynne Lacomis, Mary Lui, Anita Grewal, and Scott Geromanos for help with mass spectrometric analysis; Matthias Mann for the peptidesearch and sequencetag programs; Stephen Nimer for CMK nuclear extract; Gerald Crabtree and Ganjam Kalpana for antibodies to mammalian SWI/SNF subunits; George Stamatoyannopoulos for the pHC79-μLCRAγψβδβ cosmid and wt1 transgenic mouse line; and Richard Axel, Robert Bauchwitz, Kathryn Calame, Marian Carlson, Madalyn Castle, Srikumar Chellapan, Frank Costantini, Michael Flamm, Stephen Goff, Shankar Srinivas, Saul Silverstein, Kwok Wang, Haifeng Xue, and especially Dimitrios Thanos for helpful discussions and advice. This work was supported by Public Health Service Grants DK25274 and HL28381 from the National Institutes of Health, a grant from the Ahepa Cooley’s Anemia Foundation, National Cancer Institute Grant P30 CA08748, and National Science Foundation Grant BDI-9420123 (to P.T.). D.O. is supported by National Institutes of Health Clinical Investigator Award DK02260, and J.Y. was supported by National Institutes of Health Hematology Training Grant DK07373.
ABBREVIATIONS
- LCR
locus control region: MEL, murine erythroleukemia
- wt
wild type
- E(n)
embryonic day
References
- 1.Bank A, Mears J G, Ramirez F. Science. 1980;207:486–493. doi: 10.1126/science.7352255. [DOI] [PubMed] [Google Scholar]
- 2.Orkin S H. Eur J Biochem. 1995;231:271–281. doi: 10.1111/j.1432-1033.1995.tb20697.x. [DOI] [PubMed] [Google Scholar]
- 3.Baron M H. Biochim Biophys Acta. 1997;1351:51–72. doi: 10.1016/s0167-4781(96)00195-9. [DOI] [PubMed] [Google Scholar]
- 4.Donze D, Townes T M, Bieker J J. J Biol Chem. 1995;270:1955–1959. doi: 10.1074/jbc.270.4.1955. [DOI] [PubMed] [Google Scholar]
- 5.Perkins A C, Sharpe A H, Orkin S H. Nature (London) 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 6.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Nature (London) 1995;375:316–318. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 7.Guy L, Mei Q, Perkins A C, Orkin S H, Wall L. Blood. 1998;91:2259–2263. [PubMed] [Google Scholar]
- 8.Felsenfeld G. Cell. 1996;86:13–19. doi: 10.1016/s0092-8674(00)80073-2. [DOI] [PubMed] [Google Scholar]
- 9.Martin D I, Fiering S, Groudine M. Curr Opin Genet Dev. 1996;6:488–495. doi: 10.1016/s0959-437x(96)80072-4. [DOI] [PubMed] [Google Scholar]
- 10.Kadonaga J. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 11.Kennison J A. Annu Rev Genet. 1995;29:289–303. doi: 10.1146/annurev.ge.29.120195.001445. [DOI] [PubMed] [Google Scholar]
- 12.Pirrotta V. Curr Opin Genet Dev. 1995;5:466–472. doi: 10.1016/0959-437x(95)90050-q. [DOI] [PubMed] [Google Scholar]
- 13.Tamkun J W. Curr Opin Genet Dev. 1995;5:473–477. doi: 10.1016/0959-437x(95)90051-h. [DOI] [PubMed] [Google Scholar]
- 14.Gould A. Curr Opin Genet Dev. 1997;7:488–494. doi: 10.1016/s0959-437x(97)80075-5. [DOI] [PubMed] [Google Scholar]
- 15.Stanworth S J, Roberts N A, Sharpe J A, Sloane-Stanley J A, Wood W G. Mol Cell Biol. 1995;15:3969–3978. doi: 10.1128/mcb.15.8.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibbons R J, Picketts D J, Villard L, Higgs D R. Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 17.O’Neill D, Bornschlegel K, Flamm M, Castle M, Bank A. Proc Natl Acad Sci USA. 1991;88:8953–8957. doi: 10.1073/pnas.88.20.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dignam J D, Martin P L, Shastry B S, Roeder R G. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 19.O’Neill D, Kaysen J, Donovan-Peluso M, Castle M, Bank A. Nucleic Acids Res. 1990;18:1977–1982. doi: 10.1093/nar/18.8.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kadonaga J T. In: Strategies for Protein Purification and Characterization. Marshak D R, Kadonaga J T, Burgess R R, Knuth M W, Brennan W A, Lin S, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 130–204. [Google Scholar]
- 21.Lui M, Tempst P, Erdjument-Bromage H. Anal Biochem. 1996;241:156–166. doi: 10.1006/abio.1996.0393. [DOI] [PubMed] [Google Scholar]
- 22.Erdjument-Bromage H, Lui M, Lacomis L, Grewal A, Annan R S, Carr S A, Tempst P. J Chromatog. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- 23.Geromanos S, Philip J, Freckleton G, Tempst P. Rapid Commun Mass Spectrom. 1998;12:551–556. doi: 10.1002/(SICI)1097-0231(19980515)12:9<551::AID-RCM198>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 24.Mann M, Hojrup P, Roepstorff P. Biol Mass Spectrom. 1993;22:338–345. doi: 10.1002/bms.1200220605. [DOI] [PubMed] [Google Scholar]
- 25.Mann M, Wilm M. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 26.Enver T, Raich N, Ebens A J, Papayannopoulou T, Costantini F, Stamatoyannopoulos G. Nature (London) 1990;344:309–313. doi: 10.1038/344309a0. [DOI] [PubMed] [Google Scholar]
- 27.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 226–250. [Google Scholar]
- 28.Behringer R R, Ryan T M, Palmiter R D, Brinster R L, Townes T M. Genes Dev. 1990;4:380–389. doi: 10.1101/gad.4.3.380. [DOI] [PubMed] [Google Scholar]
- 29.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 30.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, et al. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W, Chi T, Xue Y, Zhou S, Kuo A, Crabtree G R. Proc Natl Acad Sci USA. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, Sun Z W, Iratni R, Erdjument-Bromage H, Tempst P, Hampsey M, Reinberg D. Mol Cell. 1998;1:1021–1031. doi: 10.1016/s1097-2765(00)80102-1. [DOI] [PubMed] [Google Scholar]
- 33.Qian Y W, Lee E Y. J Biol Chem. 1995;270:25507–25513. doi: 10.1074/jbc.270.43.25507. [DOI] [PubMed] [Google Scholar]
- 34.Peterson C L. Curr Opin Genet Dev. 1996;6:171–175. doi: 10.1016/s0959-437x(96)80047-5. [DOI] [PubMed] [Google Scholar]
- 35.Tsukiyama T, Wu C. Curr Opin Genet Dev. 1997;7:182–191. doi: 10.1016/s0959-437x(97)80127-x. [DOI] [PubMed] [Google Scholar]
- 36.Goodwin G H. Gene. 1997;184:27–32. doi: 10.1016/s0378-1119(96)00569-0. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 38.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nature (London) 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 39.Grosschedl R, Giese K, Pagel J. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 40.Landsman D, Bustin M. BioEssays. 1993;15:539–546. doi: 10.1002/bies.950150807. [DOI] [PubMed] [Google Scholar]
- 41.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. Nature (London) 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 42.Yoshinaga S K, Peterson C L, Herskowitz I, Yamamoto K R. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 43.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 44.Peterson K R, Stamatoyannopoulos G. Mol Cell Biol. 1993;13:4836–4843. doi: 10.1128/mcb.13.8.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanscombe O, Whyatt D, Fraser P, Yannoutsos N, Greaves D, Dillon N, Grosveld F. Genes Dev. 1991;5:1387–1394. doi: 10.1101/gad.5.8.1387. [DOI] [PubMed] [Google Scholar]