Abstract
We have identified a previously undescribed transmembrane protein, Hemese, from Drosophila melanogaster blood cells (hemocytes), by using a monoclonal pan-hemocyte antibody. Heavy glycosylation is suggested by the heterogeneous size distribution, ranging between 37 and 70 kDa. Hemese expression is restricted to the cell surfaces of hemocytes of all classes, and to the hematopoietic organs. The sequence of the corresponding gene, Hemese (He), predicts a glycophorin-like protein of 15 kDa, excluding an N-terminal signal peptide, with a single hydrophobic transmembrane region. The extracellular region consists mainly of Ser/Thr-rich sequence of low complexity, with several potential O-glycosylation sites. Hemese contains phosphotyrosine and the cytoplasmic region has potential phosphorylation sites, suggesting an involvement in signal transduction. Depletion of Hemese by RNA interference has no obvious effect under normal conditions, but the cellular response to parasitic wasps is much enhanced. This finding indicates that Hemese plays a modulatory role in the activation or recruitment of the hemocytes.
The blood cells, or hemocytes, in Drosophila participate in the humoral and cellular immune responses (1–3), apparently distinguishing self from non-self. They synthesize antimicrobial peptides (4, 5), phagocytose microbes (6) and encapsulate larger foreign bodies such as eggs of parasites (7). In contrast to the well studied humoral immune response, we know very little about the molecular mechanisms of the cellular immune reactions in Drosophila.
At least three main classes of hemocytes can be discerned in Drosophila (8). The dominating hemocytes belong to a class of small round cells with phagocytic capacity, the plasmatocytes. A second class, the crystal cells, are similar to the plasmatocytes in size and morphology but are distinguished by prominent crystal-like inclusions in the cytoplasm. The crystal cells carry phenoloxidase and they are involved in melanin deposition in wounds and around foreign objects. Finally, a class of large flat cells, the lamellocytes, appears when parasitoid wasps infest the larvae. They are believed to participate in the encapsulation of the parasites. Lamellocytes have also been described to occur spontaneously at the time of metamorphosis (8), although this is debated (9). The developmental origin of the different classes of hemocytes is not clear, though the lymph glands near the anterior end of the dorsal vessel are thought to be major sites of hematopoiesis in the larva (8).
The further study of hemocyte function would be facilitated by the identification of genes and gene products whose expression are restricted to these cells. For this purpose, we have created a library of hemocyte-specific monoclonal antibodies and used it to define specific marker molecules restricted to hemocytes. This may also identify molecules that are involved in recognition and signaling in the cellular immune response. Some antibodies recognize pan-hemocyte antigens that are expressed on all blood cells; others are restricted to hemocyte subpopulations, such as plasmatocytes, lamellocytes or crystal cells (E.K., P.V., Istvan Nagy, Robert Markus, Yves Carton, Imre Ocsovszki, D.H., Elisabeth Gateff, and I.A., unpublished data). Here we used a pan-hemocyte antibody to identify a previously undescribed gene, Hemese, which encodes a blood-cell-specific transmembrane protein.
Methods
Drosophila Stocks.
Flies were kept on cornmeal-yeast food at 25°C. We used Canton-S and Oregon-R wild-type stocks. domino is a P-element induced mutant devoid of circulating hemocytes and with melanized lymph glands in the third-instar larvae (10). The l(3)hem mutant lacks lymph glands and circulating hemocytes (11). The l(3)mbn-1 strain is a tumor suppressor mutant with enlarged lymph glands and proliferating tumorous hemocytes in the circulation (12).
Preparation of Hemocyte Extracts.
Hemocytes were collected from larvae by bleeding into Drosophila Ringer's solution (130 mM NaCl/5.0 mM KCl/1.0 mM CaCl2) on ice. After centrifugation, cells were extracted into lysis buffer (50 mM Tris⋅HCl, pH 8.0/150 mM NaCl/1.0% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS) supplemented with a protease inhibitor mixture (Boehringer Mannheim) and 1 mM phenyl-methyl-sulfonyl fluoride, at 4°C for 1 h. After centrifugation at 12,000 × g for 10 min, the supernatant was subjected to Western blot analysis or immunoprecipitation followed by Western blot analysis.
Hybridoma Production.
Monoclonal antibodies against hemocytes were raised as described elsewhere (E.K., P.V., Istvan Nagy, Robert Markus, Yves Carton, Imre Ocsovszki, D.H., Elisabeth Gateff, and I.A., unpublished data). Briefly, BALB/c mice were immunized by i.p. injection of 106 hemocytes from late third-instar lethal (3) malignant blood neoplasm [l(3)mbn-1] larvae in 1 ml of Drosophila Ringer's solution. Booster injections were given 4, 8, and 13 weeks later. Three days after the last immunization spleen cells were collected and fused with SP2/O myeloma cells by using polyethylene glycol (PEG 1450). Hybridoma culture supernatants were screened by indirect immunofluorescence on living cells. The selected hybridomas were subcloned three times by limiting dilution.
FACS Analysis of Live Hemocytes.
A 20-μl aliquot of 107 cells per ml hemocyte suspension, in insect Schneider's medium supplemented with 10% FCS, was placed in each well of a 96-well U-form multiwell plate. Hybridoma supernatant (50 μl) was added to each well, and the plates were incubated for 45 min on ice. After washing the cells three times with ice-cold Schneider's medium, and FITC-labeled anti-mouse IgG antibody (Sigma) was added at 1:100 dilution. After 45 min on ice, the hemocytes were washed three times with ice-cold Schneider's medium and the cells were analyzed with a FACSCalibur equipment (Beckton Dickinson) for fluorescence intensity.
Immunohistochemistry.
A total of 20 μl of hemocyte suspension in Schneider's medium was placed in each spot of a multispot microscope slide (SM-011, Hendley, Loughton, U.K.). The hemocytes were allowed to settle for 20 min at room temperature and then fixed in acetone for 6 min, rehydrated, and blocked in PBS containing 0.1% BSA (PBS-BSA). Samples were incubated with the primary antibody for 1 h, washed three times with PBS for 5 min each, and the bound antibody was detected by StreptABComplex/HRP-Duet kit (DAKO). The chromogen was 3-amino-9-ethylcarbasole (Sigma).
Immunoprecipitation of Antigens.
Five milliliters of hybridoma supernatant was added to 50 μl of 20% slurry of Protein G Sepharose and rotated at room temperature for 1 h. After washing the protein G Sepharose–antibody complex three times with PBS, the cell extract was added and immunoprecipitation was carried out overnight at 4°C. Finally, the immunoprecipitates were washed three times in lysis buffer and solubilized in SDS/PAGE sample buffer. The samples were subjected to SDS/PAGE and Western blotting.
Western Blot Analysis.
Protein extracts were separated by SDS/PAGE. After electrophoresis, the proteins were transferred onto nitrocellulose membrane (Hybond-C, Amersham Pharmacia) in transfer buffer (25 mM Tris, pH 8.3/192 mM glycine/20% methanol). Nonspecific binding on the nitrocellulose membrane was blocked with PBS containing 0.1% Tween 20 (PBST) supplemented with 5% nonfat dry milk for 1 h at room temperature. The blotted proteins were subjected to hybridoma supernatants for 3 h with agitation at room temperature. The blot was washed with PBST three times for 10 min each and then incubated with HRPO-conjugated anti-mouse antibody (DACO). After three washes for 10 min each in PBST, the proteins were visualized by the ECL-Plus system (Amersham Pharmacia) according to the manufacturer's instructions.
Tyrosine Phosphorylation.
Hemocyte extracts were prepared in lysis buffer containing 20 mM NaF and 5 mM NaVO4. Immunoprecipitation and Western blot analysis was carried out as described above. Signals were visualized by the ECL phosphorylation detection system (Amersham Pharmacia), with the horseradish peroxidase conjugate of the PY20 anti-phosphotyrosine antibody for direct detection of tyrosine-phosphorylated proteins.
Construction and Screening of cDNA Expression Libraries.
Total RNA was isolated from of Enterobacter cloacae-immunostimulated and nonstimulated third-instar l(3)mbn-1 mutant larvae with TRIZOL reagent (GIBCO/BRL), following the supplier's protocol. Poly(A)-enriched RNA was isolated from total RNA by chromatography on oligo(dT)-cellulose (Amersham Pharmacia) by two runs. The quality of poly(A) RNA was tested by Northern blot analysis using a labeled cDNA probe for a high molecular weight mRNA. We used 5 μg of poly(A) RNA and the ZAP Express Vector kit (Stratagene) to construct cDNA libraries according to the manufacturer's instruction. Briefly, this involved directional cloning by using a XhoI linker primer to initiate the synthesis of the first strand cDNA, and the addition of an EcoRI adapter after the synthesis of the second strand cDNA by using DNA polymerase I in combination with RNA-se H. After size selection chromatography on Sephacryl column, 100 ng of cDNA was ligated into 1 μg predigested ZAP Express Vector (Stratagene). After ligation, the DNA was packaged by using Gigapack II packaging extract (Stratagene). After packaging, the titer of the primary libraries were 5 × 105 plaque forming units (pfu)/μg vector arm.
Hemese cDNA Clones and Sequence Analysis.
We used the H2 antibody to screen ≈8 × 104 plaques from the l(3)mbn-1 larval expression libraries, transferred onto nitrocellulose filters (Amersham Pharmacia Hybond-C Extra). HRPO-conjugated secondary antibody (Amersham Pharmacia) was applied and the reaction was visualized with the ECL-Plus system (Amersham Pharmacia). Plasmid DNA was prepared from 12 independent positive clones of the pBK-CMV phagemid vector derived from the ZAP Express vector by in vivo excision using the ExAssist helper phage (Amersham Pharmacia) and sequenced on both strands. Three of them represent the same transcript, encoding a putative transmembrane protein. Database searches were performed with the blast servers at the Berkeley Drosophila Genome Project and the National Center for Biotechnology Information. Glycophorin similarity was seen with the Amino Acid Composition Search Tool at the ExPASy web site (13). The signal peptidase cleavage site was predicted by the signalp software (14), the membrane topology by TMHMM 2.0 (15), O-glycosylation by NETOGLYC 2.0 (16), all on the WWW Prediction Servers at the Center for Biological Sequence Analysis. Potential target sites for phosphorylation were found by searching prosite on the Swiss Institute for Experimental Cancer Research server.
Double-Stranded RNA (dsRNA) Synthesis for RNA Interference.
The RNA strands were synthesized from linearized plasmid templates (17). Hemese and GFP cDNAs were in BlueScript SK+ and KS+ plasmids. The plasmids were linearized with restriction endonuclease to create linear DNA templates to synthesize the sense and antisense strand with T7 polymerase (RiboMax TM Large Scale RNA Production System, Promega). The template DNA was destroyed by DNA-se I treatment and the dsRNA purified by phenol and chloroform extraction followed by ethanol precipitation and dissolved in annealing buffer (1 mM Tris, pH 7.5/1 mM EDTA/20 mM NaCl). For annealing, equimolar quantities of sense and antisense RNAs were mixed in annealing buffer to a final concentration of 1 μM each in 10 μl. The mixture was heated in a 150 ml beaker of boiling water for 1 min and left to cool at room temperature for 12 to 16 h. To monitor annealing, the RNA was electrophoresed in a 2% agarose gel in TAE buffer and stained with ethidium bromide. The dsRNA were precipitated and dissolved in injection buffer to a final concentration of 1 μg/μl.
RNA Interference.
Hemese or GFP dsRNA was precipitated and dissolved in injection buffer to a final concentration of 1 μg/μl. For injection, 0- to 30-min-old l(3)mbn-1 or Oregon-R embryos were collected and dechorionated. The embryos were desiccated and covered in Voltalef 10S oil, and the dsRNA solution was injected on the ventral side in the posterior domain by using an Eppendorf Transjector. The injection buffer was used for injection control. The injected embryos were kept at 25°C and analyzed at the second and third larval instar.
Bioassays for Encapsulation and Phagocytosis.
One-week-old females of the parasitic wasp, Leptopilina boulardi, strain G486 were used to infest second-instar Oregon-R larvae. Sixty larvae were exposed overnight to six Leptopilina females and kept at 20°C. Two days after infestation, the hemocytes were counted and the ratio of lamellocytes was determined after staining with the lamellocyte-specific antibody L1a (E.K., P.V., Istvan Nagy, Robert Markus, Yves Carton, Imre Ocsovszki, D.H., Elisabeth Gateff, and I.A., unpublished data). Phagocytosis of FITC-labeled Escherichia coli and Staphylococcus saprophyticus was carried out as described (18).
Results
Identification of Hemese, a Pan-Hemocyte Antigen.
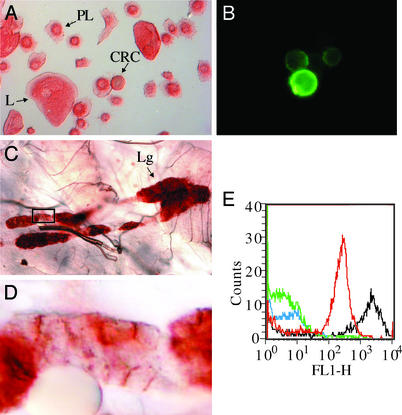
The H2 antibody defines an antigen, dubbed Hemese, which is expressed exclusively by blood cells and hematopoietic tissue. Fig. 1A shows that this antibody marks all classes of hemocytes: the plasmatocytes (PL), the lamellocytes (L), and the crystal cells (CRC). The antigen is present on the cell surface, as seen when we stain the living hemocytes by indirect immunofluorescence (Fig. 1B). Hemocytes express this antigen during all larval instars. It is also expressed on the hemocyte-like mbn-2 and Schneider-2 cell lines (data not shown), and in all lobes of the hematopoietic organ, the lymph gland (Fig. 1C). Strong staining appears on the lymph gland cells, in particular on the contact surfaces between the cells (Fig. 1D).
Figure 1.
Tissue localization of a pan-hemocyte antigen. Immunohistochemical analysis of Hemese expression in circulating hemocytes and in the lymph gland. (A–D) Isolated hemocytes (A and B) and lymph glands (C and D) from third-instar larva were stained with H2 mAb and visualized by ABC complex (A, C, and D) or by immunofluorescence of live nonpermeabilized cells (B). The lymph glands (Lg) and all hemocytes, the lamellocytes (L), the plasmatocytes (PL), and the crystal cells (CRC), express the antigen. (D) Magnified view of the boxed area in C, showing the localization of the antigen to the cell membranes between the cells in the lymph gland. (E) Flow cytometric analysis of live nonpermeabilized Drosophila hemocytes isolated from the third-instar larvae of Oregon-R (red) and l(3)mbn-1 (black) strains and stained by hybridoma supernatant containing H2 monoclonal antibody and treated with FITC-labeled anti-mouse IgG. Controls were treated with RPMI tissue culture medium only (Oregon R: green; l(3)mbn-1: blue). Over 98% of the analyzed cells express the pan-hemocyte antigen.
We also quantified the binding of the H2 antibody to live hemocytes by flow cytometry (Fig. 1E). The results confirm that Hemese is expressed on the surface of all circulating hemocytes, both in wild-type Oregon-R larvae (red curve) and in the hemocyte-overproducing tumor suppressor mutant l(3)mbn-1 (black). The mutant hemocytes express about 10 times more of the antigen than wild-type cells do.
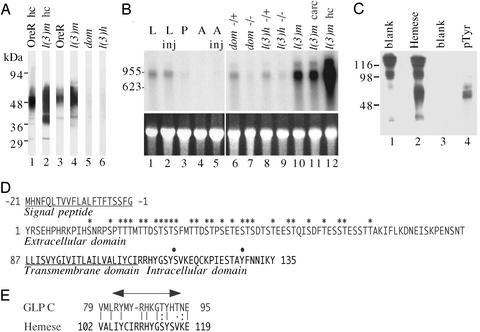
On Western blots, Hemese appears as a heterogeneous smear, with the apparent molecular mass ranging between 37 and 70 kDa (Fig. 2A). The heterogeneity suggests that the protein may be glycosylated. The protein is found in hemocyte or whole larval extracts from the wild type Oregon-R strain (Fig. 2, lanes 1 and 3) and at increased levels in the hemocyte-overproducing mutant l(3)mbn-1 (Fig. 2, lanes 2 and 4). It is essentially absent in two hemocyte-deficient mutants, domino and l(3)hem (Fig. 2, lanes 5 and 6). Thus, the expression of the antigen is strongly correlated with the presence of hemocytes.
Figure 2.
Expression and sequence of the Hemese gene. (A) Western blot analysis of the pan-hemocyte antigen on hemocyte extracts (hc, lanes 1–2) or extracts from whole larvae (lanes 3–6), of wild-type Oregon-R (OreR), the hemocyte overproducing l(3)mbn-1 strain (l(3)m), or the hemocyte-deficient mutants domino (dom) and l(3)hem (l(3)h). (B) Northern blot analysis of total RNA using the Hemese cDNA as a probe (Upper). The recovery and quantity of each RNA were assessed from the ethidium bromide loading control (Lower). Lanes 1–5: RNA from Oregon-R animals at different stages; lanes 1 and 2, larvae; lane 3, pupae; lanes 4 and 5, adults. For lanes 2 and 5, the animals were induced by an injection of Enterobacter cloacae. Lanes 6–9 show Hemese expression in larvae of hemocyte-deficient domino or l(3)hem mutants, compared with their heterozygous siblings. Lane 10 shows RNA from hemocyte-overproliferating l(3)mbn-1 larvae. Lane 12 shows RNA of l(3)mbn-1 hemocytes, and lane 11 shows the corresponding larval carcass after bleeding. (C) Tyrosine phosphorylation of the Hemese protein. The protein was immunoprecipitated by H2 antibody, bound to protein G–Sepharose, and subjected to SDS/PAGE, followed by Western blotting and detection with the H2 anti-Hemese antibody (lane 2) or with anti-phosphotyrosine antibody (lane 4). Lanes 1 and 3 are the corresponding IgG controls. (D) Deduced amino acid sequence of the Hemese gene. The predicted hydrophobic signal sequence and transmembrane region are underlined. Predicted glycosylation sites are indicated with asterisks and phosphorylation sites are indicated with dots. (E) Comparison between conserved intracellular motifs in Hemese and glycophorin C (GLP C). Arrow shows the region in glycophorin C that interacts with the Band 4.1 protein.
Expression Cloning of Hemese cDNA.
We used the H2 antibody to screen expression libraries for Hemese cDNA clones. Three independent clones were found that encode a putative transmembrane protein. Northern blot analysis shows that this transcript has a hemocyte-specific pattern of expression, as expected for the Hemese gene (Fig. 2B). There is strong expression in l(3)mbn-1 mutant larvae (Fig. 2, lane 10), and the transcript is highly enriched in hemocytes from this mutant (Fig. 2, lane 12). However, much of the Hemese mRNA remains in the carcass after bleeding (Fig. 2, lane 11), possibly because of expression in the lymph gland cells and sessile hemocytes. In contrast, Hemese expression is much reduced in hemocyte-deficient domino or l(3)hem larvae (Fig. 2, lanes 7 and 9) compared with the heterozygous controls (lanes 6 and 8). Thus, based on this pattern of expression, we could tentatively identify the isolated clones as derived from the Hemese gene. Wildtype Oregon-R larvae show a steady level of Hemese mRNA (Fig. 2, lane 1) which is not affected by injecting Enterobacter cloacae bacteria (Fig. 2, lane 2), a treatment that stimulates the humoral but not necessarily the cellular immune response. In pupae, and in uninduced or induced adults, (Fig. 2, lanes 3–5) the mRNA expression is close to or below the level of detection.
The three Hemese cDNA clones contain polyadenylated inserts of nearly identical length. The full length of the transcript is 673 nucleotides, including a short stretch at the 5′ end that we determined by 5′ RACE. This is in good agreement with the size of 0.7–0.8 kb for the polyadenylated transcript on the Northern blot. The sequence does not correspond to any of the genes predicted in the annotated version of the Drosophila genome sequence database, but the gene is represented by three EST sequences and one unpublished database entry (GenBank accession nos. AI534405, BI633401, BI636404, and AY034881) in the 34E1–2 region on chromosome 2.
The Hemese transcript contains a single ORF of 156 aa, predicted to encode a single-pass transmembrane protein of 15 kDa, excluding the signal peptide (Fig. 2D). A predicted N-terminal extracellular region of 86 amino acid residues has 29 potential O-glycosylation sites. Heavy glycosylation may account for the substantial molecular heterogeneity seen in the Western blot analysis. However, the apparent molecular mass of the purified protein, as determined by SDS/PAGE, did not change after treatment with N-glycosidase A or O-glycosidase (data not shown). It is possible that these structures are not easily accessible to our deglycosylation procedures. A putative transmembrane domain and a short cytoplasmic domain, with one potential tyrosine phosphorylation and one serine phosphorylation site, follow the extracellular domain. When a phosphotyrosine-specific antiserum was used, we confirmed the presence of constitutive tyrosine phosphorylation in a Hemese immunoprecipitate, within the same molecular mass region as the Hemese protein (Fig. 2C).
A blast search found no homologs of Hemese in the database. However, a search based on amino acid composition shows that Hemese has a striking similarity in design to the glycophorins, a family of glycoproteins in the cell membrane of human erythrocytes. Like glycophorins, Hemese is predicted to be a small single-pass transmembrane protein with an extracellular N-terminal end that is heavily O-glycosylated. Other similarities include a short sequence near the cytoplasmic face of the membrane (Fig. 2E), which resembles the docking site in glycophorin C for the Band 4.1 protein (19). Some glycophorins form SDS-resistant homodimers (20), stabilized by conserved motifs in the transmembrane segments. Hemese forms a minor band around 40 kDa and a major band in the 50- to 70-kDa range (Fig. 2A), suggesting a similar dimerization of this protein.
Hemese-Depleted Larvae Overreact to Wasp Infestation.
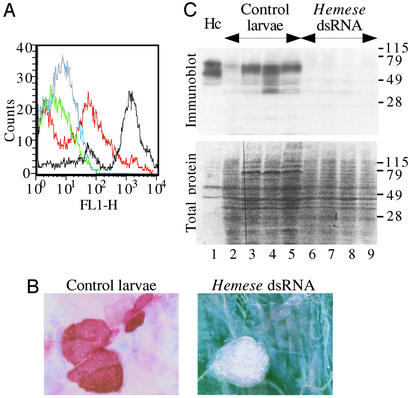
To investigate the possible function of Hemese we used RNA interference (RNAi) to suppress Hemese expression, and investigated the effect on hemocytes and hematopoietic tissue. We injected l(3)mbn-1 and Oregon-R embryos with Hemese dsRNA, or with dsRNA from the unrelated GFP gene as a negative control. As shown in Fig. 3, this treatment leads to a severe reduction of the Hemese antigen. FACS analysis of hemocytes from l(3)mbn-1 larvae (red vs. black, Fig. 3A) shows that ≈50% of the hemocytes in the treated animals lack fluorescence altogether, and the remaining cells show ≈4% of the Hemese level in control animals. RNAi also suppresses Hemese expression in the lymph glands (Fig. 3B). Furthermore, extracts from Hemese dsRNA-treated larvae show no reactivity with the H2 antibody on Western blots (Fig. 3C), confirming the absence of the gene product in the larvae. Besides demonstrating the efficiency of the dsRNA interference, these experiments provide independent evidence that the isolated cDNAs correspond to the Hemese gene. Notice that all bands disappear in the Western blot, suggesting that they are all products of the same gene, probably by posttranslational modification.
Figure 3.
Silencing of Hemese expression by RNAi. (A) FACS analysis of hemocytes derived from control l(3)mbn-1 larvae stained with the H2 antibody (black) or tissue culture medium (blue) and hemocytes from the Hemese dsRNA-treated animals stained with the H2 antibody (red) or medium (green). (B) Immunohistochemical staining by the H2 antibody of lymph glands from a control larva (bright field) and from a Hemese dsRNA-treated larva (dark field). (C) Western blot analysis of Hemese expression in protein extracts from normal hemocytes (lane 1), from individual control larvae developed from GFP dsRNA-injected embryos (lanes 2–5), and from larvae developed from Hemese dsRNA-treated embryos (lanes 6–9). Hemese was detected with the H2 antibody (Upper). (Lower) The same samples stained by Ponceau S to detect total protein content.
Silencing the Hemese gene by RNAi did not alter the development of the lymph gland as judged by morphological criteria. The size and the number of lobes was normal, and the number and morphology of the circulating hemocytes was also unaffected. As expected, injection of control GFP dsRNA had no visible effect (data not shown).
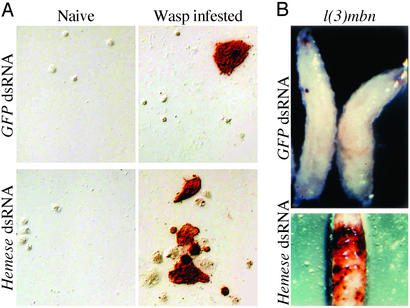
Although Hemese silencing had no dramatic consequences in untreated animals, a clear effect was seen on the response to an infestation by the parasitic wasp Leptopilina boulardi. Surprisingly, the cellular immune response to the wasp was enhanced in the Hemese-deficient animals (Fig. 4A and Table 1). Although the cellular reaction to wasp infestation is quite variable, as seen in the two experiments shown in Table 1, it is consistently much stronger when Hemese is suppressed. Both the total number of hemocytes and the number of lamellocytes are increased. No such effect was seen in control animals injected with GFP dsRNA.
Figure 4.
Effect of Hemese silencing on the hemocyte reaction. (A) Hemocyte reaction from wasp-infested larvae, injected with Hemese dsRNA, or with unrelated GFP dsRNA, at the embryonic stage. Differentiated lamellocytes could be detected with the lamellocyte-specific L1a antibody, using the ABC reaction. The formation of lamellocytes after wasp infestation was enhanced in the Hemese-deficient animals. (B) Effect of Hemese gene silencing on the formation of melanotic tumors in l(3)mbn-1 larvae.
Table 1.
Depletion of Hemese by RNAi results in an enhanced hemocyte reaction in wasp-infested larvae, as shown in two independent experiments
| No. of larvae | dsRNA | Wasp | All hemocytes | Lamellocytes | ||
|---|---|---|---|---|---|---|
| Experiment 1 | ||||||
| 6 | None | None | 889 ± 133 | NS | 0 | |
| 3 | Hemese | None | 897 ± 94 | 0 | ||
| 12 | None | G486 | 2237 ± 715 | P < 0.05 | 49 ± 37 | P < 0.001 |
| 11 | Hemese | G486 | 4894 ± 1116 | 975 ± 281 | ||
| Experiment 2 | ||||||
| 12 | None | None | 559 ± 63 | NS (P = 0.4) | 0 | |
| 12 | GFP | None | 570 ± 38 | NS (P = 0.2) | 0 | |
| 12 | Hemese | None | 498 ± 42 | 0 | ||
| 12 | None | G486 | 553 ± 50 | P < 0.05 | 101 ± 29 | P < 0.001 |
| 12 | GFP | G486 | 527 ± 43 | P < 0.05 | 97 ± 21 | P < 0.001 |
| 12 | Hemese | G486 | 758 ± 82 | 332 ± 43 |
Hemocytes were counted 2 days after infestation with Leptopilina boulardi. Lamellocytes were visualized with the lamellocyte-specific L1a antibody (E.K., P.V., Istvan Nagy, Robert Markus, Yves Carton, Imre Ocsovszki, D.H., Elisabeth Gateff, and I.A., unpublished data). Total hemocyte and lamellocyte counts per larva are given ± SEM. The significance of the difference between Hemese dsRNA-injected animals and the different controls are calculated with the t test (NS, not significant).
The phenotype of the l(3)mbn-1 mutant is reminiscent of the cellular reaction in wasp-infested larvae, with increased hemocyte counts and many lamellocytes in circulation. They also have a high incidence of so-called melanotic tumors, probably reflecting autoimmune reactions. Interestingly, this phenotype is also further enhanced in Hemese-deficient larvae. In the Hemese dsRNA-injected larvae, tumors appeared already at day 6, at a time point when no tumors were visible in the control group injected with dsRNA from the GFP gene. The tumor growth was accelerated in the Hemese dsRNA-injected group and the animals died with heavy tumors around day 12 (Fig. 4B Lower). At this time tumors just started to appear in the GFP dsRNA-injected control group (Fig. 4B Upper). Thus, like after a wasp infestation, Hemese depletion enhances the proliferation and reactivity of the l(3)mbn-1 hemocytes.
The Hemese RNAi did not affect the phagocytic capacity of the hemocytes. FITC-labeled E. coli and Staphylococcus saprophyticus bacteria were phagocytosed to the same extent in Hemese and GFP dsRNA-injected Oregon-R larvae (data not shown).
Discussion
Hemese was found in a search for hemocyte markers in Drosophila, and we conclude that it may be a very useful marker for this purpose, at least in larvae. Although Hemese expression is very low in embryos, in sessile larval hemocytes and adults, the antigen is expressed in all classes of circulating larval hemocytes, as well as in the hematopoietic organs, and it was not detected in other tissues.
The Hemese antigen is exposed on the cell surface, as shown by the fact that the H2 antibody stains live, nonpermeabilized cells. At least in the lymph glands it is enriched on the contact surfaces between the cells. In this position, Hemese may mediate attachment and/or signaling between hemocytes.
The results of the RNAi experiments give important hints about the function of the Hemese protein. Apparently, normal levels of Hemese are not important for the development of hemocytes. Even when Hemese expression is reduced by >96% there is no obvious phenotype, except in the context of a wasp infestation. However, the cellular immune reaction to the wasp is enhanced considerably in the Hemese-deficient larvae, both with respect to the proliferation of hemocytes and to the formation of lamellocytes. This finding suggests that Hemese may be important to prevent an overreaction of the cellular defense. Further support for this idea comes from the enhancement of the l(3)mbn-1 phenotype seen after Hemese silencing. The increased number of circulating hemocytes in these animals is not due to a loss of cell attachment after depletion of Hemese, because we could not observe a corresponding decrease in sessile cells (data not shown). Although the cellular reaction is enhanced in the Hemese-deficient animals, a preliminary experiment indicated that this has no major effect on the survival of infested larvae. It is likely that this is a fine-tuned response, which cannot be further improved in this way.
Although Hemese lacks significant sequence similarity to previously described proteins, the general similarity to the glycophorins is suggestive. In erythrocytes, the binding of glycophorin to the Band 4.1 protein is an important link between the cell membrane and the cytoskeleton, and it is possible that Hemese participates in a similar interaction. The tyrosine phosphorylation of the protein further suggests that Hemese could play an active role in the signaling during the cellular immune reaction in Drosophila.
Acknowledgments
This research was supported by grants from the Göran Gustafsson Foundation for Scientific Research, the Swedish Natural Science Research Council, the Hungarian National Science Foundation (Országos Tudományos Kutatási Alapprogramok Grants T035249 and T035074), and Volkswagen-Foundation Research Grant 1/71199.
Abbreviations
- FACS
fluorescence-activated cell sorting
- dsRNA
double-stranded RNA
Note Added in Proof.
In the latest release of the Genome project, the Hemese gene is now annotated in Flybase as CG31770, with the synonym Gustav.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF426744).
References
- 1.Lavine M D, Strand M R. Insect Biochem Mol Biol. 2002;32:1295–1309. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- 2.Gupta A P. Hemocytic and Humoral Immunity in Arthropods. New York: Wiley; 1986. [Google Scholar]
- 3.Vass E, Nappi A J. BioScience. 2001;51:529–535. [Google Scholar]
- 4.Samakovlis C, Kimbrell D A, Kylsten P, Engström Å, Hultmark D. EMBO J. 1990;9:2969–2976. doi: 10.1002/j.1460-2075.1990.tb07489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meister M, Hetru C, Hoffmann J A. Curr Top Microbiol Immunol. 2000;248:17–36. doi: 10.1007/978-3-642-59674-2_2. [DOI] [PubMed] [Google Scholar]
- 6.Franc N C, White K, Ezekowitz R A. Curr Opin Immunol. 1999;11:47–52. doi: 10.1016/s0952-7915(99)80009-0. [DOI] [PubMed] [Google Scholar]
- 7.Carton Y, Nappi A J. Immunogenetics. 2001;52:157–164. doi: 10.1007/s002510000272. [DOI] [PubMed] [Google Scholar]
- 8.Rizki T M, Rizki R M. In: Insect Ultrastructure. King R C, editor. Vol. 2. New York: Plenum; 1984. pp. 579–604. [Google Scholar]
- 9.Lanot R, Zachary D, Holder F, Meister M. Dev Biol. 2001;230:243–257. doi: 10.1006/dbio.2000.0123. [DOI] [PubMed] [Google Scholar]
- 10.Braun A, Hoffmann J A, Meister M. Proc Natl Acad Sci USA. 1998;95:14337–14342. doi: 10.1073/pnas.95.24.14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gateff E. Ann NY Acad Sci. 1994;712:260–279. doi: 10.1111/j.1749-6632.1994.tb33578.x. [DOI] [PubMed] [Google Scholar]
- 12.Konrad L, Becker G, Schmidt A, Klockner T, Kaufer-Stillger G, Dreschers S, Edström J E, Gateff E. Dev Biol. 1994;163:98–111. doi: 10.1006/dbio.1994.1126. [DOI] [PubMed] [Google Scholar]
- 13.Wilkins M R, Gasteiger E, Bairoch A, Sanchez J-C, Williams K L, Appel R D, Hochstrasser D F. Methods Mol Biol. 1998;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Krogh A, Larsson B, von Heijne G, Sonnhammer E L L. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 16.Hansen J E, Lund O, Tolstrup N, Gooley A A, Williams K L, Brunak S. Glycoconjugate J. 1998;15:115–130. doi: 10.1023/a:1006960004440. [DOI] [PubMed] [Google Scholar]
- 17.Kennerdell J R, Carthew R W. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- 18.Hedengren M, Åsling B, Dushay M S, Ando I, Ekengren S, Wihlborg M, Hultmark D. Mol Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 19.Marfatia S M, Leu R A, Branton D, Chishti A H. J Biol Chem. 1995;270:715–719. doi: 10.1074/jbc.270.2.715. [DOI] [PubMed] [Google Scholar]
- 20.Russ W P, Engelman D M. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]