Abstract
Influenza A virus causes widespread infection in the human respiratory tract, but existing vaccines and drug therapy are of limited value. Here we show that short interfering RNAs (siRNAs) specific for conserved regions of the viral genome can potently inhibit influenza virus production in both cell lines and embryonated chicken eggs. The inhibition depends on the presence of a functional antisense strand in the siRNA duplex, suggesting that viral mRNA is the target of RNA interference. However, siRNA specific for nucleocapsid (NP) or a component of the RNA transcriptase (PA) abolished the accumulation of not only the corresponding mRNA but also virion RNA and its complementary RNA. These siRNAs also broadly inhibited the accumulation of other viral, but not cellular, RNAs. The findings reveal that newly synthesized NP and PA proteins are required for influenza virus transcription and replication and provide a basis for the development of siRNAs as prophylaxis and therapy for influenza infection in humans.
Influenza A virus, a member of the Orthomyxoviridae family, causes the most prevalent infection of the respiratory tract in humans (1). In a typical year, 10–20% of the population in the United States is afflicted by the virus, resulting in up to 40,000 deaths (2). In what was one of the most devastating human catastrophes in history, at least 20 million people died worldwide during the 1918 influenza virus pandemic (3). The virulence of influenza A virus results from (i) its easy spread by aerosol; (ii) its ability to escape from protective immunity by frequent changes in viral antigens (antigenic drift) (4, 5); and (iii) the periodic emergence of new virulent strains by reassortment or mixing of RNA segments between viruses from two different species (antigenic shift) (6).
The threat of a new influenza pandemic persists because, despite intensive efforts, existing vaccines and therapy for influenza infection have only a limited value (7). Current vaccines, consisting of either killed virus or recombinant surface glycoproteins, induce only a weak IgG response and, as a result, protection can wane in as little as 6 months (8). In the most susceptible population, elderly and immunocompromised individuals, the efficacy of vaccination is merely 39% (2, 8). In addition, the existing vaccines have to be reformulated almost every year because the viral antigens [hemagglutinin (HA) and neuraminidase (NA)] that elicit protective antibodies usually undergo changes, rendering the previous year's vaccine ineffective against any new virus subtype. Similarly, although four antiviral drugs have been approved in the United States for treatment and/or prophylaxis of influenza, their use is limited because of severe side effects and the possible emergence of resistant viruses (9).
RNA interference (RNAi) is a process by which double-stranded RNA (dsRNA) directs sequence-specific degradation of messenger RNA (mRNA) (10–12). This phenomenon was initially observed in Caenorhabditis elegans (13), in plants (10, 14), and, recently, in mammalian cells (15). In plants, it is an evolutionarily conserved response to virus infection. Naturally occurring RNAi is initiated by the dsRNA-specific endonuclease Dicer-RDE-1, which processively cleaves long dsRNA into double-stranded fragments between 21 and 25 nt in length, termed short interfering RNA (siRNA) (15). siRNAs are then incorporated into a protein complex that recognizes and cleaves target mRNAs. Studies have shown that in mammalian cells, RNAi can be triggered by introducing synthetic 21-nt siRNA duplexes into the cells (16), bypassing the requirement for Dicer-RDE-1-mediated processing of long dsRNA. Because 21-nt siRNAs are too short to induce an IFN response in mammalian cells (16, 17), yet still able to confer transient interference of gene expression in a sequence-specific manner, they represent a previously unrecognized class of molecules that may have significant medical applications.
We report here the identification of siRNAs that can potently inhibit influenza virus production in both cell lines and embryonated chicken eggs. We show that inhibition by the most potent siRNAs is a result of both sequence-specific interference with viral mRNA accumulation and broad inhibition of all viral RNA transcription.
Materials and Methods
siRNAs.
All RNA oligonucleotides were synthesized by Dharmacon Research (Lafayette, CO). The oligonucleotides were deprotected according to the manufacturer's instructions. Equimolar amounts of complementary oligonucleotides were mixed and annealed by heating to 95°C for 5 min, then reducing the temperature slowly by 1°C every 30 sec until 35°C, then by 1°C every min until the temperature reached 5°C. The modified RNA oligonucleotides, in which the 2′-hydroxyl group was replaced with a 2′-O-methyl group in every nucleotide residue, was synthesized by Dharmacon, deprotected, and annealed to complementary strands as above. The resulting siRNA duplexes were analyzed for completion of duplex formation by gel electrophoresis. Sequences of all siRNAs tested are shown in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org.
Viruses and Assays.
Influenza viruses A/PR/8/34 (PR8) and A/WSN/33 (WSN), subtypes H1N1, were kindly provided by Peter Palese (Mount Sinai School of Medicine, New York). The viruses were grown in the allantoic cavity of 10-day-old embryonated chicken eggs (Charles River Laboratories, Wilmington, MA) at 37°C. Allantoic fluid was harvested 48 h after virus inoculation and stored at −80°C. Virus titer was measured by hemagglutination or plaque assays. The hemagglutination assay was carried out in V-bottom 96-well plates. Serial 2-fold dilutions of virus samples were mixed with an equal volume of a 0.5% suspension (vol/vol) of chicken erythrocytes (Charles River Laboratories) and incubated on ice for 1 h. Wells containing an adherent, homogeneous layer of erythrocytes were scored as positive. For plaque assays, serial 10-fold dilutions of virus samples were added onto a monolayer of Madin–Darby canine kidney (MDCK) cells in 1% semisolid agar. Two days after infection, plaques were visualized by staining with crystal violet.
Cell Culture and Virus Infection.
MDCK cells were grown in DMEM containing 10% heat-inactivated FCS, 2 mM l-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C under a 5% CO2/95% air atmosphere. For siRNA introduction, logarithmic-phase MDCK cells were trypsinized, washed, and resuspended in serum-free RPMI medium 1640 at 2 × 107 cells per ml. Cells (0.5 ml) were mixed with siRNA and electroporated at 400 V and 975 μF by using a Gene Pulser apparatus (Bio-Rad). Electroporated cells were divided into three wells of a six-well plate and cultured in DMEM for 8 h. The culture medium was then removed and 100 μl of PR8 or WSN virus in infection medium, consisting of DMEM, 0.3% BSA (Sigma), 10 mM Hepes, 100 units/ml penicillin, and 100 μg/ml streptomycin, was added to each well. After incubation for 1 h at room temperature, 2 ml of infection medium containing 4 μg/ml trypsin was added to each well and the cells were cultured at 37°C under 5% CO2. At different times after infection, supernatants were harvested from infected cultures and the virus titer was determined.
Virus and siRNA Inoculation in Chicken Embryos.
For each inoculation, 30 μl of Oligofectamine (Invitrogen) was diluted with 30 μl of Opti-MEM I (GIBCO). siRNA [2.5 nmol (10 μl)] was mixed with 30 μl of Opti-MEM I and added to diluted Oligofectamine, and the mixture was incubated at room temperature for 30 min. The mixture was then combined with 100 μl of PR8 virus [5,000 plaque-forming units (pfu)/ml] and immediately injected into the allantoic cavity of a 10-day-embryonated chicken egg. The eggs were incubated at 37°C for 17 h and allantoic fluid was harvested to measure virus titer.
RNA Extraction, Reverse Transcription (RT), and Real-Time PCR.
MDCK cells (1 × 107) were electroporated with or without siRNA, and were infected 8 h later with PR8 virus. One, 2, and 3 h after infection, culture medium was removed and the cells were lysed by using Trizol reagent (GIBCO). RNA was isolated by following the manufacturer's protocol. RT was carried out by using an Omniscript reverse transcriptase kit (Qiagen, Valencia, CA) in a 20-μl reaction mixture, containing 200 ng of total RNA and specific primers, at 37°C for 1 h. One microliter of RT reaction mixture was then used for real-time PCR by using gene-specific primers, SYBR green PCR Master Mix (Applied Biosystems), and SYBR green I dsDNA binding dye. Before the PCR, the mixture was incubated at 50°C for 2 min and 95°C for 10 min. The reaction was then performed at 95°C for 15 sec and 60°C for 1 min for 50 cycles. All reactions were done in duplicate. The levels of PCR products were monitored with an ABI PRISM 7000 sequence detection system and analyzed with ABI PRISM 7000SDS software (Applied Biosystems). Cycle times were analyzed at a reading of 0.2 fluorescence unit. Cycle times that varied by >1.0 unit between duplicates were discarded. The duplicate cycle times were averaged and normalized to the cycle time of γ-actin. Sequences of RT and PCR primers are shown in Table 3, which is published as supporting information on the PNAS web site.
Results
Design of siRNAs Specific for Influenza A Virus.
Influenza A virus has a segmented RNA genome. Three of the eight RNA segments encode three components of the RNA transcriptase (PA, PB1, and PB2) (1). Three additional RNA segments encode the major glycoproteins: HA and NA, and nucleocapsid protein (NP). Each of the remaining two RNA segments encodes two proteins, either M1 or M2, and NS1 or NS2, which function either as viral structural proteins or in the viral life cycle. Among influenza A viruses, 15 HA subtypes and 9 NA subtypes are known. Extensive differences in nucleotide sequences are also present in other genes among virus isolates from humans and different species. To design siRNAs that remain effective despite antigenic drifts and shifts, we focused on regions of the viral genome that are conserved among different subtypes and strains of virus from human, chicken, duck, horse, and swine (the influenza sequence database, www.flu.lanl.gov). Besides having no more than 1 mismatch in the 21 nucleotides among different virus subtypes and strains, the siRNAs designed did not share identity with any known human gene. We designed and tested a total of 20 siRNAs specific for NP, PA, PB1, PB2, M, and NS genes. No siRNA for HA and NA was designed because they contain no stretch of conserved 21 nucleotides, a result of extensive variations in these genes among different virus isolates from humans and various other species.
Inhibition of Influenza Virus Production in a Cell Line.
To test whether the synthetic siRNAs inhibited influenza virus production, we first examined their effect in MDCK cells. siRNAs (2.5 nmol) were introduced into MDCK cells (1 × 107) by electroporation, and 8 h later the cells were infected with either PR8 or WSN virus at a multiplicity of infection (moi) of 0.001, 0.01, or 0.1. At different times after infection, culture supernatants were harvested, serially diluted, and assayed to determine the virus titer by using an HA assay. As a control, siRNA specific for GFP (termed GFP-949) was similarly introduced into GFP-expressing MDCK cells, followed by virus infection. Virus titer was assayed as above and GFP expression was assayed by flow cytometry.
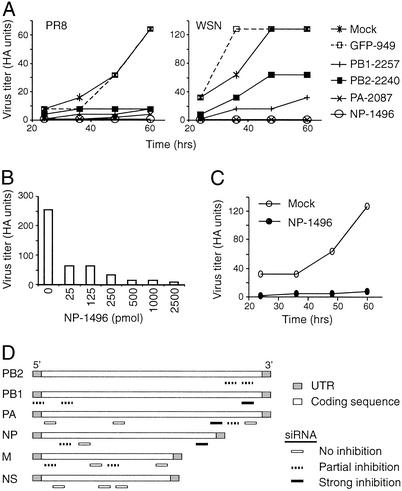
As shown in Fig. 1A, in mock transfection (no siRNA), virus titers in the infected cultures increased over time, reaching peak values between 48–60 h. Transfection of GFP-949 did not affect virus production at any time point but significantly inhibited GFP expression (data not shown), indicating that siRNA does not interfere nonspecifically with influenza virus production. Transfection of siRNAs specific for influenza virus generated three types of results (Fig. 1 A and D, and Table 1). First, ≈45% of the siRNAs had no discernable effect on the virus titer, indicating that they were not effective in interfering with influenza virus production in MDCK cells. Second, ≈40% of the siRNAs significantly inhibited virus production. The extent of inhibition varied somewhat, depending on whether PR8 or WSN virus was used. Third, ≈15% of the siRNAs potently inhibited virus production, regardless of whether PR8 or WSN virus was used. When either NP-1496 or PA-2087 siRNA was used, inhibition was so pronounced that culture supernatants lacked detectable HA activity (Fig. 1A).
Figure 1.
siRNAs interfere with influenza A virus production in MDCK cells. (A) Inhibition of influenza virus production in MDCK cells by selected siRNAs. MDCK cells were first electroporated with siRNAs and then infected 8 h later with PR8 or WSN virus at a moi of 0.01. Viral titers in the culture supernatants at different times after infection were measured by HA assay. HA units are arithmetic means based on titer endpoints of arithmetic dilutions. Virus titers (HA units) from five siRNA-treated cultures are shown over time for both PR8 (Left) and WSN (Right) infections. NP-1496, etc., are siRNAs specific for different viral genes. For example, NP-1496 indicates an siRNA specific for NP, with the beginning nucleotide at position 1496 of the NP sequence. (B) Inhibition of influenza virus production by different doses of siRNA. MDCK cells were transfected with the indicated amount of NP-1496 followed by infection 8 h later with PR8 virus at a moi of 0.1. Virus titer was measured 48 h after infection. Data shown are from one of two experiments. (C) Inhibition of influenza virus production by siRNA administered after virus infection. MDCK cells were infected with PR8 virus at a moi of 0.01 and were electroporated 2 h later with NP-1496 (2.5 nmol). Virus titer was measured at the indicated time after infection. Data shown are from one of two experiments. (D) Schematic diagrams showing the location of each siRNA in the viral genome and its relative potency in inhibiting influenza virus production in MDCK cells (based on data in Table 1). UTR, untranslated regions.
Table 1.
Effects of siRNAs on influenza virus production in MDCK cells
| Exp. | siRNA | Virus production (titer in HA units)
|
||||
|---|---|---|---|---|---|---|
| PR8 (0.001) | PR8 (0.01) | PR8 (0.1) | WSN (0.001) | WSN (0.01) | ||
| 1 | Mock | 256 | 256 | |||
| GFP-949 | 128 | 256 | ||||
| PB2-2210 | 16 | 32 | ||||
| PB2-2240 | 2 | 16 | ||||
| PB1-6 | 64 | 64 | ||||
| PB1-129 | 2 | 16 | ||||
| PB1-2257 | 1 | 4 | ||||
| 2 | Mock | 128 | 128 | |||
| GFP-949 | 64 | 128 | ||||
| PA-44 | 64 | 128 | ||||
| PA-739 | 32 | 64 | ||||
| PA-2087 | 1 | 8 | ||||
| PA-2110 | 16 | 32 | ||||
| PA-2131 | 32 | 64 | ||||
| 3 | Mock | 16 | 64 | 128 | ||
| NP-231 | 1 | 16 | 32 | |||
| NP-390 | 4 | 32 | 64 | |||
| NP-1496 | 1 | 1 | 1 | |||
| M-37 | 8 | 32 | 1 | |||
| 4 | Mock | 64 | 256 | 128 | ||
| M-37 | 32 | 256 | 1 | |||
| M-480 | 32 | 256 | 32 | |||
| M-598 | 32 | 256 | 1 | |||
| M-934 | 64 | 256 | 32 | |||
| NS-128 | 32 | 256 | 64 | |||
| NS-562 | 64 | 256 | 128 | |||
| NS-589 | 64 | 256 | 128 | |||
| NP-1496 | 1 | 16 | 1 | |||
| 5 | Mock | 64 | 128 | |||
| GFP-949 | 64 | 128 | ||||
| PB2-2240 | 8 | 64 | ||||
| PB1-2257 | 8 | 32 | ||||
| PA-2087 | 4 | 1 | ||||
| NP-1496 | 1 | 1 | ||||
| NP-231 | 8 | 64 | ||||
| NP-390 | 32 | 128 | ||||
The assays were done in the same way as in Fig. 1. Numbers in parentheses are moi values.
To estimate virus titers more precisely, we performed plaque assays with cultures that were infected with PR8 virus at a moi of 0.001 or 0.1 in the presence or absence of siRNA specific for NP (NP-1496). At a moi of 0.001, ≈6 × 105 pfu/ml was detected in the mock culture, whereas no plaques were detected in the undiluted NP-1496 culture supernatant. As the detection limit of the plaque assay is ≈20 pfu/ml, the inhibition of virus production by NP-1496 is at least 30,000-fold. Even at a moi of 0.1, NP-1496 inhibited virus production 200-fold.
To determine the potency of siRNA, a graded amount of NP-1496 was electroporated into MDCK cells, followed by PR8 infection. Virus titers in the culture supernatants were measured by HA assay. As the amount of siRNA decreased, virus titer increased in the culture supernatants (Fig. 1B). However, even when 25 pmol of siRNA was used for electroporation, an ≈4-fold decrease in virus production was detected as compared with mock transfection, indicating a high potency of NP-1496 siRNA in inhibiting influenza virus production.
For therapy, siRNA must be able to effectively inhibit an ongoing virus infection. After influenza virus infection, new virions are released in ≈4 h. To eliminate reinfection by newly released virus, MDCK cells were infected with PR8 virus for 2 h and then transfected with NP-1496. With time, virus titer increased steadily after mock transfection, whereas virus titer was only slightly increased in NP-1496-transfected cells (Fig. 1C). Thus, administration of siRNA after virus infection can also be effective.
Together, these results show that (i) some siRNAs can potently inhibit influenza virus production in MDCK cells (Fig. 1D); (ii) influenza virus production can be inhibited by siRNAs specific for different viral genes, including those encoding NP, PA, and PB1; and (iii) siRNA inhibition can still be initiated in cells with ongoing infection.
Inhibition of Virus Production in Embryonated Chicken Eggs.
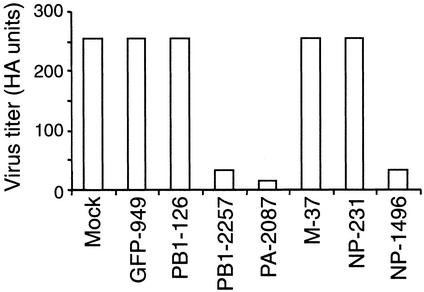
To extend the results in MDCK cells, we tested the ability of siRNAs to inhibit influenza virus production in developing chicken embryos, a widely used in vivo model of influenza virus infection. For siRNA transfection in the embryos, we used Oligofectamine, a lipid-based carrier that has been shown to facilitate intracellular uptake of DNA oligonucleotides (18). PR8 virus alone or virus plus siRNA was injected with Oligofectamine into the allantoic cavity of 10-day-old embryonated chicken eggs. Allantoic fluids were collected 17 h later for measurement of virus titers. When virus was injected alone (in the presence of Oligofectamine), high virus titers were detected (Fig. 2). As expected, coinjection of virus plus GFP-949 did not affect the virus titer. Coinjection of siRNAs specific for influenza virus, however, reduced virus titers. The results were concordant with those in MDCK cells. The same siRNAs (NP-1496, PA-2087, and PB1–2257) that potently inhibited influenza virus production in MDCK cells also inhibited virus production in chicken embryos, whereas siRNAs (NP-231, M-37, and PB1–129) that were less effective in MDCK cells (Fig. 1D and Table 1) were ineffective in chicken embryos. No significant reduction of virus titer was observed when Oligofectamine was omitted. Thus, siRNAs also effectively interfere with influenza virus production in embryonated chicken eggs.
Figure 2.
siRNAs interfere with influenza virus production in embryonated chicken eggs. A mixture of siRNAs (2.5 nmol), Oligofectamine, and PR8 virus (500 pfu) was injected into the allantoic cavity of 10-day-old embryonated chicken eggs. Allantoic fluid was collected 17 h later and assayed for virus titers as in Fig. 1. Data shown are from one of two experiments.
mRNA Is Probably the Direct Target of RNAi.
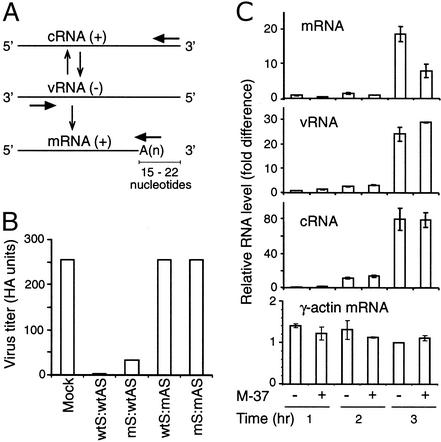
The RNA segments present in the influenza virion are called virion RNA (vRNA, −strand, Fig. 3A) (1). Transcription of vRNA by the virus-encoded transcriptase produces mRNA (+ strand), which serves as a template for synthesis of viral proteins. Transcription of vRNA also produces complementary RNA (cRNA, + strand), which serves as a template for synthesizing more vRNA for new virion production. Although siRNA is known to target mRNA for degradation (10–12), siRNA may also interfere with vRNA and cRNA. This result could occur because of the complementarity of the duplex siRNA strands: the antisense strand is complementary to mRNA and cRNA, and the sense strand is complementary to vRNA. To investigate these possibilities, we used NP-1496 siRNA in which either the sense (S or +) or antisense (AS or −) strand was modified (mS:wtAS or wtS:mAS). The modification, which replaces the 2′-hydroxyl group with a 2′-O-methyl group in every nucleotide residue, does not affect base-pairing for duplex formation, but the modified RNA strand no longer supports RNAi (M.T.M. and P.A.S., unpublished data).
Figure 3.
mRNA is the likely target for siRNA-mediated interference. (A) Schematic diagram illustrating the relationship among influenza virus vRNA, mRNA, and cRNA. Whereas cRNA is the exact complement of vRNA, mRNA contains a cap structure at the 5′ end (not shown) and a poly(A) sequence that occurs at a site 15–22 nt before the 5′ end of the vRNA segment. Arrows indicate the positions of primers used to distinguish among the various RNAs during RT. (B) Inhibition of influenza virus production requires a wild-type (wt) antisense strand in the duplex siRNA. MDCK cells were first electroporated with siRNAs formed from wt and modified (m) strands and were infected 8 h later with PR8 virus at a moi of 0.1. Virus titers in the culture supernatants were assayed 24 h after infection as in Fig. 1. Data shown are from one of two experiments. (C) M-specific siRNA inhibits the accumulation of specific mRNA. MDCK cells were electroporated with M-37, infected with PR8 virus at a moi of 0.01, and harvested for RNA isolation 1, 2, and 3 h after infection. The levels of M-specific mRNA, cRNA, and vRNA were measured by RT using RNA-specific primers as indicated in A, followed by real-time PCR. The level of each viral RNA species was normalized to the level of γ-actin mRNA (Bottom) in the same sample. The relative levels of RNAs are shown as mean value ± SD. Data shown are from one of two experiments.
MDCK cells were electroporated with wild-type or modified NP-1496 siRNAs, followed by PR8 virus infection, and virus titer in culture supernatants was measured. High virus titers were detected in cultures with mock transfection (Fig. 3B). As expected, very low virus titers were detected in cultures transfected with wild-type siRNA (wtS:wtAS), and high virus titers were detected in cultures transfected with siRNA in which both strands were modified (mS:mAS). The virus titers were high in cultures transfected with siRNA in which the antisense strand was modified (wtS:mAS), whereas the virus titers were low in cultures transfected with siRNA in which the sense strand was modified (mS:wtAS). The requirement for a wild-type antisense (−) strand in the siRNA duplex to inhibit influenza virus production suggests that the target of RNAi is either mRNA (+), cRNA (+), or both.
To further distinguish between these possibilities, we examined the effect of siRNA on the accumulation of the corresponding mRNA, cRNA, and vRNA. To follow transcription in a cohort of simultaneously infected cells, siRNA-transfected MDCK cells were harvested for RNA isolation 1, 2, and 3 h after infection (before new virion release and reinfection). The viral mRNA, vRNA, and cRNA were first independently converted to cDNA by using specific primers (Fig. 3A). The level of each cDNA was then quantified by real-time PCR. When M-specific siRNA M-37 was used, little M-specific mRNA was detected 1 or 2 h after infection (Fig. 3C). Three hours after infection, M-specific mRNA was readily detected in the absence of M-37. In cells transfected with M-37, the level of M-specific mRNA was reduced by ≈50%. In contrast, the levels of M-specific vRNA and cRNA were not significantly reduced by the presence of M-37, indicating again that viral mRNA is probably the target of siRNA-mediated interference.
Some siRNAs Inhibit Accumulation of All Viral RNAs.
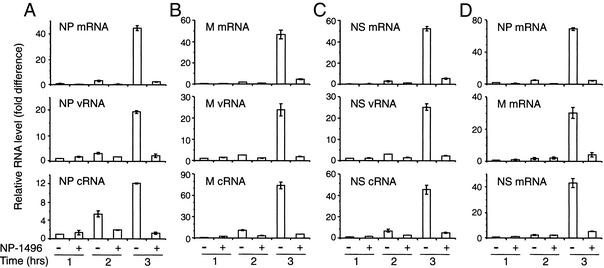
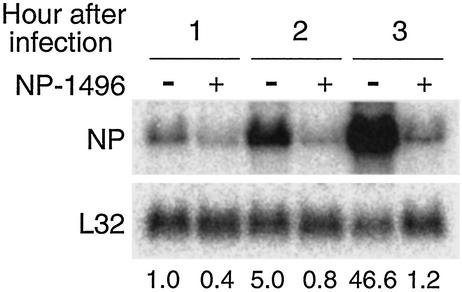
We also examined the effect of NP-1496 siRNA on the accumulation of NP-specific mRNA, vRNA, and cRNA by RT, followed by real-time PCR. With similar results as M-specific mRNA, NP-specific mRNA was low 1 or 2 h after infection (Fig. 4A Top). Three hours after infection, NP mRNA was readily detected in the absence of NP-1496, whereas in the presence of NP-1496, the amount of NP mRNA remained at the background level, indicating that siRNA inhibited the accumulation of specific mRNA. However, the NP-specific vRNA and cRNA were also abolished by the presence of NP-1496 (Fig. 4A Middle and Bottom). The same results were also obtained when Northern blotting was used to detect the viral RNA in infected cells (Fig. 5). Moreover, inhibition of other viral RNAs also occurred. In the NP-1496-treated cells, the accumulation of mRNA, vRNA, and cRNA of M, NS, PB1, PB2, and PA genes was also inhibited (Fig. 4 B and C, and data not shown). This broad inhibitory effect was also observed for PA-2087 (data not shown). As shown above, however, M-37 inhibited the accumulation of only M-specific mRNA but not M-specific vRNA or cRNA or other viral RNAs (Fig. 3C). Thus, depending on their sequence and specificity, certain siRNAs exert a global effect, inhibiting accumulation of all viral RNAs.
Figure 4.
NP-specific siRNA inhibits the accumulation of not only NP- but also M- and NS-specific mRNA, vRNA, and cRNA. MDCK (A–C) and Vero (D) cells were electroporated with NP-1496, infected with PR8 virus at a moi of 0.1, and harvested for RNA isolation 1, 2, and 3 h after infection. The levels of mRNA, cRNA, and vRNA specific for NP, M, and NS were measured by RT using RNA-specific primers, followed by real-time PCR. The level of each viral RNA species is normalized to the level of γ-actin mRNA (data not shown) in the same sample. The relative levels of RNAs are shown. Data shown are from one of two experiments.
Figure 5.
NP-specific siRNA inhibits the accumulation of all NP viral RNAs. MDCK cells were electroporated with NP-1496, infected with PR8 virus at a moi of 0.1, and harvested for RNA isolation 1, 2, and 3 h after infection. Total RNA (15 μg) was separated by a 1.2% denaturing agarose gel, transferred, and hybridized sequentially with probes specific for NP (Upper) and the ribosomal L32 gene (Lower). The numbers indicate the relative levels of NP-specific RNA normalized to the levels of L32. The level of NP-specific RNA in cells that were infected for 1 h in the absence of siRNA is arbitrarily given a value of 1. The NP-specific probe was double-stranded and hybridized to mRNA, vRNA, and cRNA.
One possible cause for the broad inhibition of viral RNA accumulation is an IFN response by the infected cells to the presence of dsRNA (17, 19, 20). Thus, we carried out the above experiments in Vero cells in which the entire IFN locus, including all α, β, and ω genes, is deleted (21, 22) (Q.G. and J.C., unpublished data). In these cells, as in MDCK cells, the accumulation of NP-, M-, and NS-specific mRNAs was inhibited by NP-1496 (Fig. 4D). In addition, we assayed the effect of siRNA on the levels of transcripts from cellular genes, including β-actin, γ-actin, GAPDH, and ribosomal L32. No significant difference in transcript levels of these genes was detected in the presence or absence of siRNA (Figs. 3C and 5 and data not shown), indicating that the inhibitory effect of siRNA is specific for viral RNAs. These results suggest that the broad inhibition of viral RNA accumulation by siRNA is not because of a cellular IFN response.
After influenza virus infection, the presence of dsRNA also activates a cellular pathway that targets RNA for degradation (17). To examine the effect of siRNA on the activation of this pathway, we assayed the levels of phosphorylated protein kinase R (PKR), the most critical component of the pathway (17). Transfection of MDCK cells with NP-1496 in the absence of virus infection did not affect the levels of activated PKR (data not shown). Infection by influenza virus resulted in an increased level of phosphorylated PKR, consistent with previous studies (19, 20, 23). However, the increase was the same in the presence or absence of NP-1496 (data not shown). Thus, the broad inhibition of viral RNA accumulation mediated by certain siRNAs is not a result of enhanced RNA degradation.
Discussion
In this report we showed that (i) siRNAs can potently inhibit influenza virus production, and (ii) some siRNAs exert their inhibitory effect by interfering with the accumulation of not only mRNA, but also other viral RNAs. These findings have significant implications for the use of siRNA for prophylaxis and therapy of influenza virus infection, and for the mechanisms underlying influenza virus transcription and replication.
Influenza virus infection is considered to have the potential to become a much more dangerous disease than at present because of easy transmission, antigenic shift and drift of the virus, and the limited efficacy of current vaccines and therapy (7). We showed that siRNAs potently inhibited influenza virus production in both cell lines and embryonated chicken eggs. Among 20 siRNAs tested, those that target NP and a component of the RNA transcriptase are especially effective, working at picomolar range and after virus infection has occurred. These results provide a basis for further development of siRNAs for prophylaxis and therapy of influenza virus infection in humans. Influenza virus naturally infects epithelial cells in the upper respiratory tract and lungs in humans. siRNAs could be conveniently administrated via intranasal or pulmonary routes. Considering that the number of virions is probably small at the beginning of a natural infection, sufficient amounts of siRNA may be taken up by epithelial cells in the upper airways and lungs to inhibit virus replication, thus, potentially achieving preventive or therapeutic effects.
After influenza virus infection, vRNA is transcribed into both mRNA and cRNA (1). Because of the complementarity of duplex siRNA strands, siRNAs may interfere directly with mRNA and with vRNA and cRNA. We showed that viral mRNA is the direct target of siRNA-mediated interference because (i) the interference requires a functional antisense strand of the siRNA duplex, and (ii) M-specific siRNA interfered with the accumulation of M-specific mRNA, but not cRNA or vRNA. These findings are consistent with previous studies showing that siRNAs target the degradation of mRNAs that are transcribed from either cellular or viral genes (10–12, 24–26). In particular, siRNA was previously shown not to interfere with vRNA of respiratory syncytial virus (RSV) (27). As with RSV, influenza vRNA and cRNA are bound by NP, which may protect these RNAs from cleavage by RNAi machinery. It is also possible that mRNAs are targeted because they are exported to the cytoplasm, whereas vRNAs and cRNAs remain in the nucleus of influenza virus-infected cells.
We found, however, that NP- or PA-specific siRNA interfered with the accumulation of not only NP- or PA-specific mRNA, but also NP- or PA-specific cRNA and vRNA. These siRNAs also inhibited the accumulation of RNAs from other viral genes. This broad inhibition was virus-specific, as it did not significantly affect RNAs transcribed from cellular genes. This virus-specific inhibition was also observed in Vero cells, which lack the IFN α, β, and ω genes, indicating that the broad inhibition is not a result of an IFN response that shuts off all transcriptions in the infected cells. In addition, the broad inhibition was not a result of a general degradation of virus-specific RNAs because activation of protein kinase R was not affected by the presence of siRNA in infected cells. Instead, these findings reveal a critical role of newly synthesized NP and PA proteins in viral transcription and replication.
The number of NP protein molecules in infected cells has been hypothesized to regulate the levels of mRNA synthesis versus genome RNA (vRNA and cRNA) replication (1). Using a temperature-sensitive mutation in the NP protein, previous studies have shown that cRNA, but not mRNA, synthesis was temperature-sensitive both in vitro and in vivo (28, 29). NP protein was also shown to be required for elongation and antitermination of nascent cRNA and vRNA transcripts (29, 30). We found that NP-specific siRNA inhibited the accumulation of all viral RNAs in infected cells. Probably, in the presence of NP-specific siRNA, the newly transcribed NP mRNA is degraded, resulting in inhibition of NP protein synthesis. Without newly synthesized NP, further viral transcription and replication are blocked, as is new virion production.
Similarly, in the presence of PA-specific siRNA, newly transcribed PA mRNA is degraded, resulting in inhibition of PA protein synthesis. Despite the presence of 30–60 copies of RNA transcriptase per influenza virion (1), without newly synthesized RNA transcriptase, further viral transcription and replication are evidently inhibited. In contrast, the matrix (M) protein is not required until a late phase of virus infection (1). Accordingly, M-specific siRNA inhibits the accumulation of M-specific mRNA but not vRNA, cRNA, or other viral RNAs. Taken together, these findings demonstrate a critical requirement for newly synthesized NP and PA proteins in influenza viral RNA transcription and replication. The broad inhibition of all viral RNA accumulation by NP- or PA-specific siRNAs probably occurs because the RNAs are not transcribed. Both the targeted mRNA degradation and the resulting global inhibition of other viral RNA transcription make the NP- and PA-specific siRNAs especially potent inhibitors of influenza virus infection.
Supplementary Material
Acknowledgments
We thank Drs. Peter Palese and Adolfo Garcia-Sastre for kindly allowing Q.G. to visit their laboratories; Drs. Christopher Basler, Astrid Flandorfer, Luis Martinez-Sobrido, and Yurie Nakaya for teaching Q.G. various techniques for handling influenza virus; and members of the Chen and Eisen laboratories for helpful discussions. This work was supported in part by National Institutes of Health grants (AI40146, AI44478, and AI50631 to J.C.; AI44477 and CA60686 to H.N.E.; and GM34277, AI32486, and CA42063 to P.A.S.) and a Cancer Center Core Grant (to Richard Hynes). M.T.M. is partially supported by a postdoctoral fellowship from the Cancer Research Institute.
Abbreviations
- vRNA
virion RNA
- cRNA
complementary RNA
- dsRNA
double-stranded RNA
- siRNA
short interfering RNA
- RNAi
RNA interference
- HA
hemagglutinin
- NA
neuraminidase
- NP
nucleocapsid protein
- MDCK cell
Madin–Darby canine kidney cell
- PR8
A/Puerto Rico/8/34 (H1N1)
- WSN
A/WSN/33 (H1N1)
- pfu
plaque-forming units
- moi
multiplicity of infection
- RT
reverse transcription
References
- 1.Lamb R A, Krug R M. In: Fundamental Virology. Knipe D M, Howley P M, editors. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 725–770. [Google Scholar]
- 2.Fukuda K, Bridges C B, Brammer T L, Izurieta H S, Cox N J. Morbid Mortal Wkly Rep. 1999;48:1–28. [Google Scholar]
- 3.Patterson K D, Pyle G F. Bull Hist Med. 1991;65:4–21. [PubMed] [Google Scholar]
- 4.Parvin J D, Moscona A, Pan W T, Leider J M, Palese P. J Virol. 1986;59:377–383. doi: 10.1128/jvi.59.2.377-383.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Webster R G, Laver W G, Air G M, Schild G C. Nature. 1982;296:115–121. doi: 10.1038/296115a0. [DOI] [PubMed] [Google Scholar]
- 6.Webster R G, Bean W J, Gorman O T, Chambers T M, Kawaoka Y. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webby R J, Webster R G. Philos Trans R Soc London. 2001;356:1817–1828. doi: 10.1098/rstb.2001.0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castle S C. Clin Infect Dis. 2000;31:578–585. doi: 10.1086/313947. [DOI] [PubMed] [Google Scholar]
- 9.Luscher-Mattli M. Arch Virol. 2000;145:2233–2248. doi: 10.1007/s007050070017. [DOI] [PubMed] [Google Scholar]
- 10.Vaucheret H, Beclin C, Fagard M. J Cell Sci. 2001;114:3083–3091. doi: 10.1242/jcs.114.17.3083. [DOI] [PubMed] [Google Scholar]
- 11.Sharp P A. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 12.Brantl S. Biochim Biophys Acta. 2002;1575:15–25. doi: 10.1016/s0167-4781(02)00280-4. [DOI] [PubMed] [Google Scholar]
- 13.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 14.Baulcombe D. Curr Biol. 2002;12:R82–R84. doi: 10.1016/s0960-9822(02)00665-6. [DOI] [PubMed] [Google Scholar]
- 15.Elbashir S, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 16.McManus M T, Sharp P A. Nat Rev Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 17.Kumar M, Carmichael G G. Microbiol Mol Biol Rev. 1998;62:1415–1434. doi: 10.1128/mmbr.62.4.1415-1434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pedroso de Lima M C, Simoes S, Pires P, Faneca H, Duzgunes N. Adv Drug Delivery Rev. 2001;47:277–294. doi: 10.1016/s0169-409x(01)00110-7. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Sastr A. Microbes Infect. 2002;4:647–655. doi: 10.1016/s1286-4579(02)01583-6. [DOI] [PubMed] [Google Scholar]
- 20.Katze M G, He Y, Gale M., Jr Nat Rev Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 21.Diaz M O, Ziemin S, Le Beau M M, Pitha P, Smith S D, Chilcote R R, Rowley J D. Proc Natl Acad Sci USA. 1988;85:5259–5263. doi: 10.1073/pnas.85.14.5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diaz M O, Pomykala H M, Bohlander S K, Maltepe E, Malik K, Brownstein B, Olopade O I. Genomics. 1994;22:540–552. doi: 10.1006/geno.1994.1427. [DOI] [PubMed] [Google Scholar]
- 23.Kim M-J, Latham A G, Krug R M. Proc Natl Acad Sci USA. 2002;99:10096–10101. doi: 10.1073/pnas.152327499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacque J-M, Triques K, Stevenson M. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gitlin L, Karelsky S, Andino R. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 26.Novina C D, Murray M F, Dykxhoorn D M, Beresford P, Riess J, Lee S-K, Collman R G, Lieberman J, Shankar P, Sharp P A. Nat Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 27.Bitko V, Barik S. BMC Microbiol. 2001;1:34–43. doi: 10.1186/1471-2180-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Medcalf L, Poole E, Elton D, Digard P. J Virol. 1999;73:7349–7356. doi: 10.1128/jvi.73.9.7349-7356.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shapiro G I, Krug R M. J Virol. 1988;62:2285–2290. doi: 10.1128/jvi.62.7.2285-2290.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beaton A R, Krug R M. Proc Natl Acad Sci USA. 1986;83:6282–6286. doi: 10.1073/pnas.83.17.6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.