Abstract
Increased levels of triglyceride-rich lipoproteins provoke lipid accumulation in the artery wall, triggering early inflammatory responses central to atherosclerosis like endothelial adhesion molecule expression. The endogenous mechanisms limiting such reactions remain poorly defined. Lipoprotein lipase (LPL) plays a central role in lipid metabolism by hydrolyzing triglyceride rich lipoproteins and releasing fatty acids. We found that LPL treatment reversed tumor necrosis factor α and very low-density lipoprotein (VLDL)-stimulated endothelial vascular cell adhesion molecule 1 (VCAM1) induction and VCAM1 promoter responses, thus recapitulating effects reported with synthetic peroxisome proliferator-activated receptor (PPAR) agonists. In fact, these LPL effects on VCAM1 were absent in endothelial cells isolated from PPARα-deficient mice. This finding suggests a novel antiinflammatory role for LPL. Further studies reveal specificity for PPAR activation through lipolysis in regards to lipoprotein substrate (VLDL ≫ LDL > HDL), PPAR isoform (PPARα ≫ PPARδ > PPARγ), and among fatty acid-releasing lipases. These PPAR responses required intact LPL catalytic activity. In vivo, transgenic mice overexpressing LPL had increased peroxisome proliferation, but not in the genetic absence of PPARα. Although human plasma possesses minimal PPARα activation despite containing abundant free fatty acids, marked PPARα activation is seen with human plasma after LPL is added in vitro or systemically released in vivo. These data suggest a previously uncharacterized pathway in which the key lipolytic enzyme LPL can act on circulating lipoproteins to generate PPARα ligands, providing a potentially important link between lipoprotein metabolism and distal PPARα transcriptional effects.
Inflammation plays a fundamental role in the development and complications of atherosclerosis (1). One early seminal event in atherogenesis is adhesion molecule expression, a response integral to inflammatory cell entry into the vessel wall (1). Elevated levels of triglyceride-rich lipoproteins (TRL), like very low-density lipoprotein (VLDL) and chylomicrons, induce vascular cell adhesion molecule-1 (VCAM1) expression (2), an effect markedly augmented by inflammatory cytokines (3). Such responses may contribute to the association between levels of TRL, the major carrier of fatty acids (FA), and atherosclerosis (4). In contrast, VCAM1 expression is typically negligible in otherwise healthy endothelium despite exposure to TRL through both dietary sources and endogenous production (4, 5), suggesting physiologic pathways that limit adhesion molecule expression. The links between endogenous lipid metabolism and pathways that might inhibit inflammatory responses, like adhesion molecule expression, remain poorly defined.
Prior reports have demonstrated that synthetic agonists for the nuclear receptor peroxisome proliferator-activated receptor α (PPARα) decreased VCAM1 expression induced by inflammatory stimuli, including tumor necrosis factor α (TNFα; refs. 6–8). Consistent with these findings, certain omega 3 FA inhibited leukocyte adhesion in vivo in WT but not in PPARα-deficient (PPARα−/−) mice (9). Such observations are of particular interest, given associations between increased dietary intake of omega 3 FA and reduced cardiovascular events, including atherosclerosis, as seen in recent large epidemiologic studies (10–12).
Lipoprotein lipase (LPL) represents an appealing candidate for an enzyme whose physiologic action might limit adhesion molecule expression. LPL exerts a proximal effect in lipid metabolism, determining catabolism of nascent chylomicrons and VLDL (4). The site of LPL action is primarily on the endothelium itself, where its activity helps determine circulating triglyceride and high-density lipoprotein (HDL) levels, known cardiovascular risk factors (13). Both genetic and functional LPL abnormalities are directly linked to vascular pathology (14, 15), although divergent effects have been reported, perhaps dependent on tissue-specific LPL responses (13). Transgenic LPL overexpression decreased lesions in both apolipoprotein E (ApoE)- and low-density lipoprotein (LDL) receptor (LDLR)-deficient mouse models of atherosclerosis (14, 16). In fact, LPL overexpression completely abrogated atherosclerosis in LDLR−/− mice despite an atherogenic diet (14). In contrast, bone marrow transplantation of macrophages (Mφ) overexpressing LPL into similar LDLR-deficient mice markedly increased atherosclerotic lesions (17). Mechanisms accounting for this differential role of LPL in atherosclerosis are not clear. LPL hydrolytic function may be decreased in Mφ (18), leaving nonenzymatic LPL-mediated lipid uptake (a result of LPL bridging lipoprotein receptors and proteoglycans) unchecked (13).
We hypothesized that physiologic triglyceride lipolysis mediated by LPL might represent a link between lipid metabolism and PPAR activation, exerting distal PPAR effects. To examine this possibility, we studied the effects of LPL on TNFα-induced VCAM1 responses in EC in the genetic presence or absence of PPARα. We further explored lipolytic activation of PPARs in vitro and in vivo, testing the role of LPL enzymatic action, different lipoprotein substrates, other FA-releasing lipases, and the PPAR isoforms involved that might form such a pathway.
Methods
Reagents and media were from Sigma or BioWhittaker, respectively, except as indicated. Fenofibric acid was a generous gift from Laboratories Fournier.
Mice with PPARα+/+ (129S3/SvImJ) genotype were obtained from The Jackson Laboratory. PPARα−/− mice were a generous gift from F. Gonzalez (National Institutes of Health; ref. 19). Mice overexpressing LPL in skeletal muscle (20, 21) were crossbred to homogeneity with PPARα−/− mice.
Cell cultures of microvascular endothelial cells (EC) were isolated from 1-mo-old PPARα+/+ and PPARα−/− mouse hearts by using intercellular adhesion molecule-2 (ICAM-2) and platelet-endothelial adhesion molecule-1 (PECAM-1) antibody (PharMingen) Dynabead selection (22). HepG2 cells and HEK293 cells [American Type Culture Collection (ATCC)] were cultured in DMEM, 10% FCS. Human EC were from saphenous veins (6). Circulating human monocytes were isolated from healthy volunteers and treated in M199 medium after differentiation (23).
Lipoproteins were isolated by using gradient ultracentrifugation of plasma pooled from multiple healthy donors. Cholesterol, triglyceride, and protein (Pierce) contents were measured by enzymatic assay. Total free FA content was determined by enzymatic assay (Roche Molecular Biochemicals), adapted for 96-well plates (see Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org). For labeling, VLDL was incubated (10 h, 23°C) with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI, final concentration, 500 ng of DiI per mg of VLDL protein). Human post-heparin samples were drawn 10 min after systemic heparin infusion (60 units/kg).
Northern analysis was done on isolated mRNA (RNEasy; Qiagen, Valencia, CA) transferred after gel electrophoresis to Hybond (Amersham Pharmacia).
Transient transfection was done by using Fugene (Roche Molecular Biochemicals) in 24-well plates containing ≈2.3 × 104 bovine aortic EC (passage 3–6) per well. Constructs were generous gifts from T. Willson [GlaxoSmithKline, ligand-binding domain (LBD)/yeast Gal4]; T. Collins (Children's Hospital, Boston; VCAM1 promoter); and H. Hobbs (University of Texas Southwestern, LDLR expression vector). LPL and LPL mutant expression vectors have been described (D.J.R., unpublished data; refs. 24 and 25). The LPL mutant generates minimal lipolysis due to a 2-bp difference (AGC/GCC) that replaces the catalytic site serine-132 with alanine.
Immunohistochemistry was performed on frozen peripheral muscle sections by staining with rabbit PMP70 antibody (1:200; Affinity BioReagents, Neshanic Station, NJ; ref. 26) and secondary tetramethylrhodamine B isothiocyanate-conjugated antibody R0156 (DAKO), and analyzed by using laser-scanning confocal microscopy (MRC 600; Bio-Rad).
Scintillation proximity assays were done using human full-length cDNA for PPARα, PPARδ, and hPPARγ2 subcloned into the pGEX-KT expression vector as described (27). The 3H2-labeled known synthetic PPAR agonists used were compound A and compound B (see Supporting Methods) with relative Kds as follows: compound A, PPARγ 2.5 nM and PPARα 5.0 nM; compound B, PPARδ 1.0 nM (27, 28). Results are expressed as the percentage inhibition with a calculated inhibitory constant (KiS).
VCAM1 cell surface ELISA on mouse PPARα+/+ and PPARα−/− EC monolayers was performed as before (6).
Results
LPL Opposes VLDL/TNFα-Induced VCAM1 Expression in a PPARα-Dependent Fashion.
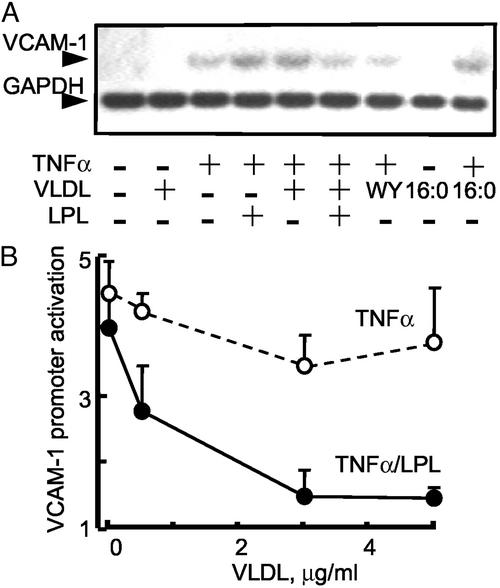
Inflammatory cytokines induce endothelial VCAM1 expression, a response known to be augmented by either TRL or saturated FA (3, 29). Consistent with these reports, VLDL and palmitic FA (16:0) both significantly increased VCAM1 mRNA in EC in the presence of TNFα (Fig. 1A, lanes 5 and 9, respectively). In contrast, LPL treatment of VLDL (LPL/VLDL) inhibited this VCAM1 response, even in the presence of TNFα (Fig. 1A, lane 6). This LPL-mediated repression of induced VCAM1 equaled that seen with the synthetic PPARα agonist WY14643 (Fig. 1A, lane 7). Neither LPL (data not shown) nor VLDL alone (Fig. 1A, lane 2) induced VCAM1 expression. This antiinflammatory response was also evident in VCAM1 promoter studies, with recombinant LPL (200 units/ml) reversing TNFα-mediated VCAM1 promoter activation in a manner dependent on VLDL substrate concentration (Fig. 1B). Thus, LPL treatment of VLDL alters transcriptional regulation of VCAM1.
Figure 1.
LPL inhibits VLDL/TNFα-mediated VCAM1 expression and promoter activity. (A) Northern analysis of VCAM1 mRNA expression in human EC treated with the PPARα agonist WY14643 (WY, 250 μM); VLDL (5 μg) with or without LPL (200 units/ml, 4 h); saturated FA (16:0) in the presence or absence of TNFα stimulation (50 ng/ml, 17 h). LPL alone had no effect (data not shown). (B) TNFα-mediated VCAM1 promoter (6) activation was tested over a concentration range of VLDL in the presence (solid line) or absence (dashed line) of LPL. All transfection experiments were performed in bovine aortic EC (DMEM, 1% Nutridoma SP). EC were pretreated with VLDL with or without LPL (200 units/ml, 4 h) and then stimulated with TNFα (2 ng/ml, 12 h). The luciferase results were normalized to β-galactosidase activity. For comparison, 250 μM WY14643 suppressed TNFα-induced VCAM1 promoter to a 2.6-fold activation. n = 3, each in triplicate.
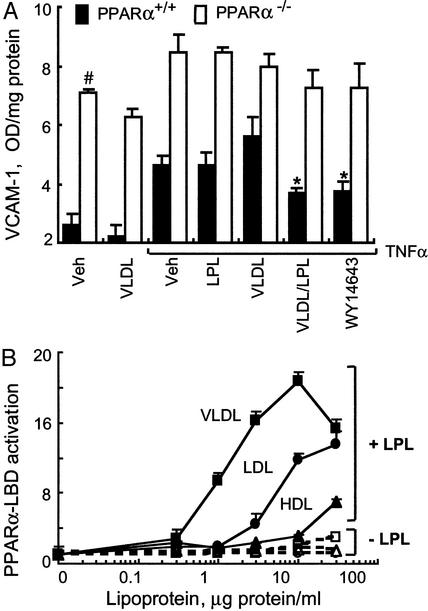
Given the possibility of PPAR-independent effects of synthetic PPAR agonists (30, 31), we next asked whether LPL/VLDL repression of VCAM1 persisted in EC isolated from PPARα−/− mice. By ELISA, PPARα−/− EC had significantly higher VCAM1 levels as compared with WT EC (Fig. 2A), consistent with the increased NF-κB activation reported in PPARα−/− mice (32). LPL/VLDL limited TNFα-induced increases in VCAM1 in WT but not in PPARα−/− EC, an effect similar to that seen with a synthetic PPARα agonist (Fig. 2A). These results suggest that LPL/VLDL effects are mediated through PPARα.
Figure 2.
LPL/VLDL inhibits VCAM1 levels through a PPARα-dependent mechanism. (A) VCAM1 surface expression in EC obtained from WT (filled bars) or PPARα−/− (open bars) mice was measured by ELISA as described (6). EC cultured in 96-well plates were treated as in Fig. 1A with murine TNFα (10 ng/ml, 18 h). Data represent absorbance at 410 nm after protein concentration normalization. In WT, but not PPARα−/−, EC both WY14643 and LPL/VLDL significantly decrease VCAM1 levels (*, P < 0.005). Basal VCAM1 levels in PPARα−/− are significantly increased as compared with WT EC (#, P < 0.001). (B) Concentration-dependent activation of PPARα-LBD by various lipoproteins in the presence or absence of LPL (30 units/ml). EC cotransfected with PPARα-LBD, the luciferase response pUASx4-TK-luc, and β-galactosidase constructs were stimulated as shown (18 h, DMEM, 1% delipidated serum). PPARα-LBD activation by fenofibric acid (100 μM) in this experiment was 16.2- ± 1.3-fold.
LPL acts on triglycerides in lipoproteins to release FA, with relative triglyceride content of common lipoproteins following the order VLDL ≫ LDL > HDL. The triglyceride content (mg triglyceride per dl) in LP studied were VLDL 264 ± 13.2, LDL 51 ± 2.6, and HDL 24 ± 1. To examine the relationship between circulating lipoproteins and LPL effects on PPARα, we performed standard chimeric PPARα-LBD/GAL4 transfection assays (33) in primary bovine aortic EC, known to lack LPL. In the absence of added LPL, these lipoproteins failed to activate the PPARα-LBD (Fig. 2B); however, in the presence of recombinant LPL, significant PPARα activation occurred in a concentration-dependent manner. Half-maximal PPARα activation values (EC50) for VLDL, LDL, and HDL were 1.5, 5, and 20 μg of protein per ml, respectively. Maximal induction was highest for VLDL (21-fold), followed by LDL (14-fold) and HDL (7-fold). Taken together, LPL treatment of TRL leads to significant PPARα activation.
PPAR Isoform-Specific Effects of VLDL Lipolysis.
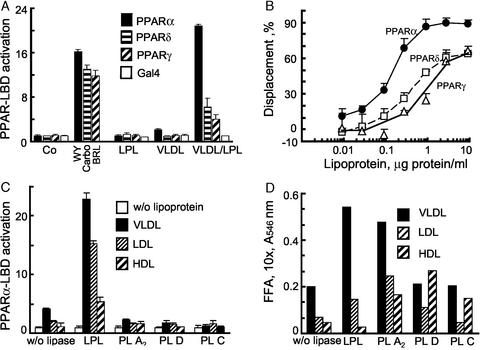
Given reports that FA in vitro can activate all PPAR isoforms, we next examined these LPL/VLDL responses in PPARγ, PPARδ, and PPARα-LBD transfection assays, and compared the responses to isoform-specific ligands (Fig. 3A). VLDL alone had no PPAR-LBD effects. In the presence of LPL, the most potent relative induction was observed for PPARα, with much less of a response for PPARδ, followed by PPARγ (P < 0.001). Control plasmid GAL4 remained unchanged.
Figure 3.
Lipolysis leads to PPARα activation in a manner selective as to PPAR isoform, lipase, and substrate. (A) PPAR-LBD activation by VLDL in EC was measured in the presence or absence of LPL by using standard pUASx4-TK-luc reporter construct and β-galactosidase normalization approaches as in Fig. 2B. Cells were stimulated with LPL (14 h, 30 units/ml) in the presence or absence of VLDL (10 μg/ml). Responses to established PPAR agonists were used for comparison: PPARγ−BRL49653 (BRL, 1 μM); PPARα−WY14643 (WY, 100 μM); PPARδ−carboprostacyclin (carbo, 10 μM). (B) Competition curves were generated by incubating the LPL/VLDL reaction mixture with specific radiolabeled PPAR activators: 5 nM 3H2 compound A with GST-hPPARα (circles) or GST-hPPARγ (triangles), or 2.5 nM 3H2 compound B with GST-hPPARδ (squares). Radioligand displacement in the presence of the indicated concentration of VLDL (0.003–10 μg of protein per ml)/LPL (200 units/ml LPL, 37°C, 1 h) is plotted. Data are from duplicate determinations. KiS values were obtained from the dose–response curves. (C) PPARα activation occurs through lipolysis mediated by LPL but not some other FA-releasing lipases. EC, transfected as in Fig. 2B, were treated (18 h) with the lipoproteins shown (10 μg of protein per ml) and either LPL (20 units/ml) or phospholipases PLA2 (20 units/ml), PLD (20 units/ml), or PLC (2 units/ml). Control (open bars), VLDL (filled bars), LDL (horizontally striped bars), and HDL (diagonally striped bars) are shown. (D) The FA generated in Fig. 3C is shown. The FA content in the media was measured spectrophotometrically as described in Methods, SD 5%; bars designated as in Fig. 3C. Spectrophotometric determinations correlated with palmitic acid standard in a concentration-dependent manner (data not shown).
As a complement to biologic LBD assays, competitive displacement of specific radioactive ligands establishes direct binding of putative ligand(s), such as LPL/VLDL-generated products, to given expressed PPAR-LBD protein in a cell-free setting (Fig. 3B; ref. 27). These assays are independent of any noncatalytic LPL effects, like bridging (13). Maximal radioligand displacement by LPL/VLDL occurred against the PPARα-LBD (99%), with responses extending to low levels of lipoprotein substrate (KiS 0.1 μg of protein per ml). Lesser effects were seen for PPARδ and PPARγ, with ≈65% maximal displacement for both (KiS 0.3 and 1.2 μg of protein per ml, respectively). LPL and VLDL added separately had no effect (data not shown). These results indicate that enzymatic hydrolysis of VLDL by LPL preferentially generates PPARα ligands.
Lipase-Specific Effects of VLDL Lipolysis.
To investigate PPAR activation through other lipases, LBD experiments were repeated using phospholipase A2, phospholipase D, phospholipase C, and LPL treatment of the major lipoprotein classes (Fig. 3C). Despite quantitative FA release (Fig. 3D) by all these enzymes, only LPL significantly activated PPARα regardless of lipoprotein substrate. Thus, PPARα activation generated from lipoproteins depends to some extent on the lipase.
LPL/VLDL Activation of PPARα Depends on Intact LPL Enzymatic Activity.
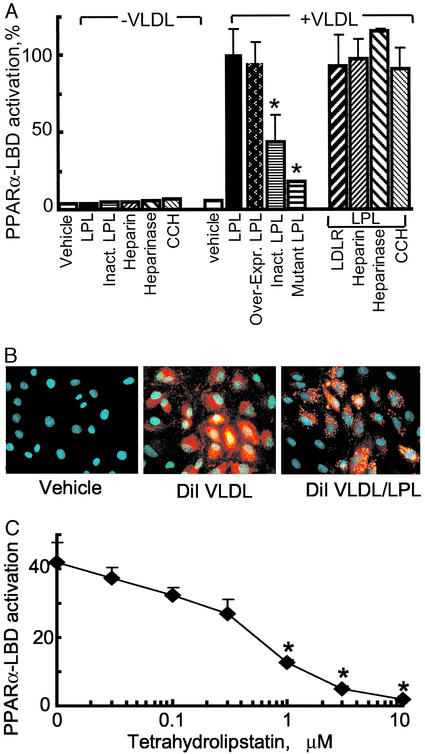
To address how LPL/VLDL activates PPARα, we asked whether PPARα-LBD responses depended on intact LPL enzymatic activity or, alternatively, on increased lipoprotein uptake through LPL's known bridging effect (13). PPARα−LBD activation experiments were performed after (i) altering bridging responses by overexpressing the LDL receptor; (ii) changing proteoglycan levels with heparin and heparinase; or (iii) impairing endosomal processing with the actin inhibitor cytochalasin. None of these treatments had an effect (Fig. 4A). The lack of PPARα responses in the absence of LPL was not due to decreased lipid uptake as shown by abundant intracellular DiI-labeled VLDL even in the absence of LPL (Fig. 4B). Thus, VLDL uptake alone is unlikely to account for PPARα-LBD activation. In contrast, addition of heat-inactivated LPL or transfection of a catalytically inactive LPL significantly decreased PPARα-LBD activation (Fig. 4A). Furthermore, the lipase inhibitor tetrahydrolipstatin decreased LPL-mediated PPARα-LBD activation in a dose-dependent fashion (Fig. 4C). Thus, multiple lines of evidence indicate that generation of PPARα activators from VLDL depends on intact LPL catalytic activity.
Figure 4.
PPARα activation by LPL/VLDL depends on intact LPL catalytic activity. (A) The role of LPL catalytic activity on PPARα-LBD activation was studied in EC by using VLDL (10 μg/ml) combined with various forms of LPL: recombinant LPL (30 units/ml) or heat-inactivated LPL (Inact. LPL); transfected expression constructs for human LPL (Over-expr. LPL) or catalytically inactive mutant LPL. Results are expressed as the percentage of standard LPL/VLDL response. Asterisk indicates a significant decrease in PPARα-LBD activation (P < 0.01, Mann–Whitney test) compared with the appropriate control. No difference was seen in EC overexpressing the LDL receptor (LDLR/LPL) or pretreated with cytochalasin D (10 μM), heparin (100 μg/ml), or heparinase (200 units/ml) for 1 h before LPL/VLDL addition. (B) Labeled VLDL uptake by EC. Cells were stimulated with VLDL (control) and DiI-labeled VLDL (5 μg of protein per ml) with or without LPL (30 units/ml, 18 h). Fluorescent microscopy (×20 magnification) of representative formalin-fixed specimens is shown. DiI VLDL is shown in red and DNA is shown in blue. (C) LPL was preincubated with the lipase inhibitor tetrahydrolipstatin at the concentrations shown (15 min at room temperature) before addition of LPL (30 units/ml)/VLDL (10 μg/ml). Significant inhibition in PPARα-LBD activation (P < 0.01, Mann–Whitney test) as compared with control is shown (*).
LPL/VLDL Induces PPARα-Regulated Genes and Responses in a PPARα-Dependent Manner in Vitro and in Vivo.
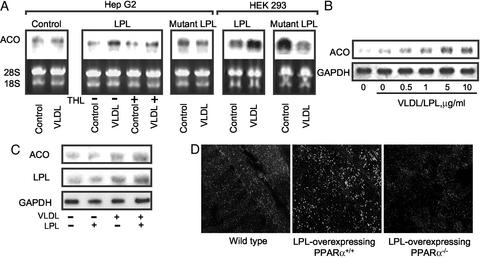
To study whether LPL/VLDL leads to known PPARα biologic responses, we examined a series of well-established PPARα effects, including induction of acyl-CoA-oxidase (ACO) (34). Transient transfection of WT LPL increases ACO expression after VLDL treatment in HepG2 cells (Fig. 5A Left). In contrast, this response was decreased by pretreatment with the lipase inhibitor tetrahydrolipstatin or abolished by transfection of a catalytically inactive LPL. Similar effects are seen in HEK293 cells (Fig. 5A Right). LPL treatment also induced ACO mRNA in a time- (data not shown) and VLDL concentration-dependent manner in both HepG2 (Fig. 5B) and primary human Mφ (Fig. 5C). Consistent with reports of LPL itself being PPARα-regulated (35), LPL/VLDL induced LPL expression in Mφ (Fig. 5C), suggesting a positive feedback loop through LPL/VLDL-activated PPARα.
Figure 5.
Enzymatically intact LPL induces PPARα target gene mRNA expression. (A Left) HepG2 cells were transfected (24 h, DMEM, 1% delipidated serum) with empty vector (PcDNA3), human WT LPL, or a catalytically inactive LPL (mutant LPL), and expression levels were confirmed with RT-PCR (data not shown). Northern analysis reveals that VLDL treatment (5 μg/ml, 3 h) induced ACO mRNA expression in LPL-treated cells, but not if they had been pretreated (15 min) with tetrahydrolipostatin (THL, 10 μM). HepG2 cells expressing the mutant LPL had no ACO induction after VLDL treatment. (Right) Similar Northern results are seen in HEK293 cells transfected and stimulated as before. (B) The concentration response of ACO mRNA expression to VLDL was tested in HepG2 cells incubated with LPL (200 units/ml, 1 h). (C) LPL/VLDL treatment induces ACO and LPL mRNA expression in human Mφ treated with LPL/VLDL as described in Fig. 5B. One representative result of three is shown. (D) In vivo analysis of skeletal muscle from WT LPL-overexpressing/PPARα+/+, and LPL-overexpressing/PPARα−/− mice reveals increased peroxisomal proliferation with LPL overexpression in the presence but not absence of PPARα. Peroxisomal expression from fasting mice (age 12 wk, n = 3) was analyzed by using PMP70 antibody.
The hallmark of PPARα activation by synthetic agonists in rodents is the induction of peroxisomes. To address LPL effects in vivo in a gain of function model, peroxisome proliferation was measured by staining for the peroxisomal protein PMP70 in the skeletal muscle of LPL-overexpressing transgenic mice bred to PPARα+/+ or PPARα−/− mice (Fig. 5D). The increased PMP70-positive granules observed in LPL-overexpressing/PPARα+/+ mice were absent in LPL-overexpressing/PPARα−/− mice. Of note, the transgenic LPL mice selected for these experiments were the “low expression” (MCK-L) variety, with three LPL transgene copies and an ≈5.5-fold increase in LPL activity (21). Thus, increased LPL expression in vivo induces peroxisome proliferation in a PPARα-dependent manner. Interestingly, rodents on high-fat diets have increased peroxisomes (36), although the mechanism for this induction has not been previously identified.
Endogenous Lipolysis in Humans Generates PPARα Activators.
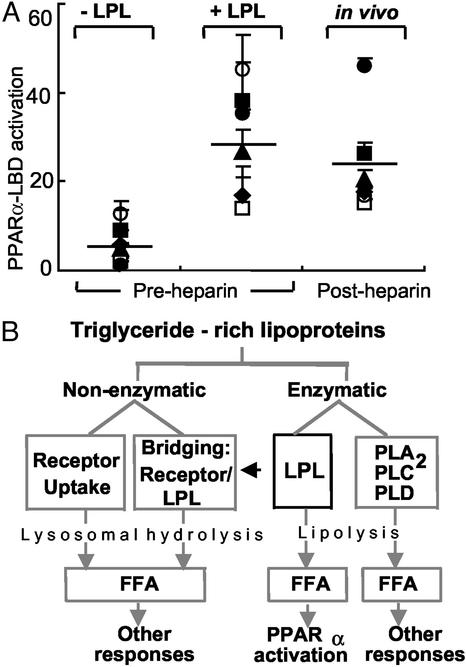
Given this evidence for LPL-meditated PPARα activation, we next asked whether similar effects could be demonstrated in humans. In plasma from six normal volunteers, minimal PPARα activity was seen despite the presence of ample free FA (207 ± 17 μM FA; Fig. 6A). The addition of LPL to these same samples led to a 29-fold increase in PPARα LBD activity (Fig. 6A), a response equivalent to the synthetic agonist WY14643 (20.3-fold ± 0.3-fold increase). Moreover, release of endogenous tissue-bound LPL through heparin infusion led to similar PPARα activation (23-fold) in the absence of adding any exogenous LPL (Fig. 6A). These results suggest that lipolytic activity is involved in generating PPARα activity of human plasma.
Figure 6.
LPL-mediated PPARα activation occurs as a unique component of triglyceride-rich lipoprotein metabolism. (A) PPARα-LBD transactivation assays were performed as in Fig. 2B by using human plasma samples (5%) obtained from six normolipidemic donors at baseline (−LPL), after addition of LPL in vitro (200 units/ml, 37°C, 1 h) to pre-heparin plasma, or after systemic heparin infusion (60 units/kg) in vivo. Symbols represent individual donors; lines indicate mean values. The relative PPARα-LBD fold activation for 10 μM WY14643 in this experiment was 20.3 ± 0.3; FA concentrations were 1.7 ± 0.8 μM (−LPL), 18.2 ± 9.3 μM (+LPL), and 18.2 ± 6.8 μM (post-heparin). (B) The metabolism of TRL occurs through both nonenzymatic and enzymatic mechanisms. Although these pathways ultimately result in FA production and their various downstream effects, PPARα activation through lipolysis seems restricted to some degree to LPL. The data presented suggest that specific mechanisms of endogenous lipolysis can lead to specific transcriptional responses through ligand generation. Thus, LPL-mediated PPARα activation may provide a link between circulating triglyceride-rich lipoproteins and distal PPARα effects such as antiinflammatory responses, FA oxidation, and lipid metabolism. The latter may include a possible positive feedback loop through PPARα leading to increased LPL expression and activity by inducing LPL and repressing ApoCIII.
Discussion
Our results reveal previously uncharacterized roles for LPL, the central enzyme in triglyceride metabolism, both in limiting inflammatory responses as well as generating PPARα ligands. LPL treatment of VLDL results in well-established indicators of PPARα activation, including LBD responses, direct ligand displacement, induction of known PPARα target genes, and peroxisome proliferation. These results were seen in vitro and in vivo in both gain of function and loss of function models. These responses suggest an endogenous metabolic pathway linking LPL enzymatic action to subsequent PPARα activation and its distal transcriptional effects. To our knowledge, there exists no previously documented evidence for an enzymatic pathway leading to PPARα ligand generation. Thus, LPL, an enzyme central to physiologic TRL metabolism, may replicate the effects of synthetic PPARα ligands. A positive feedback loop through PPARα is suggested by LPL/VLDL-mediated induction of LPL expression itself. The functional impact of this pathway may be augmented by PPARα-mediated repression of the endogenous LPL inhibitor ApoCIII (37). By examining the relationship of this lipolytic pathway to nuclear receptor activation, new insights are gained into both LPL and PPAR function.
Selectivity for the PPAR isoform targeted by LPL action is evident in vitro. PPARα likely represents the major PPAR response to physiologic lipolysis, as might occur after eating. Although minimal PPARγ responses are seen at relevant VLDL concentrations, LPL-mediated PPARδ activation does occur. This PPARδ response through LPL may be relevant under certain circumstances and in cell types where PPARδ expression predominates over PPARα. Differences are also seen among lipases, with only certain FA-releasing enzymes generating PPARα-LBD activation. This specificity may derive from various lipase-specific properties: different substrates (triglyceride vs. phospholipids, saturated vs. unsaturated FA), subsequent FA modifications, and/or the lipolytic products generated. Regardless, these results argue against the assumption based on prior in vitro data that FA in general activate PPARs (38–40) or that FA generation by any means of lipolysis would do the same. Rather, our findings suggest different transcriptional responses dependent on the specific mechanism of FA generation and uptake (Fig. 6B).
One such example is seen in divergent FA effects on VCAM1 expression. Certain FA alone, including palmitic and stearic acids common in VLDL, augmented TNFα-induced VCAM1 in EC. In contrast, LPL-treated VLDL, and the FA so produced, limit TNFα-induced VCAM1 in a PPARα-dependent fashion, recapitulating synthetic PPARα agonist effects. Thus, LPL lipolysis may represent a mechanism through which heterogeneous lipoproteins, despite containing multiple nuclear receptor ligands, including hydroxy-octadecadienoic acids (HODEs), oxidized lipids, and retinoids, can generate specific nuclear receptor responses. These results support a broader view of circulating lipoproteins as more than just transporters or resources for energy substrates, but also a source of ligands capable of generating specific transcriptional responses. Such a perspective may have clinical implications regarding the downstream effects of circulating lipids and intact lipase function.
Physiologic activation of PPARα through LPL may shed new light on the widely studied but still controversial role of LPL and PPARα in atherogenesis. Tordjman et al. (41) found higher triglyceride levels but unaltered fasting post-heparin LPL activity in mice lacking both PPARα and ApoE, as compared with ApoE−/− alone. As those authors pointed out, their studies would not account for natural PPARα ligands generated after feeding as our results would predict. Surprisingly, these ApoE−/−/PPARα−/− mice had less atherosclerosis despite evidence that PPARα activation may limit atherosclerosis and inflammation. Many factors may have contributed to the phenotype of these ApoE−/−/PPARα−/− mice: abnormal triglyceride metabolism in ApoE−/−, known interaction between ApoE and LPL (42), improved insulin sensitivity in PPARα−/− mice (41), and/or species-specific differences in PPARα responses. Recently PPARα agonist treatment was reported to decrease lesional cholesterol content in ApoE−/− mice, with a more pronounced effect in ApoE−/− mice expressing a human ApoA1 transgene (43). Cell-specific differences in PPARα levels or LPL action may also influence atherosclerotic responses, for example, as seen in Mφ (17).
In contrast, generalized LPL overexpression dramatically decreased atherosclerotic lesions in both LDLR−/− (14) and ApoE−/− mice (16). In humans, impaired LPL hydrolytic activity, typically associated with hypertriglyceridemia, may promote atherosclerosis, especially in diabetic patients (44). Similarly, genetic alterations in LPL hydrolytic activity are also associated with atherosclerosis (45). Of note, in the CARE trial, coronary heart disease risk was associated with plasma TRL levels only if combined with increased levels of the LPL inhibitor ApoCIII (46). Our data would predict that impaired LPL enzymatic activity (for example, in diseases like diabetes) might decrease endogenous PPARα activation and its subsequent downstream effects, including antiinflammatory responses. If so, fibrate therapy might be particularly important in patients with impaired LPL function, compensating for inadequate endogenous PPARα ligand generation. This “pharmacologic bypass” of LPL-mediated PPARα activation may contribute to the atherosclerotic and cardiovascular benefits seen in fibrate trials (47).
Aside from their possible relevance to pathologic states, these findings provide a candidate physiologic mechanism uniting LPL and PPAR pathways, each of which has been previously, but separately, implicated in energy balance (13, 48). The FA generated by LPL, as occurs postprandially, provides a major energy source through subsequent FA β-oxidation involving largely PPARα-regulated enzymes (48). Interestingly, fasting, which results in triglyceride catabolism, induces PPARα expression (49). Thus, PPARα is integral in energy supply through LPL, as after feeding or during states of increased demand, as occurs during fasting. Taken together, our findings suggest a mechanism linking physiologic metabolism of TRL and subsequent PPARα transcriptional effects, implicating LPL as a key enzyme in the endogenous generation of PPARα ligands.
Supplementary Material
Acknowledgments
We thank E. Schwartz, D. Sahady, N. Sharma, and S. Laclair for excellent technical support; and Drs. P. Sarraf, I. Goldberg, and U. Schoenbeck for helpful discussions. Grant support included the American Diabetes Association, International HDL, and Leducq Foundation Awards (to J.P.), National Institutes of Health Grant HL50350 (to A.S.), and Fonds zur Förderung der Wissenschaftlichen Forschung Grant P13617 (to G.H.).
Abbreviations
- TRL
triglyceride-rich lipoprotein
- VLDL
very low-density lipoprotein
- LDL
low-density lipoprotein
- HDL
high-density lipoprotein
- DiI
1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate
- ACO
acyl-CoA-oxidase
- VCAM1
vascular cell adhesion molecule 1
- FA
fatty acid
- PPARα
peroxisome proliferator-activated receptor α
- TNFα
tumor necrosis factor α
- LPL
lipoprotein lipase
- LDLR
LDL receptor
- ApoE
apolipoprotein E
- Mφ
macrophage
- EC
endothelial cell
- LBD
ligand-binding domain
References
- 1.Glass C K, Witztum J L. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 2.Huo Y, Ley K. Acta Physiol Scand. 2001;173:35–43. doi: 10.1046/j.1365-201X.2001.00882.x. [DOI] [PubMed] [Google Scholar]
- 3.Moers A, Fenselau S, Schrezenmeir J. Exp Clin Endocrinol Diabetes. 1997;105:35–37. doi: 10.1055/s-0029-1211793. [DOI] [PubMed] [Google Scholar]
- 4.Havel R J. Proc Nutr Soc. 1997;56:659–666. doi: 10.1079/pns19970065. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Cybulsky M I, Gimbrone M A, Jr, Libby P. Arterioscler Thromb. 1993;13:197–204. doi: 10.1161/01.atv.13.2.197. [DOI] [PubMed] [Google Scholar]
- 6.Marx N, Sukhova G, Collins T, Libby P, Plutzky J. Circulation. 1999;99:3125–3131. doi: 10.1161/01.cir.99.24.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson S M, Parhami F, Xi X P, Berliner J A, Hsueh W A, Law R E, Demer L L. Arterioscler Thromb Vasc Biol. 1999;19:2094–2104. doi: 10.1161/01.atv.19.9.2094. [DOI] [PubMed] [Google Scholar]
- 8.Pasceri V, Wu H D, Willerson J T, Yeh E T. Circulation. 2000;101:235–238. doi: 10.1161/01.cir.101.3.235. [DOI] [PubMed] [Google Scholar]
- 9.Sethi S, Ziouzenkova O, Ni H, Wagner D D, Plutzky J, Mayadas T N. Blood. 2002;100:1340–1346. doi: 10.1182/blood-2002-01-0316. [DOI] [PubMed] [Google Scholar]
- 10.Albert C M, Campos H, Stampfer M J, Ridker P M, Manson J E, Willett W C, Ma J. N Engl J Med. 2002;346:1113–1118. doi: 10.1056/NEJMoa012918. [DOI] [PubMed] [Google Scholar]
- 11.Hu F B, Bronner L, Willett W C, Stampfer M J, Rexrode K M, Albert C M, Hunter D, Manson J E. J Am Med Assoc. 2002;287:1815–1821. doi: 10.1001/jama.287.14.1815. [DOI] [PubMed] [Google Scholar]
- 12.Marchioli R, Barzi F, Bomba E, Chieffo C, Di Gregorio D, Di Mascio R, Franzosi M G, Geraci E, Levantesi G, Maggioni A P, et al. Circulation. 2002;105:1897–1903. doi: 10.1161/01.cir.0000014682.14181.f2. [DOI] [PubMed] [Google Scholar]
- 13.Goldberg I. J Lipid Res. 1996;37:693–707. [PubMed] [Google Scholar]
- 14.Shimada M, Ishibashi S, Inaba T, Yagyu H, Harada K, Osuga J, Ohashi K, Yazaki Y, Yamada N. Proc Natl Acad Sci USA. 1996;93:7242–7246. doi: 10.1073/pnas.93.14.7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison A C, Ballantyne C M, Bray M, Chambless L E, Sharrett A R, Boerwinkle E. Genet Epidemiol. 2002;22:233–242. doi: 10.1002/gepi.0191. [DOI] [PubMed] [Google Scholar]
- 16.Yagyu H, Ishibashi S, Chen Z, Osuga J, Okazaki M, Perrey S, Kitamine T, Shimada M, Ohashi K, Harada K, et al. J Lipid Res. 1999;40:1677–1685. [PubMed] [Google Scholar]
- 17.Van Eck M, Zimmermann R, Groot P H, Zechner R, Van Berkel T J. Arterioscler Thromb Vasc Biol. 2000;20:E53–E62. doi: 10.1161/01.atv.20.9.e53. [DOI] [PubMed] [Google Scholar]
- 18.Hendriks W L, Van Vark L C, Schoonderwoerd K, Jansen H, Havekes L M. Biochem J. 1998;330:765–769. doi: 10.1042/bj3300765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, Gonzalez F. Ann NY Acad Sci. 1996;804:524–529. doi: 10.1111/j.1749-6632.1996.tb18642.x. [DOI] [PubMed] [Google Scholar]
- 20.Hoefler G, Noehammer C, Levak-Frank S, el-Shabrawi Y, Schauer S, Zechner R, H. R. Biochimie. 1997;79:163–168. doi: 10.1016/s0300-9084(97)81509-x. [DOI] [PubMed] [Google Scholar]
- 21.Levak-Frank S, Radner H, Walsh A, Stollberger R, Knipping G, Hoefler G, Sattler W, Weinstock P, Breslow J, Zechner R. J Clin Invest. 1995;96:976–986. doi: 10.1172/JCI118145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marelli-Berg F M, Peek E, Lidington E A, Stauss H J, Lechler R I. J Immunol Methods. 2000;244:205–215. doi: 10.1016/s0022-1759(00)00258-1. [DOI] [PubMed] [Google Scholar]
- 23.Marx N, Mackman N, Schonbeck U, Yilmaz N, Hombach V, Libby P, Plutzky J. Circulation. 2001;103:213–219. doi: 10.1161/01.cir.103.2.213. [DOI] [PubMed] [Google Scholar]
- 24.Faustinella F, Smith L C, Semenkovich C F, Chan L. J Biol Chem. 1991;266:9481–9485. [PubMed] [Google Scholar]
- 25.Emmerich J, Beg O U, Peterson J, Previato L, Brunzell J D, Brewer H B, Jr, Santamarina-Fojo S. J Biol Chem. 1992;267:4161–4165. [PubMed] [Google Scholar]
- 26.Lawrence J, Wollenberg G, Frank J, DeLuca J. Toxicol Appl Pharmacol. 2001;170:113–123. doi: 10.1006/taap.2000.9098. [DOI] [PubMed] [Google Scholar]
- 27.Elbrecht A, Chen Y, Adams A, Berger J, Griffin P, Klatt T, Zhang B, Menke J, Zhou G, Smith R G, Moller D E. J Biol Chem. 1999;274:7913–7922. doi: 10.1074/jbc.274.12.7913. [DOI] [PubMed] [Google Scholar]
- 28.Berger J, Leibowitz M D, Doebber T W, Elbrecht A, Zhang B, Zhou G, Biswas C, Cullinan C A, Hayes N S, Li Y, et al. J Biol Chem. 1999;274:6718–6725. doi: 10.1074/jbc.274.10.6718. [DOI] [PubMed] [Google Scholar]
- 29.De Caterina R, Bernini W, Carluccio M A, Liao J K, Libby P. J Lipid Res. 1998;39:1062–1070. [PubMed] [Google Scholar]
- 30.Chawla A, Barak Y, Nagy L, Liao D, Tontonoz P, Evans R M. Nat Med. 2001;7:48–52. doi: 10.1038/83336. [DOI] [PubMed] [Google Scholar]
- 31.Lawrence J W, Li Y, Chen S, DeLuca J G, Berger J P, Umbenhauer D R, Moller D E, Zhou G. J Biol Chem. 2001;276:31521–31527. doi: 10.1074/jbc.M103306200. [DOI] [PubMed] [Google Scholar]
- 32.Poynter M E, Daynes R A. J Biol Chem. 1998;273:32833–32841. doi: 10.1074/jbc.273.49.32833. [DOI] [PubMed] [Google Scholar]
- 33.Forman B, Tontonoz P, Chen J, Brun R, Spiegelman B, Evans R. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 34.Dreyer C, Krey G, Keller H, Givel F, Helftenbein G, Wahli W. Cell. 1992;68:879–887. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- 35.Schoonjans K, Peinado-Onsurbe J, Lefebvre A, Heyman R, Briggs M, Deeb S, Staels B, Auwerx J. EMBO J. 1996;15:5336–5348. [PMC free article] [PubMed] [Google Scholar]
- 36.Ishii H, Fukumori N, Horie S, Suga T. Biochim Biophys Acta. 1980;617:1–11. doi: 10.1016/0005-2760(80)90218-0. [DOI] [PubMed] [Google Scholar]
- 37.Staels B, Vu Dac N, Kosykh V A, Saladin R, Fruchart J C, Dallongeville J, Auwerx J. J Clin Invest. 1995;95:705–712. doi: 10.1172/JCI117717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kliewer S A, Sundseth S S, Jones S A, Brown P J, Wisely G B, Koble C S, Devchand P, Wahli W, Willson T M, Lenhard J M, Lehmann J M. Proc Natl Acad Sci USA. 1997;94:4318–4323. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forman B M, Chen J, Evans R M. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krey G, Braissant O, L'Horset F, Kalkhoven E, Perroud M, Parker M, Wahli W. Mol Endocrinol. 1997;11:779–791. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- 41.Tordjman K, Bernal-Mizrachi C, Zemany L, Weng S, Feng C, Zhang F, Leone T C, Coleman T, Kelly D P, Semenkovich C F. J Clin Invest. 2001;107:1025–1034. doi: 10.1172/JCI11497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jong M C, Dahlmans V E, Hofker M H, Havekes L M. Biochem J. 1997;328:745–750. doi: 10.1042/bj3280745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duez H, Chao Y S, Hernandez M, Torpier G, Poulain P, Mundt S, Mallat Z, Teissier E, Burton C A, Tedgui A, et al. J Biol Chem. 2002;107:48051–48057. doi: 10.1074/jbc.M206966200. [DOI] [PubMed] [Google Scholar]
- 44.Reynisdottir S, Angelin B, Langin D, Lithell H, Eriksson M, Holm C, Arner P. Arterioscler Thromb Vasc Biol. 1997;17:2287–2292. doi: 10.1161/01.atv.17.10.2287. [DOI] [PubMed] [Google Scholar]
- 45.Nordestgaard B, Abildgaard S, Wittrup H, Steffensen R, Jensen G, Tybjaerg-Hansen A. Circulation. 1997;96:1737–1744. doi: 10.1161/01.cir.96.6.1737. [DOI] [PubMed] [Google Scholar]
- 46.Sacks F, Alaupovic P, Moye L, Cole T, Sussex B, Stampfer M, Pfeffer M, Braunwald E. Circulation. 2000;102:1886–1892. doi: 10.1161/01.cir.102.16.1886. [DOI] [PubMed] [Google Scholar]
- 47.Rubins H B, Robins S J, Collins D, Fye C L, Anderson J W, Elam M B, Faas F H, Linares E, Schaefer E J, Schectman G, Wilt T J, Wittes J. N Engl J Med. 1999;341:410–418. doi: 10.1056/NEJM199908053410604. [DOI] [PubMed] [Google Scholar]
- 48.Barger P M, Kelly D P. Trends Cardiovasc Med. 2000;10:238–245. doi: 10.1016/s1050-1738(00)00077-3. [DOI] [PubMed] [Google Scholar]
- 49.Kersten S, Desvergne B, Wahli W. Nature. 2000;405:421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.