Abstract
High-density lipoprotein (HDL) protects against atherosclerosis. Endothelial lipase (EL) has been postulated to be involved in lipoprotein, and possibly HDL, metabolism, yet the evidence has been scarce and conflicting. We have inactivated EL in mice by gene targeting. EL−/− mice have elevated plasma and HDL cholesterol, and increased apolipoproteins A-I and E. NMR analysis reveals an abundance of large HDL particles. There is down-regulation of the transcripts for phospholipid transfer protein, but up-regulation of those for hepatic lipase and lipoprotein lipase. Plasma lecithin:cholesterol acyltransferase is unchanged despite an increase in hepatic mRNA; lecithin:cholesterol acyltransferase activity toward endogenous EL−/− substrate is, however, reduced by 50%. HDL clearance is decreased in EL−/− mice; both the structure of HDL and the presence of EL are factors that determine the rate of clearance. To determine EL's role in humans, we find a significant association between a single-nucleotide polymorphism 584C/T in the EL (LIPG) gene and HDL cholesterol in a well characterized population of 372 individuals. We conclude that EL is a major determinant of HDL concentration, structure, and metabolism in mice, and a major determinant of HDL concentration in humans.
Keywords: lipoprotein metabolism‖atherosclerosis‖gene targeting
Low high-density lipoprotein (HDL) is the most common lipoprotein abnormality in patients with atherosclerotic cardiovascular disease (1). It is generally accepted that HDL protects the vessel wall against atherosclerosis development. A number of mechanisms have been proposed for the protective function of HDL. HDL may be a crucial transport vehicle that delivers cholesterol from the periphery back to the liver for disposal, a process known as “reverse cholesterol transport” (2). It may protect low-density lipoprotein (LDL) from oxidation, a process that greatly increases its atherogenicity (3). In addition, HDL may inhibit the inflammatory response and down-regulate the production of adhesion molecules by endothelial cells (4); it also has anticoagulant properties on the vessel wall (5) and facilitates the production of nitric oxide, a vasculoprotective molecule (6).
Endothelial lipase (EL) is a recently identified molecule that has been proposed to play a role in lipoprotein, and perhaps HDL, metabolism (7). EL displays substantial sequence similarity to lipoprotein lipase (LPL) and hepatic lipase (HL), the other two members of the vascular lipase gene family (8). Like LPL and HL, EL has triglyceride lipase activity; however, compared with the other lipases, it has substantially greater phospholipase activity (8, 9). EL is produced by endothelial cells as well as by the liver, lung, macrophage, testis, ovary, and placenta (8). The literature on the possible physiological function of EL is confusing and conflicting (7–10). Hirata and colleagues (8, 10) postulated that EL plays an important role in angiogenesis. On the other hand, Jaye et al. (7) showed that massive overexpression of EL in the liver by adenovirus-mediated gene transfer in mice causes marked depletion of HDL, together with a major reduction in non-HDL lipoproteins. However, McCoy et al. (9) found that EL is completely inactive in vitro in the presence of serum, and deLemos et al. (11) found no significant difference in allele frequencies of six polymorphisms in the EL (LIPG) gene between individuals with normal and those with high HDL cholesterol (HDL-c). Thus, the physiological role of EL in lipoprotein metabolism, if any, is unknown. In this communication, we have inactivated the EL gene in mice by gene targeting and examined the effect of absence of EL on lipoprotein metabolism in vivo. Our analysis indicates that EL is a major determinant of HDL concentration, structure, and metabolism. To determine whether EL plays a significant role in the determination of plasma HDL levels in humans, we analyzed the frequency of a specific single-nucleotide polymorphism (SNP) in LIPG in a well characterized population of 372 individuals and found a significant association of the SNP with HDL-c level, indicating that EL is a major determinant of HDL concentration in humans.
Materials and Methods
Generation of Endothelial Lipase Knockout Mice.
Endothelial lipase (EL) genomic DNA was isolated from a mouse strain 129 DNA BAC library (Genome Systems, St. Louis). A 12-kb XbaI fragment containing the first to the third exon was used to construct the targeting vector. A replacement-type targeting vector, containing the PGK-NEO-bpA (NeoA) and herpes simplex virus thymidine kinase (TK) cassette, was used to generate the construct. A Neo cassette was inserted to replace exon 2 of the endothelial lipase gene (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). We used R1 embryonic stem (ES) cells and generated knockout clones as described (12). Nine positive R1 ES cell clones were injected into C57BL/6J blastocysts, and chimera mice were crossed to C57BL/6J mice and germ-line transmission was confirmed in eight. Mice were weaned at 21 days and fed either a standard chow diet (7001, Teklad, Madison, WI) or a high-fat, high-cholesterol diet (960393, ICN) containing 1.23% cholesterol and 17.84% fat 6 weeks after weaning.
Plasma Lipids, FPLC, and Northern and Immunoblot Analysis.
Total plasma cholesterol and triglyceride, phospholipids, free cholesterol, and HDL-c were measured using kits from Sigma or Wako Chemicals (Richmond, GA). For FPLC analysis, plasma pooled from 7–10 mice (200 μl) was loaded onto two Superose-6 columns connected in series (Pharmacia FPLC System, Amersham Pharmacia Biotech). Fractions of 0.5 ml were collected with an elution buffer (1 mM EDTA/154 mM NaCl/0.02% NaN3, pH 8.2) as described (12): fractions 4–9, very low-density lipoprotein (VLDL); 10–20, intermediate-density lipoprotein (IDL)/LDL; and 22–32, HDL. Northern blotting and immunoblotting by using the probes and antibodies identified in the figure legends was performed as described (12). Immunoblots were developed by chemiluminescence. Goat anti-human apolipoprotein A-I, E polyclonal antibodies, and mouse antiactin monoclonal antibody were purchased from Chemicon. Rabbit anti-scavenger receptor BI (SR-BI) polyclonal antibody was purchased from Novus Biologicals (Littleton, CO). Polyclonal antibodies to mouse apoA-II and lecithin:cholesterol acyltransferase (LCAT) were gifts from Gustav Schonfeld (Washington University School of Medicine, St. Louis) and John S. Parks (Wake Forest University Medical School, Winston-Salem, NC), respectively.
NMR Lipoprotein Analysis.
Lipoprotein subclass profiles were measured on frozen aliquots (100 μl) of EDTA plasma by proton NMR spectroscopy at LipoScience as described (13). The diameter ranges of the five HDL subclasses quantified by NMR were determined by calibration using purified HDL subclasses characterized by gradient gel electrophoresis. The designations of the NMR-derived HDL subclasses and their estimated diameter ranges are as follows: H1, 7.3–7.7 nm; H2, 7.8–8.2 nm; H3, 8.3–8.8 nm; H4, 8.9–10.0 nm; and H5, 10.1–13.0 nm. In some analyses, the three largest and two smallest HDL subclasses were grouped together and called H345 and H12, respectively. Concentrations of the HDL subclasses are expressed in units of cholesterol (mg/dl), mean HDL particle size was determined by weighting the relative percentage of each subclass by its diameter, and HDL subclass particle concentrations were expressed in particles number (nmol/liter) as derived from raw NMR data as described (14).
LCAT Activity Assay.
LCAT activity was assayed using an artificial substrate proteoliposome of apoA-I:cholesterol:lecithin according to the method by Chen and Albers (15). Mouse plasma (15 μl) was incubated with the labeled substrate for 1 h at 37°C. Plasma LCAT activity against endogenous substrate (endogenous esterification rate) was measured by using the lipoproteins of the whole plasma as substrate as described (16). The percentage incorporated into cholesteryl ester (CE) was calculated and the results were expressed as the percentage of cholesterol esterified.
HDL Clearance.
The [1α,2α(n)-3H]cholesterol with specific activity of 39.0 Ci/mmol (1 Ci = 37 GBq) used for HDL labeling was purchased from Amersham Pharmacia Biotech. Pooled plasma was collected from C57/BL6 wild-type and EL−/− mice. HDL was then isolated by ultracentrifugation (49,000 rpm for 18 h on TLA110 rotor) at a density of 1.21 g/ml adjusted by solid KBr. The B > 1.21 g/ml fraction was dialyzed against at least four changes of 50 vol of 0.15 M NaCl containing 0.01% EDTA overnight. Four milliliters of the 1.12 B fraction was incubated with 200 μCi of [1α,2α(n)-3H]cholesterol at 37°C for >4 h with mild agitation. The resulting labeled fraction was subjected to TLC analysis that showed >95% of [3H]cholesterol was converted to CE by the endogenous LCAT activity, CE:FC ratio >20:1 (17). We injected 300 μl of [3H]cholesteryl ester-labeled HDL into mice fasted overnight via tail vein. We collected 50 μl of blood 3, 6, 10, 15, 30, and 60 min and 3, 6, 10, and 24 h after injection. The plasma decay curve was used to fit an exponential curve by computer analysis. The fractional catabolic rate (FCR) was calculated as the inverse of the area under the decay curves as described (18, 19).
Human Population Genotyping and Phenotypic Correlation.
The design of the Lipoprotein and Coronary Atherosclerosis Study (LCAS) is described in ref. 20. Genotyping was performed by PCR–restriction fragment length polymorphism and confirmed by cycle sequencing. LIPG genotypes were determined using mutagenic oligonucleotide primers with sequences 5′-CATGAGCTGAGATTGTTGTCAGTGC-3′ and 5′-CAGTCAACCACAACTACATTGGCGTCTTTCTCTCAT-3′. The 254-bp product was typed with two restriction enzymes. For the C allele, there was no site for NdeI, whereas NlaIII produced a 250-bp band; for the T allele, NdeI digestion produced 217- and 37-bp products and NlaIII produced 213- and 37-bp products. An investigator who had no knowledge of the clinical and angiographic data performed the genotyping. Two people read the genotypes and all ambiguous genotypes were repeated and sequenced. For phenotype–genotype correlations, continuous variables were expressed as mean ± SD. Differences among the genotypes were compared by ANOVA or Kruskal–Wallis test, as appropriate. Distribution of the categorical variables was compared using the Pearson χ2 test. Genotype-by-treatment interactions were assessed using ANOVA (General Linear Model).
Results and Discussion
Targeted Disruption of EL in Mice.
Targeted disruption of the EL gene (see Fig. 6a) was designed to delete exon 2 and parts of the flanking introns. Germ-line transmission was confirmed by tail blots (see Fig. 6b). The disruption led to an absence of detectable EL mRNA expression in liver (see Fig. 6c). Three independent knockout mouse lines were established by intercrossing heterozygous animals. They displayed identical phenotypes. Experiments were performed initially in EL+/− and EL−/− F2 mice and their wild-type littermates and confirmed in F3 and F4 animals.
EL-Deficient Mice Have Increased Plasma Cholesterol, Phospholipid, HDL Cholesterol, and Associated Apolipoproteins.
Compared with wild-type littermates, EL−/− mice fed regular chow had an elevated plasma cholesterol (42.6% increase compared with wild type), mainly because of an increase in HDL-c (55.7% increase compared with wild type; Table 1). Heterozygous mice showed an intermediate increase in these values (28.2% and 22.8%, respectively). The ratio of free cholesterol to cholesteryl ester remained similar in wild-type and knockout mice (71.4% and 71.9% cholesteryl ester/total cholesterol in −/− and +/+, respectively). Total plasma phospholipid was increased 52.8% in EL−/− mice, but EL+/− mice did not show any significant change. Plasma triglyceride was similar in all three groups. On high (1.23%)-cholesterol diet feeding, plasma lipids rose in all groups. EL−/− mice continued to display higher total cholesterol (53.2%), HDL-c (51.4%), and phospholipid (47.6%). Their plasma triglyceride level rose more than that in wild-type mice (52.3% higher). Again, EL+/− mice on the high-cholesterol diet showed an intermediate increase in plasma lipids (Table 1).
Table 1.
Plasma lipids level on EL+/+ and EL−/− mice
| Genotype | Normal chow diet, mg/dl
|
High-cholesterol diet, mg/dl
|
||||||
|---|---|---|---|---|---|---|---|---|
| CHOL | TG | PL | HDL-c | CHOL | TG | PL | HDL-c | |
| EL+/+ | 108.8 ± 11.2 | 45.9 ± 8.7 | 696.8 ± 79.0 | 79.3 ± 17.8 | 209.5 ± 72.5 | 48.2 ± 15.2 | 1109.9 ± 110.8 | 86.7 ± 10.6 |
| EL+/− | 139.4 ± 15.8** | 47.6 ± 11.9 | 773.6 ± 144.5 | 97.3 ± 22.5* | 253.3 ± 42.9** | 57.2 ± 15.2 | 1319.8 ± 132.1** | 112.0 ± 26.9* |
| EL−/− | 154.2 ± 12.8** | 43.6 ± 7.7 | 1064.9 ± 204.8** | 123.2 ± 35.4** | 320.9 ± 97.4** | 73.4 ± 6.6* | 1638.4 ± 96.8** | 131.3 ± 15.5** |
Values are expressed as mean ± SD. TG, triglyceride; CHOL, total cholesterol; PL, total phospholipids (n = 8–11).
, Significant difference between +/+ and +/− or −/− (P < 0.05);
, Significant difference between +/+ and +/− or −/− (P < 0.01).
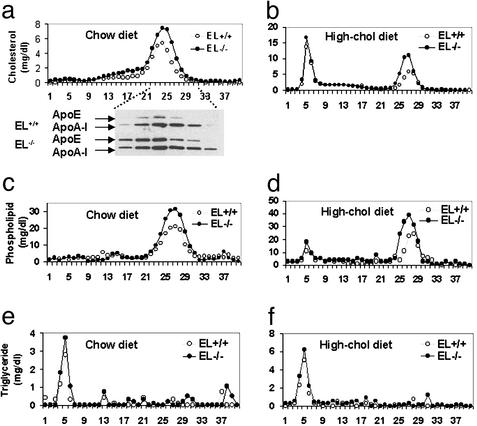
The absence of EL caused a selective rise in the HDL fraction (Fig. 1a). On FPLC the HDL peak was both broader and higher in EL−/− compared with EL+/+ mice. This change in HDL profile was reflected by higher total HDL-c, as well as a broadened distribution of apoA-I and apoE in the HDL region of the FPLC (Fig. 1a). When the mice were fed a high-cholesterol diet, there was an exaggeration of the cholesterol in the HDL fractions, but the cholesterol in the chylomicron/VLDL region was similar in the two types of mice (Fig. 1b). The same changes in phospholipid and triglyceride distribution were apparent on the two types of diet (Fig. 1 c–f); there was also a tendency for these lipids to be higher in the chylomicron/VLDL region on the high-cholesterol diet (Fig. 1 d and f).
Figure 1.
FPLC analysis of plasma lipoprotein and apolipoproteins in EL+/+ and EL−/− mice on the chow and high-cholesterol diets. (a) Total cholesterol levels in FPLC fractions on the chow diet were determined and combined immunoblot analysis of fractions 22–32 from the chromatograms shown was performed with polyclonal antibodies to apoE and apoA-I. (b) Total cholesterol levels in FPLC after the high-cholesterol diet for 6 weeks. (c and d) Phospholipid levels in FPLC fractions on the normal chow diet and on the high-cholesterol diet. (e and f) Triglyceride levels of FPLC on the normal chow and high-cholesterol diets.
EL−/− Mice Have Large HDL Particles.
The FPLC profiles on the high-cholesterol diet suggest that the HDL in EL−/− may be larger than those in EL+/+ mice. To directly test this hypothesis, we used NMR spectroscopy to measure the complete lipoprotein subclass profiles of the mice, including HDL subclass levels and mean HDL particle sizes.
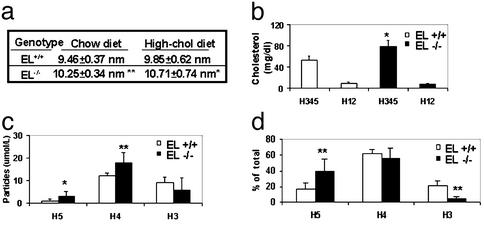
On regular chow, VLDL and LDL particles are undetectable in most of the mice in both EL−/− and EL+/+ groups. Levels of the smaller-size HDL subclasses (H12) were nearly identical but those of the larger-size HDL subclasses (H345) were much higher in the EL−/− group: 78.2 ± 11.7 mg/dl, compared with 52.9 ± 7.6 mg/dl in the EL+/+group (Fig. 2b). This is consistent with the differences seen in HDL cholesterol levels measured by the standard precipitation method (Table 1). Another significant difference between the two groups is the increased number of the larger-size HDL particles, H4 and H5 (Fig. 2c), in EL−/−. As a result, the mean HDL diameter was larger in the knockouts: 10.25 ± 0.34 nm compared with 9.46 ± 0.37 nm in wild type (Fig. 2a). On the high-cholesterol diet, the same difference in HDL size and subclass distribution was observed between the EL−/− and EL+/+ groups (Fig. 2a). When we considered the percent distribution of the three classes of HDL particles, we detected a doubling in the proportion of the largest (H5) particles accompanied by a marked 4-fold reduction in the proportion of the smallest H3 HDL particles (Fig. 2d). Thus, with the loss of function of EL, the changes in the NMR HDL subclass profiles suggest that both the increased abundance of larger HDL particles and the corresponding shift in distribution contribute to the increased HDL cholesterol level in these mice.
Figure 2.
Mean HDL particle size, subclass particle concentration, and size distribution by NMR analysis. (a) Mean HDL particle diameter on the chow diet and the high-cholesterol diet after 6 weeks in EL+/+ and EL−/− mice (n = 9 and 11, respectively). (b) Plasma concentrations of HDL subclass on the chow diet as expressed in cholesterol units (mg/dl). (c) HDL subclass particle concentrations on the chow diet. (d) Percentage of the larger HDL subclass particles, H3, H4, and H5 on the chow diet. Significant difference between EL+/+ and EL−/−: *, P < 0.05; **, P < 0.01.
Absence of EL Affects Expression of Selected Gene Products That Are Involved in HDL Structure and Metabolism.
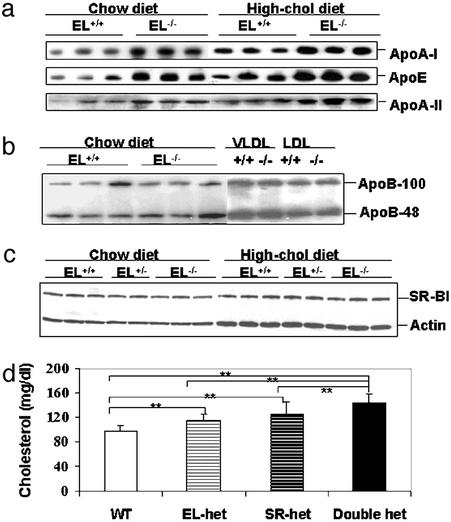
By immunoblotting, plasma level of apoB-100 andB-48, which play major roles in VLDL/LDL and chylomicron metabolism, were not different between EL−/− and EL+/+ mice (Fig. 3b). A high-fat, high-cholesterol diet raised the levels of both proteins equally in EL−/− and EL+/+ mice (data not shown). In contrast, the plasma level of apoA-I and apoE, the major apolipoprotein components of HDL, were elevated in EL−/− mice (Fig. 3a), and the difference between knockout and wild-type mice was maintained when these animals were fed a high-fat, high-cholesterol diet. Further analysis by Northern blotting revealed that the level of apoA-I or apoE transcripts in the liver was similar in EL−/− and EL+/+ mice (Fig. 4a). Therefore, the difference in plasma apoA-I and apoE was not caused by an increase in the transcription level; it was the result of a posttranscriptional regulatory mechanism.
Figure 3.
Immunoblot analysis of apolipoprotein and SR-BI expression, and plasma cholesterol level of offspring from the EL+/− SR-BI+/− cross. (a) Expression of HDL-associated apolipoproteins, apoA-I, apoE, and apoA-II expression level in 1 μl of plasma on the chow and high-cholesterol diets. (b) Non-HDL-associated apolipoproteins, apoB-100 and apoB-48 level, in plasma, VLDL, and LDL fractions on the chow diet. (c) SR-BI expression in liver tissue with actin level as an internal control. (d) Plasma cholesterol level of offspring of different genotypes obtained from the EL+/− SR-BI+/− mating pairs (n = 8∼12). Significant difference compared with respective control group as indicated: *, P < 0.05; **, P < 0.01.
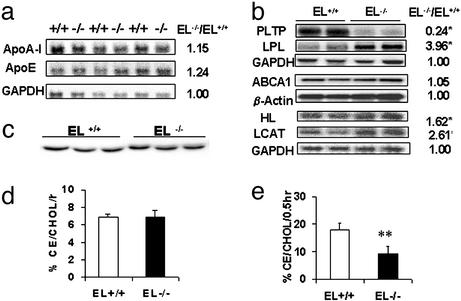
Figure 4.
Northern blot analysis of genes in HDL metabolism and LCAT activity by using exogenous and endogenous substrates. (a) Northern blot analysis of apoA-I and apoE in liver. (b) Northern blot analysis of LCAT, HL, and PLTP in liver, LPL in skeletal muscle, and RNase protection assay of ABCA1 expression in liver. Glycerol aldehyde-6-phosphate dehydrogenase (GAPDH) and β-actin levels serve as internal control. (c) LCAT protein level by Western blot analysis. (d) LCAT activity by using artificial proteoliposome substrate with 10 μl of plasma (n = 8). (e) LCAT endogenous esterification rate in 50 μl of plasma from EL+/+ and EL−/− mice (n = 8). Significant difference between EL+/+ and EL−/−: **, P < 0.01.
We next examined the level of SR-BI, the major HDL receptor, in the liver of wild-type and knockout mice (Fig. 3c). Immunoblotting detected no change in the amount of SR-BI protein in EL−/− mice. Thus, a change in SR-BI protein expression per se was not the basis for the higher plasma HDL in EL−/− mice. This was true whether the mice were on a regular chow or on a high-cholesterol diet. Furthermore, EL-deficient mice intercrossed into a SR-BI-deficient background displayed a stepwise change in plasma cholesterol level that suggests independent, additive function of the two genes on plasma cholesterol concentration (Fig. 3d). We were unable to obtain sufficient number of mice that were homozygous for both knockout genes for study because of a very high embryonic lethality rate when both genes were nonfunctional.
Another molecule involved in cholesterol transport between tissues and the plasma compartment is ATP-binding cassette transporter A1 (ABCA1). By Northern blotting, we found that the level of ABCA1 transcripts in the mouse liver was unchanged in the presence or absence of EL (Fig. 4b). Apart from SR-BI and ABCA1, HDL metabolism in mice could be modulated, directly or indirectly, by a number of enzymes, including LPL, HL, phospholipid transfer protein (PLTP), and LCAT. Northern blotting indicates that LPL and HL, the other members of the vascular lipase gene family, displayed up-regulation in the absence of EL (Fig. 4b). Because EL, LPL, and HL together form the vascular lipase gene family and share some overlapping lipase activities, overexpression of the other two members of the family is likely the result of a compensatory response to the absence of EL function in these mice. The level of PLTP was, however, significantly depressed in EL−/− mice (Fig. 4b). The role of PLTP in HDL metabolism is complex (21, 22). Both PLTP knockout and PLTP overexpression in mice have been reported to be associated with low HDL (23, 24). The marked down-regulation of PLTP in the absence of EL is intriguing. It may be a response to the change in the structure or metabolism of the HDL, a major PLTP substrate, in EL−/− mice. The contribution of PLTP down-regulation to the lipoprotein phenotype observed in these mice will be the subject of future investigations.
LCAT is a major HDL-modifying enzyme. By Northern blotting we detected increased hepatic LCAT mRNA expression in EL−/− mice (Fig. 4b). However, immunoblot analysis revealed no change in plasma LCAT mass (Fig. 4c), and LCAT activity assayed with artificial proteoliposomes was unchanged in EL−/− mice [EL−/− 6.90% vs. EL+/+ 6.91%; Fig. 4d, % CE/CHOL/0.5hr]. As LCAT activity is modulated by the structure and composition of the endogenous lipoprotein substrate (16, 25, 26), we measured endogenous plasma LCAT activity in the two types of mice against their own endogenous lipoproteins (from EL−/− or EL+/+ mice) as substrates (Fig. 4e). By this assay, LCAT endogenous esterification rate was reduced by ≈50% in EL−/− mice, indicating that, despite normal plasma LCAT concentration, the EL+/+ HDL was a better LCAT substrate than EL−/− HDL, presumably because of differences in their structure and/or composition.
Absence of EL Delays HDL Clearance in Vivo.
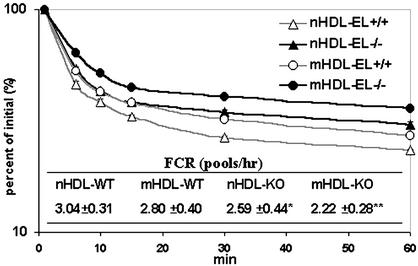
To study the effect of absence of EL function on HDL removal from the circulation, we determined the rate of disappearance of labeled HDL injected into mice of different genotypes. [3H]cholesteryl ester-labeled HDL from EL+/+ and EL−/− mice (nHDL for HDL from wild-type mice, or mHDL for HDL from EL−/− mice) was injected into EL+/+ or EL−/− mice and its rate of disappearance from the circulation was determined over a 24-h period (Fig. 5). The decay curves from this experiment displayed a decreasing rate of HDL removal from the circulation in the following order: nHDL → EL+/+, mHDL → EL+/+, nHDL → EL−/−, and mHDL → EL−/−. The FCR of 3.04 ± 0.31 pools per h was thus the highest in the nHDL → EL+/+ group, followed by FCRs of 2.80 ± 0.40, 2.59 ± 0.44, and 2.22 ± 0.28 pools per h for the mHDL → EL+/+, nHDL → EL−/−, and mHDL → EL−/− groups, respectively (Fig. 5). Therefore, wild-type HDL injected into wild-type mice had the shortest residence time in the circulation, which is similar to what was reported previously (19). In contrast, HDL from EL−/− injected into EL−/− mice was removed at the slowest rate from the circulation with an FCR that is 37% lower than that in the nHDL → EL+/+ group. The injection of mHDL into wild-type mice or of nHDL into EL−/− mice displayed intermediate rates of disappearance. These data indicate that HDL structure and systemic differences between wild-type and EL−/− mice play significant and additive roles in HDL clearance. The absence of EL in the recipient mice substantially delayed clearance; similarly, mHDL that had not been acted on by EL also was cleared at a slower rate than nHDL that had been acted on by EL.
Figure 5.
Normal and mutant HDL (nHDL and mHDL) decay curves and FCR in EL+/+ and EL−/− mice on the chow diet. Radioactivity count in the first time point, 3 min after injection, was used as 100%. The decay of radioactivity in total plasma was plotted in logarithmic scale (n = 7∼8). Table shows FCR values for each genotype and results are expressed as mean ± SD. Significant difference compared with control wild-type group injected with normal HDL (nHDL → EL+/+): *, P < 0.05; **, P < 0.01.
The elevated HDL-c and apoA-I in the absence of EL could offer protection against atherosclerosis. On the other hand, absence of EL function could have proatherogenic consequences as it interferes with HDL clearance, an important step in reverse cholesterol transport. Additional experiments are needed to determine whether the overall effect of EL inactivation is pro- or antiatherogenic.
EL Sequence Variants Are Associated with Plasma HDL Cholesterol in Humans.
We showed above that absence of EL is associated with reduced HDL removal from the circulation and an elevated HDL-c level in mice. An important issue is whether EL is a determinant of HDL-c in humans. A study by deLemos et al. (11) failed to find a difference in allele frequencies of EL (LIPG) gene variants between individuals with high HDL-c and those with normal HDL-c. Because allele frequency comparison between different populations is not a very sensitive method for detecting the contribution of a specific gene locus to a phenotype, we analyzed the genotype distribution of an SNP, 584C/T, in LIPG in a well studied population (11). This SNP produces a protein variant (T111I) with a high frequency compared with other SNPs and EL protein variants, which are extremely rare (11). T111 is conserved between mouse and human EL and HL, but is not conserved in LPL. Because I111 is not a conservative substitution, it is possible that the EL variant T111I might possess altered enzymatic activity or property compared with the more abundant “wild-type” T111. We analyzed the frequency distribution of the 584C/T SNP in a well characterized population from the LCAS (20). The LCAS and selected substudies have been published (27–29).
The complete genotype distribution of the 584C/T SNP of LIPG is shown in Table 2. The minor T allele has a frequency of 0.26 in the LCAS population. Demographic data, such as age, gender, ethnic background, height, weight, body mass index, systolic and diastolic blood pressure, waist/hip ratio, and history of smoking, diabetes, and myocardial infarction are not significantly different among the different genotypes (data not shown). We detected a significant association of the 584C/T (T111I) SNP in LIPG with mean plasma levels of HDL-c. Patients with the TT allele have a 14% higher mean HDL-c compared with those with the CC allele. In addition, there is also a strong association of the SNP with the mean plasma apoC-III concentration and the ratio of HDL-c/LDL-c and apoA-I/apoB in this population. Importantly, there is an allele-dependent variation in HDL-c, as well as these other parameters, with the rank order TT > CT > CC. The same allele-dependent rank order is evident when plasma apoA-I concentration alone is considered, although in this case the association does not reach statistical significance (P = 0.076). The LCAS patients were followed for a 2.5-year period and we detected no significant association between the SNP and progression or regression of coronary lesions during this relatively brief period (data not shown). We also detected no significant genotype-by-treatment interactions between the LIPG 584C/T SNP and response of HDL-c to fluvastatin therapy (data not shown). Taken together, the high HDL-c observed in EL−/− mice and the association of the 584C/T SNP with HDL-c in a well defined human population strongly suggest that EL is a major determinant of plasma HDL-c in mice and humans.
Table 2.
Genotypes and levels of plasma lipids and apolipoproteins
| LIPG (584C/T) (n = 372) | CC 180 | CT 167 | TT 25 | P |
|---|---|---|---|---|
| TC, mg/dl | 220.2 ± 24.9 | 220.1 ± 23.2 | 225.4 ± 27.8 | 0.584 |
| HDL-c, mg/dl | 42.9 ± 10.5 | 44.5 ± 11.7 | 48.9 ± 14.4 | 0.035 |
| LDL-c, mg/dl | 145.3 ± 20.2 | 144.0 ± 19.8 | 143.0 ± 19.9 | 0.769 |
| HDL-c/LDL-c, mg/dl | 0.31 ± 0.08 | 0.32 ± 0.10 | 0.34 ± 0.09 | 0.039 |
| TG, mg/dl | 160.4 ± 57.7 | 157.9 ± 54.6 | 167.5 ± 65.0 | 0.721 |
| apoA-I, mg/dl | 130.2 ± 26.2 | 134.5 ± 27.7 | 142.1 ± 29.0 | 0.076 |
| apoB, mg/dl | 137.0 ± 19.2 | 132.5 ± 22.4 | 133.7 ± 18.8 | 0.128 |
| apoA-I/apoB, mg/dl | 0.97 ± 0.24 | 1.05 ± 0.34 | 1.08 ± 0.26 | 0.016 |
| apoC-III, mg/dl | 36.5 ± 11.6 | 36.4 ± 11.3 | 43.6 ± 18.0 | 0.018 |
| Lp(a), mg/dl | 36.6 ± 34.0 | 37.8 ± 34.0 | 36.4 ± 32.8 | 0.940 |
P, statistical significance value among three genotypes by ANOVA.
The association between the LIPG gene SNP and plasma HDL-c in this study is at odds with that of deLemos et al. (11). The apparent discrepancy is likely the result of different study designs. The previous study population consisted of selected black and white individuals vs. a group of white individuals with HDL-c levels greater than the 90th percentile. The LCAS, which was not preselected on the basis of HDL-c, was a prospective study of 372 subjects between 35 and 75 years old who had LDL cholesterol levels of 115–190 mg/dl and ≥1 coronary lesion causing 30–75% diameter stenosis (27). The LCAS subjects included only 27 (or 7%) African Americans, as opposed to the study of deLemos et al. (11), which included 50% African Americans as the non-high HDL control for a high-HDL group consisting of 100% white subjects. Distribution of the 584C/T genotypes among the LCAS subjects displayed Hardy–Weinberg equilibrium (P = 0.91, comparing observed vs. expected genotypes), and the association between the 584C/T genotypes and plasma levels of lipid and apolipoprotein parameters was independent of race, gender, or age. The P values and the progressive allele-dependent rank order of the association with the three 584C/T genotypes were particularly strong for apoA-I/apoB ratio, apoC-III, and HDL-c levels (Table 2). It has been estimated that each 1 mg/dl increase in HDL-c level lowers cardiovascular mortality by 2–3% (30). In the Helsinki Heart Study (30), LOCAT (31), and VA-HIT (32), changes in mean HDL-c levels of 3.9, 4.0, and 1.9 mg/dl were associated with a significant decrease in the clinical event rate and cardiovascular mortality. In the LCAS population, individuals homozygous for the T584 allele have a mean plasma HDL-c level that is 6.0 mg/dl (14%) higher than individuals homozygous for the C584 allele. This difference would be considered significant in terms of vascular protection and potential outcome. Yet, we did not detect any association between cardiovascular mortality and the 584C/T SNP. The apparent lack of protection could be caused by the relatively short follow-up period of only 2.5 years in the LCAS population. Alternatively, the protective effect of a high HDL-c could be dependent on the molecular mechanism behind the higher HDL-c concentration. It is thus also possible that the high HDL-c associated with the T allele of the 584C/T SNP may not offer any protection. Additional studies are needed to resolve this important issue.
Supplementary Material
Acknowledgments
We thank Benny Chang, Kerry Ko, Hideto Kojima, Lan Li, and Leslie Wu for assistance with the study, and Dr. John S. Parks and Dr. Gustav Schonfeld for providing antibodies to mouse LCAT and apoA-II, respectively. This work was supported by National Institutes of Health Grants HL-16512 and HL-51586 and the Betty Rutherford Chair for Diabetes and Endocrinology Research (to L.C.). The LCAS was supported by Novartis Pharmaceuticals Corporation Grant B351 and National Institutes of Health General Clinical Research Center Grant 5M01RR00350, and partially by National Institutes of Health Grants HL-68884 and P50-HL-54313 and Nijad Fares and Jeff Hines.
Abbreviations
- EL
endothelial lipase
- EL−/−
homozygous EL knockout
- FCR
fractional catabolic rate
- HDL
high-density lipoprotein
- HDL-c
HDL cholesterol
- HL
hepatic lipase
- LDL
low-density lipoprotein
- LPL
lipoprotein lipase
- LCAS
Lipoprotein and Coronary Atherosclerosis Study
- LCAT
lecithin:cholesterol acyltransferase
- LIPG
human EL gene
- PLTP
phospholipid transfer protein
- SNP
single-nucleotide polymorphism
- SR-BI
scavenger receptor BI
- VLDL
very low-density lipoprotein
Note
In a recent review article, Choi et al. (33) commented on an unpublished study showing that mice homologous for EL-null alleles have elevated HDL cholesterol level, an observation that corroborates the data in the current study.
References
- 1.Genest J J, Jr, Martin-Munley S S, McNamara J R, Ordovas J M, Jenner J, Myers R H, Silberman S R, Wilson P W, Salem D N, Schaefer E J. Circulation. 1992;85:2025–2033. doi: 10.1161/01.cir.85.6.2025. [DOI] [PubMed] [Google Scholar]
- 2.Fielding C J, Fielding P E. J Lipid Res. 1995;36:211–228. [PubMed] [Google Scholar]
- 3.Mackness M I, Abbott C, Arrol S, Durrington P N. Biochem J. 1993;294:829–834. doi: 10.1042/bj2940829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barter P J. Clin Exp Pharmacol Physiol. 1997;24:286–287. doi: 10.1111/j.1440-1681.1997.tb01821.x. [DOI] [PubMed] [Google Scholar]
- 5.Griffin J H, Kojima K, Banka C L, Curtiss L K, Fernandez J A. J Clin Invest. 1999;103:219–227. doi: 10.1172/JCI5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tretjakovs P, Kalnins U, Dabina I, Dinne I, Erglis A, Kumsars I, Jurka A. Med Sci Monit. 2000;6:507–511. [PubMed] [Google Scholar]
- 7.Jaye M, Lynch K J, Krawiec J, Marchadier D, Maugeais C, Doan K, South V, Amin D, Perrone M, Rader D J. Nat Genet. 1999;21:424–428. doi: 10.1038/7766. [DOI] [PubMed] [Google Scholar]
- 8.Hirata K, Dichek H L, Cioffi J A, Choi S Y, Leeper N J, Quintana L, Kronmal G S, Cooper A D, Quertermous T. J Biol Chem. 1999;274:14170–14175. doi: 10.1074/jbc.274.20.14170. [DOI] [PubMed] [Google Scholar]
- 9.McCoy M G, Sun G S, Marchadier D, Maugeais C, Glick J M, Rader D J. J Lipid Res. 2002;43:921–929. [PubMed] [Google Scholar]
- 10.Hirata K, Ishida T, Matsushita H, Tsao P S, Quertermous T. Biochem Biophys Res Commun. 2000;272:90–93. doi: 10.1006/bbrc.2000.2747. [DOI] [PubMed] [Google Scholar]
- 11.deLemos A S, Wolfe M L, Long C J, Sivapackianathan R, Rader D J. Circulation. 2002;106:1321–1326. doi: 10.1161/01.cir.0000028423.07623.6a. [DOI] [PubMed] [Google Scholar]
- 12.Chang B H, Liao W, Li L, Nakamuta M, Mack D, Chan L. J Biol Chem. 1999;274:6051–6055. doi: 10.1074/jbc.274.10.6051. [DOI] [PubMed] [Google Scholar]
- 13.Grundy S M, Vega G L, Otvos J D, Rainwater D L, Cohen J C. J Lipid Res. 1999;40:229–234. [PubMed] [Google Scholar]
- 14.Colhoun H M, Otvos J D, Rubens M B, Taskinen M R, Underwood S R, Fuller J H. Diabetes. 2002;51:1949–1956. doi: 10.2337/diabetes.51.6.1949. [DOI] [PubMed] [Google Scholar]
- 15.Chen C H, Albers J J. J Lipid Res. 1982;23:680–691. [PubMed] [Google Scholar]
- 16.Barter P J, Hopkins G J, Gorjatschko L. Atherosclerosis. 1985;58:97–107. doi: 10.1016/0021-9150(85)90058-9. [DOI] [PubMed] [Google Scholar]
- 17.Thomas M S, Rudel L L. Anal Biochem. 1983;130:215–222. doi: 10.1016/0003-2697(83)90672-3. [DOI] [PubMed] [Google Scholar]
- 18.Hayek T, Ito Y, Azrolan N, Verdery R B, Aalto-Setala K, Walsh A, Breslow J L. J Clin Invest. 1993;91:1665–1671. doi: 10.1172/JCI116375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji Y, Wang N, Ramakrishnan R, Sehayek E, Huszar D, Breslow J L, Tall A R. J Biol Chem. 1999;274:33398–33402. doi: 10.1074/jbc.274.47.33398. [DOI] [PubMed] [Google Scholar]
- 20.West M S, Herd J A, Ballantyne C M, Pownall H J, Simpson S, Gould L, Gotto A M., Jr Control Clin Trials. 1996;17:550–583. doi: 10.1016/s0197-2456(96)00178-x. [DOI] [PubMed] [Google Scholar]
- 21.Jauhiainen M, Metso J, Pahlman R, Blomqvist S, van Tol A, Ehnholm C. J Biol Chem. 1993;268:4032–4036. [PubMed] [Google Scholar]
- 22.Tu A Y, Nishida H I, Nishida T. J Biol Chem. 1993;268:23098–23105. [PubMed] [Google Scholar]
- 23.Jiang X C, Bruce C, Mar J, Lin M, Ji Y, Francone O L, Tall A R. J Clin Invest. 1999;103:907–914. doi: 10.1172/JCI5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foger B, Santamarina-Fojo S, Shamburek R D, Parrot C L, Talley G D, Brewer H B., Jr J Biol Chem. 1997;272:27393–27400. doi: 10.1074/jbc.272.43.27393. [DOI] [PubMed] [Google Scholar]
- 25.Barter P J, Hopkins G J, Gorjatschko L, Jones M E. Biochim Biophys Acta. 1984;793:260–268. doi: 10.1016/0005-2760(84)90328-x. [DOI] [PubMed] [Google Scholar]
- 26.Sparks D L, Pritchard P H. Biochem Cell Biol. 1989;67:358–364. doi: 10.1139/o89-056. [DOI] [PubMed] [Google Scholar]
- 27.Herd J A, Ballantyne C M, Farmer J A, Ferguson J J, III, Jones P H, West M S, Gould K L, Gotto A M., Jr Am J Cardiol. 1997;80:278–286. doi: 10.1016/s0002-9149(97)00346-9. [DOI] [PubMed] [Google Scholar]
- 28.Ballantyne C M, Herd J A, Stein E A, Ferlic L L, Dunn J K, Gotto A M, Jr, Marian A J. J Am Coll Cardiol. 2000;36:1572–1578. doi: 10.1016/s0735-1097(00)00918-9. [DOI] [PubMed] [Google Scholar]
- 29.Lutucuta S, Ballantyne C M, Elghannam H, Gotto A M, Jr, Marian A J. Circ Res. 2001;88:969–973. doi: 10.1161/hh0901.090301. [DOI] [PubMed] [Google Scholar]
- 30.Frick M H, Elo O, Haapa K, Heinonen O P, Heinsalmi P, Helo P, Huttunen J K, Kaitaniemi P, Koskinen P, Manninen V, et al. N Engl J Med. 1987;317:1237–1245. doi: 10.1056/NEJM198711123172001. [DOI] [PubMed] [Google Scholar]
- 31.Frick M H, Syvanne M, Nieminen M S, Kauma H, Majahalme S, Virtanen V, Kesaniemi Y A, Pasternack A, Taskinen M R. Circulation. 1997;96:2137–2143. doi: 10.1161/01.cir.96.7.2137. [DOI] [PubMed] [Google Scholar]
- 32.Robins S J, Collins D, Wittes J T, Papademetriou V, Deedwania P C, Schaefer E J, McNamara J R, Kashyap M L, Hershman J M, Wexler L F, Rubins H B. J Am Med Assoc. 2001;285:1585–1591. doi: 10.1001/jama.285.12.1585. [DOI] [PubMed] [Google Scholar]
- 33.Choi S Y, Hirata K I, Ishida T, Quertermous T, Cooper A D. J Lipid Res. 2002;43:1763–1769. doi: 10.1194/jlr.r200011-jlr200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.