Abstract
Heart failure (HF) is the end result of progressive and diverse biological adaptations within the diseased myocardium. We used cDNA microarrays and quantitative PCR to examine the transcriptomes of 38 left ventricles from failing and nonfailing human myocardium. After identification of a pool of putative HF-responsive candidate genes by microarrays on seven nonfailing and eight failing hearts, we used quantitative PCR and a general linear statistical model in a larger sample set (n = 34) to validate and examine the role of contributing biological variables (age and sex). We find that most HF-candidate genes (transcription factors, Cebpb, Npat; signaling molecules, Map2k3, Map4k5; extracellular matrix proteins, Lum, Cola1; and metabolic enzymes, Mars) demonstrated significant changes in gene expression; however, the majority of differences among samples depended on variables such as sex and age, and not on HF alone. Some HF-responsive gene products also demonstrated highly significant changes in expression as a function of age and/or sex, but independent of HF (Ngp1, Cd163, and Npat). These results emphasize the need to account for biological variables (HF, sex and age interactions) to elucidate genomic correlates that trigger molecular pathways responsible for the progression of HF syndromes.
The prevalence and clinical manifestations of chronic heart failure (HF) in humans vary considerably with age and sex (gender). HF, although not restricted to the elderly, is primarily a syndrome manifest in the myocardium of older persons, and generally affects men more than women (1, 2). Genetic characteristics and lifestyle or environmental stressors, such as smoking, diet, obesity, and medication, also affect the development and progression of this disorder. Although molecular and genetic studies to date have identified a finite number of genes responsible for a limited number of cardiovascular diseases (3, 4), multiple genetic pathways are believed to be responsible for transducing stress-related, mechanical and neurohormonal stimuli into changes in gene expression. The balance between these pathophysiologic stresses and the ability of the myocardium to adapt ultimately determine the threshold for clinical manifestation, its severity and the prognosis for patients with HF.
The human genome project has provided clones and sequences that have led to the development of large-scale gene expression arrays that reduce the time necessary for the identification of transcriptionally regulated genes (5). The accompanying gene predictions provide a starting point for the understanding of function, genomic diversity, the complexity of interactions and the role of genes in promoting cellular and organismal phenotypes. Large-scale transcriptome analyses therefore seem well suited to the identification of disease genes that can be used to understand the underlying causes of HF. Recently several array-based studies of human HF have “identified” transcripts with altered expression (6–9). Although attempts have been made to confirm the data by independent techniques, the data represent primarily a “preliminary molecular profile” of the failing myocardium. Many of the analyses have been performed on pooled samples and most of the data exists as a ratio-based assessment (fold change) of putative “HF-regulated” genes that contain potential type I and type II statistical errors (5), generally attributable to array batch variability, array to array hybridization differences and gene sequence variance (10). An added degree of biological variability occurs in human HF studies of mixed etiology, leading Tan et al. (9) to correctly suggest that large numbers of samples that take into account absolute expression levels need to be included in statistical analyses to generate unique profiles of a particular disease etiology. Most human heart samples, however, are acquired as biopsies (fresh, frozen, or fixed), as hearts rejected for transplantation, or as explants of diseased myocardium. These samples are taken from patients of different ages and sex, making it exceedingly difficult to determine simply by arrays which gene products are responsive to HF as opposed to other sources of biological variability.
In this study, we have taken advantage of large-scale gene expression arrays to identify candidate genes potentially responsive to HF. We then examined a subset of these candidate gene products on a larger sample set by quantitative (Q)-PCR. We were able to demonstrate that many of the putative HF-responsive genes are in fact dependent on variables like age and sex, some of which are independent of HF. Only a small fraction of the total gene products examined demonstrated HF-dependent gene expression changes independent of age or sex. Complex biological interactions are therefore responsible for the breadth of alterations in gene expression generally seen in microarray analyses of failing and nonfailing human hearts.
Methods and Procedures
Human Tissue Samples.
Human ventricular myocardium was obtained from patients with severe HF secondary to idiopathic dilated cardiomyopathy and from unused donor hearts (Table 1) (11). Total RNA was extracted (12), and poly(A+) RNA was prepared by using Oligotex mRNA kits (Qiagen, Valencia, CA). Purified mRNA was quantified either by spectrophotometry or with RiboGreen (Molecular Probes), where ribosomal RNA (16S and 23S rRNA) dilutions, from 1 to 10,000 ng/ml, were used to generate standard curves.
Table 1.
Patient (Px) characteristics for samples employed for microarray and Q-PCR analyses
| Px number | Age | Sex | CHF etiology | LVEF | HW |
|---|---|---|---|---|---|
| F1 | 42 | F | DCM | 7.5 | 495 |
| F2 | 47 | F | DCM | 17.5 | 282 |
| F3* | 63 | F | DCM | 17.5 | 339 |
| F4 | 48 | F | DCM | 7.5 | 881 |
| F5 | 63 | M | DCM | 7.5 | 492 |
| F6* | 36 | M | DCM | 17.5 | 550 |
| F7 | 56 | M | DCM | 12.5 | 464 |
| F8 | 63 | M | DCM | 7.5 | 696 |
| F9 | 65 | F | DCM | 10 | 636 |
| F10 | 16 | M | DCM | 7.5 | 490 |
| F11 | 67 | M | DCM | 7.5 | 725 |
| F12 | 41 | M | DCM | 12.5 | 473 |
| F13 | 71 | M | DCM | 7.5 | 573 |
| F14 | 44 | M | DCM | 7.5 | 691 |
| F15 | 66 | M | DCM | 12.5 | 645 |
| F16 | 39 | M | DCM | 5 | 787 |
| F17 | 43 | M | DCM | 7.5 | 805 |
| F18 | 41 | M | DCM | 7.5 | 578 |
| F19 | 69 | F | DCM | 7.5 | 434 |
| F20 | 20 | M | DCM | 7.5 | 377 |
| F21 | 66 | F | DCM | 10 | 642 |
| N1 | 63 | F | N/A | 65 | 342 |
| N2 | 59 | F | N/A | 60 | 220 |
| N3 | 43 | M | N/A | 70 | ND |
| N4* | 60 | F | N/A | 55 | 434 |
| N5* | 41 | F | N/A | 35 | ND |
| N6 | 59 | F | N/A | ND | 200 |
| N7 | 56 | M | N/A | ND | 387 |
| N8 | 38 | M | N/A | ND | 344 |
| N9 | 27 | M | N/A | 60 | 447 |
| N10 | 62 | M | N/A | 52.5 | 349 |
| N11 | 75 | M | N/A | ND | 515 |
| N12 | 65 | M | N/A | ND | 633 |
| N13 | 16 | M | N/A | 52.5 | 373 |
| N14 | 66 | M | N/A | 60 | 518 |
| N15 | 79 | M | N/A | ND | 583 |
| N16 | 65 | M | N/A | 57.5 | 533 |
| N17 | 40 | M | N/A | ND | ND |
F, failing; N, nonfailing; CHF, chronic heart failure; LVEF, left ventricular ejection fraction. Boldface denotes samples employed for microarray and Q-PCR analyses. N/A, not applicable; ND, data not available.
Microarray analyses only; all other samples were utilized in Q-PCR assays.
Microarray Analyses.
We used microarrays from Incyte (Human UniGem V: 10,176 cDNAs, including yeast control cDNAs) to identify candidate genes implicated in HF. Incyte Genomics synthesized cDNA probes from 200 ng of poly(A+) RNA in the presence of Cy3 (nonfailing) or Cy5 (failing) fluorescent nucleotides and used the labeled cDNA in microarray hybridizations (10). We performed preliminary ratiometric data analyses with GEMTOOLS 2.4, and considered changes as potentially significant when a >1.8-fold change in expression was observed. To overcome type I and type II statistical problems inherent to ratiometric analyses, we Z transformed differential expression [DE-raw data normalized for fluorescence intensity (Cy3 vs. Cy5)] data to normalize differences among independent hybridizations (13).
Real-Time Q-PCR Data Analysis.
Reaction protocols and data analyses were as described (13, 14). For Q-PCR, 1 μg of total RNA (normalized conditions) was reverse transcribed with random hexonucleotides, and Q-PCRs were run in the presence of SYBR green. Each assay was performed in triplicate, and three negative controls were run for every assay: no template (sample lacking cDNA), no reverse transcriptase, and no RNA in reverse transcriptase reaction. We prepared standard curves from cDNA dilutions of target genes and endogenous references [cardiac calsequestrin (Casq2) and glyceraldehyde phosphate dehydrogenase (Gapd)], and we used one nonfailing human myocardial sample (N10) as an RNA reference throughout this study. The threshold cycle (Ct) of each sample was transformed to the log of the dilutions and normalized. Normalized data are presented as a ΔCt (Ct − Ctreference gene) value where both Casq2 and Gapd were used as reference genes (13).
Data Analysis and Statistics.
We used SAS or SPSS for all statistical analyses (SAS Institute, Cary, NC; SPSS, Chicago). For candidate gene analysis, we performed two-sample t tests on Z transformed data, using all available data, but after omitting outliers, which were identified as observations that were beyond the 1.5× interquartile range of the lower and upper quartiles (15). Because many t tests were performed (large number of target cDNAs), we took care with the interpretation of the P values, because, by chance, we would expect 5% of these results to be statistically significant. The t tests were specifically used to screen for a short list of genes that warranted further investigation by Q-PCR.
From the Q-PCR data, we modeled the relationship between gene values (ΔCt) and age, sex, and HF, using a general linear model (SAS statistical package) with age as a numerical variable and sex and HF as categorical variables. The initial model included each of the main effects as well as the three two-way interactions. Backward elimination of statistically nonsignificant terms was used to obtain the final models. Data from this reduced model is presented in the text. Data are expressed as mean ± SEM and statistical significance is taken at P < 0.05.
Results
Identification of Potential Candidate Genes.
Fifteen human heart samples (seven nonfailing and eight failing hearts) were used in the cDNA microarray analysis (Table 1). One nonfailing sample was used in two independent microarray hybridizations. The median age for nonfailing and failing myocardium was 59 and 52 years, respectively. The maximum age difference among patients was 12 years, and six of eight hybridizations were sex matched. The algorithm used for this study to identify HF-responsive gene products is shown in Fig. 1A. The primary goals of these microarray analyses were to (i) identify transcripts present in sufficient quantities to give quantifiable signals, (ii) compare expression levels between the failing and nonfailing hearts after omitting outliers, and (iii) identify putative “HF-responsive” candidate genes.
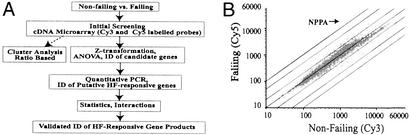
Figure 1.
(A) Algorithm used to identify, validate, and statistically analyze putative HF-candidate genes. (B) Identification of candidate genes by competitive hybridization assay using Cy3 (nonfailing)- and Cy5 (failing)-labeled probes. Data are presented as scatter plots, and each point represents the ratio of normalized Cy3/Cy5 intensities [balanced differentiation expression (BDE) value] of the gene. A BDE value >1 indicates an mRNA abundance higher in control samples, whereas a BDE <1 indicates an abundance higher in the experimental group. An absolute BDE >1.8 may imply a significant change in expression; however, the potential significance depends on the relative signal strength. The majority of the expression ratios are centered near the unity line of 1; however, some, like Nppa, show increases in expression in failing myocardium.
Microarray Analyses.
Fig. 1B shows the fluorescence plot of a representative competitive hybridization conducted with Cy3- and Cy5-labeled cDNA from nonfailing and failing human heart, respectively. We compared hybridization signals to generate a ratio-based value of BDE, which takes into account the intrinsic difference in fluorescence intensities between Cy3- and Cy5-labeled DNA (10). Most of the signals corresponding to individual cDNA elements fell on or close to the unity line (BDE ≈1), indicating that the differentiation expression of failing versus nonfailing myocardium did not change for the majority of cDNA targets. Although 2,393 individual cDNA elements showed potentially significant changes in gene expression with at least one hybridization, only 106 gene products had an absolute change in expression of ≥1.8-fold in three or more hybridizations (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org). These included many well known cardiac hypertrophy and failure associated gene transcripts, including those for natriuretic peptide precursors A (Nppa) and B, SR calcium ATPase, Na+/Ca2+ exchanger, osteoblast-specific factor 2, and numerous extracellular matrix (fibronectin) and cytoskeletal proteins (vinculin).
Candidate Genes.
We Z transformed the normalized array data, after omitting outliers, to adjust the results in proportion to the standard error to allow meaningful comparisons among independent hybridizations. Z transformation is based on the assumption that most individual signal intensities (transcript abundance) are not altered between samples (BDE ≈1). Then we used statistical analyses (ANOVA) to identify 162 gene products (Table 4, which is published as supporting information on the PNAS web site) that demonstrated significant changes (P < 0.05) in gene expression, of which 35 had P values <0.01. As with the ratio-based analyses, this list of likely HF-candidate genes included Nppa, P = 0.000001, collagen type 1, α1 (Col1a1, P = 0.0026), and lumican (Lum, P = 0.022). We also detected numerous examples of gene products that were not identified with the ratio-based approach, including 14 that were subsequently analyzed by Q-PCR.
Postarray Analyses.
We used Q-PCR to examine and validate a subset of candidate HF-responsive gene products on 34 heart samples from men and women (Table 1). Eleven of the samples used in the microarray analyses were available for use in the Q-PCR assays. The aim of Q-PCR analyses was to identify reproducible changes in gene expression, putatively due only to HF, on an expanded sample set with a high degree of biological variability. Nonfailing patients ranged in age from 16 to 79, whereas the ages for diseased myocardium ranged from 20 to 71 years. The median age was identical to those samples used for the microarray experiments: 59 and 52 years of age, respectively.
Data Normalization.
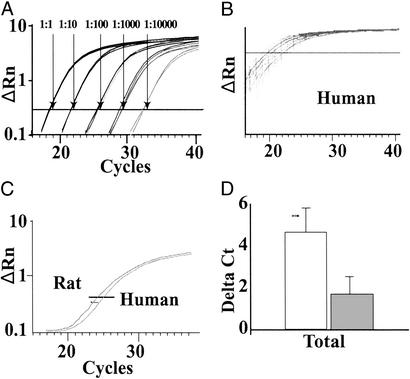
Because human cardiac samples are nonhomogeneous and contain myocytes, nonmyocytes, and potentially fibrotic tissue, we used Casq2 and Gapd as controls of RNA quality and as reference standards (ΔCt). Casq2 should be restricted to cardiomyocytes, whereas Gapd should be present in both cardiomyocytes and noncardiomyocytes. Casq2, but not Gapd, was present on the microarray, and its expression did not significantly change with HF (P > 0.05), supporting its use as an internal control. Note, from dilution curves of reverse transcriptase reactions, that every 3.6 amplification cycles, on average, reflects a 10-fold change in expression (Fig. 2A). The abundance of Casq2 and Gapd transcripts varied by 2 orders of magnitude between individual human samples (Fig. 2B). Neither Casq2 nor Gapd transcripts, however, demonstrated significantly different levels of expression either before normalization or after direct comparisons (ΔCt) with the other respective control transcript. In contrast to human samples, those from rat (or mouse) heart show very limited changes in overall expression for either transcript (Fig. 2C).
Figure 2.
Q-PCR analysis of candidate gene transcripts. (A) Casq2 amplifications with serial dilutions (1:10 to 1:100,000) of cDNA template. On average a 10-fold difference in expression was observed every 3.6 amplification cycles. ΔRn represents the difference between Rn+ [ratio of SYBR green emission intensity to that of a passive reference (ROX) containing template DNA] and Rn− (ratio of SYBR green emission intensity to that of ROX for samples lacking template or during early cycles of real-time PCR). (B) Amplification plot of Casq2, showing more than one order of magnitude variability in Casq2 expression levels among 34 human samples (failing and nonfailing). Similar results were seen with each of the control and target templates, implying either a high degree of genetic variability or the potential for differing proportions of myocytes versus nonmyocytes in the human samples. (C) Amplification plot of Casq2 from rat samples (n = 4). In contrast to that seen in humans, very little variability in the amount of Casq2 was seen in age-matched male rats. (D) Normalized Lumican data, presented as a ΔCt value, after normalization with Casq2. Both Casq2 and Gapd were more highly expressed than any of the target sequences examined. Larger ΔCt values therefore imply lower expression levels (e.g., Lum mRNA abundance decreases with HF). Open bars represent nonfailing myocardium, whereas shaded columns indicate data from failing myocardium.
HF-Responsive Gene Products.
We designed unique primer sets for 51 candidate gene products. Twenty primer sets amplified target transcripts inappropriately and have not been included in the data analysis. Thirty-one of the primer sets (Tables 5 and 6, which are published as supporting information on the PNAS web site) yielded amplification products of appropriate size at the predicted melting temperature. Next, we simultaneously amplified each target (i.e., candidate gene transcripts) and control cDNA with all 34 human samples in the same thermal cycling reaction. This effectively minimized PCR variability, and permitted calculation of ΔCt (i.e., TargetΔCtG,Gapd or TargetΔCtC,Casq2). We compared mean ΔCt values for each target in failing and nonfailing myocardial samples. From these statistical analyses, only 12 of the 31 gene products demonstrated significant changes in gene expression between the failing and nonfailing groups (Table 2 and Table 7, which is published as supporting information on the PNAS web site).
Table 2.
General linear model analysis of Q-PCR data and identification of significant interactions among sex, age, and HF
| Category | Gene name | Sex | HF | Age | HF*S | S*A | HF*A | Model |
|---|---|---|---|---|---|---|---|---|
| HF | LumΔCtC | 0.01 | 0.0117 | |||||
| Tgm2ΔCtC | 0.01 | 0.01 | 0.0031 | |||||
| Map2k3ΔCtC | 0.001 | 0.04 | 0.0024 | |||||
| LumΔCtG | 0.001 | 0.0005 | ||||||
| CebpbΔCtG | 0.03 | 0.0274 | ||||||
| Map2k3ΔCG | 0.002 | 0.05 | 0.0039 | |||||
| Pla2g2aΔCtG | 0.01 | 0.004 | 0.0013 | |||||
| Sex (S) | Ngp1ΔCtC | 0.006 | 0.0060 | |||||
| Map2k3ΔCtC | 0.001 | 0.04 | 0.0024 | |||||
| Tgm2ΔCtC | 0.01 | 0.006 | 0.0031 | |||||
| Cd163ΔCtG | 0.03 | 0.0256 | ||||||
| Map2k3ΔCtG | 0.002 | 0.05 | 0.0039 | |||||
| Age (A) | Pla2g2aΔCtG | 0.01 | 0.004 | 0.0013 | ||||
| HF*S | Cnn1ΔCtC | 0.02 | 0.01 | 0.0014 | ||||
| Igfbp4ΔCtC | 0.001 | 0.008 | 0.0040 | |||||
| Grp58ΔCtC | 0.03 | 0.003 | 0.004 | 0.0099 | ||||
| NppaΔCtC | 0.04 | 0.01 | 0.03 | 0.0500 | ||||
| CdkniaΔCtC | 0.003 | 0.0195 | ||||||
| Rasi1ΔCtC | 0.04 | 0.03 | 0.003 | 0.0181 | ||||
| Map4k5ΔCtC | 0.03 | 0.02 | 0.0331 | |||||
| Pla2g2aΔCtC | 0.04 | 0.03 | 0.0336 | |||||
| Cnn1ΔCtG | 0.04 | 0.004 | 0.0110 | |||||
| Igfbp4ΔCtG | 0.0003 | 0.001 | 0.0015 | |||||
| CdkniaΔCtG | 0.02 | 0.003 | 0.0221 | |||||
| HF*A | MarsΔCtG | 0.002 | 0.01 | 0.0030 | ||||
| HF*S/HF*A | Dscr3ΔCtC | 0.01 | 0.02 | 0.0206 | ||||
| MarsΔCtC | 0.002 | 0.04 | 0.03 | 0.0024 | ||||
| S*A | Pin1ΔCtC | 0.006 | 0.003 | 0.0050 | ||||
| CD163ΔCtC | 0.01 | 0.05 | 0.0016 | |||||
| NpatΔCtC | 0.004 | 0.01 | 0.0017 | |||||
| Ngp1ΔCtG | 0.01 | 0.03 | 0.0065 | |||||
| NpatΔCtG | 0.007 | 0.02 | 0.0076 | |||||
| HF*S/S*A | Nptx2ΔCtC | 0.03 | 0.02 | 0.05 | 0.0777 | |||
| FlncΔCtC | 0.006 | 0.009 | 0.02 | 0.0341 | ||||
| Sca1ΔCtC | 0.0009 | 0.05 | 0.004 | 0.0178 | ||||
| Colla1ΔCtG | 0.007 | 0.007 | 0.0006 | 0.005 | 0.0007 | |||
| Grp58ΔCtG | 0.009 | 0.004 | 0.003 | 0.05 | 0.0092 | |||
| Nptx2ΔCtG | 0.0004 | 0.01 | 0.05 | 0.0008 | 0.0011 | |||
| Pin1ΔCtG | 0.04 | 0.01 | 0.01 | 0.0060 | ||||
| RasI1ΔCtG | 0.002 | 0.02 | 0.0001 | 0.002 | 0.0002 | |||
| FlcnΔCtG | 0.0001 | 0.008 | 0.0001 | 0.0002 | ||||
| Sca1ΔCtG | 0.002 | 0.01 | 0.008 | 0.0279 |
Gene names are listed in text or in Table 5. Only significant P values are shown. Asterisk denotes interactions between the named variables. Boldface indicates significance within the specified category.
Next we modeled the relationship between gene values (ΔCt) and age, sex, and HF, by using a general linear model. After normalization to Casq2 or to Gapd, only three [transglutaminase 2 (TGM2), Lum, mitogen-activated protein kinase kinase 3 (Map2k3)] and four [(Lum, Map2k3, CCAAT/enhancer binding protein, β (Cebpb), phospholipase A2 (Pla2g2a)] of the 31 gene products, respectively, were significantly altered with HF, independent of age or sex (Table 3). Regardless of the normalization procedure, Lum (Fig. 2D) and Map2k3 transcript abundance increased, on average, by 8.3- or 4.7-fold, respectively. Many other significant changes in expression with HF (P < 0.05) were identified, but the extent of the change caused by HF depended on age or sex (Table 3). The observed HF changes in gene expression that varied by sex included Calponin 1, (Cnn1)ΔCtC/G, Pla2g2aΔCtC, insulin growth factor binding protein 4, (Igfbp4)ΔCtC/G, mitogen activated protein 4 kinase 5 (Mapk4k5)ΔCtC and cyclin-dependent kinase inhibitor (Cdknia)ΔCtC/G. In some cases only one sex (Cnn1, Fig. 3A) demonstrated changes with HF. In other cases, opposite effects occurred between males and females. For example, in males Grp58ΔCtC expression decreased with HF, whereas in females, expression increased (Fig. 3B), and when males and females were combined in this larger data set, no significant change in expression could be demonstrated.
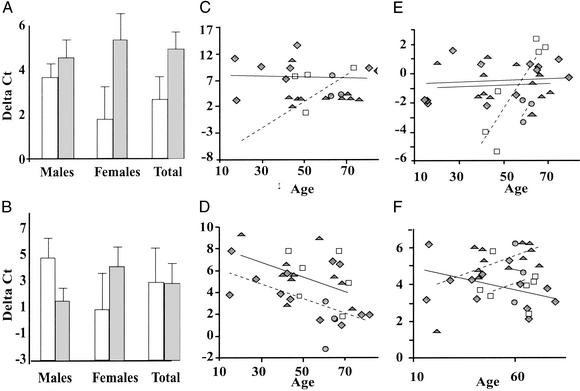
Figure 3.
General linear model statistical analyses of Q-PCR data on a subset of HF-responsive gene products, which vary by age or sex. Data are presented as ΔCt after normalization to Casq2 or Gapd. Note the greater the ΔCt value, the lower the abundance. (A) Calponin 1 (Cnn1ΔCtG), a HF-responsive gene product that affected primarily females. (B) Grp58ΔCtG, a HF-responsive gene that was differentially expressed in males and females. (C) MarsΔCtG, a HF-responsive gene with age-dependent decreases in expression (R2 = 0.36). Note that nonfailing hearts did not demonstrate any age-dependent effects. (D) Pla2g2aΔCtG, a HF-gene product with age-dependent increases in expression in both males and females (R2 = 0.36). HF patients appear to have Pla2g2a at quantities equivalent to nonfailing old hearts. (E) FlncΔCtG, a HF-responsive gene product that shows striking age-dependent decreases in females (R2 = 0.56). Males demonstrate almost no HF or age dependence. (F) Dscr3ΔCtC, age-, sex-, and HF-dependent interactions, indicative of complex biological interactions that contribute to the breadth of gene expression changes generally seen in microarray analyses of failing and nonfailing human hearts (R2 = 0.36). Diamonds, nonfailing males; triangles, failing males; circles, nonfailing females; squares, failing females.
The abundance of some transcripts was altered with HF, but the extent of change varied by age. For example, methionyl-tRNA synthetase (Mars) decreased significantly in abundance with HF as a function of age, whereas nonfailing hearts did not demonstrate any changes in the expression of this transcript with aging (Fig. 3C). Pla2g2a abundance, on the other hand, significantly decreased as a function of both age and HF, but the change in HF did not depend on age, as can be seen by the parallel, negatively sloped lines in Fig. 3D. The abundance of some other transcripts that varied with age exhibited differential changes in HF. Filamin C, γ (Flnc), for example, shows striking age- and HF-dependent changes in females, and only modest changes in males (Fig. 3E).
More complicated interactions with HF were also identified, including that for Downs syndrome critical region 3 (Dscr3)ΔCtC, which did not show any significant changes with HF, sex, or age alone. When interactions were taken into account, sex, age and HF contributed to significantly altered changes in expression of Dscr3 (Fig. 3F). The effects of HF on gene expression therefore depend on a multitude of complex genomic interactions that are affected by age and sex.
Non-HF-Responsive Gene Products.
A subset of the candidate genes evaluated by Q-PCR did not demonstrate any significant change in expression with HF, but varied as a function of sex and/or age, underscoring the biological heterogeneity of these samples (Table 3). For example Pin1, identified as a highly significant HF-candidate gene (P < 0.00001), was not HF-responsive when normalized by Casq2. The altered expression was sex and age dependent, with females having a significantly reduced abundance relative to males (P = 0.006), potentially explaining the high degree of significance in the putative candidate gene list, which contained a proportionally larger number of samples from females. Pin1 did, however, show HF- and sex-related interactions when normalized to Gapd. Other sex and age interactions could be demonstrated for Cd163 antigen (Cd163)ΔCtC, nuclear protein, ataxia-telangiectasia (Npat)ΔCtC/G, and Ngp autoantigen (Ngp1)ΔCtG. None of these age effects were significant until sex was taken into account. Sex-responsive genes include Ngp1ΔCtC, transglutamase 2 (Tgm2)ΔCtC, Cd163ΔCtG and Map2k3ΔCtC/G. Interestingly, the abundance of the control transcripts, Casq2 and Gapd demonstrated sex-related differences, which may affect the overall interpretation of these data. Females had greater abundances of Casq2 and Gapd than males (P = 0.007 and 0.006, respectively), but no further interactions could be identified.
The expression of only 9 of 31 candidates (Sas, Pggt1b, Sdcc, Rgn, Rnf4, Ap1b, Smt3h1, Cct7, and Aip1; see Table 2 for gene names) tested by Q-PCR did not significantly change with HF, sex, or age. For Sas, Sdcc, Rgn, and Rnf4, the signal intensities from the microarrays were very low (BDE values <500), and low signal intensities represent a common cause for false positives on microarrays (5). Alternatively, the statistical model only accounted for 71% of the total variance, indicating that other independent variables may be responsible for changes in transcript abundance.
Discussion
Large-scale transcriptome analyses appear ideally suited for the identification of disease-causing genes. Gene expression arrays, however, have an inherent probability of low sensitivity or specificity, and the extent to which these shortcomings are linked with sample diversity with respect to sex or age have not been addressed in previous studies of human HF. We therefore used gene expression arrays as a screening tool to quantify the expression of gene products in failing and nonfailing hearts. We identified 162 candidate “HF-responsive” gene products (P < 0.05), including many, which were not identified by using a ratio-based approach. We went on to examine a subset of these candidate genes by Q-PCR on a larger and more diverse sample population. The statistical analyses demonstrated that the majority of expression changes were subject to diverse biological inputs, involving age and sex.
Of thirty-one candidate genes examined by Q-PCR, 22 (71%) demonstrated significant and verifiable changes in abundance, but only when modeled to take into account HF, age, sex, and their interactions (Table 3). Only five putative HF-responsive transcripts were linked to HF irrespective of sex or age. Eight transcripts demonstrated HF-associated changes in expression, which varied by sex. Some of these involved only one sex (Cnn1), whereas other transcripts, like those for Grp58, demonstrated opposite effects in men and women. One of the transcripts analyzed in the present study demonstrated a HF response that varied only as a function of age (MarsΔCtG). Eight others demonstrated complex interactions, involving HF, sex, and age. Three of the 22 putative HF gene products, irrespective of normalization procedure, were not regulated as a function of HF (Ngp1, CD163, Npat).
We used Casq2 and Gapd for normalization purposes. Casq2, a sarcoplasmic reticulum-restricted protein, should be restricted to cardiomyocytes, whereas Gapd, a metabolic enzyme involved in glycoloysis, should be present in all cells within the heart. Because the microarray analyses are derived from labeling of total cardiac mRNA, data normalized with Gapd, an indicator of total cardiac mRNA, would be expected to correlate more closely with that from microarrays; however, 19 putative HF-genes transcripts were identified by both Casq2 and by Gapd. Only 16 of these putative HF-gene transcripts were common to both normalization procedures. Differences between the two normalizing control genes, however, may be indicative of sample heterogeneity (i.e., the relative contribution of myocytes and nonmyocytes in each sample), another source of biological variation.
The underlying causes for altered gene expression within functional groups (e.g., proteins of the ECM) involve multiple biological interactions that differ for individual genes. For example, Lum is a member of leucine-rich proteoglycan family that is thought to regulate the assembly and diameter of collagen fibers (16), and both Lum and Col1a1 are associated with the pathology of fibrosis in heart (17, 18). We show that Col1a1 has multivariant HF-, sex-, and age-dependent components. In contrast, Lum expression depends primarily on HF. Similarly, Rasi1 (Mmp19), a novel zinc-binding endopeptidases that degrades various components of the extracellular matrix (19), shows complex sex-, age-, and HF-dependent changes in expression, whereas Igfbp4, which is localized to the subendothelial connective tissue components of heart and mediates cell growth and metabolism (20), is controlled only by sex- and HF-dependent processes.
We identified several HF-dependent gene products that may act as potential regulators for transducing mechanical, stress and neurohormonal stimuli into changes in gene expression. These include the signal transduction proteins of Map2k3 and Map4k5. Mitogen-activated protein kinases (MAPKs) consist of at least three phosphorylation cascades that terminate in the activation of extracellular signal-regulated kinases (ERK), c-Jun NH2-terminal kinases (JNK) or p38 MAPKs (21). Map2k3 specifically activates p38, which targets a number of cardiac regulators including Mef2c, whereas Map4k5 targets kinases in the JNK signaling cascade. Both JNK and p38 cascades are activated in response to G-protein coupled receptor activation and stress stimulation and have been implicated in cardiac hypertrophy and failure (22). Novel factors that we have identified and which may regulate some human HF-mediated events include Pin1ΔCtG and CebpbΔCtG. Pin1 is an essential nuclear peptidylprolyl cis/trans isomerase involved in the regulation of mitosis, whereas Cebpb, a DNA-binding protein predominantly characterized by a leucine zipper motif, regulates plasma free fatty acid levels and insulin signal transduction events in skeletal muscle (23). Because we also identified the inhibitory insulin growth factor binding protein, Igfbp4, as a HF-responsive gene product, it is possible that Cebpb works in concert with insulin signaling pathways to regulate remodeling events.
In conclusion, biological sample variability adversely affects the interpretation of microarray studies of failing human myocardium. Gene products with altered levels of expression that are identified by microarrays do not necessarily change with HF or other diseases in the same way or to the same extent in men and women or with age. In this study, we demonstrate that sex and age contribute significantly to changes in gene expression identified by microarrays. Even when age and sex are taken into account, our model accounts for only 71% of the total variance. The remainder represents unexplained additional sources of biological diversity. Unraveling transcriptome variability in human HF therefore needs to take into account genomic diversity caused by age and sex. Other factors like diabetes, hypertension, smoking, diet, obesity, medication, HF etiology, and duration, which have not been examined in the present study, represent additional, potentially important sources of biological diversity that need to be taken into account when examining changes in gene expression with human HF. Thus, very large sample sizes, or possibly, metaanalyses of studies of smaller sample sizes are tantamount to a sufficient accounting of the diversity of altered gene expression that directs alterations in specific molecular pathways that underlie changes in cardiac structure and function in various types of human HF or disease.
Supplementary Material
Acknowledgments
This work was supported in part by National Institutes of Health Grant AG17022 (to K.M.).
Abbreviations
- BDE
balanced differentiation expression
- HF
heart failure
- Q-PCR
quantitative PCR
- ECM
extracellular matrix
- Gapd
glyceraldehyde phosphate dehydrogenase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lakatta E G. In: Principles of Geriatric Medicine and Gerontology. Hazzard W R, editor. New York: McGraw–Hill; 1999. pp. 645–660. [Google Scholar]
- 2.Lakatta E G. Clin Geriatr Med. 2000;16:419–444. doi: 10.1016/s0749-0690(05)70021-5. [DOI] [PubMed] [Google Scholar]
- 3.Seidman J G, Seidman C. Cell. 2001;104:557–567. doi: 10.1016/s0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 4.Lifton R P, Gharavi A G, Geller D S. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 5.Juhasz O, Zhu Y, Garg R, Anisimov S V, Boheler K R. Eur J Heart Failure. 2002;4:687–697. doi: 10.1016/s1388-9842(02)00169-1. [DOI] [PubMed] [Google Scholar]
- 6.Barrans J D, Stamatiou D, Liew C. Biochem Biophys Res Commun. 2001;280:964–969. doi: 10.1006/bbrc.2000.4137. [DOI] [PubMed] [Google Scholar]
- 7.Hwang J J, Allen P D, Tseng G C, Lam C W, Fananapazir L, Dzau V J, Liew C C. Physiol Genomics. 2002;10:31–44. doi: 10.1152/physiolgenomics.00122.2001. [DOI] [PubMed] [Google Scholar]
- 8.Barrans J D, Allen P D, Stamatiou D, Dzau V J, Liew C C. Am J Pathol. 2002;160:2035–2043. doi: 10.1016/S0002-9440(10)61153-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan F L, Moravec C S, Li J, Apperson-Hansen C, McCarthy P M, Young J B, Bond M. Proc Natl Acad Sci USA. 2002;99:11387–11392. doi: 10.1073/pnas.162370099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yue H, Eastman P S, Wang B B, Minor J, Doctolero M H, Nuttall R L, Stack R, Becker J W, Montgomery J R, Vainer M, Johnston R. Nucleic Acids Res. 2001;29:E41–1. doi: 10.1093/nar/29.8.e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dipla K, Mattiello J A, Jeevanandam V, Houser S R, Margulies K B. Circulation. 1998;97:2316–2322. doi: 10.1161/01.cir.97.23.2316. [DOI] [PubMed] [Google Scholar]
- 12.Chirgwin J M, Przybyla A E, MacDonald R J, Rutter W J. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- 13.Anisimov S V, Tarasov K V, Stern M D, Lakatta E G, Boheler K R. Genomics. 2002;80:213–222. doi: 10.1006/geno.2002.6821. [DOI] [PubMed] [Google Scholar]
- 14.Anisimov S V, Tarasov K V, Riordon D, Wobus A M, Boheler K R. Mech Dev. 2002;202:25–74. doi: 10.1016/s0925-4773(02)00177-6. [DOI] [PubMed] [Google Scholar]
- 15.Ott R L. An Introduction to Statistical Methods and Data Analysis. Belmont, CA: Duxbury; 1993. [Google Scholar]
- 16.Blochberger T C, Vergnes J P, Hempel J, Hassell J R. J Biol Chem. 1992;267:347–352. [PubMed] [Google Scholar]
- 17.Baba H, Ishiwata T, Takashi E, Xu G, Asano G. Jpn Circ J. 2001;65:445–450. doi: 10.1253/jcj.65.445. [DOI] [PubMed] [Google Scholar]
- 18.Weber K T. Hypertension. 2001;38:588–591. doi: 10.1161/01.hyp.38.3.588. [DOI] [PubMed] [Google Scholar]
- 19.Murphy G, Knauper V, Cowell S, Hembry R, Stanton H, Butler G, Freije J, Pendas A M, Lopez-Otin C. Ann NY Acad Sci. 1999;878:25–39. doi: 10.1111/j.1749-6632.1999.tb07672.x. [DOI] [PubMed] [Google Scholar]
- 20.Boes M, Booth B A, Sandra A, Dake B L, Bergold A, Bar R S. Endocrinology. 1992;131:327–330. doi: 10.1210/endo.131.1.1377125. [DOI] [PubMed] [Google Scholar]
- 21.Han J, Molkentin J D. Trends Cardiovasc Med. 2000;10:19–22. doi: 10.1016/s1050-1738(00)00039-6. [DOI] [PubMed] [Google Scholar]
- 22.Sugden P H, Clerk A. Circ Res. 1998;83:345–352. doi: 10.1161/01.res.83.4.345. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Shao J, Muhlenkamp P, Liu S, Klepcyk P, Ren J, Friedman J E. J Biol Chem. 2000;275:14173–14181. doi: 10.1074/jbc.m000764200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.