Abstract
Saccharomyces cerevisiae, bakers' yeast, is not a pathogen in healthy individuals, but is increasingly isolated from immunocompromised patients. The more frequent isolation of S. cerevisiae clinically raises a number of questions concerning the origin, survival, and virulence of this organism in human hosts. Here we compare the virulence of a human isolate, a strain isolated from decaying fruit, and a common laboratory strain in a mouse infection model. We find that the plant isolate is lethal in mice, whereas the laboratory strain is avirulent. A knockout of the SSD1 gene, which alters the composition and cell wall architecture of the yeast cell surface, causes both the clinical and plant isolates to be more virulent in the mouse model of infection. The hypervirulent ssd1Δ/ssd1Δ yeast strain is a more potent elicitor of proinflammatory cytokines from macrophages in vitro. Our data suggest that the increased virulence of the mutant strains is a consequence of unique surface characteristics that overstimulate the proinflammatory response.
Bakers' yeast, Saccharomyces cerevisiae, is not a pathogen in healthy individuals, but is increasingly isolated from immunocompromised patients (1). This organism, used commercially in the preparation of food and beverages, is usually found in the environment associated with rotting fruit. The use of live Saccharomyces in the treatment of diarrhea in Europe has been linked to yeast sepsis, making it clinically relevant (1, 2). In addition to its use as an oral probiotic, live S. cerevisiae shows promise as a vaccine delivery vector (3), and the predominant yeast cell wall molecule, β-glucan, is a potent immunostimulatory molecule with potential clinical use (4). The possible infectious properties and clinical utility of S. cerevisiae highlight the importance of understanding the yeast–immune system interaction.
Some clinical isolates of S. cerevisiae proliferate and cause death in DBA/2 mice (5, 6), but not other standard laboratory mouse strains. Clinical yeast isolates are able to grow at higher temperatures (41°C) than laboratory strains, and this characteristic has been correlated with their survival in mice (7). Recent work has also shown that metabolic genes required for the virulence of more traditional human pathogens such as Cryptococcus neoformans and Candida albicans are also required for survival of clinical isolates in mice (8). These experiments have established that the mouse models of S. cerevisiae virulence are relevant to a broad range of fungal pathogens and are useful for identification of new determinants of fungal virulence.
We have genetically characterized a number of Saccharomyces strains recently isolated from humans and found that in the CISC44 (CISC, clinical isolate of Saccharomyces cerevisiae) strain the SSD1 gene is heterozygous and required for high-temperature growth (Htg). In wild strains of S. cerevisiae isolated either from humans or plants, we show that knockout of the SSD1 gene, which alters the composition and cell wall architecture of the yeast cell surface, causes yeast to be more virulent in the mouse model of infection. The ssd1Δ/ssd1Δ mutant yeast is a more potent elicitor of proinflammatory cytokines from macrophages, suggesting that the hypervirulence is due to overstimulation of the proinflammatory response and septic shock.
Materials and Methods
Strains and Manipulations.
CISC44 was isolated by A. W. Fothergill at the University of Texas Health Science Center, San Antonio, and was obtained from Robert H. Rubin of Massachusetts General Hospital, Boston. It was isolated from a patient by esophageal brushing. EM93 was a gift from Robert Mortimer of the University of California, Berkeley (9). W303 (Z18) was obtained from Rick Young of the Whitehead Institute, Boston. Prototrophic S288c strains FY4 and FY5 were obtained from Fred Winston of Harvard Medical School, Boston (10). Plasmids containing the S288c allele of SSD1 were gifts of Ralph Keil of the Pennsylvania State University, Hershey. Standard yeast manipulations were performed and all media used were as described (11). Preliminary characterization of CISCs showed that the majority were bona fide diploid Saccharomyces by several criteria: they underwent meiosis, produced haploid progeny that mated well to laboratory strains (S288C, Σ1278b, and W303), and resulted in F1 heterozygotes that were fertile. In-frame knockouts of the entire ORF in S288c were made using dominant drug resistance cassettes (G418r and HYGr) as described (12). The deleted ORF with the drug resistance cassette, including 500 bp flanking each side, were amplified by PCR and sequentially transformed to make the knockouts in the diploid EM93 and CISC44 strains. Complementation of the homozygous knockout in the EM93 background was done by inserting the NATr cassette upstream of the SSD1 gene in S288C. PCR was used to amplify the marked SSD1* gene and this PCR fragment was transformed into the ssd1Δ/ssd1Δ mutant in the EM93 background to replace the ssd1Δ∷HYGr knockout allele. Temperature resistance was assayed by diluting overnight cultures and spotting to yeast extract/peptone/dextrose (YPD) plates that were incubated at the indicated temperatures for 2–3 days.
Mouse Infections.
Tail-vein infections were done essentially as described (6). A culture of each yeast strain grown in rich media (YPD) overnight at room temperature was diluted and allowed to grow for more than two generations at room temperature. Cells were harvested at OD600 = 1–2, washed three times with PBS, counted, and resuspended at 2 × 108 colony-forming units (cfu)/ml in PBS. The flocculation of the ssd1Δ/ssd1Δ mutant strains was eliminated by the washes in PBS, allowing an accurate count of cfu. Four-week-old DBA/2 or BALB/c mice (Taconic Farms) were anaesthetized with i.p. injection of Avertin (tribromoethanol in tert-amyl alcohol; dosage of 250 mg/kg). Anaesthetized mice were injected in the tail vein with 100 μl of yeast suspension (2 × 107 yeast). Mice were monitored after 2 h to ensure return of the righting reflex. Mice were then monitored for morbidity twice daily for 14 days. In each experiment, infections of DBA/2 mice with live EM93 and the ssd1Δ/ssd1Δ in the EM93 background were used as controls. Where applicable, significance of differences in survival were calculated by Kaplan–Meier survival analysis using log-rank scores.
Cell Wall Analysis.
Cells from cultures grown for seven to eight generations in YPD medium containing [U14C]glucose were harvested from log-phase cells grown at room temperature (to assure homogeneous labeling of all carbon containing molecules and equal 14C incorporation for all strains). Cells were homogenized with glass beads and cell walls were isolated by centrifugation. Walls were enzymatically fractionated in a three-step procedure previously described (13). Each of the cell wall polysaccharides were quantified as the percentage of total wall label. Chromatographic profiles are on the digestion products obtained after step 1 of the serial enzymatic cell wall fractionation scheme (13). The relative amount of L5 and L4 was calculated by adding the radioactivity in all fractions of each peak, relative to the total of both peaks, after background subtraction. The fractions taken for calculation are those between the lines (Fig. 3 a and b). L5 (laminaripentaose) and L2 (GlcNAc and laminaribiose) were both analyzed by TLC to confirm the polysaccharide content of the peaks. Statistical significance was measured using one-tailed Student's t test, assuming two-sample equal variance.
Figure 3.
The cell surface is quantitatively and qualitatively altered in ssd1Δ/ssd1Δ mutants. (a and b) Profiles of β-glucanase digestion products of β1,3-glucan. L5 represents laminaripentaose from linear, unsubstituted portions of the chains. L4 represents laminaritetraose from substituted portions of the glucan chain. The glucanase also releases laminaribiose (L2) from glucan chains and GlcNAc from chitin, which coelute on the column in higher number fractions. Peaks were quantitated as described in Materials and Methods. (a) Profile of CISC44. (b) Profile of CISC44 ssd1Δ/ssd1Δ mutant. (c) The EM93 ssd1Δ/ssd1Δ mutant is more flocculent than the WT and heterozygous strains. Strains were grown overnight, vortexed, and photographed within 5 min. Flocculation causes the cells to clump together and fall out of suspension. This flocculation was found to depend on Ca2+ (data not shown). (d) The EM93 ssd1Δ/ssd1Δ mutant is more hydrophobic than the WT and heterozygous strains in the octane bipartitioning assay. Strains were grown overnight and assayed as described in Materials and Methods. Greater retention in the octane phase indicates an increase in hydrophobicity.
Assays of Flocculation and Hydrophobicity.
Cells were grown overnight in SC media (minimal media containing glucose and all amino acids). To test flocculation, culture tubes were vortexed vigorously for 1 min, stood on end, and photographed within 5 min. The hydrophobicity test was done exactly as described (14). The amount of cells in the aqueous layer before and after vortexing vigorously with octane was quantitated by spectrophotometer.
Macrophage Stimulation with Yeast.
Overnight cultures of yeast strains grown in rich media (YPD) were washed three times with sterile PBS (GIBCO/BRL), counted, and resuspended at 1.5 × 108 yeast per ml. Murine RAW 264.7 macrophages (American Type Culture Collection) were grown in RPMI-10 [RPMI (GIBCO/BRL) + 10% heat-inactivated FCS (Intergen, Purchase, NY) + penicillin (100 units/ml)-streptomycin (0.1 mg/ml)]. They were plated to 24-well plates at 2.5 × 105 macrophages per well and grown overnight. Peritoneal macrophages were elicited from 7- to 10-week-old DBA/2 mice with Biogel-10 beads (Bio-Rad) and harvested as described (15). Macrophages were plated in 24-well plates at 2.5 × 105 macrophages per well and grown overnight in RPMI-10 before the infection. Before infection, macrophages were washed three times with fresh RPMI-10. Yeast were then added at a yeast:macrophage ratio of 30:1 and allowed to bind and be phagocytosed for 30 min at 37°C in 5% CO2. Macrophages were then washed extensively to remove unbound yeast and incubated in 0.5 ml of medium for 3–6 h at 37°C in 5% CO2. All infections were done at least three times independently, and duplicate or triplicate wells were used each time.
Cytokine Measurements.
Murine tumor necrosis factor α (TNFα), IL-1β, and IL-6 levels in the infection supernatants were measured by ELISA according to the manufacturer's instructions (R & D Systems), with the exception that antibody binding was detected using the Supersignal ELISA substrate (Pierce) and quantified on a Luminoskan luminometer (Labsystems, Franklin, MA). Standard curves were calculated using DELTASOFT3 imaging software (Biometallics, Princeton) or excel (Microsoft). Infections were done in duplicate or triplicate and ELISA measurements were taken in duplicate or triplicate. Five independent experiments were performed, and results from a single representative experiment are presented as the averages and standard deviations of four to nine measurements.
Results and Discussion
A number of clinical isolates, dubbed CISCs, were subjected to genetic analysis and shown to be authentic diploids of S. cerevisiae (see Materials and Methods). One of these diploids, CISC44, showed a 2:2 meiotic segregation for the ability to grow at high temperatures (Fig. 1 a and b), suggesting that Htg+ in this strain depends on a single genetic locus. We investigated the molecular nature of the Htg+ phenotype because temperature growth has been associated with virulence in S. cerevisiae (7, 16). Crosses of an Htg+ haploid segregant by the laboratory W303 strain (Htg−) also segregated 2 Htg+:2 Htg− (Fig. 1 c and d). The Htg+ locus was cloned by transforming a single-copy library (from an Htg+ haploid segregant, 44-8c) into the Htg− W303 strain and isolating a plasmid that conferred the Htg+ phenotype to W303.
Figure 1.
The SSD1 gene is heterozygous in a clinical isolate and regulates Htg in three strain backgrounds. (a–d) A single locus regulates temperature resistance in CISC44 clinical isolate. The four products of one meiosis are in each vertical column. The meiotic products of CISC44 (a and b) or a cross between CISC Htg+ spore 44-8c and W303 (c and d) were replica-plated to YPD plates at 30°C (a and c) or 41°C (b and d). (e and f) W303 (row 3) is converted to Htg+ by transformation with a centromeric plasmid containing the SSD1 gene from CISC44 (row 4) or from S288C (rows 5 and 6) or a high copy (2 μ) plasmid containing the S288C allele (row 7), but not with vector alone (row 3); CISC44 is included as an Htg+ control (row 1) and CISC44 ssd1Δ/ssd1Δ (row 2) is included as an Htg− control. Strains were spotted in 10-fold dilution series (left to right) on YPD plates at 30°C (e) or 41°C (f). (g and h) Homozygous knockout of both alleles of SSD1 yields Htg− strains in both EM93 and CISC44 backgrounds. EM93 (row 1), EM93 ssd1Δ/ssd1Δ (row 2), CISC44 (row 3), and CISC44 ssd1Δ/ssd1Δ (row 4) were spotted in a 10-fold dilution series to YPD plates at 37°C (g) or 41°C (h). Linkage of Htg+ to SSD1 was further validated by crosses of Htg+ spore 44-8c to strains with neutral markers inserted at the SSD1 locus (data not shown).
Sequence analysis showed that the cloned gene was SSD1, a polymorphic gene in laboratory strains that is functional in the fully sequenced S288c strain but nonfunctional in W303 (17). Either the CISC44-derived SSD1 gene or the S288c-derived SSD1 confers the Htg+ phenotype to the Htg− W303 strain (Fig. 1 e and f). SSD1 has been implicated in temperature resistance in several laboratory strains (17–19). To establish that SSD1 regulates temperature resistance in environmental isolates, we constructed homozygous mutants (ssd1Δ∷G418r/ssd1Δ∷HYGr) by sequential gene disruption in two different strains, CISC44 and EM93. EM93, a feral diploid strain that was isolated from a rotting fig, is the progenitor to the fully sequenced S288c lab strain and contributes ≈88% of the S288c gene pool (9). As shown in Fig. 1 g and h, SSD1 is required for Htg on YPD media in both the EM93 and CISC44 backgrounds.
Loss of SSD1 function in laboratory strains results in a number of phenotypes associated with alterations in the cell wall: dramatic increases in sensitivity to osmotin, a plant-derived antifungal compound; changes in the composition of the cell wall polysaccharides (20); and sensitivity to the cell wall damaging agent calcofluor white (18). Both ssd1Δ/ssd1Δ mutants constructed in the feral strain backgrounds exhibit similar cell wall changes and cell integrity phenotypes to those described in lab strains: a partial deficiency in β1,3- and β1,6-glucan (≈10% less) accompanied by concomitant increases in mannan and chitin (Table 1), as well as sensitivity to calcofluor white.
Table 1.
The cell wall composition of ssd1Δ/ssd1Δ mutants in wild isolates
| Strain | Chitin | β-1,6-glucan | β-1,3-glucan | Mannan |
|---|---|---|---|---|
| EM93 WT | 3.4 ± 1.7 | 12.4 ± 0.3 | 42.9 ± 1.1 | 41.4 ± 1.8 |
| EM93 ssd1Δ/ ssd1Δ | 4.1 ± 1.3 | 11.0 ± 0.2* | 39.8 ± 0.7* | 45.6 ± 1.2** |
| CISC44 WT | 3.1 ± 1.1 | 12.4 ± 0.7 | 39.4 ± 2.0 | 45.1 ± 0.9 |
| CISC44 ssd1Δ/ ssd1Δ | 5.7 ± 1.5** | 10.9 ± 0.5* | 33.4 ± 1.4* | 50.1 ± 0.9* |
All results are presented as the percentage of total cell wall. Results are presented as average ± SD of duplicates from three independent experiments.
and
indicate, respectively, P < 0.001 and P < 0.01 for the difference between WT and ssd1Δ/ssd1Δ.
The fact that selective pressures have kept CISC44 heterozygous at the SSD1 locus raised the possibility that homozygous mutation may affect the survival and/or virulence of the yeast in the mammalian host, counterbalancing the clearly detrimental ex vivo fitness of such strains. To test the contribution of SSD1 to virulence we infected DBA/2 mice i.v., a standard model of S. cerevisiae infection (6, 21). The EM93 strain was much more virulent than the clinical isolate CISC44, which caused no lethality in mice even 4 weeks after infection (Fig. 2a). Consistent with previous studies, we found that the prototrophic S288C lab strain was avirulent (data not shown).
Figure 2.
Homozygous ssd1Δ/ssd1Δ mutants are more virulent in the mouse model of infection. DBA/2 mice were infected with the indicated yeast strains (see Materials and Methods) and monitored twice daily. (a) Comparison of infections with WT (×) and ssd1Δ/ssd1Δ (○) yeast in the CISC44 (dotted line) and EM93 (solid line) backgrounds. (b) Reintroduction of SSD1 into the ssd1Δ/ssd1Δ mutant in the EM93 background restores virulence to WT levels. Two ssd1Δ/ssd1Δ strains transformed with WT SSD1 were assayed and had indistinguishable virulence (data not shown). Only one transformed strain (ssd1Δ/SSD1*; dotted line) is shown for the sake of clarity. In addition, the heterozygote used to obtain the homozygous deletion was found to have essentially the same virulence as the transformed strains (data not shown). (c) Comparison of infections of DBA/2 and BALB/c mice shows that lethality is not through aggregation and mechanical obstruction. Infections of DBA/2 mice (solid lines) resulted in 100% lethality, whereas infections of BALB/c mice (dashed lines) caused no lethality. EM93 and the ssd1Δ/ssd1Δ mutant in the EM93 background were used in all experiments as controls, and the survival curves represent the combination of four experiments. Survival curves with CISC44 and the ssd1Δ/ssd1Δ mutant in the CISC44 background are the combination of three independent experiments.
The homozygous ssd1Δ/ssd1Δ strains of yeast constructed in both the CISC44 and EM93 backgrounds were dramatically more virulent than the corresponding WT strains. The CISC44 comparison is striking because the ssd1Δ/ssd1Δ mutant was lethal, whereas the CISC44 WT strain caused no death. Mice infected with the EM93 ssd1Δ/ssd1Δ strain had an accelerated onset of lethality as compared with the isogenic WT strain; a significant proportion of the mice died even 1 day after infection. This accelerated death is a consequence of the ssd1Δ/ssd1Δ mutations in EM93, because the SSD1/ssd1Δ heterozygote and an ssd1Δ/ssd1Δ strain with a functional SSD1 gene reintegrated at the SSD1 locus (ssd1Δ/SSD1*) had virulence indistinguishable from that of the EM93 WT strain and less than that of the ssd1Δ/ssd1Δ mutant (Fig. 2b).
Several experiments suggest that the ssd1Δ/ssd1Δ mutant yeast are viable in the mouse and that their increased virulence requires proliferation in vivo. Although the ssd1Δ/ssd1Δ mutants do not grow at 41°C, they still grow well at mammalian body temperature, 37°C (Fig. 1g). Mice that have died after tail-vein injections of either the ssd1 mutant or WT have large numbers of live yeast (>106 yeast) in their kidneys. Moreover, neither mutant nor WT yeast that have been killed by mild heating to 65°C for 15 min cause any lethality, suggesting that the increased virulence of the mutant cells is not a consequence of lysis and release of toxins. These data suggest that the mutant yeast are viable in vivo and cause mortality due to their growth and interaction with the host, rather than killing the mice through toxicity, as is the case for lipopolysaccharide (LPS).
We investigated the architecture of β-glucan in the cell wall of WT and ssd1Δ/ssd1Δ strains because alterations in the composition of fungal cell walls are known to alter the immune response (22). The two primary components of the S. cerevisiae cell wall (β-glucan and mannan) are recognized by specialized innate immune receptors, termed pattern recognition receptors (23, 24), which are thought to direct innate and adaptive immune responses to these fungal antigens. To probe the β-glucan structure, we analyzed the chromatographic profile of products released from WT and ssd1Δ/ssd1Δ mutant cell walls by the combined action of a recombinant β1,3 endoglucanase (Quantazyme) and chitinase (see Materials and Methods). Because of the cleavage specificity of Quantazyme (25), the 5-glucose (L5):4-glucose (L4) products ratio indicates the structural features of the glucan chains: L5 is diagnostic of linear chains and L4 is indicative of branching or chain substitutions, which affect enzyme accessibility. The cell walls from the hypervirulent CISC44 ssd1Δ/ssd1Δ and EM93 ssd1Δ/ssd1Δ mutants show an increase in L5 as compared with L4, indicating a relative increase in linear, unsubstituted β-glucan chains (Fig. 3 a and b, Table 2). These experiments show that the quantitative changes in cell wall composition are accompanied by significant qualitative changes in β-glucan structure.
Table 2.
The β1,3-glucan structure (linear vs. substituted) of ssd1Δ/ssd1Δ mutants
| Strain | Laminaripentaose (L5WT/L5ssdΔssdΔ) | Laminaritetraose (L4WT/L4ssdΔssdΔ) |
|---|---|---|
| EM93 | 0.95 ± 0.01* | 1.34 ± 0.13** |
| CISC44 | 0.91 ± 0.02* | 1.83 ± 0.20** |
All results are presented as the ratios of the areas under the curve for each peak and are the average ± SD of three experiments.
and
indicate, respectively, P < 0.001 and P < 0.005 for the difference between WT and mutant.
These differences in cell wall architecture between the ssd1Δ/ssd1Δ mutant and parent strain are accompanied by physical changes in the yeast cell surface. The ssd1Δ/ssd1Δ mutant in the EM93 background shows much greater flocculence (cell–cell adherence) and greater hydrophobicity than the isogenic WT and heterozygous strains (Fig. 3 c and d). Hydrophobicity is a property of cell wall composition and is positively correlated with virulence in C. albicans (26). The flocculence of the ssd1Δ/ssd1Δ strains does not appear to contribute to virulence by causing mechanical obstruction of the vascular system, because the WT and ssd1Δ/ssd1Δ mutant strains are both avirulent in BALB/c mice (Fig. 2c). BALB/c mice are resistant to lethal infection with S. cerevisiae strains but should be equally susceptible to mechanical obstruction (6). The fact that neither WT nor ssd1Δ/ssd1Δ mutant yeast were virulent in BALB/c mice suggests that aggregation and associated embolism are not responsible for the hypervirulence of ssd1Δ/ssd1Δ strains.
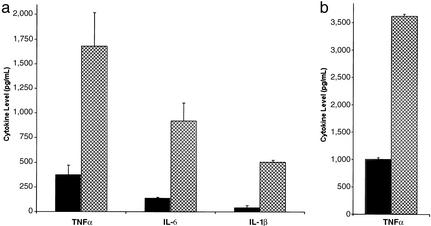
These striking cell wall changes suggested that the mutants may be more virulent because their altered cell surface leads to misrecognition by the innate immune system. The rapidity of death in all cases (within 1 week) also implied that the lethality was due to septic shock induced through overstimulation of the proinflammatory arm of the innate immune system. Analysis of the production of proinflammatory cytokines by primary peritoneal macrophages exposed to WT and mutant yeast provided strong support for this hypothesis. In comparison to the WT CISC44 yeast, the ssd1Δ/ssd1Δ mutant yeast stimulated 3-fold more TNFα, 3-fold more IL-6, and 7-fold more IL-1β (Fig. 4a). The enhancement of TNFα production stimulated by the mutant yeast was also seen when a murine macrophage cell line was exposed to the mutant yeast (Fig. 4b). Overproduction of TNFα, IL-1β, and IL-6 is associated with septic shock (27), suggesting that the hypervirulent yeast is indeed a more efficient inducer of sepsis in the mice.
Figure 4.
The ssd1Δ/ssd1Δ mutant yeast elicits greater proinflammatory cytokine induction in both primary and cultured macrophages. (a) Peritoneal macrophages were exposed to WT and mutant yeast for 6 h. (b) Macrophages from the RAW 264.7 murine cell line were exposed to WT and mutant yeast for 6 h. TNFα, IL-6, and IL-1β in culture supernatants were measured by duplicate ELISAs of duplicate wells. Gray bar, CISC44 stimulation; crosshatched bar, stimulation with the ssd1Δ/ssd1Δ mutant. The ssd1Δ/ssd1Δ mutant in the EM93 background also causes a significant, although smaller, increase of cytokine production in RAW264.7 and primary macrophages. For both a and b, more than four independent experiments were performed and a single representative experiment is shown.
The established association between fungal cell surface polysaccharides and immune recognition of fungi (22–24, 28, 29) suggests that the changes in the cell surface properties of the ssd1Δ/ssd1Δ mutants are responsible for their increased virulence. The marked difference in virulence between the two feral isolates suggests that there are other differences between these strains that contribute to virulence in the DBA/2 mouse model. The fact that EM93, a feral strain from a rotting fig, is more virulent than the CISC44 strain and as virulent as other characterized CISC strains (6) suggests that proliferation of S. cerevisiae in humans may not require any special modification of strains found in the environment. Indeed, fermenting plant material could be the source of the Saccharomyces strains found in immunocompromised patients.
Acknowledgments
We thank Robert Mortimer, Dr. Robert Rubin, and Rick Young for providing strains; Ralph Keil for providing plasmids with the S288c allele of SSD1; the Hacohen and Young labs for providing key ELISA reagents, equipment, and expertise; Gordon Brown and the Gordon lab for providing invaluable expertise in innate immunity and assistance in completion of the initial TNFα experiment; members of the Fink Lab and Nir Hacohen for comments on the manuscript; and Michael Lorenz for assistance with tail-vein injections. This work was supported by National Institutes of Health RO1 Grants GM 40266 (to G.R.F.), GM 59773 (to C.A.), and GM 31318 (to P. W. Robbins). R.T.W. is a Burroughs Wellcome Fellow of the Life Science Research Foundation.
Abbreviations
- CISC
clinical isolate of Saccharomyces cerevisiae
- TNFα
tumor necrosis factor α
- Htg
high-temperature growth
- YPD
yeast extract/peptone/dextrose
References
- 1.Piarroux R, Millon L, Bardonnet K, Vagner O, Koenig H. Lancet. 1999;353:1851–1852. doi: 10.1016/S0140-6736(99)02001-2. [DOI] [PubMed] [Google Scholar]
- 2.Lherm T, Monet C, Nougiere B, Soulier M, Larbi D, Le Gall C, Caen D, Malbrunot C. Intensive Care Med. 2002;28:797–801. doi: 10.1007/s00134-002-1267-9. [DOI] [PubMed] [Google Scholar]
- 3.Stubbs A C, Martin K S, Coeshott C, Skaates S V, Kuritzkes D R, Bellgrau D, Franzusoff A, Duke R C, Wilson C C. Nat Med. 2001;7:625–629. doi: 10.1038/87974. [DOI] [PubMed] [Google Scholar]
- 4.Sutherland I W. Trends Biotechnol. 1998;16:41–46. doi: 10.1016/S0167-7799(97)01139-6. [DOI] [PubMed] [Google Scholar]
- 5.McCusker J H, Clemons K V, Stevens D A, Davis R W. Genetics. 1994;136:1261–1269. doi: 10.1093/genetics/136.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byron J K, Clemons K V, McCusker J H, Davis R W, Stevens D A. Infect Immun. 1995;63:478–485. doi: 10.1128/iai.63.2.478-485.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCusker J H, Clemons K V, Stevens D A, Davis R W. Infect Immun. 1994;62:5447–5455. doi: 10.1128/iai.62.12.5447-5455.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldstein A L, McCusker J H. Genetics. 2001;159:499–513. doi: 10.1093/genetics/159.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mortimer R K, Johnston J R. Genetics. 1986;113:35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 11.Guthrie C, Fink G R. In: Guide to Yeast Genetics and Molecular Biology. Abelson J N, Simon M I, editors. Vol. 194. San Diego: Academic; 1991. pp. 3–21. [Google Scholar]
- 12.Goldstein A L, McCusker J H. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 13.Magnelli P, Cipollo J F, Abeijon C. Anal Biochem. 2002;301:136–150. doi: 10.1006/abio.2001.5473. [DOI] [PubMed] [Google Scholar]
- 14.Reynolds T B, Fink G R. Science. 2001;291:878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- 15.Haworth R, Gordon S. In: Immunology of Infection. Kaufmann S H E, Kabelitz D, editors. Vol. 25. Boston: Academic; 1998. pp. 287–310. [Google Scholar]
- 16.Steinmetz L M, Sinha H, Richards D R, Spiegelman J I, Oefner P J, McCusker J H, Davis R W. Nature. 2002;416:326–330. doi: 10.1038/416326a. [DOI] [PubMed] [Google Scholar]
- 17.Sutton A, Lin F, Arndt K T. Cold Spring Harbor Symp Quant Biol. 1991;56:75–81. doi: 10.1101/sqb.1991.056.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Kaeberlein M, Guarente L. Genetics. 2002;160:83–95. doi: 10.1093/genetics/160.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uesono Y, Toh-e A, Kikuchi Y. J Biol Chem. 1997;272:16103–16109. doi: 10.1074/jbc.272.26.16103. [DOI] [PubMed] [Google Scholar]
- 20.Ibeas J I, Yun D J, Damsz B, Narasimhan M L, Uesono Y, Ribas J C, Lee H, Hasegawa P M, Bressan R A, Pardo J M. Plant J. 2001;25:271–280. doi: 10.1046/j.1365-313x.2001.00967.x. [DOI] [PubMed] [Google Scholar]
- 21.McCullough M J, Clemons K V, McCusker J H, Stevens D A. J Clin Microbiol. 1998;36:2613–2617. doi: 10.1128/jcm.36.9.2613-2617.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mansour M, Levitz S. Curr Opin Microbiol. 2002;5:359. doi: 10.1016/s1369-5274(02)00342-9. [DOI] [PubMed] [Google Scholar]
- 23.Janeway C A, Jr, Medzhitov R. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 24.Underhill D M, Ozinsky A. Annu Rev Immunol. 2002;20:825–852. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 25.Jeffries T W, Macmillan J D. Carbohydr Res. 1981;95:87–100. [Google Scholar]
- 26.Hazen K C, Glee P M. Curr Top Med Mycol. 1995;6:1–31. [PubMed] [Google Scholar]
- 27.Astiz M E, Rackow E C. Lancet. 1998;351:1501–1505. doi: 10.1016/S0140-6736(98)01134-9. [DOI] [PubMed] [Google Scholar]
- 28.Brown G D, Taylor P R, Reid D M, Willment J A, Williams D L, Martinez-Pomares L, Wong S Y, Gordon S. J Exp Med. 2002;196:407–412. doi: 10.1084/jem.20020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraser I P, Koziel H, Ezekowitz R A. Semin Immunol. 1998;10:363–372. doi: 10.1006/smim.1998.0141. [DOI] [PubMed] [Google Scholar]