Abstract
The opportunistic bacterial pathogen Pseudomonas aeruginosa colonizes airways of individuals with cystic fibrosis (CF) with resultant chronic destructive lung disease. P. aeruginosa adaptation to the CF airway includes biofilm formation and antibiotic resistance. Isolates from asymptomatic individuals in the first 3 years of life have unique characteristics, suggesting that adaptation occurs before clinical symptoms. One defined early adaptation is expression of a specific proinflammatory lipopolysaccharide (LPS) that is associated with antimicrobial peptide resistance. This CF-specific LPS is induced when P. aeruginosa is grown in medium that is limited for magnesium. Therefore, qualitative and quantitative proteomic approaches were used to define 1,331 P. aeruginosa proteins, of which 145 were differentially expressed on limitation of magnesium. Among proteins induced by low magnesium were enzymes essential for production of 2-heptyl 3-hydroxy 4-quinolone, the Pseudomonas quinolone signal (PQS), which interacts with the homoserine lactone signaling pathway. Measurement of PQS in P. aeruginosa isolates from asymptomatic children with CF indicated that strains with increased synthesis of PQS are present during early colonization of CF patient airways.
Magnesium is an essential cofactor in many biochemical reactions, and depletion of magnesium destabilizes the Gram-negative bacterial outer membrane (1). Growth in medium limited for magnesium promotes Gram-negative virulence gene expression (2–4) suggesting that, at least in part, it simulates environmental conditions that bacteria encounter on colonization of host tissues. Many of these virulence genes function to remodel the bacterial envelope (5–10). Both the outer membrane lipopolysaccharide (LPS) structure and the protein content are significantly changed on growth of Salmonella enterica (8, 9) and P. aeruginosa (6, 11) in medium with low magnesium concentration. These changes result in increased resistance to the vertebrate host's antimicrobial peptides (4, 9) and modulation of bacterial recognition by Toll-like receptors (12), both important strategies to combat hosts' innate immunity and to establish the infection.
P. aeruginosa lung infection is a major cause of mortality in patients with cystic fibrosis (CF), the most common genetic disease of Caucasians (13). Airways of almost 100% of CF patients are infected with P. aeruginosa by 3 years of age (14). The inability of their immune systems to clear the bacteria results in chronic infection and an associated high level of airway inflammation (13). Bacterial adaptation to the CF airway includes various phenotypic changes such as mucoidy, which is a result of production of a complex polysaccharide alginate at the bacterial surface (15). Another bacterial phenotype that develops in the course of infection is resistance to various antimicrobial therapy agents (16). One of the earliest reported adaptations of P. aeruginosa isolates from airways of infants with CF is the synthesis of specific LPS at the bacterial surface. The result of such envelope modification is increased bacterial resistance to antimicrobial peptides (4, 6) and increased proinflammatory signaling through the human Tlr4 receptor (12). Similar LPS structure is also produced by P. aeruginosa grown in medium limited for magnesium (6). P. aeruginosa was also isolated from CF patients' airways in the form of biofilms, antibiotic-resistant bacterial communities (17). Expression of genes that contribute to formation of P. aeruginosa biofilms is, at least in part, regulated through quorum sensing, intercellular bacterial communication via small molecules (18). Quorum sensing also regulates a number of P. aeruginosa secreted virulence and nutrient-scavenging factors (19). Therefore, it has been suggested that gene regulation through quorum sensing is essential for the establishment of P. aeruginosa infection and for bacterial adaptation to the CF patients' airways (17, 19, 20). In this study, proteomic analysis revealed that the synthesis of Pseudomonas quinolone signal (PQS), which interacts with quorum sensing, is increased during growth of a laboratory strain in low magnesium and increased in P. aeruginosa isolates from airways of infants with CF.
Materials and Methods
Bacterial Strains.
P. aeruginosa PAO-1 Lo was obtained from Stephen Lory (Harvard Medical School, Boston). PAO-1 Ig (obtained from Barbara Iglewski, University of Rochester, Rochester, NY) and PQS-null mutant [PAO-1 Ig with insertion of ISphoA/hah-Tc transposon in ORF PA0999 (21)] were generous gifts of David D'Argenio (University of Washington, Seattle). P. aeruginosa CF clinical isolates were collected as a part of a multicenter study on infection and inflammation in young infants with CF, whereas isolates from blood and urinary tract infections were obtained from Stephen Lory. Bacterial strain clonality was determined by restriction fragment length polymorphism analysis of a region upstream of exoA and whole chromosome digests (Jane Burns and Maynard Olson, personal communication).
Proteomic Analysis of P. aeruginosa.
P. aeruginosa PAO-1 was grown at 37°C with aeration (200 rpm) in N medium (2) supplemented with 38 mM glycerol, 0.1% casamino acids, and either 8 μM or 1 mM MgCl2. Bacteria were harvested for protein analysis in the late logarithmic phase of growth. The doubling time was 180 min in medium containing 8 μM MgCl2 and 120 min in medium containing 1 mM MgCl2. The final density of P. aeruginosa culture grown in 8 μM MgCl2 was at 70% of the density of the culture grown in 1 mM MgCl2, although both cultures reached a stationary phase of growth after 12 h. Bacterial protein was labeled with either light (dO; for cultures grown in 8 μM MgCl2) or heavy (d8; for cultures grown in 1 mM MgCl2) isotope-coded affinity tag (ICAT) and ICAT peptides were analyzed by microcapillary liquid chromatography–electrospray ionization-tandem MS (see supporting information on the PNAS web site, www.pnas.org, for a detailed protocol). Automated data processing for protein identification and quantitation was achieved by using sequest and express software tools (22). Uninterpreted tandem mass spectra were searched against the P. aeruginosa database (www.pseudomonas.com). Peptides that showed sequest scores >1.5 were further analyzed manually by detailed spectral analysis as described (22). Protein abundance ratios larger than +1.5 or smaller than –1.5 were set as a threshold indicating significant changes (23). Relative abundance ratios between –1.5 and +1.5 were considered to represent the steady state (23, 24).
Analysis of PQS Production.
P. aeruginosa PAO-1 Lo was grown overnight at 37°C in N minimal medium supplemented with either 8 μM or 1 mM MgCl2 or in tryptic soy broth (TSB). PAO-1 Ig and various clinical P. aeruginosa were grown overnight at 37°C in TSB. PQS was extracted from bacterial cultures by acidified ethyl acetate and analyzed by thin-layer chromatography as described (25, 26).
Results
Proteomic Analysis Defined 1,331 P. aeruginosa Proteins, of Which 145 Were Regulated by Growth in Low Magnesium.
Qualitative and quantitative proteomic approaches were used to determine the whole cell and envelope proteome during growth of the Gram-negative bacterial pathogen P. aeruginosa in varying magnesium concentrations [growth in presence of low (8 μM) vs. growth in high (1 mM) magnesium]. Quantitative proteomic analysis of P. aeruginosa was performed on differential labeling of proteins with ICAT (27) (see Materials and Methods). Protein identification and quantification were achieved in silico (22).
DNA-microarray transcriptional profiling has defined that ≈2,250 P. aeruginosa ORFs are transcribed during growth in low and high magnesium (R. K. Ernst and S.I.M., unpublished results), suggesting that ≈40% of the potential P. aeruginosa proteome is expressed under these conditions. Furthermore, 681 (30%) of these genes were regulated by growth in 8 μM Mg2+ (R. K. Ernst and S.I.M., unpublished results). A similar ratio of regulated vs. expressed proteins was determined in this study. One thousand three hundred and thirty-one proteins that represent 59% of the expressed P. aeruginosa proteome as estimated from transcriptional profiling were detected in low- and high-magnesium growth conditions by using qualitative and quantitative proteomic analysis. The relative abundance of 546 proteins (486 from the whole cell and 163 from the membrane fraction) or 24% of expressed proteome was determined by quantitative ICAT analysis (see Tables 3 and 4, which are published as supporting information on the PNAS web site). This analysis defined 145 magnesium stress-response proteins, of which 76 proteins were induced and 69 were repressed on growth in low magnesium. Regulated proteins included 34 metabolic enzymes, 31 putative enzymes, 37 other previously characterized proteins, and 43 hypothetical proteins (see Tables 5 and 6). The overlap between transcriptional profiling and proteomic analysis results has been indicated for 23 regulated proteins (see Tables 5 and 6).
A number of conserved Gram-negative magnesium stress-response proteins involved in bacterial virulence were among the most abundant proteins induced in low magnesium (Table 1). These included transcriptional regulator PhoP and magnesium transporter homologue MgtA (Table 1). PhoP orthologues regulate expression of genes essential for virulence and magnesium acquisition in several Gram-negative bacteria (2, 4, 28). Some of the PhoP-induced enzymes promote increased bacterial resistance to antimicrobial peptides, a key component of host's innate immunity (5, 6, 8, 10). For example, PhoP-activated LPS modifications, including the addition of aminoarabinose and palmitate to lipid A, promote resistance to the antibiotic polymyxin and other cationic antimicrobial peptides (6, 9, 10). Consistent with the addition of aminoarabinose to LPS, homologues of S. enterica enzymes necessary for aminoarabinose addition and resistance to polymyxin (PA3552–3554) were among the most highly regulated proteins in this study (Tables 1 and 5).
Table 1.
Selected proteins differentially expressed in P. aeruginosa during growth in low magnesium
| Gene no. | Protein | n | u-ICAT | Fold abundance | SD |
|---|---|---|---|---|---|
| Conserved magnesium stress response | |||||
| 1179 | Two-component response regulator PhoP | 6 | 1 | 10.34 | 0.96 |
| 3552 | Conserved hypothetical protein, PmrH homologue | 7 | 1 | 2.84 | 0.25 |
| 3553 | Probable glycosyl transferase, PmrF homologue | 1 | 1 | 2.33 | na |
| 3554 | Conserved hypothetical protein, PmrI homologue | 34 | 7 | 6.09 | 1.16 |
| 4635 | Conserved hypothetical protein, MgtC homologue | 13 | 1 | 3.99 | 0.56 |
| 4825 | Magnesium transport ATPase MgtA | 85 | 3 | 5.80 | 1.75 |
| Quorum sensing and adaptation | |||||
| 0934 | GTP pyrophosphokinase RelA | 2 | 1 | 1.53 | 0.00 |
| 0996 | Probable coenzyme A ligase | 1 | 1 | 1.57 | na |
| 0997 | Hypothetical protein | 16 | 2 | 1.57 | 0.26 |
| 0998 | Hypothetical protein | 6 | 2 | 2.04 | 0.24 |
| 0999 | 3-Oxoacyl-[acyl-carrier-protein] synthase III FabH1 | 4 | 1 | 1.63 | 0.14 |
| 1432 | Autoinducer synthesis protein LasI | 2 | 1 | 3.10 | 0.05 |
| Secreted factors | |||||
| 3478 | Rhamnosyltransferase chain B RhlB | 4 | 1 | −1.57 | 0.15 |
| 1899 or 4210* | Probable phenazine biosynthesis protein PhzA | 2 | 2 | −2.69 | 0.00 |
| 1900 | Probable phenazine biosynthesis protein PhzB | 8 | 2 | −1.70 | 0.10 |
| 1903 or 4214* | Phenazine biosynthesis protein PhzE | 1 | 1 | −2.70 | na |
| 1904 or 4215* | Probable phenazine biosynthesis protein PhzF | 2 | 1 | −1.85 | 0.01 |
| 4211 | Probable phenazine biosynthesis protein PhzB | 2 | 1 | −1.93 | 0.07 |
| 4224 | Hypothetical protein PchG | 21 | 2 | −5.23 | 0.25 |
| 4225 | Pyochelin synthetase PchE | 4 | 3 | −1.97 | 0.14 |
| 4226 | Dihydroaeruginoic acid synthetase PchE | 15 | 3 | −4.23 | 0.19 |
| 4228 | Pyochelin biosynthesis protein PchD | 30 | 5 | −4.07 | 0.19 |
| 4230 | Salicylate biosynthesis protein PchB | 17 | 2 | −4.42 | 0.36 |
| 4231 | Salicylate biosynthesis isochorismate synthase PchA | 1 | 1 | −2.21 | na |
n, number of independent identifications and quantification events for each protein; u-ICAT, number of unique peptide sequences; fold abundance, average ratios of all quantified peptides for each protein representing fold increase in protein abundance during growth of P. aeruginosa in 8 μM Mg2+; na, not applicable. Positive values represent increased relative abundance on growth in low magnesium; negative values represent decreased relative abundance.
Identified peptides are identical in both proteins.
Growth of P. aeruginosa in Low Magnesium Results in Altered Subcellular Compartmentalization of Large Enzyme Complexes.
Significant Gram-negative envelope remodeling is part of bacterial adaptation to magnesium limitation (4, 6, 9). To define proteins that contribute to the envelope remodeling either structurally or by providing an enzymatic function to this process, membrane protein of P. aeruginosa was also analyzed by ICAT. In this experiment, relative abundance of 163 P. aeruginosa proteins during growth in varying magnesium concentrations was determined (see Table 4). Seventy of these proteins were induced, and 16 were repressed in the membrane fraction when P. aeruginosa was grown in low magnesium. Abundance ratios of 106 of membrane-associated proteins were also determined by ICAT analysis of the whole cell protein as described above. However, 19 proteins that were apparently induced at the membrane were also found to be at steady-state levels (relative abundance ratios of –1.5 to +1.5) when whole cell protein was analyzed (Table 2). These results suggested that, although the overall cellular concentration of these 19 proteins remained constant, they were more concentrated at the membrane during growth in low magnesium. In addition, although the cellular concentration of the heat-shock chaperone GroEL was reduced by growth in low magnesium, GroEL level was at the “steady state” at the membrane (Table 2). In contrast, relative abundance of protein PA2069 was decreased at the membrane when compared with its concentration in the whole cell during growth in low magnesium. These results suggested that increased abundance of these 21 proteins at the membrane fraction is not a result of increase in transcription of the corresponding genes or protein regulation by posttranslational processing. Instead, changes in protein abundance ratios at the membrane reflected changes in their increased subcellular compartmentalization at the envelope during growth of P. aeruginosa in magnesium-limiting condition.
Table 2.
Differential P. aeruginosa protein fractionation on growth in low magnesium
| Gene no. | Protein | Membrane protein quantification
|
Whole cell protein quantification
|
M/WC abundance ratio | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | u-ICAT | Fold abundance | SD | n | u-ICAT | Fold abundance | SD | |||
| Energy metabolism | ||||||||||
| 1583 | Succinate dehydrogenase (A subunit) SdhA | 3 | 2 | 2.24 | 0.40 | 25 | 4 | 1.36 | 0.22 | 1.65 |
| 1584 | Succinate dehydrogenase (B subunit) SdhB | 7 | 3 | 2.20 | 0.28 | 2 | 1 | −1.14 | 0.09 | 2.50 |
| 1585 | 2-Oxoglutarate dehydrogenase (E1 subunit) SucA | 10 | 3 | 2.97 | 0.59 | 9 | 1 | 1.04 | 0.03 | 2.90 |
| 1770 | Phosphoenolpyruvate synthase PpsA | 6 | 1 | 3.41 | 1.07 | 40 | 3 | 1.12 | 0.23 | 3.04 |
| 5300 | Cytochrome c5 | 5 | 1 | 2.18 | 0.17 | 5 | 1 | 1.43 | 0.11 | 1.52 |
| 5554 | ATP synthase beta chain | 4 | 1 | 1.74 | 0.18 | 45 | 1 | 1.14 | 0.13 | 1.53 |
| 5555 | ATP synthase gamma chain | 2 | 1 | 1.98 | 0.03 | 11 | 1 | 1.33 | 0.08 | 1.48 |
| 5556 | ATP synthase alpha chain | 4 | 1 | 2.00 | 0.32 | 45 | 1 | 1.47 | 0.12 | 1.36 |
| 5557 | ATP synthase delta chain | 5 | 1 | 2.15 | 0.08 | 5 | 1 | 1.17 | 0.17 | 1.84 |
| Translation | ||||||||||
| 3656 | 30S ribosomal protein S2 | 6 | 1 | 2.01 | 0.32 | 15 | 1 | 1.27 | 0.05 | 1.58 |
| 4239 | 30S ribosomal protein S4 | 9 | 2 | 2.19 | 0.22 | 70 | 6 | 1.43 | 0.28 | 1.53 |
| 4241 | 30S ribosomal protein S13 | 5 | 1 | 2.42 | 0.05 | 74 | 3 | 1.32 | 0.23 | 1.83 |
| 4246 | 30S ribosomal protein S5 | 6 | 1 | 2.18 | 0.24 | 60 | 1 | 1.46 | 0.16 | 1.50 |
| 2071 | Elongation factor G | 2 | 1 | 2.17 | 0.30 | 11 | 1 | 1.29 | 0.12 | 1.68 |
| 4266 | Elongation factor G | 9 | 1 | 2.41 | 0.22 | 21 | 2 | 1.21 | 0.11 | 1.99 |
| Other | ||||||||||
| 4385 | GroEL protein | 3 | 1 | −1.16 | 0.02 | 3 | 1 | −3.03 | 0.64 | 2.61 |
| 2991 | Soluble pyridine nucleotide transhydrogenase | 1 | 1 | 1.64 | na | 3 | 1 | 1.00 | 0.05 | 1.64 |
| 4670 | Ribose-phosphate pyrophosphokinase | 23 | 2 | 2.13 | 0.22 | 10 | 2 | 1.31 | 0.12 | 1.63 |
| 3068 | Conserved hypothetical protein | 1 | 1 | 2.13 | na | 6 | 1 | 1.01 | 0.05 | 2.11 |
| 3263 | Conserved hypothetical protein | 3 | 1 | 1.66 | 0.10 | 1 | 1 | 1.06 | na | 1.56 |
| 2069 | Probable carbamoyl transferase | 3 | 1 | −9.10 | 0.02 | 2 | 2 | −1.06 | 0.11 | −8.58 |
n, number of independent identifications and quantification events for each protein; u-ICAT, number of unique peptide sequences; fold abundance, average ratios of all quantified peptides for each protein representing fold increase in protein abundance during growth of P. aeruginosa in 8 μM Mg2+; M/WC abundance ratio, ratio of relative protein abundances determined on ICAT analysis of membrane fraction and ICAT analysis of whole cell protein. Positive values represent increased relative abundance on growth in low magnesium; negative values represent decreased relative abundance.
Regulation of P. aeruginosa Quorum Sensing by Growth in Low Magnesium Concentration.
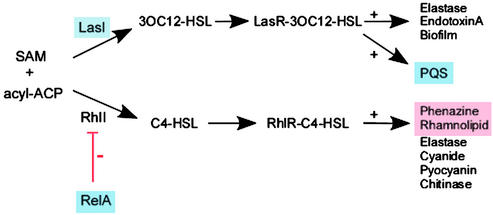
A number of P. aeruginosa virulence factors are regulated by quorum sensing, bacterial communication via small molecule signals N-(3-oxododecanoyl)-l-homoserine lactone (3OC12-HSL) and N-butyryl-l- homoserine lactone (C4-HSL) (17, 18, 29, 30). ICAT analysis indicated that magnesium limitation induced several quorum-sensing proteins including the 3OC12-HSL synthase LasI (31) and the starvation and general stress-response regulator RelA, which negatively regulates synthesis of C4-HSL quorum-sensing signal (32, 33) (Table 1, Fig. 1). Furthermore, proteins of the quorum-sensing RhlR regulon, such as rhamnosyltransferase and enzymes of both P. aeruginosa phenazine antibiotic biosynthesis operons, were repressed during growth in low magnesium, consistent with the negative regulation of C4-HSL (Table 1, Fig. 1).
Figure 1.
Regulation of P. aeruginosa quorum sensing by growth in low magnesium. Proteins highlighted blue are induced; those highlighted red are repressed. SAM, S-adenosylmethionine; ACP, acyl-carrier protein; LasI, 3OC12-HSL synthase; RhlI, C4-HSL synthase; C4-HSL, N-butyryl-l-homoserine lactone; 3OC12, N-(3-oxododecanoyl)-l-homoserine lactone; LasR and RhlR, transcriptional regulators. Lines marked + or − denote positive or negative regulation, respectively.
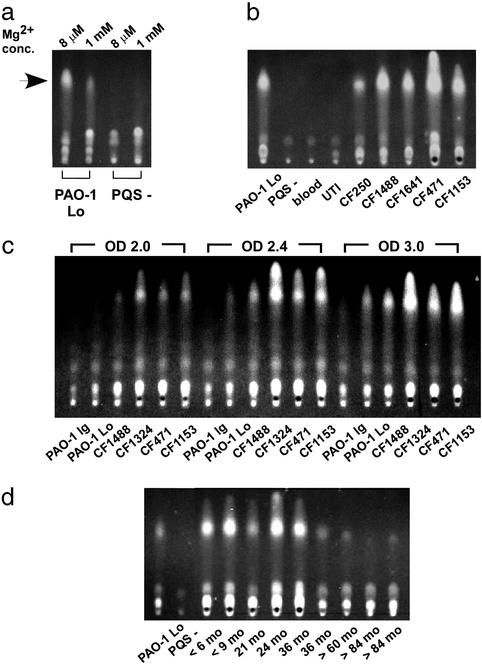
Recently, a non-HSL P. aeruginosa intercellular signal, 2-heptyl 3-hydroxy 4-quinolone (PQS), which is a part of the P. aeruginosa quorum-sensing hierarchy, has been described (25). A chromosomal region encoding five proteins (PA0996–1000) was recently shown to be essential for the production of PQS (21, 34). Four proteins encoded by this operon (PA0996-PA0999) were synthesized at increased levels when P. aeruginosa was grown in low magnesium (Table 1). This result suggested that levels of PQS could be increased by growth in low magnesium. To test this hypothesis, PQS produced by P. aeruginosa was extracted and analyzed by thin-layer chromatography. The amount of PQS was increased ≈5-fold when P. aeruginosa was grown in medium of low magnesium concentration (Fig. 2a). This result provided strong correlation between synthesis of PQS and abundance of proteins essential for its production.
Figure 2.
PQS production by P. aeruginosa. (a) PQS produced by wild-type PAO-1 Lo and a PQS-null mutant (PQS−) grown in N-minimal medium containing either 8 μM or 1 mM Mg2+. PQS is indicated by the arrow. (b) PQS production by P. aeruginosa isolates from five infants with CF and non-CF isolates from blood or urinary tract infection (UTI) grown in rich medium (TSB). (c) PQS production during growth in TSB by two laboratory P. aeruginosa strains (PAO-1 Ig and PAO-1 Lo) and isolates from four patients with CF. Absorbance of bacterial culture at 600 nm (OD) at which PQS was extracted is indicated. All strains had similar growth rates. Bacteria remained in logarithmic growth phase until OD 4.0, doubling every 120 min. (d) PQS production by sequential clonal P. aeruginosa isolates from a patient with CF grown in TSB. The patient's age in months (mo) is indicated.
Production of PQS Is Increased and Dysregulated in PA Isolates from Infant Patients with CF.
The P. aeruginosa quorum-sensing signals 3OC12-HSL and C4-HSL have been detected in CF patients' sputum, suggesting that corresponding regulatory mechanisms are active in CF lung-adapted bacteria (17, 29, 30). In that study, P. aeruginosa isolated from airways of patients of age 18 and older were in biofilm-like structures. The regulation of PQS synthesis by magnesium observed in this study suggested that, similarly to P. aeruginosa LPS modifications, PQS production might be increased in isolates from young children with CF. In addition, PQS might be important for early adaptation to the CF airway. Therefore, P. aeruginosa isolates from five 24- to 36-mo-old CF individuals and from patients with blood or urinary tract infections were compared when grown to stationary phase in rich medium (Fig. 2b). This analysis demonstrated that all five isolates from individuals with CF produced more PQS than isolates from other diseases. Furthermore, in four of five CF isolates, PQS production was increased relative to the laboratory strain PAO-1. These results suggested that the CF strains had accumulated mutations that led to dysregulated production of PQS irrespective of environmental conditions. To examine this possibility, PQS production during growth of P. aeruginosa in rich medium was analyzed. Although two laboratory strains of P. aeruginosa produced significant amounts of PQS only on entry into stationary phase, clinical P. aeruginosa isolates from all four CF patients tested produced 7- to 15-fold more PQS during logarithmic growth (Fig. 2c).
To further investigate PQS production by clinical P. aeruginosa, sequential clonal isolates collected over the first 8 years of life from a patient with CF were analyzed. In isolates from age <36 mo, the production of PQS was significantly increased when compared with the laboratory P. aeruginosa strain. In isolates from 36 mo and older, a significant reduction in amount of PQS was observed (Fig. 2d). These findings suggested that production of PQS could change in the course of bacterial persistence in the CF airway.
Discussion
This study describes a highly sensitive quantitative proteomic analysis of whole bacteria. Unlike more conventional proteome analysis, quantitative analysis such as ICAT enhances the possibility for identification of low abundance and membrane proteins (22, 35). In this study, abundance ratios of 546 proteins of P. aeruginosa grown in varying magnesium concentration were determined. Among 145 proteins regulated by growth in low magnesium, conserved magnesium-stress response proteins such as LPS modification enzymes were highly regulated. These results provide a strong functional correlation between LPS modification enzyme levels and the LPS structural modifications seen in this environmental condition, confirming the validity of proteomic analysis. Furthermore, although 72 of 145 regulated proteins showed relatively modest changes in their relative abundance (1.5- to 2.0-fold change), these shifts in abundance were apparently biologically significant. For example, proteins essential for PQS production were induced ≈1.5- to 2-fold (Table 1), but this increase in protein abundance had a significant impact on the amount of PQS produced when bacteria were grown in presence of low magnesium (Fig. 2a).
Although magnesium limitation slowed the growth of P. aeruginosa, our results show that the observed changes in protein abundance were more likely a consequence of this specific growth condition, rather than a result of nonspecific stress onto the bacterial cell. Both proteomic and transcriptional analyses showed that conserved magnesium-stress response factors such as regulator PhoP, the magnesium transporter homologue MgtA, and LPS modification enzymes were induced on magnesium limitation (Tables 1 and 5). Furthermore, steady-state levels of several stress and heat-shock response proteins such as catalase KatA and heat-shock proteins ClpA, ClpB, ClpP, and DnaJ were detected by ICAT and not demonstrated to be changed in abundance. In addition, the nutritional response to iron limitation was not induced, because enzymes of pyoverdine synthesis were at the steady-state levels, whereas pyochelin synthesis enzymes were repressed (see Table 7, which is published as supporting information on the PNAS web site).
Comparative ICAT analysis of P. aeruginosa whole cell and envelope protein revealed that 20 proteins were increased in abundance in the membrane fraction when P. aeruginosa was grown in magnesium-limited medium (Table 2). Nine of these proteins are soluble components of large membrane-associated enzyme complexes participating in energy metabolism pathways, whereas six proteins are components of the protein translation machinery. In the bacterial cell, gene expression and protein synthesis are temporally and spatially coupled with protein export. Escherichia coli membrane-associated polysomes almost exclusively translate membrane and transported proteins, whereas cytoplasmic proteins are typically synthesized by the cytoplasmic polysomes (36). The intracellular “shift” of ribosomal components to the envelope fraction suggests that the majority of protein synthesis occurs at the cytoplasmic membrane during P. aeruginosa growth in low magnesium. This likely enables the extensive envelope remodeling characteristic of the response to this environmental stress (6, 11). Furthermore, increased abundance of elongation factor G (Ef-G) in the Pseudomonas envelope fraction was observed in this study. Ef-G is one of several GTPases that are essential to dislodge the 50S ribosomal subunits from the 70S-translation termination complex (37). Thus, increased abundance of Ef-G at the membrane likely reflects increased polysome “recycling,” allowing for an increased rate of membrane protein synthesis during growth in low magnesium. Therefore, these data suggest that polysomes, components of general and specialized secretory pathways, cellular energy generators, and other enzymes form large membrane-associated supramolecular complexes during cellular adaptation to magnesium limitation.
Another interesting observation of this study was regulation of P. aeruginosa quorum-sensing proteins by low magnesium. Increased abundance of LasI and repression of Rhl regulon members such as enzymes for production of secreted factors suggested that the balance of synthesis of the quorum-sensing signals is shifted toward increased production of 30C12-HSL. Signaling via 3OC12-HSL is essential for development of P. aeruginosa biofilms (18), bacterial communities found within the CF airway. Therefore, it is possible that growth of P. aeruginosa under magnesium limitation simulates early stages of the lung invasion, before establishment of mature biofilms.
Growth in low magnesium also induced enzymes essential for production of a third, recently described quinolone signal PQS (25, 38, 39). PQS is integrated into the quorum-sensing regulons, because its production was shown to depend on LasR/3O-C12-HSL and it was shown to regulate expression of lasB encoding elastase (25). It has been proposed that PQS may act as an additional connecting signal between the Las and Rhl quorum-sensing systems (40). PQS concentrations were found to be highest in late stationary phase, suggesting that this molecule is not involved in cell-density sensing (40). However, several recent studies indicated that bacterial stress or starvation could induce quorum-sensing genes during earlier stages of P. aeruginosa growth (33). Results of this study suggest that PQS synthesis is induced during the logarithmic phase when P. aeruginosa is grown under magnesium limitation. Seemingly, P. aeruginosa quorum sensing in low magnesium is integrated with starvation/stress response through induction of regulators such as RelA. This study also demonstrated that synthesis of PQS signal by P. aeruginosa isolates from airways of infant CF patients was also induced during logarithmic phase of growth, even though bacterial cultures were grown in rich medium. These results suggest that an early induction of quorum sensing through the PQS signal might be selected for in P. aeruginosa isolates from CF patients in the first 3 years of life, similarly to what has been observed for magnesium-regulated proinflammatory LPS modifications.
Although our results provide evidence that some of the low magnesium-induced response factors are dysregulated in CF, it is unlikely that the bacteria adapted to the airway are expressing all low magnesium modulated responses in a dysregulated fashion. Growth in low magnesium promotes significant stress to the Gram-negative outer membrane (1) and, as shown by this study, induces general nutrient-starvation and stress responses that are likely integrated with quorum-sensing regulatory pathways in P. aeruginosa. These processes likely promote slower bacterial growth under magnesium limitation. Likewise, on host colonization, bacterial pathogens are subjected to nutritional stress and exposure to the host innate immune defense mechanisms. Among these are antimicrobial peptides that cause damage to the bacterial membrane. These and other factors likely promote induction of Gram-negative virulence genes both within the host and during growth of bacteria in the medium limited for magnesium. All of the properties induced during bacterial growth in low magnesium medium are not likely to be advantageous in the unique environment of the CF airway in which clonal isolates live for up to 30–40 years. The CF airway environment should select for a set of bacterial characteristics important for resistance to innate immune killing. Definition of these bacterial characteristics, such as production of PQS and LPS modifications, should provide valuable insight into chronic infectious processes and the pathogenesis of CF airway disease.
Supplementary Material
Acknowledgments
We thank Kimberly Lee, Greg Niemi, and Richard Newitt for help with assembling a functional microcapillary liquid chromatography–electrospray ionization-tandem MS system; Samuel Donohue and Michael Wright for help with protein labeling protocol; Priska Von Haller for help with data organization and analysis; Robert Ernst for communicating unpublished data; and David D'Argenio, Cammie Lesser, Robert Ernst, Martin Bader, Adeline Hajjar, Chris Wilson, Chandra Tucker, and Visvanathan Ramamurthy for critical reading of the manuscript. This work was supported by Cystic Fibrosis Foundation Grant MILLER00P0 and National Institutes of Health Grants R01 AI47938 (to S.I.M.) and R33CA93302-01 (to D.R.G. and R.A.).
Abbreviations
- CF
cystic fibrosis
- LPS
lipopolysaccharide
- PQS
Pseudomonas quinolone signal
- ICAT
isotope-coded affinity tag
- TSB
tryptic soy broth
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Nikaido H, Vaara M. Microbiol Rev. 1985;49:1–32. doi: 10.1128/mr.49.1.1-32.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia Vescovi E, Soncini F C, Groisman E A. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 3.Martinez de Tejada G, Miller J F, Cotter P A. Mol Microbiol. 1996;22:895–908. doi: 10.1046/j.1365-2958.1996.01538.x. [DOI] [PubMed] [Google Scholar]
- 4.Macfarlane E L, Kwasnicka A, Ochs M M, Hancock R E. Mol Microbiol. 1999;34:305–316. doi: 10.1046/j.1365-2958.1999.01600.x. [DOI] [PubMed] [Google Scholar]
- 5.Guo L, Lim K B, Poduje C M, Daniel M, Gunn J S, Hackett M, Miller S I. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 6.Ernst R K, Yi E C, Guo L, Lim K B, Burns J L, Hackett M, Miller S I. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- 7.Young M L, Bains M, Bell A, Hancock R E. Antimicrob Agents Chemother. 1992;36:2566–2568. doi: 10.1128/aac.36.11.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guina T, Yi E C, Wang H, Hackett M, Miller S I. J Bacteriol. 2000;182:4077–4086. doi: 10.1128/jb.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo L, Lim K B, Gunn J S, Bainbridge B, Darveau R P, Hackett M, Miller S I. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 10.Gunn J S, Lim K B, Krueger J, Kim K, Guo L, Hackett M, Miller S I. Mol Microbiol. 1998;27:1171–1182. doi: 10.1046/j.1365-2958.1998.00757.x. [DOI] [PubMed] [Google Scholar]
- 11.Bell A, Hancock R E. J Bacteriol. 1989;171:3211–3217. doi: 10.1128/jb.171.6.3211-3217.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajjar A M, Ernst R K, Tsai J H, Wilson C B, Miller S I. Nat Immunol. 2002;3:354–359. doi: 10.1038/ni777. [DOI] [PubMed] [Google Scholar]
- 13.Davis P B, Drumm M, Konstan M W. Am J Respir Crit Care Med. 1996;154:1229–1256. doi: 10.1164/ajrccm.154.5.8912731. [DOI] [PubMed] [Google Scholar]
- 14.Burns J L, Gibson R L, McNamara S, Yim D, Emerson J, Rosenfeld M, Hiatt P, McCoy K, Castile R, Smith A L, Ramsey B W. J Infect Dis. 2001;183:444–452. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- 15.Boucher J C, Yu H, Mudd M H, Deretic V. Infect Immun. 1997;65:3838–3846. doi: 10.1128/iai.65.9.3838-3846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mouton J W, den Hollander J G, Horrevorts A M. J Antimicrob Chemother. 1993;31:919–926. doi: 10.1093/jac/31.6.919. [DOI] [PubMed] [Google Scholar]
- 17.Singh P K, Schaefer A L, Parsek M R, Moninger T O, Welsh M J, Greenberg E P. Nature. 2000;407:762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 18.Davies D G, Parsek M R, Pearson J P, Iglewski B H, Costerton J W, Greenberg E P. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 19.Van Delden C, Iglewski B H. Emerg Infect Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whiteley M, Lee K M, Greenberg E P. Proc Natl Acad Sci USA. 1999;96:13904–13999. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.D'Argenio D A, Calfee M W, Rainey P B, Pesci E C. J Bacteriol. 2002;184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han D K, Eng J, Zhou H, Aebersold R. Nat Biotechnol. 2001;19:946–951. doi: 10.1038/nbt1001-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffin T J, Gygi S P, Ideker T, Rist B, Eng J, Hood L, Aebersold R. Mol Cell Proteom. 2002;1:323–333. doi: 10.1074/mcp.m200001-mcp200. [DOI] [PubMed] [Google Scholar]
- 24.von Haller P D, Donohoe S, Goodlett D R, Aebersold R, Watts J D. Proteomics. 2001;1:1010–1021. doi: 10.1002/1615-9861(200108)1:8<1010::AID-PROT1010>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 25.Pesci E C, Milbank J B, Pearson J P, McKnight S, Kende A S, Greenberg E P, Iglewski B H. Proc Natl Acad Sci USA. 1999;96:11229–11234. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calfee M W, Coleman J P, Pesci E C. Proc Natl Acad Sci USA. 2001;98:11633–11637. doi: 10.1073/pnas.201328498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gygi S P, Rist B, Gerber S A, Turecek F, Gelb M H, Aebersold R. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 28.Johnson C R, Newcombe J, Thorne S, Borde H A, Eales-Reynolds L J, Gorringe A R, Funnell S G, McFadden J J. Mol Microbiol. 2001;39:1345–1355. doi: 10.1111/j.1365-2958.2001.02324.x. [DOI] [PubMed] [Google Scholar]
- 29.Erickson D L, Endersby R, Kirkham A, Stuber K, Vollman D D, Rabin H R, Mitchell I, Storey D G. Infect Immun. 2002;70:1783–1790. doi: 10.1128/IAI.70.4.1783-1790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Middleton B, Rodgers H C, Camara M, Knox A J, Williams P, Hardman A. FEMS Microbiol Lett. 2002;207:1–7. doi: 10.1111/j.1574-6968.2002.tb11019.x. [DOI] [PubMed] [Google Scholar]
- 31.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B H, Greenberg E P. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whiteley M, Parsek M R, Greenberg E P. J Bacteriol. 2000;182:4356–4360. doi: 10.1128/jb.182.15.4356-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Delden C, Comte R, Bally A M. J Bacteriol. 2001;183:5376–5384. doi: 10.1128/JB.183.18.5376-5384.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallagher L A, McKnight S L, Kuznetsova M S, Pesci E C, Manoil C. J Bacteriol. 2002;184:6472–6480. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gygi S P, Corthals G L, Zhang Y, Rochon Y, Aebersold R. Proc Natl Acad Sci USA. 2000;97:9390–9395. doi: 10.1073/pnas.160270797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Randall L L, Hardy S J. Eur J Biochem. 1977;75:43–53. doi: 10.1111/j.1432-1033.1977.tb11502.x. [DOI] [PubMed] [Google Scholar]
- 37.Karimi R, Pavlov M Y, Buckingham R H, Ehrenberg M. Mol Cell. 1999;3:601–609. doi: 10.1016/s1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- 38.Pearson J P. J Bacteriol. 2002;184:2569–2571. doi: 10.1128/JB.184.10.2569-2571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diggle S P, Winzer K, Lazdunski A, Williams P, Camara M. J Bacteriol. 2002;184:2576–2586. doi: 10.1128/JB.184.10.2576-2586.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKnight S L, Iglewski B H, Pesci E C. J Bacteriol. 2000;182:2702–2708. doi: 10.1128/jb.182.10.2702-2708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.