Abstract
RNA interference represents an exciting new technology that could have therapeutic applications for the treatment of viral infections. Hepatitis C virus (HCV) is a major cause of chronic liver disease and affects >270 million individuals worldwide. The HCV genome is a single-stranded RNA that functions as both a messenger RNA and replication template, making it an attractive target for the study of RNA interference. Double-stranded small interfering RNA (siRNA) molecules designed to target the HCV genome were introduced through electroporation into a human hepatoma cell line (Huh-7) that contained an HCV subgenomic replicon. Two siRNAs dramatically reduced virus-specific protein expression and RNA synthesis to levels that were 90% less than those seen in cells treated with negative control siRNAs. These same siRNAs protected naive Huh-7 cells from challenge with HCV replicon RNA. Treatment of cells with synthetic siRNA was effective >72 h, but the duration of RNA interference could be extended beyond 3 weeks through stable expression of complementary strands of the interfering RNA by using a bicistronic expression vector. These results suggest that a gene-therapeutic approach with siRNA could ultimately be used to treat HCV.
RNA interference (RNAi) is a phenomenon in which small double-stranded RNA molecules induce sequence-specific degradation of homologous single-stranded RNA (1). In plants and insects, RNAi activity plays a role in host-cell protection from viruses and transposons (2, 3). From a practical perspective, RNAi is proving to be a very powerful technique to “knock down” specific genes to evaluate their physiological roles in Caenorhabditis elegans (1, 4), Drosophila melanogaster (5), and humans (6).
In plants and invertebrates, RNAi can be induced through transfection or microinjection of long double-stranded RNA (1, 7). The double-stranded RNA is cleaved into 19- to 23-nt RNA fragments known as small interfering RNAs (siRNAs) (8). siRNAs are incorporated into a ribonuclease enzyme complex known as the RNA-induced silencing complex (RISC). The antisense strand of siRNA within the RISC serves as a guide for sequence-specific degradation of homologous messenger RNAs. Only RNA molecules <30 bases in length can be used to exclusively induce RNAi in mammalian cells because longer molecules also activate the nonspecific double-stranded RNA-dependent response (9, 10). In plants and nematodes, RNAi activity is long-term and disseminates throughout the organism via an uncharacterized amplification mechanism. In mammalian cells, amplification activity seems absent, and interference activity is transient, lasting for only 3–5 days. More recently, DNA expression vectors have been developed to express hairpin or duplex siRNAs. These vectors employ the type III class of RNA polymerase promoters to drive the expression of siRNA molecules (11–14). In addition, stable cell lines containing siRNA expression plasmids have been produced to induce RNAi over longer durations (13, 15).
The potential of using RNAi activity for treatment of viral diseases and cancer has aroused a great deal of interest in the scientific community. Other laboratories have reported the use of RNAi activity in cultured cells infected with HIV, human papillomavirus, and polio or containing a variety of cancer genes (16–21). Hepatitis C virus (HCV) is a major health concern, and an estimated 3% of the world's population (270 million individuals) is chronically infected with this viral pathogen. It is estimated that 40–60% of infected individuals progress to chronic liver disease, and many of these patients ultimately require liver transplantation (22). Currently, the only treatment available for patients with chronic HCV infections consists of combination therapy with IFN and ribavirin. The standard therapy has a poor response rate (23), and thus there is a great need for the development of new treatments for HCV infections. Our laboratory has investigated the effect of RNAi activity on the replication of HCV by using the recently established replicon system (24–26). We have identified two siRNAs capable of dramatically reducing viral protein and RNA synthesis. In addition, we also have shown that RNAi can protect naive Huh-7 cells from challenge with replicon RNA. Finally, the duration of protective interference activity was extended beyond 3 weeks by expressing siRNAs from a bicistronic expression vector that replicates as an episome.
Materials and Methods
Cell Culture.
The cell line Huh-7 (27) was kindly provided by Stanley M. Lemon (University of Texas Medical Branch, Galveston) and were routinely grown in DMEM supplemented with 1× nonessential amino acids, 100 units/ml penicillin, 100 μg/ml streptomycin, 10% FCS (Wisent, Montreal). Cell lines carrying HCV replicons were grown in medium containing 800 μg/ml G418-active ingredient (geneticin, GIBCO/Invitrogen, Carlsbad, CA).
Construction of HCV Replicons and pCEP4-H1/H1 Expression Vector and Synthesis of siRNAs.
Plasmids pHCVrep1b BB7 (25) and p90/HCV FL-long pU (28) were provided by Charles M. Rice (Center for the Study of Hepatitis C, The Rockefeller University, New York). The plasmid pHCVrepAB12 was made by adding two additional adaptive mutations, E1202G and T1280I, (26) to the NS3-coding region and an additional 12 nucleotides to the HCV internal ribosome entry site (29). Sequence changes were made by using the QuikChange mutagenesis kit (Stratagene). One strand of each complementary pair of mutagenic primer is shown. Adaptive mutations E1202G and T1280I were introduced through mutagenesis of nucleotides A2330G and C2564T of the replicon sequence by using primers (5′-CCTGTGGAGAACCTAGGGACACCATGAGATCC-3′) and (5′-CCTAATATCAGGATCGGGGTGAGAACAATT-3′). The 12-nt insert was added by using the primer 5′-CCTCAAAGAAAAACCAAACGTAACACCAACGGGCGCGCCATGATTGAAC-3′. The negative control replicon pHCVrepAB12mut contain an aspartic acid-to-asparagine mutation in the NS5b polymerase coding sequence was made by using the primer 5′-CGATGCTCGTATGCGGAAACGACCTTGTCGTTATCTG-3′. pHCVrepAB12Luc was made by removing the neomycin gene from pHCVrepAB12 by digestion with AscI and PmeI and inserting the luciferase gene, which had been amplified from the plasmid pGL2 (Promega) by using standard techniques. The plasmid pCEP4d was made by digesting pCEP4 (Invitrogen) with PvuII and SnaBI and religating to remove the cytomegalovirus immediate early promoter element. A DNA insert encoding tandem H1 promoters driving the sense and antisense siRNAs 6367 and 6367 mismatch (mm) was made by PCR. A detailed description of the cloning method is available on request. All plasmid constructs were sequenced for confirmation. Synthetic siRNA duplexes described in Table 1 were obtained from Dharmacon. siRNAs 6188 mm and 6367 mm are negative control duplexes, each containing six nucleotide mismatches in the target sequence.
Table 1.
siRNAs used in this study
| siRNA | Target gene | siRNA sequences (sense strand only) (5′ to 3′) |
|---|---|---|
| 1,743 | EMCV-IRES | CGUCUAGGCCCCCGAACCACTT |
| 2,365 | NS3 | CUCGUCCCCUCCGGCCGUACCTT |
| 2,886 | NS3 | GGGGGGGAGGCACCUCAUUUUTT |
| 6,188 | NS5b | GGAGAUGAAGGCGAAGGCGUCTT |
| 6,367 | NS5b | GACACUGAGACACCAAUUGACTT |
| 6,793 | NS5b | GGGCAGAACUGCGGCUAUCGCTT |
| 6,188 mm | NA | GGACAUCUAGCGGUAGCCCACTT |
| 6,367 mm | NA | GAGAGUCAGUCAGCUAAUCACTT |
| DDB1 S | NA | AACAAGUCUCGUAUGUAGUGGTT |
EMCV, encephalomyocarditis virus; IRES, internal ribosome entry sites; NA, not applicable.
Production of Monoclonal Antibodies Against HCV 1a H77 NS4a/3 and NS5b.
The plasmids pETNS4A/NS3 and pETNS5B, containing the coding sequences for NS4A/NS3 and NS5B, were transformed into BL21 bacteria. The His-tagged proteins were purified (Amersham Pharmacia) and injected into mice. Hybridoma cell lines were produced and screened by using standard methods (30).
In Vitro Transcription.
HCV replicon RNAs were transcribed in vitro by using the T7-Megascript in vitro transcription kit (Ambion, Austin, TX) according to manufacturer instructions. After RNA synthesis, the DNA template was removed by three repeated digests with 0.2 units/μl DNase I enzyme at 37°C for 30 min.
Electroporation of HCV Replicon and siRNA and Selection with G418.
Cells were electroporated by using the protocol described by Lohmann et al. (24). Either 10 ng of HCVrepAB12neo/HCVrepAB12neomut replicon RNA or 10 μg of HCVrepAB12Luc/HCVrepAB12Lucmut RNA were electroporated into naive Huh-7 cells alone or with 100 nM siRNA. AB12-A2 cells were electroporated with 1 μM siRNA. Plasmid pcDNAluc (1 μg) was added to each sample to determine electroporation efficiency. If the cells were to be assayed for colony formation, they were transferred to 8 ml of DMEM and seeded into one 10-cm-diameter tissue-culture dish. Twenty-four hours later and every 3–4 days subsequently, the medium was replaced with fresh DMEM supplemented with 800 μg/ml G418 until colonies were visible. Colonies were fixed and stained with 0.1% gentian violet. To screen for luciferase expression, three 35-mm plates were seeded, each with 5% of the electroporated cells. At 3, 48, and 72 h postelectroporation, the cells were harvested and assayed for luciferase activity (Promega). The luciferase levels at 3 h postelectroporation were used to correct for transfection efficiency.
Transfection of Plasmid DNA into Huh-7 Cells.
Huh-7 cells were transfected with pCEP4d plasmids expressing siRNA 6367, siRNA 6367 mm, or with no insert. The plasmids were transfected into Huh-7 cells by using Lipofectamine 2000 (Invitrogen) and the suggested method. Medium containing 75 μg/ml hygromycin (Invitrogen) was added to the cells 24 h posttransfection.
RNA Purification and Northern Blot Analysis.
RNA samples were purified from Huh-7 cells by using Trizol reagent (Life Technologies, Invitrogen). Total RNA (5 μg) was treated with glyoxal and subjected to electrophoresis in a 0.9% agarose gel by using standard techniques (31). The gels were transferred to Hybond n+ nylon membrane (Amersham Pharmacia) and probed with 32P-labeled neomycin-resistance gene DNA that had been labeled by using the Ready-To-Go DNA-labeling kit (Amersham Pharmacia). An HCV sense strand-specific riboprobe was made by using HindIII linearized pHCVrepAB12 replicon plasmid as a template for use in the Riboprobe T7 system with [α-32P]UTP (Promega).
SDS/PAGE and Western Blot Analysis.
Equal numbers of naive or replicon-containing Huh-7 cells were lysed in SDS sample buffer 72 h after electroporation with siRNAs. Protein was electrophoresed on a polyacrylamide gel (Novex Invitrogen) and transferred to Hybond-C Extra-supported nitrocellulose membrane (Amersham Pharmacia). The blots were probed with monoclonal antibodies specific for NS3, NS5b, and actin by using standard methods. Proteins were visualized by using enhanced chemiluminescence (ECL, Amersham Pharmacia).
Results
Construction of the HCV Replicon Used in the Study.
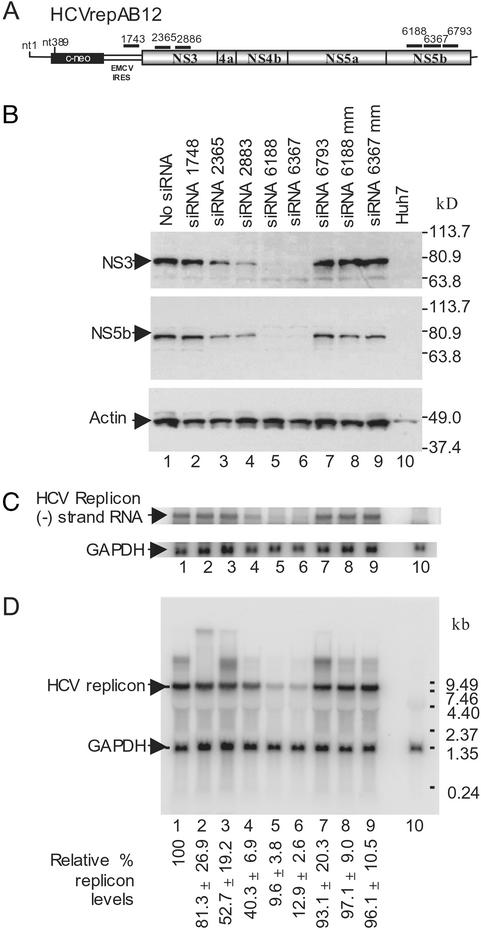
The design of the bicistronic HCV replicon used in this study is shown in Fig. 1A. The HCVrepBB7 replicon construct was obtained from Charles M. Rice and contained the adaptive mutation S2204I (25). We constructed an enhanced replicon construct by introducing two additional adaptive mutations, E1202G and T1280I (26), and extending the HCV internal ribosome entry site by 12 nucleotides (29). The enhanced replicon construct, HCVrepAB12, had a colony-forming efficiency of 1 × 105 colonies per μg of RNA, which is a 1,700-fold improvement over the efficiency of HCVrepBB7 in our hands. A G418-resistant cell clone, AB12-A2, was isolated, amplified, and screened for the presence of the replicon RNA and absence of replicon DNA by PCR.
Figure 1.
Synthetic siRNA directed inhibition of HCV replicon in Huh-7 cells. (A) Schematic diagram of the HCVrepAB12neo replicon RNAs showing the approximate locations of the siRNA target sequences. EMCV, encephalomyocarditis virus; IRES, internal ribosome entry site. (B) Western blot analysis of HCV nonstructural protein levels in AB12-A2 cells 72 h after electroporation. Samples were electroporated in the absence of siRNA (lane 1, no siRNA), with 1 of 6 HCV sequence-specific siRNAs (lanes 2–6), or with a control siRNA containing mismatched nucleotides (lanes 8 and 9, siRNA 6188 mm and siRNA 6367 mm). A sample from the parental Huh-7 cells is shown (lane 10). Blots were probed with monoclonal antibodies to either NS3 (Top) or NS5b (Middle). A third blot was probed with antiactin (Bottom) to control for protein loading. Protein size markers are shown on the right side. (C) Northern blot analysis of negative-strand HCV replicon RNA levels in siRNA-treated replicon cells 48 h after induction of RNAi. RNA was purified from a portion of the samples described for B. The Northern blot was probed with 32P-labeled (−) strand-specific riboprobe to detect negative strand HCV replicon RNA and a 32P-labeled GAPDH DNA to control for RNA loading. The locations of the HCV replicon (−) strand RNA and control GAPDH RNA are indicated. (D) Northern blot analysis of HCV replicon RNA levels in AB12-A2 cells 48 h after electroporation of siRNAs. The samples analyzed by Northern blot are identical to those described for B. RNA size markers are shown on the right side, and the RNA bands corresponding to HCV replicon RNA and GAPDH mRNA are indicated on the left side. HCV replicon and GAPDH RNA levels on Northern blots were quantitated by PhosphorImager (Molecular Dynamics) analysis. The average level of HCV RNA present in each sample is given below each lane as a percentage relative to the levels seen in cells not treated with siRNA. Percent SDs are given based on the average of three independent experiments.
RNAi Silences HCV Subgenomic Replication and Gene Expression.
Six siRNAs were designed to trigger RNAi through homology with specific regions of the HCV subgenomic replicon (Fig. 1A; Table 1). The effect of each siRNA on HCV protein and RNA levels was examined by Western and Northern blot analyses of samples of the HCV replicon cell line (AB12-A2) after electroporation. Of the six triggers tested, siRNAs 6188 and 6367 elicited the most potent effect. At 72 h postelectroporation, HCV nonstructural proteins NS3 and NS5b levels were below the detection limit by Western blot analysis (Fig. 1B, lanes 5 and 6), and the levels of replicon RNA were 9.6% and 12.9%, respectively, when compared with the levels of replicon RNA in control cells electroporated in the absence of siRNA (Fig. 1D, lane 1 vs. lanes 5 and 6). Electroporation of negative control siRNAs containing six mismatched nucleotides did not have any effect on the levels of HCV replicon RNA or HCV nonstructural proteins in AB12-A2 cells. These results indicate that the effects of the siRNAs on HCV replicon protein and RNA levels were sequence-specific and not caused by induction of nonspecific host-defense pathways (Fig. 1 B and C, lanes 8 and 9). Thus the effects of siRNAs on HCV protein and RNA levels seem to be the result of siRNA-directed degradation of the HCV replicon RNA by RISC. The other four siRNAs used in this experiment were less effective or had no significant effect (Fig. 1 B and D, lanes 2–4 and 7). The relative effectiveness of the siRNA trigger sequences was confirmed by quantification of HCV replicon RNA levels by real-time RT-PCR (data not shown). By using this method, the relative reduction in HCV replicon RNA levels was more dramatic, with a 99% and 94% decrease in replicon RNA levels after treatment with siRNA 6188 and siRNA 6367. RNAi also reduced the levels of negative-strand HCV replicon RNA (Fig. 1C), indicating that RISC endonuclease activity also may target and degrade the (−) strand of the HCV-replication intermediate.
HCV-Specific siRNA Protects Cells from Challenge with the HCV Subgenomic Replicon.
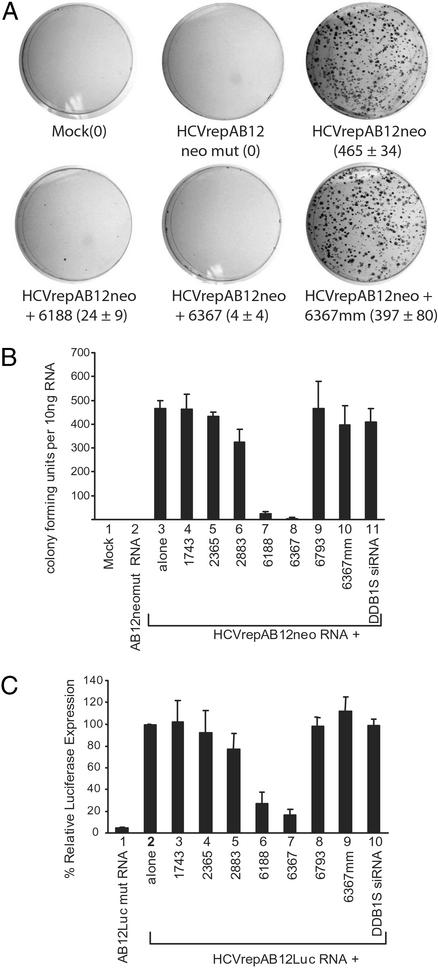
Electroporation of 10 ng of HCVrepAB12neo replicon RNA into Huh-7 cells resulted in the growth of ≈465 G418-resistant colonies (Fig. 2 A and B, lane 3). Triggering HCV-specific gene silencing by coelectroporation of siRNAs with replicon RNA caused a dramatic decrease in the number G418-resistant colonies when siRNAs 6188 and 6367 were used (Fig. 2 A and B, lanes 7 and 8). The efficacy of individual siRNAs mirrored the results seen in the previous experiments. Small interfering RNAs 6188 and 6367 were the most potent inhibitors and caused a 95% and 99% reduction, respectively, in the numbers of G418-resistant colonies formed. The siRNAs 1748, 2365, 2883, and 6793 had only marginal effects on colony formation (Fig. 2B, lanes 4–6 and 9). Control siRNAs, siRNA 6367 mm and DDB1S (a nonspecific siRNA), gave no significant reduction in G418-resistant colony formation (Fig. 2B, lanes 10 and 11). A control replicon in which the NS5B gene was mutated and rendered nonfunctional (HCVrepAB12neo-mut) could not produce G418-resistant colonies after electroporation into Huh-7 cells (Fig. 2 A and B, lane 2). Similar results were seen in a transient assay designed to measure the stability and replication of the HCV subgenomic replicon through the use of a luciferase assay (Fig. 2C). A luciferase reporter gene was inserted into the replicon RNA in place of the neomycin-resistance gene to produce HCVrepAB12Luc. Stability of the replicon was determined by measuring luciferase expression levels at 72 h postelectroporation of the various siRNAs. Again, the effects of the siRNA triggers on luciferase expression levels were similar to those seen in the previous assays (Fig. 2C). Small interfering RNAs 6188 and 6367 were the most potent inhibitors and led to a reduction in relative luciferase expression levels to 27% and 16%, respectively, compared with controls in which siRNA was absent (Fig. 2C, lanes 7 and 8 vs. lane 1). Control siRNA 6367 mm and DDB1S had no effects on luciferase expression (Fig. 2C, lanes 10 and 11). These data suggest that coelectroporation of replicon RNA together with certain siRNAs can induce strong RNAi activity. In the case of the most effective siRNA sequences, HCV replicon colony growth was abolished almost completely.
Figure 2.
RNAi protects cells from HCV replicon RNA replication. (A) Huh-7 cells were electroporated with 10 ng of HCVrepAB12neo alone (HCVrepAB12neo) or 10 ng of HCVrepAB12neo in a solution containing 100 nM of the indicated siRNA molecules (HCVrepAB12neo + siRNA). Negative control samples were electroporated with 10 ng of nonreplicating replicon RNA (HCVrepAB12neomut) or in the absence of RNA (Mock). Cells were selected in G418 until colonies were visible. The colonies were enumerated after staining. (B) The experiment described for A was repeated three times, and the average number of colonies that grew in each sample was plotted as a histogram. Error bars show the SDs of the average of three independent experiments. (C) The effect of siRNA on transient luciferase expression from an HCV replicon carrying the luciferase reporter gene. Huh-7 cells were electroporated with 10 μg of control nonreplicating luciferase replicon RNA (HCVrepAB12Lucmut), replicating replicon RNA (HCVrepAB12Luc) alone (alone), or in a solution containing the indicated siRNA. Luciferase levels were measured at 3 and 72 h postelectroporation. The levels at 3 h postinfection were used to estimate electroporation efficiencies. The luciferase levels measured at 72 h from the HCVrepAB12Luc RNA alone were defined as 100%, and the luciferase levels measured in the other samples are expressed as relative percentages. The data represent the average of three independent experiments, and error bars represent the SDs.
Duration of RNAi Activity on HCV Subgenomic Replicon Triggered by Synthetic siRNAs.
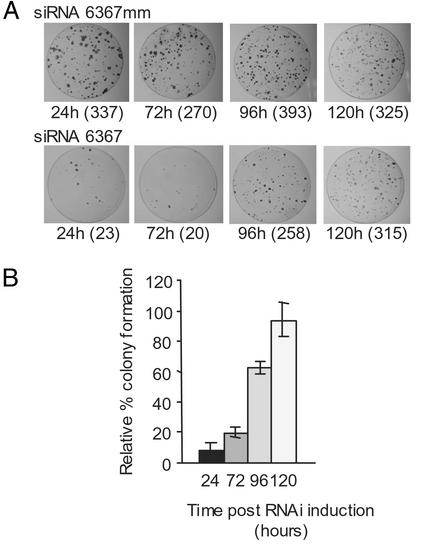
We investigated the duration of RNAi activity on the HCV replicon in Huh-7 cells by first introducing siRNA 6367 or control siRNA 6367 mm into Huh-7 cells by electroporation to induce RNAi, and then at various times after electroporation the cells were reelectroporated with 10 ng of HCVrepAB12 replicon RNA to assess the potency of interference activity at that particular time. Negative control cells were electroporated with siRNA 6367 mm (Fig. 3A Upper), and the numbers of colonies that formed (ranging between 270 and 393) did not change significantly over the duration of the experiment. The small variation in the colony numbers between each experiment reflected differences in electroporation efficiencies. However, in cells electroporated with siRNA 6367 RNA, the interference activity was strong at early times after induction and became weaker over time, as evidenced by the increase in numbers of G418-resistant colonies that grew when replicon RNA was electroporated 96 and 120 h after induction of interference (Fig. 3A Lower). The combined results of three replicate experiments are shown graphically (Fig. 3B). When HCV replicon RNAs were electroporated 24 or 72 h after induction of RNAi with siRNA 6367, the effect of gene silencing on the HCV subgenomic replicon was potent and caused a 92% and 80% reduction in the number of G418-resistant colonies (Fig. 3B, 24 and 72 h). However, at 96 h after introduction of siRNA 6367 to the cells, there was considerably less RNAi activity, and activity was insignificant by 120 h postinduction (Fig. 3B, 96 and 120 h). As has been seen in other mammalian systems, the effect of RNAi mediated by exogenously added synthetic siRNAs is short-lived and seemed to extend to 96 h in our experiments.
Figure 3.
The duration of RNAi activity triggered by synthetic siRNAs in Huh-7 cells. Huh-7 cells were electroporated with 100 nM of either siRNA 6367 or negative control siRNA 6367 mm. At 24, 72, 96, or 120 h after induction of interference, each population of cells was challenged with 10 ng of HCVrepAB12 replicon RNA and then grown in medium containing G418 until colonies were visible. (A) The colonies that grew after a time-course experiment are shown. (Upper) Time course of colony formation in cells electroporated with the negative control siRNA 6367 mm. (Lower) Time course of colony formation by cells electroporated with HCV-specific siRNA 6367. (B) A histogram showing the relative percentage of colony formation when HCV replicon RNA was electroporated 24, 72, 96, and 120 h after induction of RNAi. The number of G418-resistant colonies that grew from cells that had been electroporated with control siRNA 6367 mm was defined as 100%, and the number of colonies that formed on plates with cells treated with siRNA 6367 are plotted relative to this value. Data represent the averages of three experiments for each time point. Error bars reflect the SD from the average.
Prolonged Duration of RNAi by Bicistronic Plasmids Expressing Complementary siRNAs.
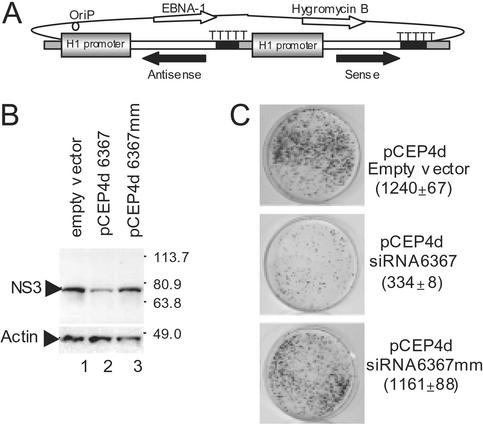
RNAi was induced in Huh-7 liver cells by transfecting cells with a vector that expressed complementary strands of an siRNA under control of two separate H1 promoters (Fig. 4). The plasmid pCEP4 (Invitrogen) was chosen to express the siRNA molecules because of its ability to replicate as a multicopy episome in mammalian cells. The cytomegalovirus promoter was removed from the pCEP4 to produce the plasmid pCEP4d to eliminate possible competition between the endogenous cytomegalovirus promoter and the H1 promoter. Tandem H1 promoters driving expression of sense and antisense siRNA sequences were inserted into pCEP4d as depicted in Fig. 4A. To test transient RNAi activity by pCEP4d expressing siRNA 6367, the plasmid pCEP4d6367 was transfected into AB12-A2 cells, and the level of viral NS3 protein was assessed at 72 h posttransfection by Western blot analysis (Fig. 4B). The level of NS3 was lower in cells containing pCEP4d6367 (Fig. 4B, lane 2) when compared with cells that were transfected with pCEP4d empty vector (Fig. 4B, lane 1) or with a plasmid expressing negative control siRNA, pCEP4d6367 mm (Fig. 4B, lane 3). This result indicated that the H1 promoter expressed complementary siRNA strands that induced RNAi directed against HCV. To demonstrate stable siRNA activity against HCV in Huh-7 cells, the plasmid pCEP4d6367, pCEP4d6367 mm, or empty vector was introduced into cells by transfection. Cell lines that contained the episomal expression vector were selected by using hygromycin for 3 weeks. After selection and expansion of the hygromycin-resistant cell lines, cells were electroporated with 100 ng of HCV subgenomic replicon RNA. Colonies that were resistant to G418 because of the presence of replicon were counted. HCV-specific RNAi activity was observed in the cell line containing pCEP4d6367 as evidenced by 70% fewer G418-resistant colonies (Fig. 4C) compared with control cell lines. Control cells contained the empty vector pCEP4d or the vector expressing siRNA pCEP4d6367 mm (Fig. 4C). Thus, potent HCV-specific RNAi activity was induced for an extended period of time by using cells that constitutively expressed siRNA molecules.
Figure 4.
Prolonging HCV-specific RNAi by using plasmid siRNA expression vectors. (A) A schematic diagram of the pCEP4d plasmid, which replicates in the cell as a multicopy episome and contains dual H1 promoters to drive expression of the complementary siRNA strands. (B) Western blot evaluation of the levels of HCV nonstructural protein NS3 in AB12-A2 cells. At 72 h posttransfection, AB12-A2 cells containing pCEP4d, pCEP4d6367, or pBS6367 mm were harvested for analysis by Western blot. Identical blots were probed with either anti-NS3 (Upper) or antiactin (Lower) that served as a control for protein loading. (C) Stable cell lines that expressed siRNA yielded 75% less HCV replicon-dependent colony growth. Huh-7 cells were transfected with pCEP4d empty vector (Top), pCEP4d6367 (Middle), or pCEP4d6367 mm (Bottom) and grown for 21 days in medium containing hygromycin. After selection, the cells were electroporated with 100 ng of HCVrepAB12neo replicon RNA. Colonies were grown in medium containing 800 μg/ml G418 for 14–20 days, fixed, and stained with gentian violet. The numbers of colonies on each plate are shown in parentheses. In each case, the number of colonies represents the average of two independent experiments with the SDs.
Discussion
RNAi represents an exciting new technology that could have applications in the treatment of viral diseases. Previous reports have shown that siRNAs directed against the HIV genome can effectively inhibit virus production in model cell-culture systems (1, 19, 20, 32). In addition, RNAi activity directed toward the major HIV receptor protein, CD4, led to decreased entry of HIV into cells (19). However, replication of HIV occurs through an integrated DNA genome, representing a situation where RNAi is ineffective in clearing the virus. On the other hand, the HCV genome is a (+) sense single-stranded RNA that functions as both the viral messenger RNA and a template for RNA replication via a negative-strand intermediate (33). This situation suggests that HCV could be a particularly attractive target for RNAi therapy that could eliminate viral RNA from the infected cell and potentially cure a patient of hepatitis. We have demonstrated that HCV replicon RNA is susceptible to RNAi in a human hepatoma cell line (Huh-7). Introduction of two different siRNAs into target cells that contained HCV replicon RNA caused a dramatic decrease in the levels of viral proteins and RNA. This effect was likely due to the degradation of HCV messenger RNA by the RISC endonuclease. HCV-specific RNAi activity also led to a reduction in the levels of HCV (−) strand replication intermediate RNA and allows for the possibility that replicating HCV RNA may also be susceptible to degradation by RISC. We do not know the effect of RNAi on HCV immediately after virus entry into cells, because an efficient cell-culture system for growth of HCV is not available at this time. However, we have shown that up to a 99% reduction in the efficiency of HCV replicon colony formation when interference activity was induced concurrent with replicon RNA entry into cells. Thus RNAi protects cells from “infection” by HCV replicon RNA. Because the early events of an HCV infection include translation of the newly uncoated genomic RNA, it is likely that the viral RNA will also be susceptible to RNAi at this time. However, this remains to be determined.
The efficacy of each of the six siRNAs that were designed to target different regions of the HCV replicon RNA varied greatly, which is in agreement with siRNAs targeted to other genes (19). The reasons that certain siRNAs did not induce HCV-specific RNAi are not known, but one could speculate several possibilities. SiRNAs that are inefficient in RNAi response may target regions of RNA that are inaccessible to RISC because of secondary structure, protein binding, or both. Alternatively, these siRNAs may not form RISCs that are productive in eliciting RNAi.
Because of the great variability in RNA sequences between different quasispecies and genotypes of HCV, for therapeutic applications it may be necessary to include several different combinations of siRNA to target a particular region of the genome. In addition, the high mutation rate of HCV that is apparent during replication makes the appearance of escape mutants from RNAi a distinct possibility, as was seen for poliovirus (16). However, the development of viral resistance to RNAi may not merely be limited to the production of escape mutants through sequence divergence. Many plant viruses (2, 34) and at least one animal virus (35) synthesize gene products that seem to block RNAi activity. Whether HCV possesses such an activity remains to be determined (35).
The utility of siRNA as a therapy against HCV infection will depend on the development of efficient delivery systems that induce long-lasting RNAi activity. HCV is an attractive target for its localization in the liver, an organ that can be readily targeted by nucleic acid molecules and viral vectors. In the future, chemically modified synthetic siRNAs, with improved resistance to nucleases coupled with enhanced duration of RNAi, may become a possibility for therapeutic applications. On the other hand, gene therapy offers another possibility to express siRNAs that target HCV in a patient's liver. Our laboratory has produced cells that exhibit stable RNAi directed against a virus. We constructed a self-contained episomal expression vector that contains the oriP origin of replication, a coding sequence for EBNA1 protein that is required for episome maintenance, and two H1 tandem promoters that drive the synthesis of each of the siRNA strands. This expression vector extended the duration of RNAi activity to 3 weeks. Others laboratories have observed long-acting RNAi through the establishment of stable cell lines that constitutively express specific siRNAs (13, 15, 36). Two recent reports have described the use of recombinant adenoviruses and retroviruses to deliver and express siRNA in culture. The adenovirus was also used to deliver siRNAs to the livers of mice (37, 38). Similar vectors could eventually be used from a prophylactic or therapeutic standpoint to evaluate the effects of siRNA on HCV replication in model systems such as chimpanzees and mice with chimeric human livers (39). Based on the experiments presented in this article, the use of siRNA as a treatment for HCV infections has great potential for use alone or in combination with conventional IFN/ribavirin therapy as a means to decrease virus loads and eventually clear the persistent virus from its host.
Acknowledgments
We thank Dr. Charles M. Rice for providing the plasmids pHCVrep1bBB7 and p90/HCV FL-long pU. This work was supported by Canadian Institutes of Health Research Grant EOP-38155 and CanVac (Canadian National Centres of Excellence).
Abbreviations
- RNAi
RNA interference
- siRNA
small interfering RNA
- RISC
RNA-induced silencing complex
- HCV
hepatitis C virus
- mm
mismatch
References
- 1.Hannon G J. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 2.Vance V, Vaucheret H. Science. 2001;292:2277–2280. doi: 10.1126/science.1061334. [DOI] [PubMed] [Google Scholar]
- 3.Cullen B R. Nat Immunol. 2002;3:597–599. doi: 10.1038/ni0702-597. [DOI] [PubMed] [Google Scholar]
- 4.Kim S K. Nat Rev Genet. 2001;2:681–689. doi: 10.1038/35088523. [DOI] [PubMed] [Google Scholar]
- 5.Clemens J C, Worby C A, Simonson-Leff N, Muda M, Maehama T, Hemmings B A, Dixon J E. Proc Natl Acad Sci USA. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McManus M T, Sharp P A. Nat Rev Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- 7.Zamore P D. Science. 2002;296:1265–1269. doi: 10.1126/science.1072457. [DOI] [PubMed] [Google Scholar]
- 8.Elbashir S M, Lendeckel W, Tuschl T. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ui-Tei K, Zenno S, Miyata Y, Saigo K. FEBS Lett. 2000;479:79–82. doi: 10.1016/s0014-5793(00)01883-4. [DOI] [PubMed] [Google Scholar]
- 10.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 11.Yu J Y, DeRuiter S L, Turner D L. Proc Natl Acad Sci USA. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sui G, Soohoo C, Affar el, B, Gay F, Shi Y, Forrester W C. Proc Natl Acad Sci USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyagishi M, Taira K. Nat Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- 14.Paul C P, Good P D, Winer I, Engelke D R. Nat Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- 15.Paddison P J, Caudy A A, Hannon G J. Proc Natl Acad Sci USA. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gitlin L, Karelsky S, Andino R. Nature. 2002;418:430–434. doi: 10.1038/nature00873. [DOI] [PubMed] [Google Scholar]
- 17.Jiang M, Milner J. Oncogene. 2002;21:6041–6048. doi: 10.1038/sj.onc.1205878. [DOI] [PubMed] [Google Scholar]
- 18.Jacque J M, Triques K, Stevenson M. Nature. 2002;418:435–438. doi: 10.1038/nature00896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novina C D, Murray M F, Dykxhoorn D M, Beresford P J, Riess J, Lee S K, Collman R G, Lieberman J, Shankar P, Sharp P A. Nat Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 20.Coburn G A, Cullen B R. J Virol. 2002;76:9225–9231. doi: 10.1128/JVI.76.18.9225-9231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borkhardt A. Cancer Cell. 2002;2:167–168. doi: 10.1016/s1535-6108(02)00129-0. [DOI] [PubMed] [Google Scholar]
- 22.Kim W R. Hepatology. 2002;36:S30–S34. doi: 10.1053/jhep.2002.36791. [DOI] [PubMed] [Google Scholar]
- 23.McHutchison J G. J Gastroenterol Hepatol. 2002;17:431–441. doi: 10.1046/j.1440-1746.2002.02777.x. [DOI] [PubMed] [Google Scholar]
- 24.Lohmann V, Korner F, Dobierzewska A, Bartenschlager R. J Virol. 2001;75:1437–1449. doi: 10.1128/JVI.75.3.1437-1449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blight K J, Kolykhalov A A, Rice C M. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 26.Krieger N, Lohmann V, Bartenschlager R. J Virol. 2001;75:4614–4624. doi: 10.1128/JVI.75.10.4614-4624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 28.Kolykhalov A A, Agapov E V, Blight K J, Mihalik K, Feinstone S M, Rice C M. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 29.Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 30.Kohler G, Milstein C. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 31.Brown T, Mackey K. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Vol. 1. Hoboken, NJ: Wiley; 1997. pp. 4.9.1–4.9.16. [Google Scholar]
- 32.Lee N S, Dohjima T, Bauer G, Li H, Li M J, Ehsani A, Salvaterra P, Rossi J. Nat Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 33.Major M E, Rehermann B, Feinstone S M. In: Fields Virology. Knipe D M, Howely P M, editors. Vol. 1. Philadelphia: Lippincott; 2001. pp. 1127–1161. [Google Scholar]
- 34.Li W X, Ding S W. Curr Opin Biotechnol. 2001;12:150–154. doi: 10.1016/s0958-1669(00)00190-7. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Li W X, Ding S W. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 36.Brummelkamp T R, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 37.Barton G M, Medzhitov R. Proc Natl Acad Sci USA. 2002;99:14943–14945. doi: 10.1073/pnas.242594499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia H, Mao Q, Paulson H L, Davidson B L. Nat Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 39.Mercer D F, Schiller D E, Elliott J F, Douglas D N, Hao C, Rinfret A, Addison W R, Fischer K P, Churchill T A, Lakey J R, et al. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]