Abstract
Helicobacter pylori elicits an oxidative stress during host colonization. This oxidative stress is known to cause lesions in the host DNA. Here we addressed the question as to whether the pathogen DNA is subject to lethal or mutational damage by the host-generated oxidative response. H. pylori Hpnth mutants unable to repair oxidized pyrimidines from the bacterial DNA were generated. H. pylori strains lacking a functional endonuclease III (HpNth) showed elevated spontaneous and induced mutation rates and were more sensitive than the parental strain to killing by exposure to oxidative agents or activated macrophages. Although under laboratory conditions the Hpnth mutant strain grows as well as the wild-type strain, in a mouse infection the stomach bacterial load gradually decreases while the population in the wild-type strain remains stable, showing that endonuclease III deficiency reduces the colonization capacity of the pathogen. In coinfection experiments with a wild-type strain, Hpnth cells are eradicated 15 days postinfection (p.i.) even when inoculated in a 1:9 wild-type:mutant strain ratio, revealing mutagenic lesions that are counterselected under competition conditions. These results show that the host effectively induces lethal and premutagenic oxidative DNA adducts on the H. pylori genome. The possible consequences of these DNA lesions on the adaptability of H. pylori strains to new hosts are discussed.
The well recognized pathogen Helicobacter pylori chronically infects up to 50% of the world's human population. It is a Gram-negative, microaerophilic bacterium associated with gastritis, peptic ulcer, and gastric cancer (1, 2). H. pylori induces an oxidative stress that enhances DNA damage in the host's gastric mucosa (2–5). Although postulated, oxidative host-generated lethal or premutagenic DNA damage on H. pylori DNA has not been demonstrated (6–8). Superoxide dismutase-deficient mutants have an impaired ability to colonize the mouse stomach, confirming that this pathogen is exposed to the toxic effect of oxygen-derived products during infection (6). The fact that superoxide dismutase is associated with the bacterial surface is consistent with its action as a primary barrier for exogenous reactive molecules. These results, however, do not allow the identification of the targets on which host generated reactive oxygen species (ROS) act to interfere with the colonization process. The hypothesis that DNA bacterial damage constitutes a host defense mechanism has not, to our knowledge, been experimentally tested with any host–pathogen couple. Legionella pneumophila and Salmonella typhimurium strains deficient in DNA recombination enzymes present an impaired mice colonization capacity (9, 10, 11). Given the pleiotropic effects of the mutations tested, it is not possible to rule out an intrinsic impaired growth capability of the deficient strains or to determine the type of DNA lesions that cause the pathogen lethality during infection.
The most abundant kind of DNA lesions resulting from oxidative stress are base derivatives (12). Guanines are especially susceptible to damage by ROS yielding mainly 8-oxoguanine, a strongly premutagenic lesion (13). In contrast, pyrimidine residues are chemically more resistant, but some of their oxidation products, such as 5,6-dihydroxydihydrothymine [known as thymine glycol], can entail lethal consequences for the cell (13). This lesion can block both DNA and RNA polymerases (14, 15). Other common pyrimidine derivatives are 5,6-dihydrothymine and urea (13). In Escherichia coli, endonuclease III (EndoIII) is the enzyme responsible of excising these lethal or mutagenic pyrimidine lesions (16, 17). In addition to its DNA glycosylase activity, all of the EndoIII enzymes described possess an abasic (AP) lyase activity that cleaves the DNA backbone 3′ of the AP site. EndoIII from E. coli is coded by the nth gene. nth single mutants exhibit a weak mutator phenotype but are not hypersensitive to DNA-damaging agents able to produce the substrates for this enzyme (17). However, nth nei double mutants show a strong mutator phenotype and are hypersensitive to H2O2 and x-rays (18, 19). The product of nei is EndoVIII, a DNA glycosylase that shares a common range of substrates with Nth (19).
From the analysis of the complete genomic sequence of H. pylori, two ORFs were assigned as coding for Nth homologs, hp0585 and hp0602 (20, 21). Both gene products have the same level of similarity to other EndoIII proteins. We have previously demonstrated (22) that hp0602 actually codes for a 3-methyladenine-DNA glycosylase, MagIII. Here we show that the product from ORF hp0585 codes for the H. pylori Nth. Its specific and essential role in the maintenance of the genetic information and the clearance of the products of genotoxic agents was established by the analysis of H. pylori strains in which endoIII was disrupted. The impaired ability of these strains to remove oxidized pyrimidines from their DNA led to a reduced colonization capacity in a mouse infection model. These results show that the environment in the host during the colonization process is generating oxidative DNA lesions that can affect the survival and genetic information of H. pylori.
Materials and Methods
Materials and Reference Compounds.
All chemicals were, unless otherwise stated, from Sigma. [32P]ATP (3,000 Ci/mmol; 1 Ci = 37 GBq) and Nensorb-20 cartridges were from New England Nuclear. T4 polynucleotide kinase, T4 DNA ligase, and restriction enzymes were from New England Biolabs.
Bacterial Strains, Media, and Plasmids.
E. coli strain KL16 (thi, relA1, Hfr, zed∷Tn10) and its derivative, SW2-38 (KL16 nth∷Kan, Δnei∷Cm) (19), were grown in LB broth or agar supplemented with 200 μg/ml ampicillin and/or 100 μg/ml rifampicin. The H. pylori strains used were X47-2AL (23), its derivative, X47-2AL endoIII (this work), ADM1, and 13/5 (24). H. pylori strains were grown on blood or horse serum Brucella agar plates (HBAP) at 37°C in a 5% CO2/95% humidity atmosphere. Disruption mutant H. pylori strains were cultured with 20 μg/ml kanamycin. Mutagenesis assays were made on HBAP supplemented with 20 μg/ml rifampicin. Plasmid pGEX-4T-1 was obtained from Amersham Pharmacia Biotech.
Plasmid Constructions.
The Hpnth genes with their own start codons were amplified by PCR from two independent H. pylori isolates (ADM1 and 13/5), using oligonucleotide primers deduced from the 26695 genome sequence (hp0585). The amplified DNA products were cloned in pGEX-4T-1 previously digested with the restriction enzymes BamHI and SalI, resulting in plasmids pGEX-HP585A and pGEX-HP585T, respectively. In this system, the HP0585 polypeptide is produced as a fusion protein with GST at its N terminus.
Complementation Assays of E. coli SW2-38.
SW2-38 was electrotransformed with pGEX-4T-1, pGEX-HP585A, or pGEX-HP585T, and wild-type KL16 with pGEX-4T-1. For mutagenesis assays, overnight cultures were diluted to start ten 2-ml cultures from each genotype with inoculae of 1,000–5,000 cells per ml. The cultures were grown at 37°C to mid-log phase and isopropyl β-d-thiogalactoside (IPTG) was added to 50 μM. After overnight incubation, 200 μl of the cultures were plated onto LB rifampicin plates and serial dilutions were plated onto LB plates. For survival assays, five overnight cultures in HBAP, of SW2-38pGEX, SW2-38 pGEX-HP585A, SW2-38 pGEX-HP585T, and KL16pGEX were pelleted, washed in PBS, resuspended, and divided in five aliquots. Each aliquot was treated for 10 min with 0, 10 50, 250, or 500 mM H2O2. The suspensions were serially diluted and plated onto LB-agar.
Overproduction and Purification of EndoIII.
E. coli SW2-38 carrying the pGEX or pGEX-HP585 plasmids were used to inoculate 500 ml of LB medium supplemented with ampicillin and were incubated at 37°C with shaking until the A600 reached 1.5. IPTG was added to 50 μM, and growth was continued overnight at 18°C with gentle shaking. Cells were harvested by centrifugation, and HpNth was purified by GST affinity chromatography. The eluted protein was dialyzed against 20 mM Tris⋅HCl (pH 8)/5 mM β-mercaptoethanol/10% glycerol.
DNA Substrates.
The 34-mer oligodeoxyribonucleotides used in this study have the following sequence: 5′-GGCTTCATCGTTGTC[X]CAGACCTGGTGGATACCG-3′ with X being dihydrothymine (dHT) or AP residues. The dHT oligonucleotide was a kind gift from J. Cadet (Commissariat à l'Energie Atomique, Grenoble, France). Uracil-containing oligonucleotide and its complementary strand with each of the four bases opposite the lesion in the duplex were purchased from Oligo Express (Paris). The AP site was obtained by treatment of the uracil-containing oligonucleotide with uracil DNA glycosylase.
Enzymatic Activity Assays.
For the cleavage of lesion-containing DNA duplexes, the oligonucleotide carrying the modified base was 32P-labeled at the 5′ end and annealed to its complementary strand. In a standard reaction (10-μl final volume), 50 fmol of labeled duplex were incubated in the reaction buffer (25 mM Tris⋅HCl, pH 7.6/2 mM Na2EDTA/50 mM NaCl) with the indicated protein fraction at 37°C. Reactions were stopped by addition of 0.2 M NaOH, unless otherwise stated. After addition of 6 μl of formamide dye the products were separated by 7 M urea/20% PAGE. Gels were analyzed by autoradiography.
Construction of an H. pylori Mutant Deficient in EndoIII.
The hp0585 ORF with its flanking sequences was amplified by PCR from the genomic DNA from strain 26695 by using the following primers: F-585 5′-CAUCAUCAUGAAAGCGTGTGAAAATGGGTT-3′ and R-585 5′-CUACUACUACCACTACGCTTTAAAGCTAGC-3′. The cloned ORF was disrupted in E. coli by insertion into the recombinant plasmid of a transposable element (MiniTn3-Km) as described (24). Twenty-six independent disruptions of the pILL570-hp0585 plasmid were pooled and introduced by natural transformation into H. pylori strain X47. Insertions in hp0585 were checked by PCR using the F-585 primer together with the Km-1 or Km-2 from the kanamycin cassette (24). Each PCR fragment was directly sequenced. Independent mutants with TnKm insertions in hp0585 were further used as individual mutants or pooled.
Frequency of Spontaneous Mutations to Rifampicin Resistance.
Ten independent cultures each of wild-type and Hpnth H. pylori were initiated with 2 × 104 cells per culture and grown for 48 h in HBAP agar. After resuspension in peptone broth, serial dilutions on the suspensions were performed. One hundred-microliter aliquots were plated HBAP with or without rifampicin. Colonies on both selective and nonselective plates were counted after incubation for 4 days. The frequency of resistant mutants was calculated using the median method.
H. pylori Sensitivity to Chemical Oxidative Stress.
Five independent, 24-h cultures of wild-type and Hpnth-deficient H. pylori were harvested, washed, and resuspended in PBS to give a final concentration of 2.5 × 109 cells per ml [estimation, OD600 ≅ 0.1 as 2.5 × 108 colony-forming units (cfu)]. Ten microliters of cell suspension was serially diluted and plated to obtain the viable cfu. Subsequently, menadione and fresh hydrogen peroxide were added to yield the indicated final concentrations. The cells were washed 15 min later, diluted, and plated on HBAP. Plates were incubated for 5 days and colonies were counted.
Macrophages Experiments.
The mouse macrophage J774A.1 cells (TIB-67, American Type Culture Collection) were grown in RPMI medium supplemented with 10% heat-inactivated FCS, 2 mM l-glutamine, and 0.1% NaHCO3 at 37°C in a humidified 5% CO2 atmosphere for 2–3 days. Cells were harvested by trypsin-EDTA treatment. Macrophages (2.5–5 × 106) in 1 ml of medium were placed in each of the 24 wells of a flat-bottom plate. After overnight growth, macrophages were activated with 1 μg/ml phorbol 12-myristate 13-acetate (PMA) and 0.1 μg/ml lipopolysaccharide. After 36–48 h, wild-type and Hpnth-deficient H. pylori cultures were harvested and PBS washed, and the number of bacteria was estimated as above. Wild-type and endoIII-deficient cells were resuspended in supplemented RPMI medium at 2.5–5 × 108 cfu/ml. Cocultures were initiated by addition of 1 ml of bacterial suspension. The actual bacterial load was determined by plating. After 2 and 4 h, the H. pylori-containing medium was harvested, wells were washed twice with PBS, and the washes were added to the harvest. H. pylori cells were centrifuged, washed, diluted if necessary, and plated in duplicate basic, kanamycin, and/or rifampicin plates.
Experimental Infections.
Six- to 8-week-old Swiss specific pathogen-free mice (R. Janvier, Centre d'Elevage, Le Genest St. Isle, France) were fed a commercial pellet diet and water ad libitum. Mice were inoculated intragastrically once with 100 μl of suspensions of 108 cfu/ml H. pylori consisting of strain X47-2AL (n = 15), strain X47Hpnthpool (n = 15), a mix of strains X47-2AL and X47Hpnthpool in a 1:9 ratio (n = 30), or a mix of strains X47-2AL and X47Hpnthpool in a 9:1 ratio (n = 30). Control groups of mice (n = 6) were given peptone trypsin broth alone. Mice were killed 15, 30, and 60 days after inoculation [noninfected (n = 2) and H. pylori-infected (n = 5 and 10 for mono- and coinfection, respectively) at each time point], and stomachs were removed for assessment of colonization by H. pylori. The presence of H. pylori bacteria was determined by a biopsy urease test (25) performed on half of each stomach and by quantitative culturing of the remaining gastric tissues. Viable counts of H. pylori were estimated by serial dilutions of the homogenized tissues in peptone broth and plating in parallel on 2.0% agar standard selective plates (26) and selective plates supplemented with kanamycin.
Results
Expression of H. pylori hp0585 Complements the E. coli nth nei DNA Repair Phenotypes.
Two ORFs, hp0602 and hp0585, were annotated as putative EndoIII coding genes in the complete genomic sequence of H. pylori (21, 22). We have recently shown (22) that hp0602 actually codes for a 3-methyladenine glycosylase activity, Mag III. hp0585 from two independent isolates, 13/5 and ADM1, was amplified and cloned in the E. coli expression vector pGEX. Sequencing of the hp0585 ORF and comparison of the putative proteins from 13/5 and ADM1 and that from the sequenced 26695 strain show that, for any pair, there are at least five amino acid differences among the 218 residues. Moreover, there is a two-codon deletion in hp0585 from strain 13/5. Therefore, the analysis of both clones was carried out in parallel. The cloned genes derived from H. pylori 13/5 and from ADM1 were named hp585T and hp585A, respectively.
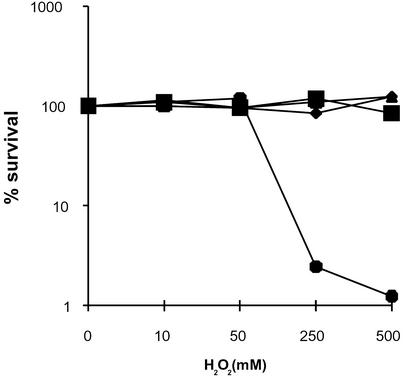
E. coli SW2-38 is hypersensitive to oxidizing agents because of null mutations in both nth and nei genes (19). Independent transformants of SW2-38 for each plasmid (pGEX, pGHP585A, or pGHP585T) and the isogenic wild-type strain KL16 transformed with pGEX were grown on horse serum-containing plates. After harvesting, wild-type and mutant E. coli strains were challenged with increasing concentrations of hydrogen peroxide before spreading on LB plates to calculate the survival fraction. Given the level of resistance to H2O2 observed, it is clear that, even after washing, horse serum provides an extra protection against reactive oxygen species (ROS). Fig. 1 shows that both alleles of hp0585 are able to restore the wild-type resistance to H2O2 in the nei, nth double mutant. Moreover, production of either of the hp0585-encoded proteins partially complements the mutator phenotype characteristic of the SW2-38 strain (Table 1). These results suggest that ORF hp0585 encodes a protein with an EndoIII-like activity.
Figure 1.
Sensitivity to H2O2 of a nei, nth E. coli mutant strain expressing hp0585. Survival curves for E. coli SW2-38 (nei, nth) harboring pGEX-4T1 (●), pGEXHP585A (♦), or pGEXHP585T (▴) and the parental wild-type strain transformed with pGEX-4T1 (■). Bacteria were precultured in H. pylori medium and incubated for 10 min with the indicated amounts of hydrogen peroxide. Values are the mean of n = 5 for each genotype and H2O2 concentration and are representative of two independent experiments.
Table 1.
Mutation frequencies of wild-type and SW2-38 (nei, nth) E. coli expressing hp0585
| Strain | Spontaneous mutation frequency of
rifR per 108cfu
|
|
|---|---|---|
| Experiment 1 | Experiment 2 | |
| Wild-type pGEX | 2.0 | 1.6 |
| SW2-38 pGEXHP585A | 6.7 | 5.8 |
| SW2-38 pGEXHP585T | 6.9 | 6.3 |
| SW2-38 pGEX | 16.3 | 19.9 |
Each culture was initiated by inoculating with 2,000–10,000 cells. rifR, rifampicin-resistant colonies.
hp0585 Codes for a Protein with EndoIII Activity.
Although the overall identity between the predicted protein product of hp0585 and E. coli Nth is low, it is possible to identify in the putative HpNth the pattern of cysteines giving rise to the iron–sulfur cluster and the helix–hairpin–helix motif, characteristic of the EndoIII family of proteins (27, 28). To examine whether H. pylori hp0585 codes for a protein with EndoIII activity, hp585A or hp585T fused to GST were expressed in E. coli SW2-38. A DNA glycosylase activity on dHT-containing oligonucleotides was detected in the lysates of cells expressing GST fused to either of the hp0585 variants, but was absent in extracts from the same strain transformed with the vector pGEX (data not shown).
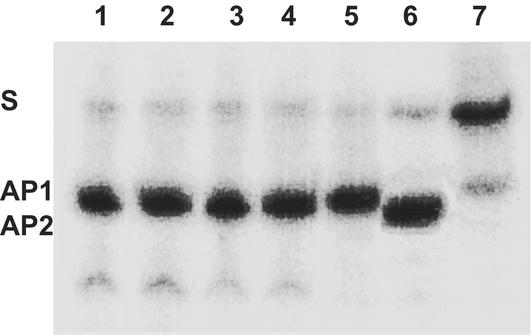
Both GST-HP585 fusion proteins overproduced in SW2-38 were purified to yield a single 53-kDa Coomassie blue band in SDS/PAGE (data not shown). The fusion proteins can efficiently cleave the dHT-containing oligonucleotide substrate at the lesion site even without a NaOH treatment of the reaction products (Fig. 2), showing that, in addition to a dHT DNA glycosylase activity, the hp0585 products have an associated AP lyase activity. By comparison to an EndoIV (Nfo) hydrolyzed tetrahydrofuran-containing oligonucleotide, it was confirmed that the hp0585 product is that of an AP lyase. These results, together with the genetic complementation assays, showed that hp0585 indeed codes for the H. pylori Nth.
Figure 2.
H. pylori HpNth cleavage activity on a dihydrothymine 34-mer. A dHT-containing 34-mer (lanes 1–5 and 7) was incubated with 10 ng of HpNth from 13/5 (lanes 1 and 2) or ADM1 (lanes 3 and 4), Nth from E. coli (lane 5), or reaction buffer alone (lane 7) and subjected to NaOH treatment (lanes 2 and 4) and/or stopped and loaded in 7 M urea/20% PAGE. Lane 6 shows the tetrahydrofuran 34-mer cleavage by Nfo. S, substrates; AP-l and AP-e, the positions of the AP lyase and AP endonuclease products, respectively.
EndoIII-Deficient H. pylori Strains Fail to Repair DNA Lesions.
Disruption mutants were generated by insertion of a kanamycin transposon into hp0585 from H. pylori X47, a model strain used in mouse infection experiments. The disruption of this gene was confirmed by PCR amplification and sequencing of the genomic DNA. Two mutants from independent transformation events, with insertions at positions 315 and 385, were selected and used for the subsequent experiments, either individually or pooled to avoid possible secondary mutations effects. Mutation frequencies were determined by quantifying the appearance of rifampicin-resistant colonies (rifR). As shown in Table 2, cells lacking HpNth show a 4-fold-higher spontaneous mutation frequency compared with the wild-type strain. Thus, in the absence of HpNth, a significant number of premutagenic pyrimidine lesions are left unrepaired.
Table 2.
Mutation frequencies of wild-type and Hpnth-deficient X47 H. pylori
| Genotype | Spontaneous mutation frequency,
rifR per 108cfu
|
|
|---|---|---|
| Experiment 1 | Experiment 2 | |
| Parental strain | 6 | 6 |
| Hpnth315 | 23 | 26 |
| Hpnth385 | 17 | 22 |
Cultures were done on HBAP inoculated with 2 × 104 cfu and grown 48 h.
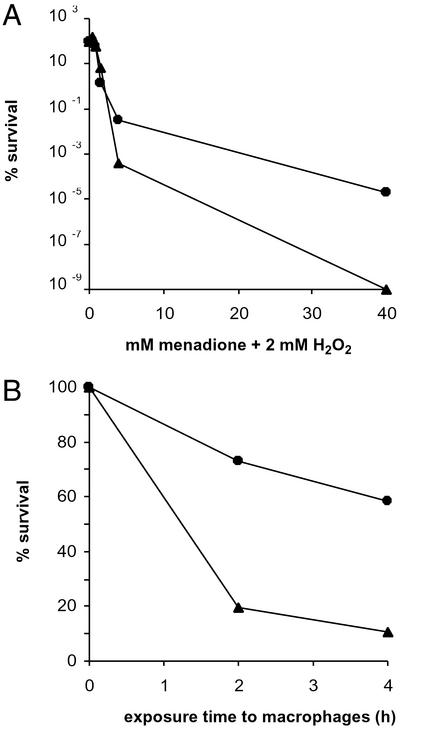
Some of the free-radical-modified DNA bases are not mutagenic but can block RNA and DNA polymerases (14, 15). Consequently, HpNth could provide H. pylori with a defense against the lethal effect of exogenous oxidizing agents. Fig. 3A shows that X47Hpnthpool is more sensitive than the parental strain to the exposure to a mixture of menadione and H2O2. To analyze whether this hypersensitivity of H. pylori Hpnth mutants to a chemically generated oxidizing stress reflected a hypersensitivity in a more physiological context, coculture experiments were performed in the presence of activated mouse J774.A macrophages. Macrophages were preactivated by exposure to lipopolysaccharide and phorbol 12-myristate 13-acetate (PMA) and subsequently mixed with either wild-type X47 or mutant X47Hpnthpool. Bacteria were harvested and plated 2 and 4 h later. Fig. 3B shows the viability of the wild-type and mutant derivatives, relative to that of the cells that survived under the same culture conditions but without macrophages. When the mutation frequency was measured before and after exposure to macrophages, both genotypes showed a marked increase in rifampicin-resistant colonies after exposure to activated macrophages (Table 3). However, the frequencies obtained show that the macrophage-induced mutation frequencies are 5-fold higher in X47endoIII than in the wild-type X47. Taken together, these results show that the HpNth-deficient strain is less effective in removing lethal and premutagenic DNA lesions generated by the exposure to macrophages.
Figure 3.
Sensitivity of H. pylori wild type or Hpnth to oxidative stress generators. (A) Survival curves for H. pylori X47Hpnth (▴) and the parental wild-type strain (●) incubated for 15 min with increasing amounts of menadione and 2 mM H2O2. Data show the surviving fraction relative to the initial cfu and are representative of two independent experiments. (B) Influence of HpNth deficiency on the resistance to the bacteriocidal activity of J774.1A macrophages. Bacteria were cocultured with preactivated macrophages for the times indicated (see Materials and Methods and Results). The survival fraction of the wild-type (●) and endoIII-derivative H. pylori (▴) compared with the surviving bacteria in RPMI without macrophages is shown. Data are representative of two independent experiments.
Table 3.
Spontaneous and induced mutation frequencies of wild-type and Hpnth-deficient X47 H. pylori
| Genotype | Experiment | Mutation frequency,
rifR per 108cfu
|
Induced mutations (×108) | |
|---|---|---|---|---|
| Before macrophages | After macrophages | |||
| Wild-type | 1 | 4 | 24 | 20 |
| 2 | 2 | 21 | 19 | |
| Hpnthpool | 1 | 21 | 116 | 95 |
| 2 | 16 | 121 | 105 | |
Stomach Colonization Is Impaired in H. pylori Lacking EndoIII.
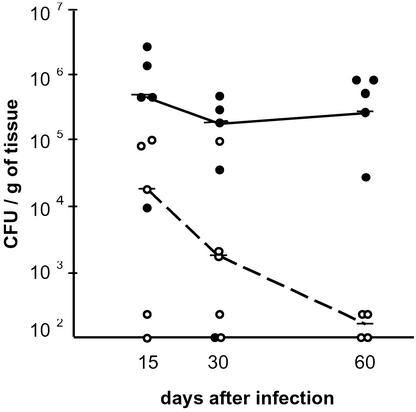
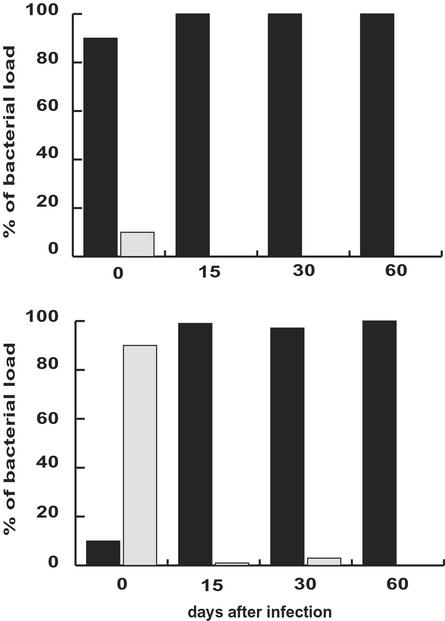
To determine whether a deficiency in HpNth activity is important for H. pylori host colonization and, at the same time, whether DNA damage on the pathogen genome is specifically generated during infection, the relative abilities of wild-type and HpNth-deficient strains to colonize the stomach were evaluated in a mouse infection model. For the Hpnth-deficient group, a pool of six independent mutant clones [insertions at positions 315 (x2), 385 (x2), 428, and 440] was used. Mice were inoculated with 1 × 107 cfu of the mouse-adapted X47 H. pylori strain or its Hpnth derivative. Fifteen, 30, and 60 days p.i., 10 infected mice from each group were killed and the H. pylori content of their gastric tissue was determined by plating (Fig. 4). Fifteen days p.i., wild-type-infected animals had 10-fold more cfu than mutant-infected mice. Whereas the bacterial load in the wild-type population remained high, the mutant bacterial load decreased gradually, yielding 102 and 103 less cfu per gram of tissue than the wild-type 30 and 60 days p.i., respectively. The impaired colonization capability of HpNth-deficient cells was confirmed in coinfection experiments. Groups of 30 mice were infected with either 1 × 106 wild-type and 1 × 107 mutant mix, or the inverse ratio mixture. Fifteen, 30, and 60 days p.i., 10 infected mice from each group were killed and their stomach bacterial loads were quantified by plating on plates containing (for counting Hpnth− H. pylori) or not containing kanamycin (for counting total H. pylori). After 30 days only one mouse, inoculated with the predominantly mutant mix, harbored Hpnth bacteria (KmR) and these mutant cells represented less than 1% of the population harvested. At 60 days p.i. none of the stomachs of the mice infected had detectable HpNth-deficient cells (Fig. 5). To rule out the possibility that cells lacking EndoIII had an intrinsic impaired growth capability, the survival in vitro (on HBAP) of X47 vs. X47Hpnth was measured during 2 weeks (≈50 generations). No reduction in population was observed in Hpnth mutant cells when compared with the wild-type strain in the same conditions (Table 4). These results show that (i) the host-generated response to infection induces an oxidative stress that damages the pathogen DNA and (ii) that H. pylori HpNth provides a bacterial defense against the host response.
Figure 4.
Colonization ability of Hpnth vs. wild-type H. pylori. On day 0, mice were inoculated with 1 × 107 of either wild-type (●) or Hpnth-deficient H. pylori (○). Mice were killed after 15, 30, or 60 days and the cfu content of the stomach tissue was determined on normal or kanamycin-containing HBAP. Points correspond to individual animals. Horizontal bars represent the median of the population per gram of stomach tissue. Lines indicate the trends of wild-type (—) and HpNth-deficient (- - -) bacterial loads.
Figure 5.
Colonization competition between Hpnth-deficient and wild-type H. pylori. On day 0, germ-free mice (n = 10 for each time and genotype) were inoculated with a ratio 9:1 wild-type (■):Hpnth-deficient H. pylori (░⃞) (Upper) or the reverse proportion (Lower). Mice were killed after 15, 30, or 60 days and cfu in the stomach were counted on normal and kanamycin-containing HBAP to determine the wild-type and mutant bacterial load, respectively.
Table 4.
In vitro survival of wild-type and Hpnth X47 H. pylori
| Passage | Genotype
|
|
|---|---|---|
| X47 | X47endoIII | |
| 1 | 1.9 × 108 | 4.2 × 108 |
| 2 | 4.4 × 1010 | 3.5 × 1010 |
| 3 | 3.3 × 1010 | 2.8 × 1010 |
| 4 | 3.8 × 1010 | 3.2 × 1010 |
| 5 | 4.1 × 1010 | 2.5 × 1010 |
| 6 | 1.9 × 1010 | 2.3 × 1010 |
Discussion
In H. pylori, two ORFs, hp0585 and hp0602, display similar levels of homology to EndoIII family members (20, 21). In a previous study (22) we demonstrated that hp0602, although labeled as endoIII, codes for a 3-methyladenine DNA glycosylase. Here we show that the protein encoded by hp0585 is indeed a DNA glycosylase that recognizes oxidized pyrimidines as substrates and has an AP lyase activity. We therefore named this protein HpNth. It is interesting to note that, even when HpNth from ADM1 and 13/5 H. pylori strains depict multiple amino acid substitutions, no differences were noted between their enzymatic activities.
H. pylori cells deficient in HpNth do not show a growth phenotype under normal laboratory conditions. As is the case for E. coli nth mutants (17), H. pylori cells deficient in HpNth show a weak mutator phenotype. However, the H. pylori Hpnth-deficient cells differ from the E. coli nth mutants in that they are more sensitive to the killing effect of oxidative agents. In E. coli, the impairment of a second DNA glycosylase, EndoVIII encoded by the nei gene, is required to obtain a hypersensitivity to H2O2 (18, 19). The genomic sequence of H. pylori does not show an ortholog of nei, coding for an EndoVIII-like protein, suggesting that in this pathogen HpNth is the only DNA glycosylase capable of removing toxic oxidized pyrimidines (20, 21). Interestingly, the same kind of difference between E. coli and H. pylori gene content exists for another base excision repair pathway. The removal of methylated adenines from DNA can be performed by either of two 3-methyladenine glycosylases in E. coli (Tag and AlkA), whereas in H. pylori only one DNA glycosylase, Mag III, has been found to protect against killing by DNA-methylating agents (22). This limited redundancy in H. pylori gene content when compared with E. coli can be extended to other gene families as cell division proteins (hp1159), thioredoxin reductase (hp1164), and carboxynorspermidine decarboxylase (hp0020), as discussed by Chalker et al. (29).
Because the H. pylori Hpnth mutants are only affected in their viability when exposed to a specific environmental stress [no growth defect was observed under normal culture conditions, even when exposed to pH 3 and 4, with or without urea (ref. 7; data not shown)], they provide a tool for evaluating whether the host reaction induces a stress capable of damaging the bacterial DNA during infection. Experiments with E. coli and S. typhimurium had shown that mutagenesis is enhanced when bacterial cells are exposed to phagocytes (30, 31). Also, recombination mutants of L. pneumophila and S. typhimurium are impaired in their ability to colonize mice (9–11). However, mutations in Rep, RecBC, or RecA could have pleiotropic effects on basic DNA metabolism. Furthermore, both of these species are intracellular pathogens, and the phagolysosome is considered the most challenging environment that invading bacteria encounter within the host and could therefore be considered a special case (32). In contrast, H. pylori resides in the mucus layer overlying the gastric epithelium of the stomach (2). For host-generated oxidative stress to reach the pathogen DNA, it needs to overcome several barriers. Some of these are the stomach mucus layer itself, membrane lipids, and bacterial antioxidant enzymes such as superoxide dismutase, catalase, and thioredoxin reductase (33, 34). Because the coculture with activated macrophages generates oxidative adducts in the bacterial DNA, we conclude that the bacterial antioxidant defenses are not sufficient to avoid oxidative DNA damage generated by a hostile environment. Furthermore, during monoinfections in Hpnth mutant strains, the bacterial load gradually decreases, whereas in wild-type the population remains stable. Therefore, the gradual decrease of the bacterial load in mice infected with Hpnth bacteria indicates that the host is playing an active role in the accumulation of oxidative lethal lesions, most likely by blocking pyrimidine lesions such as thymine glycol, on the pathogen DNA.
The macrophages experiments show that, besides genotoxic damage, mutations can be induced in H. pylori by oxidative DNA damage. The fact that Hpnth strains, when forced to compete with wild type, in coinfection experiments are eradicated much faster than in monoinfections, even when inoculated in a 1:9 wild-type:mutant strain ratio, suggests that secondary, nonlethal genetic changes induced by the host reaction are counterselected. The mutations are most likely the consequence of premutagenic pyrimidine modifications generated in pathogen DNA by the oxidative environment associated with the infection.
It has been proposed that the high genetic variability found in H. pylori could reflect a homeostatic mechanism by which this pathogen would adapt to stress situations (35–37). For other pathogens, a high mutation rate increases the fitness of the pathogen when facing a new host (38). Although our results show that mutagenic lesions reduce the colonization ability of the mutant strain, it is important to note that infections described were performed with a strain that had been previously selected for its good colonization capacity, and therefore, it is expected that most mutagenic lesions would be deleterious. It would then be interesting to analyze whether the mutations induced in a nonadapted strain by the oxidative environment created during infection could help H. pylori to overcome the genotoxic effect of the stress and allow its adaptation to a new host.
In conclusion, the identification of a mutant that, by the lack of a DNA repair activity, is more susceptible to killing by oxidative agents in vitro provided a tool for assessing the impact of the oxidative stress reaching the DNA of H. pylori during stomach infection. The reduced survival of Hpnth H. pylori in the host shows that once adapted and in a stable environment, deleterious DNA lesions induced during the infection lead to the disappearance of the bacterial population. This underscores the importance of the capacity to repair DNA for microbial persistence during infection.
Acknowledgments
We thank Jimena Ortega (Instituto Leloir) for sequencing, Marta Bravo (Instituto Leloir) for her help with FPLC experiments, Jean Cadet Commissariat à l'Energie Atomique (CEA) for modified oligonucleotides, and Susan Wallace (University of Vermont, Burlington) for bacterial strains. We are grateful to Cristian Danna for his critical reading of the manuscript and especially to Serge Boiteux for his continuous support and advice. This work was supported in France by the Program pour le Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitologie, the CEA, and the Centre National de la Recherche Scientifique, and in Argentina by grants from Laboratorios Bagó SA and the Argentine Federal Government (Agencia Nacional de Promocion Cientifica y Tecnologica PICT 01-09016 and Carrillo Oñativia 2001). The collaboration between the Argentine and French laboratories was made possible by the ECOS-Sud program. E.J.O. was the recipient of a doctoral fellowship from the University of Buenos Aires (FOMEC) and of the Carrillo-Oñativa program. L.I. is a Research Career Investigator of the Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina.
Abbreviations
- AP
abasic
- cfu
colony-forming units
- dHT
dihydrothymine
- Endo
endonuclease
- HBAP
horse serum Brucella agar plates
- p.i.
postinfection
References
- 1.Ge Z, Taylor D E. Annu Rev Microbiol. 1999;53:353–387. doi: 10.1146/annurev.micro.53.1.353. [DOI] [PubMed] [Google Scholar]
- 2.Ernst P B, Gold B D. Annu Rev Microbiol. 2000;54:615–640. doi: 10.1146/annurev.micro.54.1.615. [DOI] [PubMed] [Google Scholar]
- 3.Baik S C, Youn H S, Chung M H, Lee W K, Cho M J, Ko G H, Park C K, Kasai H, Rhee K H. Cancer Res. 1996;56:1279–1282. [PubMed] [Google Scholar]
- 4.Farinati F, Cardin R, Degan P, Rugge M, Mario F D, Bonvicini P, Naccarato R. Gut. 1998;42:351–356. doi: 10.1136/gut.42.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Obst B, Wagner S, Sewing K F, Beil W. Carcinogenesis. 2000;21:1111–1115. [PubMed] [Google Scholar]
- 6.Seyler R W, Jr, Olson J W, Maier R J. Infect Immun. 2001;69:4034–4040. doi: 10.1128/IAI.69.6.4034-4040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson S A, Blaser M J. Infect Immun. 1995;63:2185–2193. doi: 10.1128/iai.63.6.2185-2193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson S A, Latch R L, Blaser J M. Gene. 1998;209:113–122. doi: 10.1016/s0378-1119(98)00028-6. [DOI] [PubMed] [Google Scholar]
- 9.Merino D, Reglier-Poupet H, Berche P, Charbit A. Mol Microbiol. 2002;44:877–887. doi: 10.1046/j.1365-2958.2002.02929.x. [DOI] [PubMed] [Google Scholar]
- 10.Buchmeier N A, Lipps C J, So M Y, Heffron F. Mol Microbiol. 1993;7:933–936. doi: 10.1111/j.1365-2958.1993.tb01184.x. [DOI] [PubMed] [Google Scholar]
- 11.Cano D A, Pucciarelli M G, Garcia-del Portillo F, Casadesus J. J Bacteriol. 2002;184:592–595. doi: 10.1128/JB.184.2.592-595.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imlay J A, Linn S. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 13.Laval J, Jurado J, Saparbaev M, Sidorkina O. Mutat Res. 1998;402:93–102. doi: 10.1016/s0027-5107(97)00286-8. [DOI] [PubMed] [Google Scholar]
- 14.Clark J M, Beardsley G P. Nucleic Acids Res. 1986;14:737–749. doi: 10.1093/nar/14.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tornaletti S, Maeda L S, Lloyd D R, Reines D, Hanawalt P C. J Biol Chem. 2001;276:45367–45371. doi: 10.1074/jbc.M105282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laspia M F, Wallace S S. J Bacteriol. 1988;170:3359–3366. doi: 10.1128/jb.170.8.3359-3366.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham R P, Weiss B. Proc Natl Acad Sci USA. 1985;82:474–478. doi: 10.1073/pnas.82.2.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito Y, Uraki F, Nakajima S, Asaeda A, Ono K, Kubo K, Yamamoto K. J Bacteriol. 1997;179:3783–3785. doi: 10.1128/jb.179.11.3783-3785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang D, Hatahet Z, Blaisdell J O, Melamedeol R J, Wallace S S. J Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, et al. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 21.Alm R A, Ling L S, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, et al. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 22.O'Rourke E J, Chevalier C, Boiteux S, Labigne A, Ielpi L, Radicella J P. J Biol Chem. 2000;275:20077–20083. doi: 10.1074/jbc.M001071200. [DOI] [PubMed] [Google Scholar]
- 23.Londono-Arcila P, Freeman D, Kleanthous H, O'Dowd A M, Lewis S, Turner A K, Rees E L, Tibbitts T J, Greenwood J, Monath T P, Darsley M J. Infect Immun. 2002;70:5096–5106. doi: 10.1128/IAI.70.9.5096-5106.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clayton C L, Mobley H L T. Helicobacter pylori Protocols. Totowa, NJ: Humana Press; 1997. [Google Scholar]
- 25.Ferrero R L, Thiberge J M, Huerre M, Labigne A. Infect Immun. 1998;66:1349–1355. doi: 10.1128/iai.66.4.1349-1355.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chevalier C, Thiberge J M, Ferrero R L, Labigne A. Mol Microbiol. 1999;31:1359–1372. doi: 10.1046/j.1365-2958.1999.01271.x. [DOI] [PubMed] [Google Scholar]
- 27.Hollis T, Ichikawa Y, Ellenberger T. EMBO J. 2000;19:758–766. doi: 10.1093/emboj/19.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thayer M M, Ahern H, Xing D, Cunningham R P, Tainer J A. EMBO J. 1995;14:4108–4120. doi: 10.1002/j.1460-2075.1995.tb00083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chalker A F, Minehart H W, Hughes N J, Koretke K K, Lonetto M A, Brinkman K K, Warren P V, Lupas A, Stanhope M J, Brown J R, Hoffman P S. J Bacteriol. 2001;183:1259–1268. doi: 10.1128/JB.183.4.1259-1268.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlosser-Silverman E, Elgrably-Weiss M, Rosenshine I, Kohen R, Altuvia S. J Bacteriol. 2000;182:5225–5230. doi: 10.1128/jb.182.18.5225-5230.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weitzman S A, Stossel T P. Science. 1981;212:546–547. doi: 10.1126/science.6259738. [DOI] [PubMed] [Google Scholar]
- 32.Mahan M J, Tobias J W, Slauch J M, Hanna P C, Collier R J, Mekalanos J J. Proc Natl Acad Sci USA. 1995;92:669–673. doi: 10.1073/pnas.92.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mori M, Suzuki H, Suzuki M, Kai A, Miura S, Ishii H. Helicobacter. 1997;2:100–105. doi: 10.1111/j.1523-5378.1997.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 34.Windle H J, Fox A, Ni Eidhin D, Kelleher D. J Biol Chem. 2000;275:5081–5089. doi: 10.1074/jbc.275.7.5081. [DOI] [PubMed] [Google Scholar]
- 35.Wang G, Humayun M Z, Taylor D E. Trends Microbiol. 1999;7:488–493. doi: 10.1016/s0966-842x(99)01632-7. [DOI] [PubMed] [Google Scholar]
- 36.Janvier B, Grignon B, Audibert C, Pezennec L, Fauchere J L. FEMS Immunol Med Microbiol. 1999;24:27–33. doi: 10.1111/j.1574-695X.1999.tb01261.x. [DOI] [PubMed] [Google Scholar]
- 37.Akopyants N S, Eaton K A, Berg D E. Infect Immun. 1995;63:116–121. doi: 10.1128/iai.63.1.116-121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, Taddei F. Science. 2001;291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]