Abstract
Toxigenic Vibrio cholerae cause cholera, a severe diarrheal disease responsible for significant morbidity and mortality worldwide. Two determinants, cholera enterotoxin (CT) and toxin coregulated pilus (TCP) are critical factors responsible for this organism's virulence. The genes for these virulence determinants belong to a network of genes (the ToxR regulon) whose expression is modulated by transcriptional regulators encoded by the toxRS, tcpPH, and toxT genes. To define the ToxR regulon more fully, mutants defective in these regulatory genes were transcriptionally profiled by using V. cholerae genomic microarrays. This study identified 13 genes that were transcriptionally repressed by the toxT mutation (all involved in CT and TCP biogenesis), and 27 and 60 genes that were transcriptionally repressed by the tcpPH and toxRS mutations, respectively. During the course of this analysis, we validated the use of a genomic DNA-based reference sample as a means to standardize and normalize data obtained in different microarray experiments. This method allowed the accurate transcriptional profiling of V. cholerae cells present in stools from cholera patients and the comparison of these profiles to those of wild-type and mutant strains of V. cholerae grown under optimal conditions for CT and TCP expression. Our results suggest that vibrios present in cholera stools carry transcripts for these two virulence determinants, albeit at relatively low levels compared with optimal in vitro conditions. The transcriptional profile of vibrios present in cholera stools also suggests that the bacteria experienced conditions of anaerobiosis, iron limitation, and nutrient deprivation within the human gastrointestinal tract.
The causative agent of cholera, Vibrio cholerae, has provided an excellent model system for identifying bacterial virulence factors and for understanding the regulatory networks governing their expression (1, 2). Biochemical and genetic analysis, coupled with studies in experimental animals and human volunteers, has revealed many details of cholera pathogenesis. In brief, orally ingested bacteria survive passage through the stomach and then use motility and chemotaxis functions to adhere to and penetrate the mucus coat of the upper intestinal epithelium. Vibrios must then coordinately express two critical sets of genes encoded by the tcp and ctx operons, which encode the toxin coregulated pilus (TCP) and cholera enterotoxin (CT), respectively (2). TCP pili are essential for colonization in mice (3, 4) and humans (5, 6), whereas CT is the main factor responsible for generating the severe diarrhea that is the hallmark of the disease. Expression of both the tcp and ctx operons is regulated by two membrane proteins, ToxR and ToxS (7). ToxR can directly affect gene expression by binding to promoters, including those for the ctx operon, the genes encoding two outer membrane proteins OmpU and OmpT, and the gene encoding ToxT, a transcriptional regulator belonging to the AraC family of proteins (8). ToxT is the most downstream regulator of the ToxR regulon in that it can activate ctx and tcp promoters independently once ToxT expression has occurred (9, 10). However, two other membrane proteins, TcpP and TcpH, are also required to work synergistically with ToxR and ToxS to activate toxT transcription (11). Finally, several other factors, including CRP (12), AphAB (13), Nqr (14), HapR (15), and PepA (16), fine-tune this regulatory hierarchy by controlling expression of tcpPH in response to various environmental signals in the laboratory. These signals include physical parameters such as temperature and pH as well as physiological signals such as cell density, growth phase, and motility. Thus, in vitro analysis of the ToxR regulon has demonstrated the existence of an extremely complex regulatory network controlling the expression of the TCP and CT virulence genes. However, it remains largely unknown how these regulatory responses are coordinated in vivo during infection. In a series of elegant experiments, Camilli and colleagues provided evidence that CT expression during experimental infection of mice is largely dependent on early temporal expression of TCP (17). This result suggests that only a minority of cells (i.e., those that successfully colonize the intestinal mucosa) experience the in vivo regulatory signals that lead to maximal CT expression. Although various studies have identified additional factors that may contribute significantly to disease or that affect the organism's behavior during different stages of infection, it is clear that our understanding of V. cholerae gene expression patterns during infection is still poor.
The determination of the DNA sequence of the two chromosomes of V. cholerae strain N16961 (18) has provided an exceptional opportunity to define gene content in various strains (19) and gene expression across the entire bacterial genome under given experimental conditions by using the powerful technique of microarray analysis (20, 21). For example, V. cholerae microarrays displaying all of the annotated genes of this organism have been successfully used to compare the transcriptional profiles of specific mutant versus wild-type cells (20), exponential-phase bacteria recovered from animal infections versus those grown to exponential phase in vitro (21), and vibrios present in human cholera stool versus those grown to stationary phase in vitro (22). Such transcriptome analyses are useful but are frequently limited in value because of technical reasons related to variability in experimental conditions, RNA preparation, internal reference standards, and microarray manufacturing processes. Of these, selection of the appropriate internal reference is of critical importance. For example, profiling the expression pattern of bacteria during infection to identify “in vivo induced genes” is problematic. Care must be taken to select a reference in vitro condition that at least provides cells in the same physiological state as the in vivo grown cells being analyzed, because there are dramatic differences in gene expression between exponential and stationary phase in any growth medium. Selection of the appropriate reference is also critical for comparing results between different experiments, researchers, and laboratories. Microarrays manufactured by spotting oligonucleotides or PCR products onto glass slides (i.e., spotted arrays) are particularly susceptible to variation from slide to slide and from spot to spot. Therefore, spotted array results from different experiments can only be reliably compared by using an internal reference that can address the quality of all of the individual features on the array. These considerations have led several investigators to conclude that the use of a universal reference standard, such as genomic DNA (gDNA), may be the single best way to standardize the analysis of spotted arrays (23, 24).
In this study, we compared two approaches for transcriptional profiling of bacterial mutants and of bacteria contained in clinical materials. Specifically, we hoped to determine the degree of variation arising from the use of a gDNA-derived reference, compared with that arising from direct comparison of RNA preparations converted to cDNA samples. We chose the well-characterized ToxR regulon of V. cholerae as an experimental system to address these questions. Our studies of toxRS, tcpPH, and toxT mutants validated the use of a gDNA-derived reference and also provided a clear, genome-wide transcriptional profile of the ToxR regulon. Finally, the use of a gDNA-derived reference allowed us to profile expression of V. cholerae cells recovered from human cholera stool, providing a new view of the V. cholerae transcriptome during human infection.
Materials and Methods
Strains, Strain Construction, and Growth Conditions.
V. cholerae strain N16961 (El Tor, O1, StrR) was used in this study. In-frame deletions in the N16961 genes encoding toxRS and tcpPH were constructed by crossover PCR (25, 26) to yield strains M461 and M462, respectively. The N16961 toxT deletion mutant (strain M460) was constructed in a similar manner using pMD1 (27). For routine growth, all strains were cultured at 37°C in Luria broth. For the induction of the ToxR regulon, strains were cultured under AKI conditions at 37°C (28). Briefly, a saturated overnight culture grown in LB was inoculated into AKI medium to a starting density of ≈105 colony-forming units (cfu)/ml, followed by 4 h of static growth (low aeration; 10 ml in a 15-ml tube), and then transfer to shaking growth conditions (high aeration; 10 ml in a 125-ml flask shaken at 250 rpm in a G10 Gyrotory shaker (New Brunswick Scientific).
Nucleic Acid Isolation and Labeling.
V. cholerae total RNA was isolated as previously described (20), with the addition of a DNase treatment before purification with the RNeasy kit (Qiagen). The resulting RNA was reverse-transcribed to produce either Cy5- or Cy3-labeled hybridization probes as previously described (29). gDNA was prepared from overnight cultures of V. cholerae using the Easy-DNA kit (Invitrogen) according to the manufacturer's directions. gDNA was used in a Klenow reaction to produce hybridization probes as described (19).
V. cholerae Whole Genome Microarray.
The construction of the whole-genome V. cholerae microarray has been described (19) and consists of 3,890 full-length PCR products representing the annotated ORFs from the initial release of the N16961 genomic sequence (18). The procedures for hybridization of samples to the microarray, washing of arrays, and data collection were as described (19).
Data Analysis.
Gene expression levels for all microarray experiments were calculated by averaging the background-subtracted fluorescent intensities of the sample and control channels among technical replicates. Unless otherwise noted, the data for each experiment resulted from a minimum of two technical replicates for each condition or mutant that was analyzed. Microarray data normalization was performed as described (19, 21). Microarray data analysis was facilitated by the use of the GeneSpring microarray analysis software program (Silicon Genetics, Redwood City, CA). Genes showing a ≥2-fold difference in expression between the mutant strain and reference strain were classified as regulated genes.
Stool Collection, Processing, and Analysis.
Stool samples were collected from two cholera patients at the International Centre for Diarrhoeal Disease Research in Dhaka, Bangladesh. Freshly collected stool samples, containing ≈108 cfu/ml V. cholerae O139, were divided into two aliquots. One aliquot, which was frozen and maintained at −80°C, was used for the purification of total RNA as described in Merrell et al. (22). The second aliquot was used for V. cholerae enumeration, serotyping,and biotyping. The purified RNA from each patient stool sample (test sample) was used as a template to generate a pool of Cy5-labeled cDNA, which was then split and cohybridized to two microarray slides with a Cy3-labeled gDNA reference sample. The background-corrected fluorescence intensities for each feature were averaged and used to calculate a ratio for each feature (test sample/reference sample) that represents the expression level for each gene. The data set was rank-ordered from highest ratio to lowest ratio to generate a list of expression levels representing the transcriptome for that specific condition.
Results
Transcriptional Analysis of tcpPH, toxRS, and toxT Mutants.
V. cholerae O1 El Tor and O139 strains require growth under specific in vitro conditions, known as “AKI conditions,” to induce the expression of virulence factors (28, 30). As a marker for virulence gene expression, we used a CT ELISA to detect toxin production. CT production by El Tor O1 strain N16961 was consistently detected at 2 h after transfer of the static AKI culture to shaking growth conditions (data not shown). This time point was therefore selected for RNA purification for the transcriptional profiling of V. cholerae N16961 and various isogenic regulatory mutants. Growth curves for these strains indicated that the mutant strains grow at the same rate as the wild-type under AKI conditions (data not shown).
For our initial analysis, we used cDNAs derived from RNA purified from wild-type and mutant cells. In brief, the cDNAs from two strains being compared were differentially labeled with Cy3 and Cy5, mixed, hybridized to microarrays, scanned, and analyzed. For a given microarray feature, a statistically reliable change of 2-fold or more in the ratio of fluorescent signals was selected as our cut-off for reporting a gene as altered in expression in a regulatory mutant. It should be emphasized that expression ratios for various genes derived from microarray data are more of a qualitative than quantitative measure of relative gene expression. Empirically, we have found that a difference in ratio of 2- to 3-fold is a useful cut-off for identifying changes in gene content (19) or expression (21), but we suspect that smaller differences may also be biologically significant.
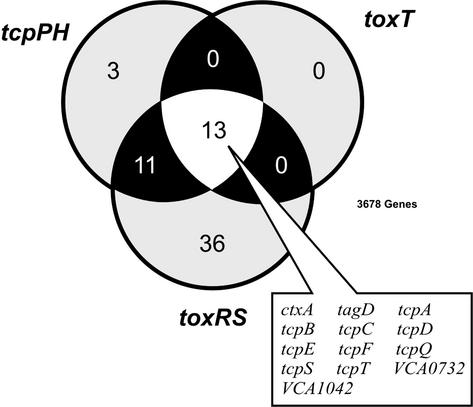
Virulence gene expression in V. cholerae is subject to the hierarchy of the ToxR regulon, where both ToxT-dependent and ToxT-independent branches require expression of the toxR gene (1, 31). Because toxT expression is known to depend on tcpPH and toxR, we did not expect to identify ToxT-regulated genes that were not also dependent on toxR and/or tcpPH. Indeed, our microarray analysis of RNA extracted from wild-type and toxT mutant cells identified only 13 genes whose expression was at least 2-fold repressed in the toxT mutant (Fig. 1 and Tables 5–8, which are published as supporting information on the PNAS web site, www.pnas.org). As expected, these 13 genes were a subset of those identified in the expression profiles of toxRS and tcpPH mutants (Fig. 1). With the exception of two genes, VCA0732 (conserved hypothetical gene) and VCA1042 (an unknown gene immediately downstream of tagE-2), all of these genes were previously identified as members of the ToxR-regulon (2). The gene encoding the cholera toxin B subunit (ctxB) was not among the 13 because the ctxB expression ratio in two samples was slightly below our cutoff (i.e., 1.81 and 1.97).
Figure 1.
Genes showing reduced expression in N16961 toxRS, toxT, and tcpPH mutant strains. Venn diagram populated with genes showing 2-fold or greater transcriptional repression in AKI growth conditions for each respective mutant relative to wild type. Numbers in the overlapping regions of the Venn diagram represent genes that were coregulated among the three mutant strains. The 13 genes whose transcription was found to be toxRS-, toxT-, and tcpPH-dependent are listed.
Our analysis indicated that tcpPH are not regulated by ToxR or ToxT, consistent with published results (32). Deletion of tcpPH resulted in decreased expression of 27 genes relative to the wild-type strain, and increased expression of 31 genes (Fig. 1 and Table 1). As predicted, tcpPH were required for transcription of the genes in the TCP and CT operons (Fig. 1). Most of the 58 genes whose expression was affected in the tcpPH mutant were also affected by ToxRS (53 genes; see Tables 5–8). The expression of five genes depended solely on tcpPH including the redox control response regulator arcA (VC2368). Loss of TcpPH decreased the expression of three porins (VC0972, ompT and ompW) and six genes involved in glycerol metabolism (VC0136, VC0137, VCA0744, VCA0745, VCA0747, and VCA0748).
Table 1.
Distribution and functional classification of genes belonging to the ToxR regulon
| toxR | tcpPH | toxT | |
|---|---|---|---|
| Total no. of genes identified | 154 | 58 | 18 |
| Total no. of genes induced | 60 | 27 | 13 |
| Total no. of genes repressed | 94 | 31 | 5 |
| Chromosome location | |||
| Large chromosome | 102 (66.2) | 39 (67.2) | 14 (77.8) |
| Small chromosome | 52 (33.8) | 19 (32.8) | 4 (22.2) |
| Functional class | |||
| Adaptation to atypical conditions | 1 (0.6) | 1 (1.7) | 0 |
| Amino acid biosynthesis | 4 (2.6) | 0 | 0 |
| Chemotaxis and motility | 6 (3.9) | 1 (1.7) | 0 |
| Cofactor | 1 (0.6) | 0 | 0 |
| Conserved, conserved hypothetical, and unknown | 33 (20.4) | 10 (17.2) | 2 (11.1) |
| Energy metabolism | 31 (20.1) | 12 (20.7) | 1 (5.6) |
| Pathogenicity | 22 (14.3) | 19 (32.8) | 11 (61.1) |
| Protein synthesis | 7 (4.5) | 0 | 0 |
| Purines, pyrimidines, nucleosides, and nucleotides | 6 (3.9) | 1 (1.7) | 1 (5.6) |
| Regulatory | 8 (5.2) | 4 (6.9) | 0 |
| Transport and cell envelope | 35 (22.7) | 10 (17.2) | 3 (16.7) |
No. of genes (% of total genes identified) showing a ≥2-fold change in expression in the indicated strain. All strains were grown under AKI conditions to promote expression of virulence genes.
The toxR mutant showed the most dramatic transcriptional changes compared with the wild type. A total of 154 genes showed a ≥2-fold change in transcription (Table 1); 60 genes showed increased expression in the mutant strain (corresponding to “ToxR-repressed genes”) and 94 genes showed decreased expression in the mutant strain (corresponding to “ToxR-activated genes”). Consistent with published reports, many of the ToxR-activated genes were involved in the production of TCP and CT. The genes encoding the ToxR-repressed OmpT porin and ToxR-activated OmpU porin showed the expected reciprocal changes in expression (i.e., ≈19-fold increase and an ≈23-fold decrease in expression in the toxR mutant, respectively).
Our analysis identified numerous genes belonging to the toxT-independent branch of the ToxR regulon. These included a large number of genes involved in cellular transport, energy metabolism, motility, iron uptake and outer membrane proteins (Tables 1 and 5–8).
Use of a Genomic DNA Reference for Transcriptional Analysis.
Having shown that our microarray-based analysis yielded the expected expression profiling results for genes known to belong to the ToxR regulon, we conclude that directly comparing cDNAs derived from a mutant and wild-type RNAs was a valid way to identify changes in gene expression between different strains. However, such an approach requires one to perform many additional microarray experiments when comparing different strains or different conditions. Because the use of a universal reference promises considerably more economy and flexibility for such comparisons (23, 24), we tested the accuracy of using a gDNA reference standard for comparing the toxT mutant to its wild-type parent. As before, total RNA was isolated from each strain and used to generate cDNA probes. We then performed two different types of hybridization experiments. The first was similar to our initial analyses and used RNA isolated from the wild-type strain to produce a cDNA reference sample that was then cohybridized with a cDNA test sample from the toxT mutant on one slide. The second experiment used chromosomal DNA to produce a gDNA reference sample that was then cohybridized on two different slides with the cDNA test samples derived from the toxT mutant and the wild-type strain. The first experiment directly generated a relative expression ratio for CT and TCP genes in the wild-type versus the toxT mutant. To obtain expression ratios using the gDNA reference, we first calculated the individual ratios of the wild-type and toxT cDNA probes to the gDNA reference, and then calculated the ratio of the two individual ratios to each other to produce the relative expression ratio (see Table 2).
Table 2.
Analysis of N16961 toxT using cDNA and gDNA reference samples
| Gene | Repression ratio*
|
|
|---|---|---|
| cDNA reference† | gDNA reference‡ | |
| tcpA | 40.8 | 36.9 |
| tcpB | 25.2 | 20.4 |
| tcpQ | 28.2 | 16.7 |
| tcpC | 20.2 | 16.9 |
| tcpR | 7.8 | 7.7 |
| tcpD | 8.0 | 3.7 |
| tcpS | 22.7 | 8.4 |
| tcpT | 19.3 | 13.8 |
| tcpE | 10.2 | 4.8 |
| tcpF | 8.3 | 7.1 |
| ctxB | 12.7 | 11.3 |
| ctxA | 15.4 | 28.7 |
Repression ratio in toxT mutant relative to the wild-type strain in AKI conditions. Reported results were calculated from one experiment.
Fluorescent intensity ratio of (wt-cDNA)/(toxT-cDNA).
Fluorescent intensity ratio of (wt-cDNA/gDNA)/(toxT-cDNA/gDNA).
We compared the results of these two methods for selected TCP and CT genes because the expression of these genes is known to be regulated by ToxT. In fact, these were the only genes whose expression was reduced 2-fold or more by the toxT mutation in this experiment. As shown in Table 2, the use of a gDNA-derived reference sample produces expression ratios for ToxT-regulated genes that are very similar to those obtained by using the direct cohybridization analysis of cDNA derived from the two strains. Therefore, using a gDNA reference for transcriptional profiling is apparently as accurate as using a cDNA reference. Based on these data and those of others (23, 24, 33), we conclude that use of a gDNA-derived reference is a convenient approach to obtaining transcriptional profiling data sets that can be accurately compared with each other to predict relative changes in gene expression.
Transcriptome of V. cholerae Present in Stools Derived from Cholera Patients.
Having established the utility of a gDNA reference for microarray analysis of mutant strains, we used this method to characterize the transcriptome of V. cholerae shed during human infection. In these experiments, we purified total RNA from rice water stools of two cholera patients and generated labeled cDNAs. The labeled cDNAs were cohybridized to microarrays in the presence of a differentially labeled N16961 gDNA reference, and the resulting data were analyzed as described in Materials and Methods. We then selected the top one-third most highly expressed genes in the cholera stool vibrios for further informatic analysis to determine their likely function (Table 3).
Table 3.
Summary of the top one-third most highly expressed genes in humans shed V. cholerae
| No. of genes (%) | |
|---|---|
| Chromosome location | |
| Large chromosome | 1,169 (90.27) |
| Small chromosome | 126 (9.73) |
| Functional class | |
| Adaptations to atypical conditions | 5 (0.39) |
| Biosynthesis of amino acids, cofactors, and prosthetic groups | 71 (5.48) |
| Cell division | 7 (0.54) |
| Cell envelope | 50 (3.86) |
| Central intermediary metabolism | 24 (1.85) |
| Chemotaxis and motility | 36 (2.78) |
| DNA metabolism | 28 (2.15) |
| Energy metabolism | 61 (4.71) |
| Fatty acid and phospholipid metabolism | 18 (1.39) |
| Hypothetical, conserved hypothetical, and unknown proteins | 713 (55.06) |
| Pathogenesis | 15 (1.16) |
| Protein fate | 27 (2.08) |
| Protein synthesis | 65 (5.02) |
| Purines, pyrimidines, nucleosides, and nucleotides | 16 (1.24) |
| Regulatory functions | 65 (5.02) |
| Toxin production and resistance | 1 (0.08) |
| Transcription | 19 (1.47) |
| Transport and binding proteins | 60 (4.63) |
The vast majority of the genes most highly expressed by vibrios present in the cholera stools resided on the large chromosome (≈90%), and 50% of the genes fell into the hypothetical, conserved hypothetical, and unknown functional category. Fifteen genes were classified by TIGR as V. cholerae pathogenicity genes, including eight genes associated with the ToxR regulon (ctxAB, tcpP, tcpH, tcpR, tcpQ, tagA, and tagD). Besides ctxAB, two genes involved in the morphogenesis of CTXΦ were also highly expressed (ace and cep), suggesting that this phage may be replicating in stool vibrios. CTXΦ replication is induced by “SOS” signals associated with DNA damage (34) and several other SOS-inducible genes were also noted as being expressed by vibrios present in the stools, including lexA, dinF, radC, and radO. Alternatively, increased expression of ace and cep may reflect RstC-mediated antirepression of the CTXΦ (39). Three genes involved in RTX toxin production (35) were highly expressed (rtxA, rtxC, and rtxD), consistent with a role for this toxin in vivo (36). A number of genes involved in the production of adhesins or type IV related pili were also expressed (e.g., pilT, mshK, VC1236, and VC1998) and included two clusters of in vivo-expressed genes that were organized in potential operons (e.g., VC0857, VV0858, and VC0861; VC2630, VC2631, and VC2632). Additional virulence-related genes that were identified included 36 genes involved in motility and chemotaxis, 8 genes involved in iron transport, and 5 genes associated with anaerobic metabolism (Tables 3 and 5–8).
Previous microarray analysis of stool vibrios did not detect induction of CT or TCP transcripts in vibrios present in cholera stools relative to an in vitro grown V. cholerae strain (22). To further understand the absolute expression levels of these genes in stool-derived vibrios, we compared the transcriptomes of N16961 wild-type and N16961 toxT grown under AKI conditions to the transcriptome of the cholera stool-derived vibrios (Table 4). We reasoned that this approach might permit a qualitative assessment of changes in gene expression levels relative to the expression of all of the genes in the genome. We rank-ordered gene expression levels across these three transcriptomes (an approach made possible by using a gDNA-derived reference sample). These data show that in N16961 grown under AKI conditions, the genes in the TCP and CT operons are expressed at a very high level, with the majority of these genes present in the top 20% of the rank-ordered list. These same genes were much lower on the rank-ordered list generated from the toxT mutant grown in AKI (Table 4), consistent with their diminished expression in the toxT mutant background (Table 2). In comparison, the TCP and CT genes appear at an intermediate position in the rank-ordered list for the transcriptome of vibrios derived from human cholera stool (Table 4). These results suggest that vibrios present in cholera stool do contain significant levels of CT and TCP gene transcripts.
Table 4.
TCP and CT expression in stool
| Gene | Expression rank
|
||||
|---|---|---|---|---|---|
| WT* | toxT* | Stool† | Loop‡ | LB‡ | |
| tcpP | 29 | 79 | 1,265 | 152 | 1,150 |
| tcpH | 33 | 44 | 989 | 191 | 1,092 |
| tcpA | 7 | 3,041 | 2,342 | 3,562 | 3,533 |
| tcpB | 28 | 3,389 | 2,743 | 3,720 | 3,831 |
| tcpQ | 26 | 3,145 | 1,207 | 3,495 | 2,684 |
| tcpC | 49 | 3,434 | 3,026 | 3,611 | 3,783 |
| tcpR | 40 | 1,878 | 890 | 2,877 | 2,759 |
| tcpD | 389 | 3,298 | 1,535 | 3,278 | 3,640 |
| tcpT | 50 | 2,481 | 1,613 | 3,472 | 3,333 |
| tcpE | 44 | 3,243 | 1,557 | 3,498 | 3,738 |
| tcpF | 192 | 1,696 | 1,590 | 3,600 | 3,676 |
| ctxB | 45 | 2,990 | 1,069 | 1,783 | 1,925 |
| ctxA | 9 | 2,927 | 678 | 2,452 | 2,151 |
There was substantial overlap between the top one-third most highly expressed V. cholerae genes in stool and the top one-third most highly expressed genes in V. cholerae grown to mid-exponential phase in rabbit ileal loops (21) (507 shared genes, see Tables 5–8). The largest group of shared genes were those classified as hypothetical, conserved hypothetical, or unknown genes (245 genes). Genes encoding the translational machinery of the cell were also highly represented, with 37 of the 59 genes for ribosomal proteins appearing on both lists (Tables 5–8). Additional overlaps included genes involved in flagellin synthesis, chemotaxis, iron transport, anaerobic metabolism, and several regulatory genes including tcpPH. The overlap in these groups of highly expressed genes suggests that vibrios growing in rabbit ileal loops and human intestine encounter similar environments. For instance, the two groups include two genes encoding subunits of fumarate reductase (VC2658 and VC2659) and the gene encoding ArcA, a repressor of aerobic respiration genes. As discussed earlier (21), the expression of these genes suggests that bacteria in both rabbit ileal loops and human cholera stool have experienced an anaerobic environment where use of fumarate as an alternative electron acceptor and the repression of aerobic metabolism genes would be beneficial. Two genes involved in biotin biosynthesis (bioC and bioD) were also highly expressed in cells harvested from human cholera stool and rabbit ileal loops, consistent with the conclusion that biotin is not available in vivo (37). Three genes that are induced by iron limitation and that are involved in uptake of ferric and ferrous iron (feoA, viuB, and irgB) were also present in both high-expression groups, suggesting that iron limitation is common to both rabbit and human gastrointestinal environments. Finally, both tcpP and tcpH were on the overlap list, indicating that these positive regulators of TCP and CT expression are expressed strongly in vibrios growing intraintestinally in both humans and rabbits.
Discussion
DNA microarray technology is revolutionizing the field of bacterial pathogenesis by allowing researchers to monitor the expression of thousands of genes during the course of an in vitro or in vivo experiment. In this report, we have applied this technology to conduct a genome-wide search for V. cholerae genes belonging to the ToxR regulon, the key group of genes responsible for the virulence properties of this organism in humans. We also used microarrays to analyze the transcriptional state of vibrios shed from cholera patients.
We first compared the gene expression profiles of V. cholerae toxRS, tcpPH, and toxT mutants that were grown under in vitro conditions that are optimal for the expression of CT by El Tor O1 and O139 strains of V. cholerae. The transcriptional profile of the toxT mutant revealed the presence of few new ToxT-regulated genes. Newly identified genes include VC1091 (oligopeptide periplasmic binding protein), VC1835 (pal); VC2766 (atpA); VCA0059 (lpp); VCA0732 (conserved hypothetical); VCA1042 (unknown gene). Among these, VC1835, VCA0059, VCA0732, and VCA1042 showed similar expression profiles in the toxRS and tcpPH mutants, supporting their identification as ToxT-regulated genes. Because VCA1042 is apparently in an operon and downstream of tagE-2 (VCA1043), its transcription may be a result of expression from the tagE-2 promoter. VC2766 had a modest expression ratio near our cutoff, and its expression was not altered in either the toxRS or tcpPH mutants. VC2766 may therefore be a false positive. The high specificity of ToxT for TCP genes is consistent with the current model of horizontal acquisition of toxT as part of the TCP island (2).
TcpPH have been shown to work cooperatively with ToxRS in activating toxT transcription. Consistent with this was the finding that the TCP and CT operons show reduced transcription in the tcpPH mutant. Interestingly, 53 of the 58 genes that showed 2-fold transcriptional changes in the tcpPH mutant also showed corresponding, and usually greater, transcriptional changes in the toxRS mutant (Supporting Data). Our analysis therefore extends the role of TcpPH in cooperating with ToxR to co-regulate many new genes. Nevertheless, it should be noted that the average difference in expression of the CT and TCP genes in the various regulatory mutants was 3.6-fold. Thus, our 2-fold cutoff may have led us to designate some genes as being ToxRS-, TcpPH-, or ToxT-regulated that actually are not. More work is needed to confirm that these newly identified genes are indeed controlled by these regulatory proteins.
The toxRS mutation resulted in the most dramatic transcriptional changes. Our analysis revealed an expanded role for ToxRS in regulating the expression of other outer membrane proteins (e.g., OmpW) and lipoproteins in addition to the previously identified OmpT and OmpU. Our results also suggest a role for ToxRS in regulating metabolic responses. For example, the expression of 23 genes involved in maltose and glycerol metabolism, and peptide transport were altered in the toxRS mutant. Interestingly, we found a role for toxRS in regulating the expression of several genes involved in motility and chemotaxis, a phenotype that is inversely correlated with virulence gene expression (38). ToxRS was also found to repress the expression of two copies of acetate kinase, which is in involved in synthesis of acetyl phosphate. Another gene involved in acetyl phosphate synthesis (pta) was previously implicated in in vivo survival and virulence (37).
In this study, we also explored the use of two microarray normalization strategies: direct comparison to a cDNA reference and the use of a universal gDNA reference. The function of the reference sample in microarray analysis is to provide a measurable expression intensity for each microarray feature. The use of a cDNA reference can make comparative analysis of microarray data problematic because most RNA samples do not contain representative transcripts for all of the features on the microarray. Thus, if the intensity value of any microarray feature in the reference sample is not significantly above background (or threshold, because it is poorly expressed or not represented in the reference sample), then that feature cannot be accurately measured in the test sample (23, 24). In addition, the inherent variability in the preparation of a cDNA reference complicates comparative analysis of microarray data. Experimental conditions that may be routinely replicated in one laboratory may not be replicated in another laboratory because of variables such as different growth conditions, handling and processing of mRNA, and microarray performance.
As an alternative to cDNA references, we investigated the use of a gDNA reference. A gDNA-derived reference addresses many of the shortcomings of a cDNA reference. A gDNA-derived reference is easier to produce, contains full genomic representation, and exhibits superior template stability and more consistent synthesis. In addition, a gDNA reference is universally available and thus allows researchers directly to compare microarray-derived expression data from different experiments and laboratories. In the case of spotted arrays, a gDNA reference provides a better control for individual microarray variation because it addresses all features, thus controlling for feature loading, spot morphology and hybridization inconsistencies. In this report, we took advantage of our knowledge of the ToxR regulon to test the utility of a gDNA reference in microarray analysis. Our results were quite clear and established that the same genes were scored as ToxT-regulated and nearly the same expression ratios were obtained when data were compared using a gDNA-derived reference (Table 2).
Validation of the gDNA-derived reference allowed us to compare the expression profile of vibrios present in the stools of cholera patients to other microarray data sets generated by using the same gDNA-derived reference. Specifically, we were able to examine whether vibrios present in cholera stools showed a degree of expression of the CT and TCP genes that corresponded more closely to wild-type or toxT bacteria grown under optimal in vitro conditions for the expression of these virulence genes (i.e., AKI conditions). Our results suggest that CT and TCP are likely expressed by vibrios in cholera stool but at a lower level than under optimal in vitro growth conditions. The TCP and CT genes appear at an intermediate position in the rank-ordered list for the transcriptome of stool-derived vibrios, indicating that vibrios present in cholera stool do contain significant levels of CT and TCP gene transcripts. The two simplest explanations are that either CT and TCP genes are expressed at an intermediate level in all vibrios present, or they are expressed at a very high level by a small subpopulation of vibrios. It may be that only a small population of V. cholerae reach an intestinal microenvironment where CT and TCP expression are maximally induced (e.g., the intestinal crypts or close proximity to the epithelial cell brush border), and the resultant diarrheal purge provides growth substrate that allows expansion of the rest of the vibrio population under in vivo growth conditions that are suboptimal for CT and TCP expression.
Additionally, vibrios present in cholera stools show a pattern of gene expression that is both different and similar to mid-exponential phase bacteria harvested from either LB medium or rabbit ileal loops (Tables 4–8). Notable similarities between the transcriptome of vibrios harvested from human stools versus rabbit ileal loops are the expression of many conserved hypothetical, hypothetical and unknown proteins in both populations, as well as the expression of many genes involved in growth under anaerobic conditions and nutrient limitation (particularly iron and biotin). Notable differences between the transcriptome of vibrios harvested from rabbit ileal loops versus human stools are that a larger fraction of the most highly expressed genes were located on the large chromosome for vibrios present in human cholera stool. In this study, we found that only 9.73% of the top one-third genes most highly expressed in human stool were located on the small chromosome, whereas we previously found that 18% of the top one-third most highly expressed genes in rabbit ileal loops resided on the small chromosome (21). The overlap between the top one-third most highly expressed genes in rabbit ileal loops and human cholera stool was significant and consisted of 507 genes. Thirty-seven of these genes encoded ribosomal proteins, and 8 genes are involved in LPS biosynthesis (Tables 5–8). Because ribosomal proteins and LPS biosynthetic genes are among the most highly expressed genes in exponential-phase cells (21, 29), we tentatively conclude that vibrios present in our cholera stools may have been actively dividing when samples were frozen for later analysis. This result suggests that growth phase may be at least in part responsible for the unique “hyperinfectious” phenotype of stool vibrios compared with stationary phase, in vitro grown vibrios as reported by Merrell et al. (22).
This report is the second in a likely series of papers that will analyze transcriptome data from V. cholerae present in clinical materials from human cholera patients. The first paper (22) used a cDNA internal reference that was derived from mid-exponential RNA purified from cells grown under laboratory conditions to compare stool vibrios to in vitro-grown stationary-phase V. cholerae. These data cannot be compared directly to our data because of the use of different reference samples. Our decision to use a gDNA-derived reference sample was based on the rationale that the generation of a reference sample from any single, arbitrarily selected in vitro condition will not contain sufficient genomic representation to develop a complete transcriptome. Furthermore, a universal gDNA reference sample facilitates comparative analysis of microarray data between different experiments, which allowed us to make direct comparisons to the transcriptome of vibrios harvested from rabbit ileal loops (21) or strains grown under in vitro conditions such as AKI. Nonetheless, Merrell et al. (22) concluded that the expression of certain signature genes suggested that low iron and anaerobiosis are physiological states experienced by vibrios in the human gastrointestinal tract. We have reached similar conclusions for vibrios harvested from rabbit ileal loops (21) and from cholera stool samples. Thus, some consistent conclusions are emerging from the analysis of V. cholerae harvested from in vivo environments, and these will guide further genetic analysis of this interesting pathogen.
Supplementary Material
Acknowledgments
We thank Su Chiang for manuscript editing, Emmy Balon for technical assistance, Betty Guo for methodological suggestions, and Scotty Merrell and Andy Camilli for helpful discussions. This work was supported by National Institutes of Health Grant AI18045 (to J.M.). J.B. was supported by a Postdoctoral Fellowship from the Cystic Fibrosis Foundation. J.Z. was supported by a National Service Research Award from the National Institutes of Health.
Abbreviations
- CT
cholera enterotoxin
- TCP
toxin coregulated pilus
- gDNA
genomic DNA
References
- 1.Cotter P A, DiRita V J. Annu Rev Microbiol. 2000;54:519–565. doi: 10.1146/annurev.micro.54.1.519. [DOI] [PubMed] [Google Scholar]
- 2.Faruque S M, Albert M J, Mekalanos J J. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thelin K H, Taylor R K. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tacket C O, Taylor R K, Losonsky G, Lim Y, Nataro J P, Kaper J B, Levine M M. Infect Immun. 1998;66:692–695. doi: 10.1128/iai.66.2.692-695.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiRita V J. Mol Microbiol. 1992;6:451–458. doi: 10.1111/j.1365-2958.1992.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 8.Higgins D E, Nazareno E, DiRita V J. J Bacteriol. 1992;174:6874–6980. doi: 10.1128/jb.174.21.6974-6980.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiRita V J, Parsot C, Jander G, Mekalanos J J. Proc Natl Acad Sci USA. 1991;88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiRita V, Neely M, Taylor R, Bruss P. Proc Natl Acad Sci USA. 1996;93:7991–7995. doi: 10.1073/pnas.93.15.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Häse C C, Mekalanos J J. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skorupski K, Taylor R K. Proc Natl Acad Sci USA. 1997;94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovacikova G, Skorupski K. J Bacteriol. 1999;181:4250–4256. doi: 10.1128/jb.181.14.4250-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Häse C C, Mekalanos J J. Proc Natl Acad Sci USA. 1999;96:3183–3187. doi: 10.1073/pnas.96.6.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovacikova G, Skorupski K. Mol Microbiol. 2002;46:1135–1147. doi: 10.1046/j.1365-2958.2002.03229.x. [DOI] [PubMed] [Google Scholar]
- 16.Behari J, Stagon L, Calderwood S B. J Bacteriol. 2001;183:178–188. doi: 10.1128/JB.183.1.178-188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee S H, Hava D L, Waldor M K, Camilli A. Cell. 1999;99:625–634. doi: 10.1016/s0092-8674(00)81551-2. [DOI] [PubMed] [Google Scholar]
- 18.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, et al. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dziejman M, Balon E, Boyd D, Fraser C M, Heidelberg J F, Mekalanos J J. Proc Natl Acad Sci USA. 2002;99:1556–1561. doi: 10.1073/pnas.042667999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J, Miller M B, Vance R E, Dziejman M, Bassler B L, Mekalanos J J. Proc Natl Acad Sci USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Q, Dziejman M, Mekalanos J J. Proc Natl Acad Sci USA. 2003;100:1286–1291. doi: 10.1073/pnas.0337479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merrell D S, Butler S M, Qadri F, Dolganov N A, Alam A, Cohen M B, Calderwood S B, Schoolnik G K, Camilli A. Nature. 2002;417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Talaat A M, Howard S T, Hale W T, Lyons R, Garner H, Johnston S A. Nucleic Acids Res. 2002;30:e104. doi: 10.1093/nar/gnf103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dudley A M, Aach J, Steffen M A, Church G M. Proc Natl Acad Sci USA. 2002;99:7554–7559. doi: 10.1073/pnas.112683499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bina J E, Mekalanos J J. Infect Immun. 2001;69:4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imai Y, Matsushima Y, Sugimura T, Terada M. Nucleic Acids Res. 1991;19:2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Champion G A, Neely M N, Brennan M A, DiRita V J. Mol Microbiol. 1997;23:323–331. doi: 10.1046/j.1365-2958.1997.2191585.x. [DOI] [PubMed] [Google Scholar]
- 28.Iwanaga M, Yamamoto K, Higa N, Ichinose Y, Nakasone N, Tanabe M. Microbiol Immunol. 1986;30:1075–1083. doi: 10.1111/j.1348-0421.1986.tb03037.x. [DOI] [PubMed] [Google Scholar]
- 29.Wei Y, Lee J M, Richmond C, Blattner F R, Rafalski J A, LaRossa R A. J Bacteriol. 2001;183:545–556. doi: 10.1128/JB.183.2.545-556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waldor M K, Mekalanos J J. Infect Immun. 1994;62:72–78. doi: 10.1128/iai.62.1.72-78.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reidl J, Klose K E. FEMS Microbiol Rev. 2002;26:125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 32.Murley Y M, Carroll P A, Skorupski K, Taylor R K, Calderwood S B. Infect Immun. 1999;67:5117–5123. doi: 10.1128/iai.67.10.5117-5123.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weil M R, Macatee T, Garner H R. BioTechniques. 2002;32:1310–1314. doi: 10.2144/02326mt01. [DOI] [PubMed] [Google Scholar]
- 34.Faruque S M, Asadulghani, Alim A R, Albert M J, Islam K M, Mekalanos J J. Infect Immun. 1998;66:3752–3757. doi: 10.1128/iai.66.8.3752-3757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fullner K J, Mekalanos J J. EMBO J. 2000;19:5315–5323. doi: 10.1093/emboj/19.20.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fullner K J, Boucher J C, Hanes M A, Haines G K, III, Meehan B M, Walchle C, Sansonetti P J, Mekalanos J J. J Exp Med. 2002;195:1455–1462. doi: 10.1084/jem.20020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chiang S L, Mekalanos J J. Mol Microbiol. 1998;27:797–806. doi: 10.1046/j.1365-2958.1998.00726.x. [DOI] [PubMed] [Google Scholar]
- 38.Gardel C, Mekalanos J J. J Bacteriol. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis B M, Kimsey H H, Kane A V, Waldor M K. EMBO J. 2002;21:4240–4249. doi: 10.1093/emboj/cdf427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.