Abstract
Bacteriophytochrome photoreceptors (BphPs) are a family of phytochrome-like sensor kinases that help a wide variety of bacteria respond to their light environment. In Agrobacterium tumefaciens, a unique pair of BphPs with potentially opposing roles in light sensing are present. Both AtBphPs contain an N-terminal chromophore-binding domain that covalently attaches a biliverdin chromophore. Whereas AtBphP1 assumes a Pr ground state, AtBphP2 is unusual in that it assumes a Pfr ground state that is produced nonphotochemically after biliverdin binding through a transient Pr-like intermediate. Photoconversion of AtBphP2 with far-red light then generates Pr but this Pr is also unstable and rapidly reverts nonphotochemically to Pfr. AtBphP1 contains a typical two-component histidine kinase domain at its C terminus whose activity is repressed after photoconversion to Pfr. AtBphP2 also functions as a histidine kinase but instead uses a distinct two-component kinase motif that is repressed after photoconversion to Pr. We identified sequences related to this domain in numerous predicted sensing proteins in A. tumefaciens and other bacteria, indicating that AtBphP2 might represent the founding member of a family of histidine phosphorelay proteins that is widely used in environmental signaling. By using these mutually opposing BphPs, A. tumefaciens presumably has the capacity to simultaneously sense red light-rich and far-red light-rich environments through deactivation of their associated kinase cascades.
Almost all organisms use a variety of photoreceptors to help them respond and adapt to their ambient light environment. One of the most influential is the phytochrome (phy) superfamily, a large and diverse group of photochromic photoreceptors recently found to be widely distributed in plants, fungi, and bacteria (1, 2). These homodimeric photoreceptors sense red light (R) and far-red light (FR) through photointerconversion between two stable conformations, a R-absorbing Pr form and a FR-absorbing Pfr form. Typically R triggers phy-regulated responses by converting the initially synthesized Pr to Pfr, whereas FR represses these responses by reverting Pfr back to Pr. By this mechanism, phys act as reversible R/FR “switches” in many aspects of photomorphogenesis (3).
Members of the phy superfamily have a similar protein organization that consists of a signature N-terminal chromophore-binding domain (CBD) that autocatalytically binds an assortment of bilin chromophores, followed by a divergent C-terminal module involved in signal transduction and homodimerization. Based on several criteria, this superfamily can be divided into three families: plant phys, cyanobacterial phys (Cphs), and bacteriophytochrome photoreceptors (BphPs) (1, 2). The phys and Cphs use the bilins 3E-phytochromobilin and phycocyanobilin (PCB) as chromophores, respectively, which are synthesized from multistep pathways beginning with a heme precursor. These bilins are attached to the apoprotein through a thiolether linkage to a specific cysteine in the CBD. The C-terminal transduction domain of Cphs is strongly related to known two-component sensor kinases, and as expected they behave as histidine kinases in vitro (4). Likewise, phys have a C-terminal domain related to sensor kinases (5, 6). However, this domain is missing critical residues essential for catalysis including the positionally conserved histidine essential for phosphotransfer, implying that phys descended from a Cph progenitor but have since acquired a new type of kinase activity. In vitro phys appear to behave as regulated serine/threonine kinases (6) but how this activity relates to phy action remains unclear (7).
The BphPs are common among photosynthetic and nonphotosynthetic eubacteria and present in some fungi (1, 8). Like Cphs, they often contain the canonical two-component histidine kinase motif at their C termini and act as histidine kinases in vitro. The main distinction is that they use the bilin biliverdin (BV) as the chromophore (8). BV is synthesized directly from heme by a heme oxygenase that is sometimes encoded within the BphP operons, indicating a tight relationship between apoprotein and chromophore synthesis. How BV is autocatalytically attached to the apoprotein is unresolved; it could involve either a histidine adjacent to the cysteine used by phys and Cphs (8) or an alternative cysteine near the N terminus of the CBD (9). BphPs act as photoregulated kinases that presumably initiate their phosphorelay cascade by phosphotransfer to an associated response regulator (RR). In some cases, this RR is translationally appended to the C-terminal end of the BphP (1, 8).
To better understand how BphPs help bacteria and fungi monitor their light environments, we have begun to dissect their sensory transduction chains at the biochemical and genetic levels. One interesting pair, AtBphP1 and AtBphP2, was detected in the bacterium Agrobacterium tumefaciens, a soil-borne pathogen of plants (9–11). Whereas the sequence of AtBphP1 suggested that it behaved as a typical BphP, the sequence of AtBphP2 was unusual. Following the CBD, AtBphP2 contains a domain only weakly related to orthodox two-component histidine kinases and ends in a RR domain, suggesting that AtBphP2 participates in a distinct type of phosphotransfer. Here, we show that AtBphP2 is indeed novel among defined members of the phy superfamily with the unique characteristics that it assumes a Pfr and not a Pr ground state and that it uses a two-component histidine kinase domain different from those previously described. Using this kinase domain as a query, we subsequently found related sequences in a variety of bacterial proteins of unknown activity but predicted to have roles in environmental signaling. As such, this domain appears to participate in a distinct type of two-component phosphorelay cascade that may be prevalent among particular families of bacteria.
Materials and Methods
Identification and Expression of AtBphPs.
AtBphP1 and AtBphP2 were identified in the A. tumefaciens genomic database (www.ddbj.nig.ac.jp/e-mail/homology.html) by blast (12). Alignmentswere created by using CLUSTALX MAC V.1.8, and searches for known proteins motifs were made by smart (http://smart.embl-heidelberg.de). The coding regions for AtBphP1 and AtBphP2, AtBphP2-N731, the C-terminal RR domain of AtBphP2, and AtRR1 were PCR-amplified from A. tumefaciens strain C58 by using primers designed to introduce NdeI and XhoI sites before the ATG and designated stop codons, respectively. The NdeI–XhoI-digested PCR products were cloned into pET21b (Novagen) that was similarly digested, resulting in the addition of a His-6 tag at the C terminus. GenBank accession numbers for sequences described herein are: AtBphP1 (NP_354963), AtBphP2 (NP_355125), AtRR1 (NP_532668), ExsG (NP_354961), Xanthomonas axonopodis (NP_644587), and Xanthomonas campestris (NP_639488).
Recombinant proteins were expressed in Escherichia coli strain BL21-Codon Plus (DE3)-RIL (Stratagene) and purified by nickel chelate affinity chromatography as described (8). For AtBphPs assembled in vitro, the apoproteins were incubated in darkness in a 10-fold molar excess of BV or PCB (Porphyrin Products, Logan, UT). Absorbance spectra of AtBphPs were determined after saturating irradiations with light at 690 nm (R) and 775 nm (FR) provided by interference filters. Covalent attachment of bilins to AtBphP1 and AtBphP2 was monitored by zinc-induced fluorescence of the chromoproteins subjected to SDS/PAGE (8).
Protein Kinase Assays.
Protein kinase assays were performed as described (8). BV adducts of AtBphP1 and AtBphP2 were either kept in the dark or irradiated with saturating R or FR and with the kinase reactions performed at 25°C with 0.2 μM [γ-32P]ATP. The reactions were quenched by addition of SDS/PAGE sample buffer. For phosphotransfer from AtBphP1 to AtRR1, the 32P-AtBphP1 intermediate was first generated by a 15-min autophosphorylation reaction with the photoreceptor in the Pr form. AtBphP1 was then left as Pr or photoconverted to Pfr and incubated for an additional 5 min with AtRR1. 32P-labeled products were quantified by using a GS-525 PhosphorImager (Bio-Rad).
Results
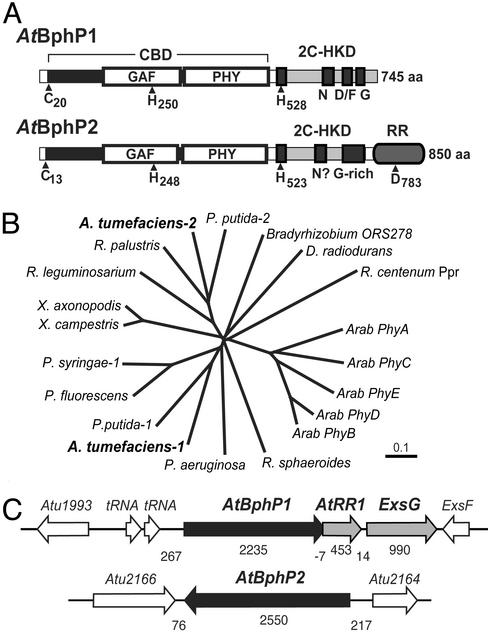
In searches of various bacteria genomic sequences for new BphPs, we and others (9–11) discovered a potential pair (AtBphP1 and AtBphP2) in the α-proteobacterium A. tumefaciens C58, a pathogen commonly used as a vehicle for plant gene transfer. They are encoded by separate operons in opposing orientations within the circular chromosome. Like phys, CphPs, and other BphPs (1, 2), both AtBphP1 and AtBphP2 contain an N-terminal region CBD that includes both the GAF and PHY subdomains (Fig. 1A). Both of these domains are essential for binding bilins and creating the striking Pr/Pfr spectral properties particular to the phy superfamily (2). Included in the CBD are the positionally conserved cysteine (Cys-20 and Cys-13) and histidine (His-250 and His-248) residues that could serve as the chromophore-binding site, by using either a thiolether or Schiff-base-type linkage, respectively (8, 9, 13). To place the AtBphPs within the phy superfamily, we generated an unrooted phylogenetic tree by comparing their GAF domain sequences to those from representative members of the phy and BphP families. As can be seen in Fig. 1B, AtBphP1 and AtBphP2 differ phyletically, with AtBphP2 clustering on a branch with Pseudomonas putida BphP2, Rhizobium leguminosarium BphP, and Rhodopseudomonas palustris BphP (14), suggesting that these four represent a distinct BphP type (Fig. 1B).
Figure 1.
Molecular characterizations of AtBphP1 and AtBphP2. (A) Structural organization of the AtBphP proteins. The CBD is identified by the bracket; the two-component histidine kinase domain (2C-HKD) by gray boxes, and the GAF and PHY domains by white boxes. Within the 2C-HKD, the H-, N-, D/F-, and G-boxes for AtBphP1, and the H-box, possible N-box, and glycine-rich domain for AtBphP2 are shown. The N-terminal cysteine and histidine residues important for BV ligation and the C-terminal histidines and aspartate residues involved in phosphotransfer are located by arrowheads. (B) Phyletic comparison of the GAF domains from AtBphP1 and AtBphP2 and representative members of the phy, Cph, and BphP families. (C) Organization of the predicted AtBphP1 and AtBphP2 operons (gray and black arrows). Numbers below indicate the size of the ORFs and the intergenic regions.
AtBphP1 terminates in a canonical two-component histidine kinase domain, which contains the H-, N-, D/F-, and G-boxes predicted to direct autophosphorylation of the chromoprotein (15–17). Often the BphP operons include other ORFs that likely encode additional component(s) of their sensory cascades (1, 8). AtBphP1 appears to be part of a three-gene operon (Fig. 1C and below). Immediately downstream is the coding region for a 151-aa protein related to the CheY superfamily of bacterial RRs (AtRR1). This RR has the positionally conserved aspartate involved in phosphotransfer but is missing an obvious effector domain (e.g., DNA binding), indicating that additional phosphorelay proteins (e.g., histidine phosphotranferases) may be needed for signal output (15). Immediately after AtRR1 is another ORF, designated ExsG, that encodes a 330-aa protein bearing an N-terminal RR domain.
The organization of AtBphP2 is distinct from that of AtBphP1. The region after the CBD bears little homology to known histidine kinases, except for the presence of a positionally conserved histidine (His-523) in a H-box-like motif [EL−HRVNK (16)] that could participate in phosphotransfer. Together with the fact that AtBphP2, like the three other BphPs in its clade (Fig. 1B), terminates in a RR domain, suggested to us that AtBphP2 participates in a histidine phosphorelay chemically similar to the two-component system (Fig. 1A). The sequence of the RR domain is related to members of the CheY superfamily, including the aspartate phosphoaccpetor site (D783), but differs phyletically from the AtRR1 present with the AtBphP1 operon. This distinction implies that AtBphP1 and AtBphP2 proteins may operate in nonoverlapping phosphorelay cascades by using distinct sets of RRs. No other ORFs appear to be associated with the AtBphP2 sequence. Although some bacteria encode within their BphP operon(s) a heme oxygenase protein responsible for synthesizing BV (8), the gene for this protein is outside of both BphP operons in A. tumefaciens.
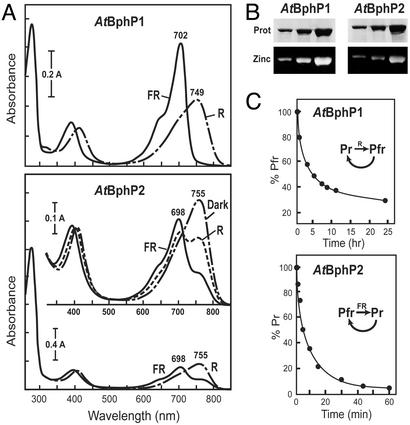
To demonstrate that both A. tumefaciens sequences function as BphPs, we assembled each with BV and measured their R/FR photochromic spectral properties. His-6-tagged versions were expressed in E. coli, purified by nickel chelate affinity chromatography, and incubated with BV in the dark. Similar to previous studies (9), recombinant AtBphP1 efficiently bound BV, as determined by fluorescence of the chromoprotein in the presence of zinc and UV light, and generated a chromoprotein with R/FR photoreversible absorbance spectra typical of BphPs (Fig. 2 A and B). The form initially synthesized was Pr with an absorbance maximum at 702 nm; it could then be converted by R to a photoequilibrium mixture containing mostly Pfr with an absorbance maximum at 749 nm (Fig. 2A). However, the Pfr form of AtBphP1 was unstable, reverting to Pr during prolonged incubations in the dark (Fig. 2C and ref. 9).
Figure 2.
Spectral properties of AtBphP1 and AtBphP2. (A) Absorbance spectra of the BV-holoproteins after saturating irradiations with R and FR or after a prolonged incubation of AtBphP2 in the dark. (B) Covalent binding of BV to AtBphP1 and AtBphP2. Apoproteins were incubated with BV in vitro, and the reaction products were subjected to SDS/PAGE and either stained for protein with Commassie blue (Upper) or assayed for the bound bilin by zinc-induced fluorescence (Lower). (C) Dark reversion of the BphPs. AtBphP1 and AtBphP2 were photoconverted to Pfr and Pr, respectively, and then assayed for their dark reversion back to the other spectral form.
AtBphP2 also bound BV but with less efficiency than AtBphP1 as determined by reduced zinc-induced fluorescence and a lower 280-nm/698-nm absorbance ratio for Pr (Fig. 2 A and B). Surprisingly, the spectral form initially observed for AtBphP2 resembled Pfr with an absorbance maximum at 755 nm and not the expected Pr. Prolonged irradiations of the BV-AtBphP2 chromoprotein with FR led to the formation of a Pr-enriched mixture at equilibrium. Its absorbance spectrum contained a peak at 698 nm, which is characteristic of Pr, plus a shoulder at ≈750 nm that likely represented residual Pfr. Subsequent saturating irradiation with R generated a Pfr-enriched mixture with two absorbance maxima at ≈700 and ≈750 nm. Regardless of the final irradiation, AtBphP2 rapidly converted (t1/2 ≈5 min) in the dark to a preparation containing almost exclusively Pfr as judged by the absence of a Pr shoulder at ≈700 nm in the spectrum (Fig. 2A). Taken together, AtBphP2 appears to assume a Pfr and not a Pr conformation as the ground state after assembly with BV.
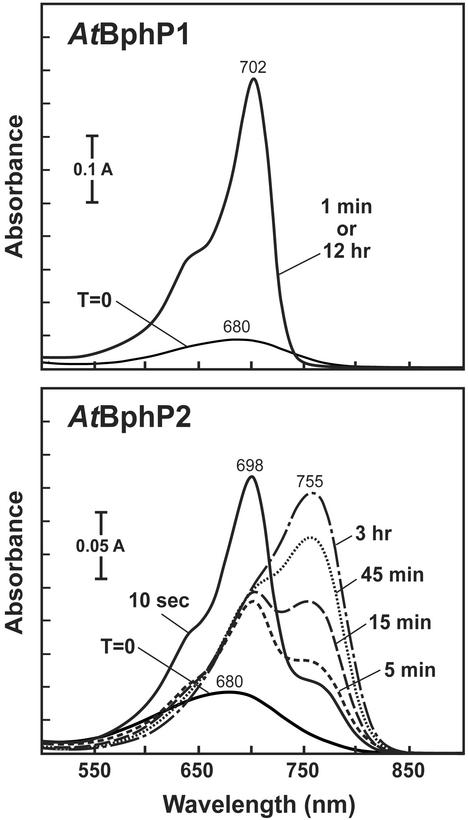
To examine the kinetics of BV assembly in more detail, recombinant AtBphP1 and AtBphP2 apoproteins were added to BV and the absorbance spectra of the mixtures were continually monitored (Fig. 3). Like previous observations with phys and Cphs (18, 19), AtBphP1 rapidly assembled with BV to generate a Pr conformation that was highly stable even after prolonged incubations in the dark. AtBphP2 also rapidly assembled with BV to generate Pr, but this form was transient and immediately began converting nonphotochemcially to Pfr (t1/2 ≈ 15 min) (Fig. 3). In fact, we could detect this transformation soon after adding BV; although most of the holoprotein after 10 sec was in the Pr form, the shoulder at ≈750 nm indicative of Pfr was readily apparent in the absorbance spectrum. After a 3-hr incubation in the dark, almost all of the holoprotein was transformed to Pfr. Thus although both BphPs initially assembled with BV as Pr, this form was unstable in AtBphP2 and rapidly converted nonphotochemically to Pfr. A truncation of AtBphP2 containing just the CBD (amino acids 1–505) retained these spectral characteristics, whereas a smaller version (1–314) did not even though it still bound BV with high efficiency, indicating that the entire CBD may be required for the unique spectral properties of AtBphP2 (data not shown).
Figure 3.
Absorbance spectrum changes of AtBphP1 and AtBphP2 immediately after their assembly with BV. The recombinant apoproteins were added to BV and incubated in the dark. Absorbance spectra were recorded at the indicated times.
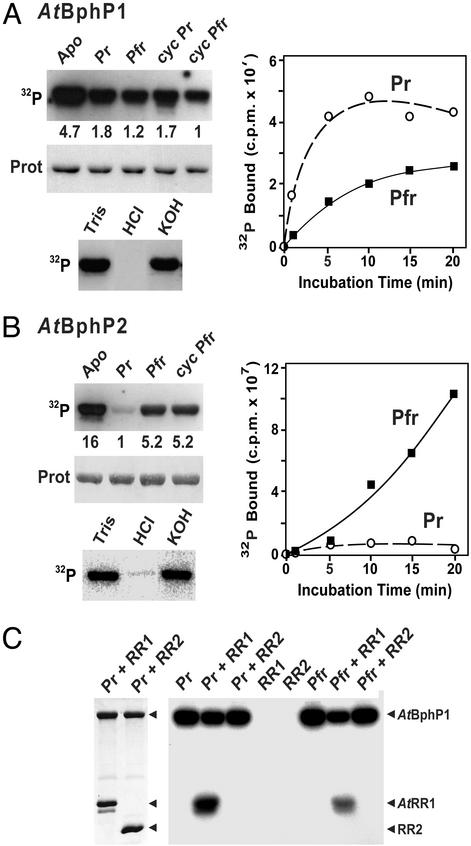
We considered it likely that AtBphP1 functions as a light-regulated kinase based on the presence of the two-component histidine kinase domain at its C terminus and an adjacent RR in the AtBphP1 operon (Fig. 1 and ref. 9). However, the activity for AtBphP2 was uncertain even with the possible H-box and appended RR given that the C-terminal region was not significantly similar to known kinases. To test for kinase activity, the purified BV-assembled holoproteins were incubated in the presence of [32P]ATP and examined for autophosphorylation activity. As can be seen in Fig. 4, both chromoproteins had an intrinsic protein kinase activty that could generate an autophosphorylated form in vitro. Their stability in 3 M KOH but not in 1 M HCl indicated that phosphohistidine intermediates were produced (4, 8, 15). They were also more active as apoproteins than as BV-holoproteins, suggesting that the chromophore represses their kinase activities. For AtBphP1, the kinase activity was ≈2 times greater as Pr than Pfr and could be photoreverted by R/FR cycles (Fig. 4A). Incubation of phosphorylated AtBphP1 with its associated AtRR1 allowed the transfer of the bound phosphate to the RR, with Pr being 10 times more efficient than Pfr (Fig. 4C). This transfer was not detected when an equal amount of the RR domain from AtBphP2 was used as the acceptor, suggesting that the phosphorelay was RR-specific. In contrast, AtBphP2 had the opposite regulation of its kinase activity (Fig. 4B). Its activity was photoreversible with the Pfr form being ≈5 times more active than Pr. A truncated form of AtBphP2 missing the RR domain (AtBphP2-N731) retained this kinase activity, suggesting that the region responsible lies between the CBD and RR domains.
Figure 4.
AtBphP1 and AtBphP2 act as light-regulated histidine kinases. Autophosphorylation of purified AtBphP1 (A) and AtBphP2 (B) without chromophore (Apo) or assembled with BV and photoconverted to Pr, Pfr, or photocycled back to Pr (cycPr) and Pfr (cycPfr). The reaction products were separated by SDS/PAGE, and the gels were either subjected to autoradiography (Top) or stained for protein with Ponceau red (Middle). Normalized relative phosphorylation levels are indicated between the panels. The stability of the autophosphorylated form of each after incubations in 50 mM Tris (pH 7.0), 1 M HCl, or 3 M KOH is shown (Bottom). The kinetics of 32P incorporation over time either as Pr (○) or Pfr (■) are shown (Right). (C) Specific transfer of phosphate from AtBphP1 to AtRR1. AtBphP1 was autophosphorylated as Pr, left either as Pr or photoconverted to Pfr, and then incubated for 5 min with equal amounts of AtRR1 or the 119-aa RR domain derived from AtBphP2 (RR2). The products were visualized as in A. (Left) Staining for protein with Commassie blue. (Right) Autoradiogram.
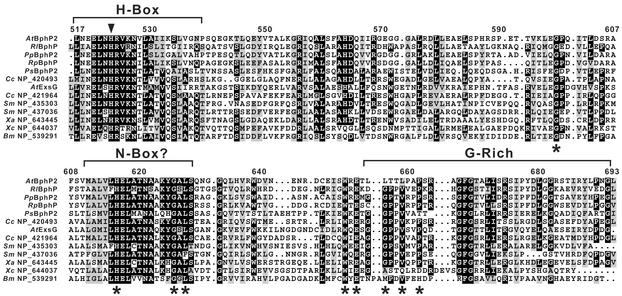
To identify the kinase domain within AtBphP2, we searched the area between the CBD and the RR domains for sequences related to motifs conserved among two-component histidine kinases (16). Whereas a potential H-box harboring the histidine critical for phosphotransfer was evident, the distil ≈200 residues aligned poorly with previously described two-component histidine kinases, suggesting that the domain was distinct. Using this domain as a query, we searched by blast for other proteins with a related domain. Surprisingly, we detected a number of hypothetical proteins in a variety of bacteria that also contain this sequence near their C termini (Fig. 5 and data not shown). Alignment of these sequences detected several islands of conservation that could represent an N-box and a glycine-rich region that could form an ATP-binding site similar to those in the histidine kinase/Bergerart-fold ATPase superfamily (16, 17, 20) (Fig. 5). Important differences from other two-component histidine kinase families were the apparent absence of a D/F-box and the presence of several conserved residues, including Gly-598, His-616, Trp-651, and Glu-653, that may define this family (Fig. 5). Site-directed mutagenesis of H616 showed that this residue is essential for the kinase activity of AtBphP2 (data not shown).
Figure 5.
Amino acid sequence alignments of the proposed two-component histidine kinase motif present in AtBphP2 with similar motifs from a variety of other bacteria proteins. Representatives include BphPs from R. leguminosarium (Rl), P. putida (Pp), R. palustris (Rp), and P. syringae (Ps), ExsG from the AtBphP1 operon, and predicted sensory proteins from a variety of bacteria, including Caulobacter crescentus (Cc), S. meliloti (Sm), Xanthomonas axonopodis (Xa), Xanthomonas campestris (Xc), and B. melitensis (Bm). Reverse type and gray boxes denote identical and similar amino acids, respectively. Brackets identify the H-box, a potential N-box, and a Gly-rich domain, and the arrowhead locates the conserved histidine presumed to form the phosphorylated intermediate. Asterisks show conserved residues that help distinguish this family from others within the two-component kinase superfamily.
To date, we identified >70 proposed proteins from a variety of α- and γ-proteobacteria that have this presumed two-component histidine kinase domain (Fig. 5 and B.K., unpublished work). In A. tumefaciens alone, seven proteins in addition to AtBphP2 were among this collection, including ExsG in the AtBphP1 operon (GenBank accession nos. NP_420493, NP_532667, NP_534647, NP_534648, NP_535462, NP_535747, NP_535749, and NP_535898). The three BphPs from R. leguminosarium, R. palustris, and P. putida that phyletically cluster based on their GAF domains also have this signature, further supporting their relatedness. Many of the proteins were found in sequenced members of the Rhizobiaceae that includes Sinorhizobium meliloti, Mesorhizobium loti, R. leguminosarium, R. palustris, and Brucella melitensis in addition to A. tumefaciens, indicating a preferential expansion of the motif in this family. Often the hypothetical proteins were also predicted by smart to contain various types of N-terminal sensor modules like GAF, PAS, and chemotactic methyltransferase, suggesting that this histidine kinase domain plays a pervasive role in environmental signaling. However, it does not appear to be universally present. We were unable to find related motifs in any of the eukaryotic DNA sequence databases or in the complete genomic sequence of a number of cyanobacteria, archaebacteria, and other eubacteria (e.g., E. coli and Bacillus subtilis).
Discussion
In agreement with first pass annotations of the genome (10, 11) and the recent report of Lamparter et al. (9), we identified a pair of BphPs in A. tumefaciens. Here, we show by detailed spectral analysis of recombinant holoproteins that AtBphP1 and AtBphP2 have contrasting photobiological properties and thus could function as opposing light sensors. The most interesting distinction is that AtBphP2 assumes the Pfr and not the Pr form as the ground state, a property opposite of that used by almost all other known members of the phy superfamily (1–3). As a consequence, AtBphP2 would be expected to function as a FR and not a R sensor in the bacterium.
AtBphP1 behaves as a typical BphP bearing a BV-binding domain followed by an obvious two-component histidine kinase domain. It assembles with BV to generate a Pr ground state that has high histidine autophosphorylation activity, which can then selectively transfer the bound phosphate to an aspartate within the RR, AtRR1 encoded within its operon. On photoconversion to Pfr by R, this activity is immediately repressed but slowly restored as Pfr reverts back to Pr in the dark. In contrast, AtBphP2 assembles with BV and first generates a transient Pr-like intermediate, which then transforms nonphotochemically to a stable Pfr form with high histidine kinase activity. On photoconversion to Pr, this kinase activity drops but is quickly restored as the unstable Pr form rapidly reverts to Pfr. Presumably AtBphP2 then phosphorylates its appended RR to initiate a phosphorelay. Because the RRs expected to participate in each of the phosphorelays are missing obvious output domains, additional factors are required for continued signal transmission. For AtBphP1, one possible candidate is ExsG, which contains a RR followed by the newly discovered histidine kinase domain.
Obviously, the photobiological differences between AtBphP1 and AtBphP2 reflect the unique way that each of the apoproteins associates with the BV chromophore. This could involve differences in how the bilin is covalently linked and/or differences in the noncovalent interactions that confer the photoreversible spectral properties characteristic to the phy superfamily. We note that AtBphP2, as compared with other phys, has a substantially reduced Pr absorbance at its peak at 698 nm versus its Soret peak at ≈400 nm. Such a reduction is characteristic of bilins that assume a more helical conformation within their protein binding pockets (19), implying that the molecular environment of BV when bound to AtBphP1 and AtBphP2 differs. Further support that the chromophore-binding pockets are different comes from attempts to assemble the AtBphPs with PCB. Whereas PCB-AtBphP1 adduct is spectrally similar to AtBphP1 assembled with BV (9), the PCB-AtBphP2 adduct acquired aberrant spectral properties, including a near absence of R/FR photoreversibility, suggesting that the “fit” is different (data not shown). We also observed that AtBphP2 binds BV less efficiently than AtBphP1 when BV is added to the apoprotein either in vitro or in vivo by coexpression of the apoprotein with a heme oxygenase (8). Whether this reflects a problem with expression/folding of the recombinant protein or intrinsic differences in holoprotein assembly is not yet known.
Are there other BphPs with photochemical properties analogous to AtBphP2? A recent report by Giraud et al. (14) suggests that the two BphPs from Bradyrhizobium ORS278 and R. palustris also use Pfr as the ground state. By exploiting such opposite BphPs, these anoxygenic photosynthetic bacteria may more easily photoregulate assembly of its photosynthetic apparatus by using a photoreceptor whose absorbance spectrum does not overlap with that of chlorophyll (14). As for the other two within the AtBphP2 clade, preliminary analysis suggests that at least one (R. leguminosarium BphP) is not photochemically related to AtBphP2 (data not shown). Hopefully, by comparative sequence analyses of the CBDs from the A. tumefaciens, Bradyrhizobium and R. palustris BphPs with those of typical phys, we may be able to identify sequence motifs that are relevant to their unique spectral characteristics.
The other novel feature of AtBphP2 is its use of a two-component kinase domain distinct from those described (16, 17). As judged by the sequence alignments for representative members of the family, the kinase domain appears to be missing a D/F-box but contains highly divergent H- and N-boxes and a C-terminal glycine-rich region that may function as the ATP-binding site (16, 17). Given the numerous predicted bacterial signaling proteins with this sequence, it appears to be widely used as a distinct type of phosphorelay signaling system in bacteria. Intriguingly, the three BphPs from R. leguminosarium, R. palustris, and P. putida that are most closely related to AtBphP2, Pseudomonas syringae BphP2, and ExsG within the AtBphP1 operon have this domain, suggesting that it plays a particularly central role in light signaling. It is interesting to note that many proteins with this kinase domain also bear an appended RR domain (e.g., BphP2s from A. tumefaciens, R. leguminosarium, R. palustris, and P. putida), implying that the associated transduction chains use histidine to aspartate phosphotransfers similar to the route used by the more common two-component pathways (15).
Why does A. tumefaciens have such opposing photoreceptor systems? The downstream targets of these two photoregulated kinases are not yet known nor is it known if the two sensory cascades converge to a common response. Given that AtBphP1 would be less active in R-enriched environments, whereas AtBphP2 would be less active in FR-enriched environments, it is tempting to speculate that these opposing photoreceptors work together to sense a broader range of wavelengths thus enabling better detection of “light versus dark.” For higher plants, simultaneous sensing of R and FR is important for their ability to respond to shade by other plants (3). By measuring the R/FR ratio, plants adjust their stem growth to improve light capture under competitive conditions. In a similar manner, it is also possible that A. tumefaciens uses this ratio as a way to detect its location within the soil strata. For example by adjusting its motility with the R/FR ratio, Agrobacterium could optimize its position within the rhizosphere to promote contact with a host root (21). It is possible that other bacteria also use such an opposing pair of BphPs. For example, P. putida like A. tumefaciens also has two BphPs. One is more related by amino acid sequence to AtBphP1 whereas the other is more related to AtBphP2, including the presence of the unique histidine kinase domain and a linked RR.
Whatever the physiological reasons, the photobiological properties of AtBphP2 and possibly Bradyrhizobium and R. palustris BphP indicate that a second type of phy photoreceptor exists in bacteria that we designate here as bathyphytochromes based on their use of the longer-wavelength Pfr form as the ground state. Given that they are capable of working antagonistically to that of Pr ground-state phys, bathyphytochromes may enhance the ability to monitor light environments with fluctuating spectral quality. With regard to understanding how phys function biochemically, photoreceptors like AtBphP2 should be useful tools because they allow a way to study Pfr without significant Pr contamination.
Acknowledgments
We thank Bosl Noh, Allison Thompson, and an anonymous reviewer for helpful suggestions and Dr. Pill-Soon Song for supplying PCB. This work was supported by grants from the U.S. Department of Energy and the National Science Foundation (to R.D.V.) and Binational Research Development Fellowship FI-316-2001 (to B.K.).
Abbreviations
- BphP
bacteriophytochrome photoreceptor
- AtBphP
Agrobacterium tumefaciens BphP
- BV
biliverdin
- Cph
cyanobacterial phytochrome
- CBD
chromophore-binding domain
- PCB
phycocyanobilin
- phy
phytochrome
- RR
response regulator
- R
red light
- FR
far-red light
Footnotes
References
- 1.Vierstra R D. In: Histidine Kinases in Signal Transduction. Inouya M, Dutta R, editors. New York: Academic; 2002. pp. 273–295. [Google Scholar]
- 2.Montgomery B, Lagarias J. Trends Plant Sci. 2002;7:357–366. doi: 10.1016/s1360-1385(02)02304-x. [DOI] [PubMed] [Google Scholar]
- 3.Smith H. Nature. 2000;407:585–591. doi: 10.1038/35036500. [DOI] [PubMed] [Google Scholar]
- 4.Yeh K C, Wu S H, Murphy J T, Lagarias J C. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- 5.Schneider-Poetsch H A. Photochem Photobiol. 1992;56:839–846. doi: 10.1111/j.1751-1097.1992.tb02241.x. [DOI] [PubMed] [Google Scholar]
- 6.Yeh K C, Lagarias J C. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quail P H. Nat Rev Mol Cell Biol. 2002;3:85–93. doi: 10.1038/nrm728. [DOI] [PubMed] [Google Scholar]
- 8.Bhoo S H, Davis S J, Walker J, Karniol B, Vierstra R D. Nature. 2001;414:776–779. doi: 10.1038/414776a. [DOI] [PubMed] [Google Scholar]
- 9.Lamparter T, Michael N, Mittmann F, Esteban B. Proc Natl Acad Sci USA. 2002;99:11628–11633. doi: 10.1073/pnas.152263999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman B S, Cao Y, Askenazi M, Halling C, et al. Science. 2001;294:2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- 11.Wood D W, Setubal J C, Kaul R, Monks D E, Kitajima J P, Okura V K, Zhou Y, Chen L, Wood G E, Almeida N F, Jr, et al. Science. 2001;294:2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 12.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis S J, Vener A V, Vierstra R D. Science. 1999;286:2517–2520. doi: 10.1126/science.286.5449.2517. [DOI] [PubMed] [Google Scholar]
- 14.Giraud E, Fardoux J, Fourrier N, Hannibal L, Genty B, Bouyer P, Dreyfus B, Vermeglio A. Nature. 2002;417:202–205. doi: 10.1038/417202a. [DOI] [PubMed] [Google Scholar]
- 15.West A H, Stock A M. Trends Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 16.Grebe T W, Stock J B. Adv Microb Physiol. 1999;41:139–227. doi: 10.1016/s0065-2911(08)60167-8. [DOI] [PubMed] [Google Scholar]
- 17.Kim D, Forst S. Microbiology. 2001;147:1197–1212. doi: 10.1099/00221287-147-5-1197. [DOI] [PubMed] [Google Scholar]
- 18.Elich T D, McDonagh A F, Palma L A, Lagarias J C. J Biol Chem. 1989;264:183–189. [PubMed] [Google Scholar]
- 19.Lamparter T, Mittmann F, Gartner W, Borner T, Hartmann E, Hughes J. Proc Natl Acad Sci USA. 1997;94:11792–11797. doi: 10.1073/pnas.94.22.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutta R, Inouye M. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 21.Chesnokova O, Coutinho J B, Khan I H, Mikhail M S, Kado C I. Mol Microbiol. 1997;23:579–590. doi: 10.1046/j.1365-2958.1997.d01-1875.x. [DOI] [PubMed] [Google Scholar]