Abstract
Melatonin is produced nocturnally by the pineal gland and is a neurochemical representation of time. It regulates neuroendocrine target tissues through G-protein-coupled receptors, of which MT1 is the predominant subtype. These receptors are transiently expressed in several fetal and neonatal tissues, suggesting distinct roles for melatonin in development and that specific developmental cues define time windows for melatonin sensitivity. We have investigated MT1 gene expression in the rat pituitary gland. MT1 mRNA is confined to the pars tuberalis region of the adult pituitary, but in neonates extends into the ventral pars distalis and colocalizes with luteinizing hormone β-subunit (LHβ) expression. This accounts for the well documented transient sensitivity of rat gonadotrophs to melatonin in the neonatal period. Analysis of an upstream fragment of the rat MT1 gene revealed multiple putative response elements for the transcription factor pituitary homeobox-1 (Pitx-1), which is expressed in the anterior pituitary from Rathke's pouch formation. A Pitx-1 expression vector potently stimulated expression of both MT1-luciferase and LHβ-luciferase reporter constructs in COS-7 cells. Interestingly, transcription factors that synergize with Pitx-1 to trans-activate gonadotroph-associated genes did not potentiate Pitx-1-induced MT1-luciferase activity. Moreover, the transcription factor, early growth response factor-1, which is induced by gonadotrophin-releasing hormone (GnRH) and trans-activates LHβ expression, attenuated Pitx-1-induced MT1-luciferase activity. Finally, pituitary MT1 gene expression was 4-fold higher in hypogonadal (hpg) mice, which do not synthesize GnRH, than in their wild-type littermates. These data suggest that establishment of a mature hypothalamic GnRH input drives the postnatal decline in pituitary MT1 gene expression.
The secretion of pineal melatonin is under the control of the circadian clock, and acts as a hormonal representation of circadian and seasonal time (1). Multiple neuroendocrine target tissues interpret this daily melatonin signal through the expression of high affinity G protein-coupled melatonin receptors. There are two major melatonin receptor subtypes in mammals, of which MT1, previously termed Mel1a, is predominantly expressed (2). Melatonin receptors have a highly restricted distribution in adult mammals, but are more widely expressed in fetal and neonatal tissues, suggesting a particular role for melatonin in development (3).
Within the rodent pituitary gland, melatonin-binding sites appear shortly after the formation of Rathke's pouch (4, 5). As this region differentiates into the adenohypophysis, melatonin receptors are transiently expressed in the pars distalis (PD) during late fetal development and then disappear over the initial 2–3 weeks of postnatal life (6). In contrast, MT1 melatonin receptors remain expressed in the pars tuberalis (PT) into adulthood (7). Hence, there is developmental and tissue-specific control of melatonin receptor expression in the pituitary. In the newborn rat, the loss of PD melatonin receptors occurs in parallel with a reduced ability of melatonin to inhibit gonadotrophin-releasing hormone (GnRH)-induced luteinizing hormone (LH) secretion from gonadotroph cells (8, 9). It has therefore been speculated that the decreased inhibition of the pituitary-gonadal axis by melatonin may contribute to the timing of puberty in mammals (9). The molecular mechanisms that determine the marked developmental and tissue-specific profiles of melatonin receptor expression have not been investigated in any tissue, however.
The anterior pituitary provides an ideal model in which to address this question because there has recently been considerable progress in identifying molecular mechanisms that control the differentiation of endocrine cell lineages in this tissue (10). The transcription factor pituitary homeobox-1 (Pitx-1) is expressed in both the developing and adult pituitary gland, from the formation of Rathke's pouch (11–13), and stimulates promoter activity of many pituitary-specific genes (14). Interestingly, the strongest expression of Pitx-1 occurs in the PT and in PD cells of ventral origin (13), and therefore overlaps with the expression profile of pituitary melatonin receptors. Furthermore, mice bearing a genetic lesion of the Pitx-1 gene exhibit a selective decrease in ventral PD thyrotroph and gonadotroph cells, and also in “PT-specific” thyrotrophs (15). Because these cell types are associated with melatonin receptor expression in rodents (16, 17), the possibility arises that Pitx-1 may be an important factor supporting melatonin receptor expression in the adenohypophysis.
The development and function of gonadotroph cells is also dependent on lineage-specifying transcription factors, including early growth response factor 1 (Egr-1), steroidogenic factor-1 (SF-1), and GATA binding protein-2 (GATA-2) (18–22), which enhance the trans-activation of gonadotroph-associated genes, such as LHβ (14, 18–24). In addition, the expression of Egr-1 in gonadotrophs is directly induced by pulsatile stimulation with GnRH and is thought to be a key element in the GnRH-induced stimulation of LHβ transcription (20, 24). We have recently identified putative cis-elements for Pitx-1 and these other gonadotroph-associated transcription factors in the rat MT1 promoter (25), suggesting that these factors might also interact to regulate melatonin receptor expression in the developing pituitary gland.
To test this hypothesis, we first characterized MT1 melatonin receptor expression in the developing pituitary by in situ hybridization. Further, we have studied the interactive effects of the pituitary transcription factors described above on trans-activation of a 1.5-kb fragment of the rat MT1 promoter. These studies suggest a previously unsuspected role for GnRH in the developmental loss of MT1 from the rat PD, which is supported by a comparison of the levels of pituitary MT1 expression in wild-type and GnRH-deficient mice.
Materials and Methods
Tissue Collection and in Situ Hybridization.
All experiments were performed in accordance with the Animals (Scientific Procedures) Act, 1986. Newborn rats and adult mice were obtained from established breeding colonies at the Rowett Research Institute and University of Nottingham, respectively, and killed by cervical dislocation. Rats were decapitated and whole heads frozen on dry ice. Mouse brains and pituitaries were dissected together, keeping the pituitary stalks intact, and frozen on dry ice. Sagittal sections (20 μm) were cut through the pituitary region of each tissue for in situ hybridization analysis.
cDNAs for murine Pitx-1 (corresponding to nucleotides 544-1140 of GenBank accession no. NM_011097) and rat MT1 (nucleotides 30–466 of GenBank accession no. U14409) were cloned into the vector pGEM-T-easy. Riboprobes were transcribed from cDNA by using T7 RNA polymerase in the presence of 35S-UTP. Hybridization, posthybridization washes and film autoradiography, and densitometric analyses (performed “blind” to animal identity) were carried out as described (26). Selected sections were coated with Ilford K5 liquid emulsion (diluted 1:1 with water) and exposed for 2–4 weeks. These sections were immersed in Ilford Phenisol developer (10 min), water (3 × 1 min) and Ilford Hypam fixer (10 min), counterstained with 1% toludine blue, and dehydrated in graded ethanol solutions. Dual in situ hybridizations combining the use of 35S- and digoxygenin-labeled riboprobes were carried out as described (27).
Cloning of the Rat MT1 Promoter and Preparation of Reporter Plasmids.
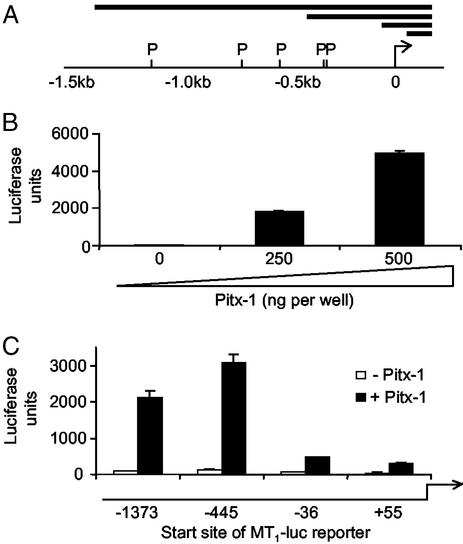
A 1.7-kb upstream fragment of the rat MT1 gene was isolated by screening a genomic library cloned in λ-phage, and subcloned into pGEM-Teasy. Restriction fragments were prepared with lengths as indicated in Fig. 2, and subcloned into the luciferase reporter vector pGL3-Basic (Promega), according to the manufacturer's protocols.
Figure 2.
Pitx-1 stimulates the rat MT1 promoter. (A) Cloned region of the rat MT1 gene. 0, Transcription start site; P, Pitx-1 consensus sequences; horizontal bars, fragments cloned into pGL3 for reporter assays. (B) Dose-dependent stimulation of MT1 promoter activity at the full-length (−1,373 to +139) promoter construct. (C) Strong stimulation of promoter activity is retained in a shortened construct (−445 to +139) that contains two proximal Pitx-1 sites, but is greatly reduced in constructs that lack these sites. (B and C) The dose of Pitx-1 expression vector used as 250 ng per well, unless otherwise stated.
Luciferase Assay.
COS-7 cells, grown in DMEM supplemented with 10% FCS, were seeded in six-well plates at a density of 150,000 cells per well. After 24 h, cells were transfected by using FuGene6 reagent (Roche), according to the manufacturer's protocol. Each well received DNA containing 100 ng of MT1-luciferase or LHβ-luciferase reporter plasmid, 100 ng of β-galactosidase reporter plasmid (as internal control), and appropriate expression vectors, made up to a total of 1.1 μg with pcDNA3. Forty-eight hours after transfection, cells were washed twice in PBS, detached by using trypsin, and centrifuged (200 × g, 10 min). Cells were then resuspended in DMEM, and aliquots were assayed for β-galactosidase activity or lysed in Steady-Glo reagent (Promega). Luciferase activity was measured by using a Packard LumiCount luminometer. Each treatment was performed in triplicate wells per experiment. Data shown are from representative experiments.
Results
Expression of MT1 mRNA in the Neonate Rat Pituitary.
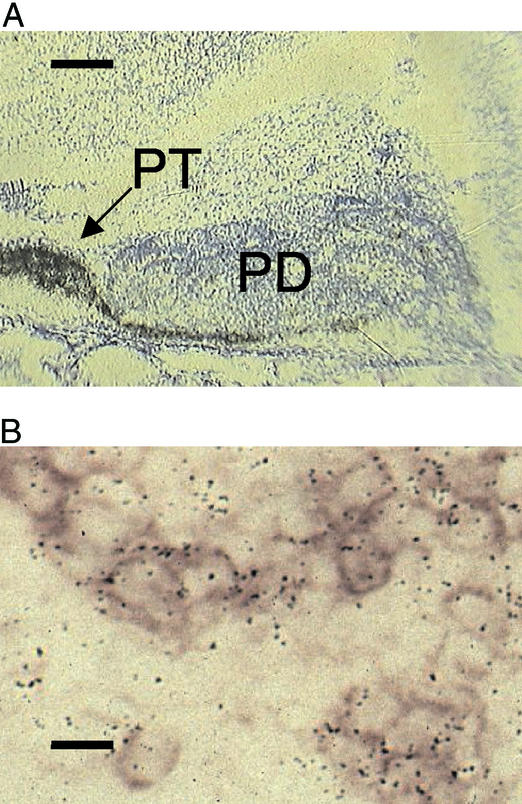
We mapped the expression of the MT1 gene within the neonatal rat pituitary gland by in situ hybridization. Strong MT1 gene expression was observed over the PT and in the adjacent rostral PD region, extending caudally over the ventral surface of the PD (Fig. 1A). This localization of MT1 expression is similar to that for the gonadotroph cells, which are concentrated in the rostral/ventral region of the PD (10). Expression of MT1 receptors in neonatal gonadotrophs was confirmed by colocalization studies, which revealed coexpression of MT1 and LHβ mRNA (Fig. 1B).
Figure 1.
Expression of MT1 mRNA in the neonate rat pituitary. (A) Strong expression occurs in the PT and extends caudally to the ventral PD. (Scale bar = 1 mm.) (B) Colocalization of LHβ (purple staining) and MT1 (black silver grains) mRNA. (Scale bar = 50 μm.)
Stimulation of the Rat MT1 Promoter by Pitx-1.
To determine the relationship between the mechanisms controlling expression of the melatonin responsive and gonadotrophic phenotypes, we have cloned a fragment of the rat MT1 promoter extending 1.5 kb upstream of the transcription start site, identified by alignment with the mouse MT1 gene (25, 28). Primary sequence analysis revealed multiple putative cis-elements for the homeobox transcription factor, Pitx-1 (Fig. 2A) and for the gonadotroph-associated transcription factors Egr-1, SF-1, and GATA-2 (25). The ability of these factors to regulate MT1 promoter activity was determined by using transient transfection assays in COS-7 cells. Cotransfection with Pitx-1 resulted in a large, dose-dependent increase in MT1 promoter activity (Fig. 2B), indicating that Pitx-1 may be a key regulator of MT1 expression in vivo. We next performed deletion studies to map Pitx-1-responsive region(s) of the rat MT1 promoter. Strong stimulation of promoter activity was retained in a truncated (−445 to +139 bp) construct that contains two adjacent proximal Pitx-1-binding sites (Fig. 2C). Deletions that removed these sites severely attenuated the ability of Pitx-1 to stimulate promoter activity (Fig. 2C).
Interaction Between Pitx-1 and Gonadotroph-Associated Transcription Factors at the Rat MT1 Promoter.
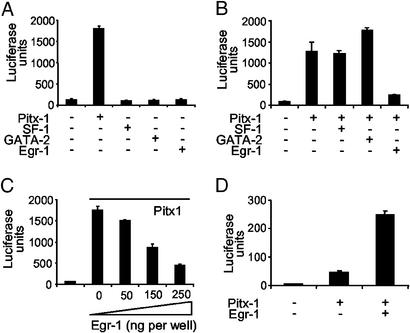
In situ hybridization studies revealed that, as in the mouse (11–13), Pitx-1 is expressed throughout the neonatal and adult rat pituitary (ref. 25 and data not shown). This finding suggests that other factor(s) act in concert with Pitx-1 to restrict MT1 expression. We therefore determined whether the other gonadotroph-associated factors, for which putative cis-elements are present in the rat MT1 promoter, also regulated promoter activity. In marked contrast to Pitx-1, no other factor was able to stimulate basal MT1-luciferase activity (Fig. 3A). Pitx-1-stimulated MT1-luciferase activity was also unaffected by cotransfection with SF-1 or GATA-2, but was greatly reduced by cotransfection with Egr-1 (Fig. 3B). The inhibition of Pitx-1-induced MT1 expression by Egr-1 was dose-dependent (Fig. 3C), and was in sharp contrast to the synergistic Pitx-1:Egr-1 interaction observed in LHβ-luciferase promoter–reporter experiments (Fig. 3D) and reported previously (20).
Figure 3.
Promoter-specific interactions with Pitx-1. (A) Pitx-1, but not gonadotroph-associated transcription factors, stimulates basal MT1 promoter activity. (B) Pitx-1 selectively interacts with gonadotroph-associated transcription factors at the MT1 promoter. (C) Egr-1 inhibits Pitx-1-stimulated promoter activity in a dose-dependent manner. (D) Pitx-1 and Egr-1 synergistically stimulate the LHβ promoter. All experiments used the full-length (−1,373 to +139) promoter construct. The dose of each expression vector was 250 ng per well unless otherwise stated.
Expression of MT1 mRNA in hypogonadal (hpg) Mice.
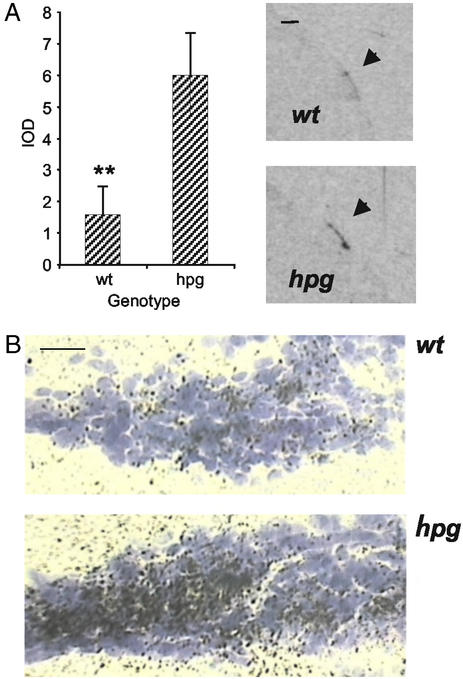
The induction of Egr-1 expression by GnRH (20, 24) suggested to us that maturation and activation of the GnRH pulse generator might be the causal factor resulting in the postnatal loss of melatonin sensitivity in gonadotrophs. To test this hypothesis we examined MT1 expression in adult mice homozygous for the naturally occurring hpg mutation, and thus unable to synthesize GnRH (29, 30). Expression of MT1 was observed in anterior pituitaries of both hpg homozygotes and wild-type littermates, in the region of the PT running back to the anterior-ventral surface of the PD (Fig. 4A). Integrated optical density measurements of MT1 expression over the whole of this region were ≈4-fold higher in the hpg group than in the wild-type (P < 0.01, Fig. 4A). Visualization of MT1 expression by liquid emulsion autoradiography showed that this difference between hpg and wild-type animals was most apparent in the caudal PT and the rostral tip of the PD (Fig. 4B).
Figure 4.
Melatonin receptor expression is increased in the pituitary of adult GnRH-deficient mice. (A) Expression of MT1 mRNA is significantly higher in hpg vs. wt mouse pituitaries. Shown are mean integrated optical density (IOD) measurements (±SEM) taken from film autoradiograms of five to seven animals per genotype (**, P < 0.01 vs. hpg, independent t test); representative film images of wt and hpg pituitaries used to calculate IOD values are shown on the right. (Scale bar = 1 mm.) (B) Expression of MT1 mRNA in the PT/PD interface of wt and hpg mouse pituitaries. (Scale bar = 50 μm.)
Discussion
Melatonin receptor expression is lost in multiple neuroendocrine tissues during development, which suggests that previously unidentified developmental cues may regulate melatonin sensitivity. Here we have used the pituitary gland as a model to study the molecular and cellular mechanisms underlying melatonin receptor expression. Our data support the hypothesis that the maturation of the GnRH neurosecretory pathway drives changes in pituitary Egr-1 expression to regulate MT1 melatonin receptor promoter activity.
Although two subtypes of melatonin receptor have been isolated in mammals (MT1 and MT2), all evidence to date suggests that the MT1 subtype is of predominant importance (31), and 2-iodomelatonin binding and responsiveness to melatonin are ablated in the PT of MT1 knockout mice (32, 33). We found that MT1 expression localizes to regions of enriched gonadotroph expression in the neonatal rat pituitary and colocalizes with LHβ mRNA expression. This result is consistent with the view that the ability of melatonin to inhibit GnRH-stimulated LH secretion in gonadotroph-enriched cultures (16) is mediated by MT1. We also observed cells that were MT1 positive but LHβ negative in the rostral PD/PT, and suspect that these represent other endocrine cell types, notably the PT-specific thryotroph, which has been previously identified in this region (34–36) and expresses melatonin receptors (17). The function of this cell type remains unclear.
To identify candidate transcription factors that may drive pituitary MT1 expression, we cloned a 1.7-kb upstream fragment of the rat MT1 gene. Primary sequence analysis revealed multiple putative cis-elements for the homeobox transcription factor Pitx-1 (25), which is highly expressed in the PT and PD cells of ventral origin, trans-activates many pituitary-specific genes, and is involved in gonadotroph development (13–15). Transient transfection assays clearly demonstrated that Pitx-1 strongly stimulates MT1 promoter activity, and suggest that the proximal doublet of Pitx-1 sites is critical to this effect. Surprisingly, reporter activity observed for the two shortest promoters was also stimulated by Pitx-1, albeit to a lesser extent than constructs containing Pitx-1 cis-elements. However, low levels of Pitx-1-induced activation have been previously reported for negative control vectors, and probably therefore represent a nonspecific effect on reporter activity (20).
Pitx-1 is expressed in the pituitary from fetal development through to adulthood (11–13, 25). It is therefore likely that other factor(s) combine with Pitx-1 to down-regulate MT1 expression in the neonatal gonadotrophs. We therefore reexamined the MT1 promoter sequence and identified putative cis-elements for the gonadotroph-associated transcription factors Egr-1, SF-1, and GATA-2 (25). Surprisingly, none of these factors enhanced basal or Pitx-1-stimulated MT1 promoter activity, even though MT1 and LHβ colocalized in the neonatal PD. Furthermore, we found that Egr-1 markedly inhibited Pitx-1-induced MT1 promoter activity, while confirming its previously reported synergism with Pitx-1 at the LHβ promoter (20), by using our experimental paradigm. Hence, these factors interact with one another to control gene expression in the ventral PD in a manner that differs qualitatively depending on promoter structure.
In gonadotroph cells, Egr-1 acts as an immediate-early gene that is induced by pulsatile GnRH stimulation, and forms a key part of the mechanism by which GnRH increases LHβ transcription (20, 24). Furthermore, the onset of pulsatile GnRH secretion in rodents occurs toward the end of gestation, after migration of GnRH neurons from the olfactory placode to the median eminence (37). This maturation coincides with the initial decrease in pituitary melatonin receptor expression, leading us to predict that, in animals that are completely unable to synthesize GnRH, pituitary MT1 expression should remain high into adult life. We therefore compared MT1 expression in adult GnRH-deficient hpg mice (29, 30) with that in their wild-type littermates. We observed MT1 labeling over the tuberal region of the PT, extending caudally along the ventral surface of the PT to its interface with the PD. Overall, MT1 mRNA expression in this region was ≈4-fold higher in hpg mice, and liquid emulsion autoradiography showed a strong increase in expression around the PT–PD interface. Because gonadotrophs are present throughout this region (17, 38–40), these data are consistent with the hypothesis that GnRH down-regulates pituitary MT1 expression in vivo. Based on this result, we suggest that melatonin receptor expression is a feature of a quiescent phase in the development of the hypothalamo-pituitary-gonadal axis, after the gonadotrophic phenotype has become established, but before the GnRH system is in place. The subsequent onset of pulsatile GnRH secretion from the hypothalamus then, through the induction of Egr-1, both activates LHβ expression and suppresses melatonin receptor expression. The maintained expression of MT1 in the PT of wild-type animals is probably largely attributable the presence of a population of PT-specific thyrotroph cells in this region that are insensitive to both GnRH and thyrotropin-releasing hormone (17, 34, 35); indeed, although Klosen and colleagues found LHβ positive cells in the rat PT, they were unable to detect LHβ coexpression with MT1 (17). Although explant experiments suggest that a small population of gonadotroph cells may remain melatonin-sensitive within the PT in adults (38, 41), the receptor subtype responsible for this phenomenon has not been defined.
The functional role of melatonin receptors in the fetal and neonatal PD is poorly understood. The ability of melatonin to inhibit GnRH-stimulated gonadotrophin secretion from neonatal PD cells (8, 9, 16) suggests a possible role in the activation of the pituitary-gonadal axis and the subsequent timing of puberty. However, daily administration of melatonin to newborn rats does not delay puberty, despite significantly reducing plasma LH titres in the perinatal period (42). In the present study, we have provided evidence that melatonin sensitivity is shut down by activation of the GnRH system around birth. This finding suggests that melatonin's function in this tissue may depend on effects exerted during fetal life, before the GnRH system becomes established.
In adult mammals, the suprachiasmatic nucleus (SCN) acts as a circadian synchronizer of peripheral tissues, thereby enhancing temporal coordination a broad spectrum of physiological activities (43). In the fetus, the SCN depends on transplacental maternal signals for entrainment to the environmental light/dark cycle (3), and efferent communication between the SCN and peripheral tissues is not established. The maternal melatonin signal acts to synchronize the fetal SCN (3), and, in adult mammals, it has been demonstrated that melatonin drives rhythmic clock gene expression in the PT (26, 33, 44). Based on these findings, a plausible hypothesis is that the maternal melatonin signal serves to synchronize both central (SCN) and peripheral components of the fetal circadian system. Hence, transient MT1 expression in the fetal pituitary may benefit the organism by maintaining circadian synchrony between this and other elements of the neuroendocrine system during development. This concept may form a useful paradigm for understanding the transient expression of melatonin receptors in multiple fetal tissues, and the highly restricted expression of MT1 in the brain and pituitary of adult animals.
Acknowledgments
We are grateful to J. Drouin (Pitx-1), D. Gordon (GATA-2), J. Milbrandt (Egr-1), and K. Parker (SF-1) for the donation of vectors, and to J. Nilson for the LHβ-luciferase reporter. We also thank K. Brown for technical assistance and J. Drouin for helpful comments at an earlier stage of these experiments. This work was supported by Biotechnology and Biological Sciences Research Council Grant 1/S11297 and by the Scottish Executive Environment and Rural Affairs Department.
Abbreviations
- Egr-1
early growth response factor-1
- GnRH
gonadotrophin-releasing hormone
- hpg
hypogonadal
- LHβ
luteinizing hormone β-subunit
- Pitx-1
pituitary homeobox-1
- PD
pars distalis
- PT
pars tuberalis
- SCN
suprachiasmatic nucleus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence of the 1.70-kb rat MT1 promoter fragment has been deposited in the GenBank database (accession no. AY228510).
References
- 1.Goldman B D. J Biol Rhythms. 2001;16:283–301. doi: 10.1177/074873001129001980. [DOI] [PubMed] [Google Scholar]
- 2.Reppert S M, Weaver D R, Godson C. Trends Pharmacol Sci. 1996;17:100–102. doi: 10.1016/0165-6147(96)10005-5. [DOI] [PubMed] [Google Scholar]
- 3.Davis F C. J Biol Rhythms. 1997;12:498–508. doi: 10.1177/074873049701200603. [DOI] [PubMed] [Google Scholar]
- 4.Rivkees S A, Reppert S M. Brain Res. 1991;568:345–349. doi: 10.1016/0006-8993(91)91424-y. [DOI] [PubMed] [Google Scholar]
- 5.Williams L M, Martinoli M G, Titchener L T, Pelletier G. Endocrinology. 1991;128:2083–2090. doi: 10.1210/endo-128-4-2083. [DOI] [PubMed] [Google Scholar]
- 6.Vanecek J. Neuroendocrinology. 1988;48:201–203. doi: 10.1159/000125008. [DOI] [PubMed] [Google Scholar]
- 7.Hazlerigg D G. J Endocrinol. 2001;170:493–501. doi: 10.1677/joe.0.1700493. [DOI] [PubMed] [Google Scholar]
- 8.Martin J E, Klein D C. Science. 1976;191:301–302. doi: 10.1126/science.1108199. [DOI] [PubMed] [Google Scholar]
- 9.Martin J E, Sattler C. Endocrinology. 1979;105:1007–1012. doi: 10.1210/endo-105-4-1007. [DOI] [PubMed] [Google Scholar]
- 10.Scully K M, Rosenfeld M G. Science. 2002;295:2231–2235. doi: 10.1126/science.1062736. [DOI] [PubMed] [Google Scholar]
- 11.Lamonerie T, Tremblay J J, Lanctot C, Therrien M, Gauthier Y, Drouin J. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 12.Szeto D P, Ryan A K, O'Connell S M, Rosenfeld M G. Proc Natl Acad Sci USA. 1996;93:7706–7710. doi: 10.1073/pnas.93.15.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanctot C, Gauthier Y, Drouin J. Endocrinology. 1999;140:1416–1422. doi: 10.1210/endo.140.3.6549. [DOI] [PubMed] [Google Scholar]
- 14.Tremblay J J, Lanctot C, Drouin J. Mol Endocrinol. 1998;12:428–441. doi: 10.1210/mend.12.3.0073. [DOI] [PubMed] [Google Scholar]
- 15.Szeto D P, Rodriguez-Esteban C, Ryan A K, O'Connell S M, Liu F, Kioussi C, Gleibermann A S, Izpisua-Belmonte J C, Rosenfeld M G. Genes Dev. 1999;13:484–494. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin J E, McKeel D W, Sattler C. Endocrinology. 1982;110:1079–1084. doi: 10.1210/endo-110-4-1079. [DOI] [PubMed] [Google Scholar]
- 17.Klosen P, Bienvenue C, Demarteau O, Dardente H, Guerrero H, Pevet P, Masson-Pevet M. J Histochem Cytochem. 2002;50:1647–1657. doi: 10.1177/002215540205001209. [DOI] [PubMed] [Google Scholar]
- 18.Lee S L, Sadovsky Y, Swirnoff A H, Polish J A, Goda P, Gavrilina G, Milbrandt J. Science. 1996;273:1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- 19.Topilko P, Schneider-Maunoury S, Levi G, Trembleau A, Gourdji D, Driancourt M-A, Rao C V, Charnay P. Mol Endocrinol. 1998;12:107–122. doi: 10.1210/mend.12.1.0049. [DOI] [PubMed] [Google Scholar]
- 20.Tremblay J J, Drouin J. Mol Cell Biol. 1999;19:2567–2576. doi: 10.1128/mcb.19.4.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingraham H A, Lala D S, Ikeda Y, Luo X, Shen W-H, Nachtigal M W, Abbud R, Nilson J H, Parker K L. Genes Dev. 1994;8:2302–2312. doi: 10.1101/gad.8.19.2302. [DOI] [PubMed] [Google Scholar]
- 22.Dasen J S, O'Connell S M, Flynn S E, Treier M, Gleibermann A S, Szeto D P, Hooshmand F, Aggarwal A K, Rosenfeld M G. Cell. 1999;97:587–598. doi: 10.1016/s0092-8674(00)80770-9. [DOI] [PubMed] [Google Scholar]
- 23.Quirk C C, Lozada K L, Keri R A, Nilson J H. Mol Endocrinol. 2001;15:734–746. doi: 10.1210/mend.15.5.0628. [DOI] [PubMed] [Google Scholar]
- 24.Dorn C, Ou Q, Svaren J, Crawford P A, Sadovsky Y. J Biol Chem. 1999;274:13870–13876. doi: 10.1074/jbc.274.20.13870. [DOI] [PubMed] [Google Scholar]
- 25. Johnston, J. D., Messager, S., Barrett, P. & Hazlerigg, D. G. (2003) J. Neuroendocrinol., in press. [DOI] [PubMed]
- 26.Messager S, Ross A W, Barrett P, Morgan P J. Proc Natl Acad Sci USA. 1999;96:9938–9943. doi: 10.1073/pnas.96.17.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mercer J G, Hoggard N, Williams L M, Lawrence C B, Hannah L T, Morgan P J, Trayhurn P. J Neuroendocrinol. 1996;8:733–735. doi: 10.1046/j.1365-2826.1996.05161.x. [DOI] [PubMed] [Google Scholar]
- 28.Roca A L, Godson C, Weaver D R, Reppert S M. Endocrinology. 1996;137:3469–3477. doi: 10.1210/endo.137.8.8754776. [DOI] [PubMed] [Google Scholar]
- 29.Cattanach B M, Iddon C M, Charlton H M, Chiappa S A, Fink G. Nature. 1977;269:338–340. doi: 10.1038/269338a0. [DOI] [PubMed] [Google Scholar]
- 30.Mason A J, Hayflick J S, Zoeller R T, Young W S, Phillips H S, Nikolics K, Seeburg P H. Science. 1986;234:1366–1371. doi: 10.1126/science.3024317. [DOI] [PubMed] [Google Scholar]
- 31.Weaver D R, Liu C, Reppert S M. Mol Endocrinol. 1996;10:1478–1487. doi: 10.1210/mend.10.11.8923472. [DOI] [PubMed] [Google Scholar]
- 32.Liu C, Weaver D R, Jin X, Shearman L P, Pieschl R L, Gribkoff V K, Reppert S M. Neuron. 1997;19:91–102. doi: 10.1016/s0896-6273(00)80350-5. [DOI] [PubMed] [Google Scholar]
- 33.von Gall C, Garabette M L, Kell C A, Frenzel S, Dehghani F, Schumm-Draeger P M, Weaver D R, Korf H W, Hastings M H, Stehle J H. Nat Neurosci. 2002;5:234–238. doi: 10.1038/nn806. [DOI] [PubMed] [Google Scholar]
- 34.Bockmann J, Bockers T M, Vennemann B, Niklowitz P, Muller J, Wittkowski W, Sabel B, Kreutz M R. Endocrinology. 1996;137:1804–1813. doi: 10.1210/endo.137.5.8612518. [DOI] [PubMed] [Google Scholar]
- 35.Bockmann J, Bockers T M, Winter C, Wittkowski W, Winterhoff H, Deufel T H, Kreutz M R. Endocrinology. 1997;138:1019–1028. doi: 10.1210/endo.138.3.5007. [DOI] [PubMed] [Google Scholar]
- 36.Sakai T, Sakamoto S, Matsubara K, Kato Y, Inoue K. J Neuroendocrinol. 1999;11:187–193. doi: 10.1046/j.1365-2826.1999.00301.x. [DOI] [PubMed] [Google Scholar]
- 37.Ebling F J P, Cronin A S. NeuroReport. 2000;11:R23–R33. doi: 10.1097/00001756-200011090-00002. [DOI] [PubMed] [Google Scholar]
- 38.Nakazawa K, Marubayashi U, McCann S M. Proc Natl Acad Sci USA. 1991;88:7576–7579. doi: 10.1073/pnas.88.17.7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gross D W, Turgeon J L, Waring D W. Endocrinology. 1984;114:2084–2091. doi: 10.1210/endo-114-6-2084. [DOI] [PubMed] [Google Scholar]
- 40.Skinner D C, Herbison A E, Robinson J E. J Neuroendocrinol. 1992;4:659–662. doi: 10.1111/j.1365-2826.1992.tb00216.x. [DOI] [PubMed] [Google Scholar]
- 41.Skinner D C, Robinson J E. Neuroendocrinology. 1997;66:263–270. doi: 10.1159/000127247. [DOI] [PubMed] [Google Scholar]
- 42.Aubert M L, Rivest R W, Lang U, Sizonenko P C. In: Development of Circadian Rhythmicity and Photoperiodism in Mammals. Reppert S M, editor. New York: Perinatology; 1989. pp. 155–191. [Google Scholar]
- 43.Reppert S M, Weaver D R. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 44.Lincoln G A, Messager S, Andersson H, Hazlerigg D G. Proc Natl Acad Sci USA. 2002;99:13890–13895. doi: 10.1073/pnas.212517599. [DOI] [PMC free article] [PubMed] [Google Scholar]