Abstract
The mediator complex is essential for regulated transcription in vitro. In the yeast Saccharomyces cerevisiae, mediator comprises >15 subunits and interacts with the C-terminal domain of the largest subunit of RNA polymerase II, thus forming an RNA polymerase II holoenzyme. Here we describe the molecular cloning of the MED1 cDNA encoding the 70-kDa subunit of the mediator complex. Yeast cells lacking the MED1 gene are viable but show a complex phenotype including partial defects in both repression and induction of the GAL genes. Together with results on other mediator subunits, this implies that the mediator is involved in both transcriptional activation and repression. Similar to mutations in the SRB10 and SRB11 genes encoding cyclin C and the cyclin C-dependent kinase, a disruption of the MED1 gene can partially suppress loss of the Snf1 protein kinase. We further found that a lexA-Med1 fusion protein is a strong activator in srb11 cells, which suggests a functional link between Med1 and the Srb10/11 complex. Finally, we show that the Med2 protein is lost from the mediator on purification from Med1-deficient cells, indicating a physical interaction between Med1 and Med2.
Keywords: cyclin C, RNA polymerase II, transcription
There is mounting biochemical and genetic evidence identifying the mediator complex as a key factor in regulating RNA polymerase II-dependent transcription. In vitro transcription performed with pure, 12-subunit core polymerase and essential general transcription factors does not respond to activator proteins, but addition of the mediator complex to this minimal system both stimulates basal transcription, enhances TFIIH-dependent phosphorylation of the polymerase C-terminal domain (CTD), and enables transcriptional regulation (1–3). Mediator purified to homogeneity is a complex of >15 polypeptides, several of which are encoded by known genes, such as SRB2, -4, -5, -6, and -7, GAL11, SIN4, RGR1, and ROX3 (3–5). However, eight mediator polypeptides were not identified previously and were designated Med1–8 according to their apparent molecular weight in SDS/PAGE. Six of them, Pgd1 (Med3) and Med2, -4/5, -6, -7, and -8 recently were identified and cloned (6, 7).
Mediator binds to the CTD of the largest subunit of the RNA polymerase II, thus forming the holo-RNA polymerase II. A similar higher molecular weight form of RNA polymerase II also was identified by Young and coworkers (8) in studies of the CTD-interacting proteins encoded by the suppressor of RNA polymerase B (SRB) genes (9, 10). An important difference between the two complexes is the presence in the latter of additional proteins, e.g., the general transcription factors TFIIB, TFIIF, and TFIIH, the SWI/SNF complex, and the Srb8–11 proteins (11–14). TFIIH contains the cyclin H-Kin28 kinase that converts the polymerase into its elongating form by phosphorylating the CTD. According to the mediator cycle model, the mediator then is released from the core polymerase and can participate in another round of initiation (15). Srb11 and Srb10 is another cyclin-kinase complex that also phosphorylates the CTD, but this phosphorylation is thought to occur before binding to the promoter, thus preventing initiation (16).
Several subunits of the yeast mediator complex originally were identified by genetic screens for mutations that would relieve transcriptional repression (17, 18). Thus, ssn (19) or gig (20) mutations cause a partial relief of glucose repression, rox mutations (21) cause a relief of oxygen repression, and are mutations (22) cause a relief of mating type repression. Subsequent work revealed that many of the corresponding genes are identical. For example, SRB10 is identical to SSN3, GIG2, and ARE1. It should be emphasized that most of the corresponding mutations also affect transcriptional activation. For example, GAL gene induction, which is reduced in med2 and med6ts cells, is also affected in srb10 cells (7, 12). Thus, it appears that mutations in these genes may result in either loss of repression or loss of induction, depending on the circumstances. In this paper, we describe the cloning of a cDNA encoding Med1, the 70-kDa subunit of the mediator complex, and functional analysis of the gene in yeast. Our data show that Med1 is required for proper regulation of transcription and is also important for the stability of the RNA polymerase II holoenzyme.
MATERIALS AND METHODS
Yeast Strains.
Saccharomyces cerevisiae strain BJ926 was used to purify the RNA polymerase II holoenzyme for amino acid sequence determination of Med1. All other experiments were performed in W303–1A congenic strains (23). The mig1, snf1, and med2-Δ1∷HIS3 disruptions have been described (7, 24, 25). The med1-Δ2∷HIS3 disruption was made by cloning the HIS3 BamHI fragment between NsiI and XbaI sites in MED1. The med1-Δ2∷LEU2 and med1-Δ2∷URA3 disruptions have the LEU2 HpaI-SalI fragment and the URA3 HindIII fragment, respectively, cloned into the same position. The srb11-Δ1∷LEU2 disruption was made by cloning the LEU2 HpaI-SalI fragment between the BsaBI and BglII sites of SRB11. The GAL1-lacZ reporter has an EcoRI-Sau3AI fragment of the GAL1 promoter fused to an SphI-AgeI fragment of pLGΔ312 (26) containing the CYC1-lacZ fusion. The his3∷(TRP1,GAL1-lacZ) integrating reporter gene was made by cloning a SnaBI-NcoI fragment of pJO116 (see below) containing the TRP1 marker and the GAL1-lacZ reporter between the MscI and NheI sites of HIS3. The construction then was targeted to the HIS3 marker previously used to disrupt the SNF1 gene (20), thus creating an snf1-Δ1∷his3∷(TRP1,GAL1-lacZ)-integrated marker in the recipient strain.
Plasmids.
The MED1 ORF, including the N-terminal His6 epitope tag, was excised as a PstI-BamHI fragment from plasmid pET6xhisMed1 (see below) and then was cloned between the EcoRI and BamHI sites of the two-hybrid vector pEG202, thus generating pDB176. An SphI fragment of pDB176 encoding the entire lexA-Med1 fusion expressed from the ADC1 promoter then was cloned into the SphI sites of plasmids pFL45 and pFL39 (27), thus generating plasmids pDB177 and pDB181, respectively. The negative control plasmid pDB185 was made similarly by cloning an SphI fragment of pRFHM1 encoding a lexA-bicoid fusion into the SphI site of pFL39. The positive control plasmid pDB198 was made in two steps. First, the lexA expression cassette from pEG202 was cloned as an SphI fragment into the SphI site of pFL39, thus generating pDB193. pDB198 then was obtained by cloning a KpnI-BamHI fragment encoding the VP16 activating domain into the XhoI site of pDB193. pDB187, finally, was made by cloning the same VP16 fragment between the two XhoI sites in pDB181. Plasmid pJO116 has the GAL1-lacZ reporter (see above) inserted into the polylinker of the CEN4 TRP1 vector pFL39 (27).
Cloning of the Med1 cDNA, Expression of Recombinant Med1 Protein, and Immunization of Rabbits.
An oligonucleotide complementary to nucleotides 1–25 in the Med1 ORF containing an NdeI site and six histidine codons in frame with the ORF and a noncoding 3′ oligonucleotide complementary to nucleotides 1,721–1,702 at the end of the Med1 ORF, including a BamHI restriction site to facilitate cloning, were used to amplify the MED1 ORF in a PCR. The resulting DNA fragment was ligated to the vector pET3a, resulting in the plasmid pET6xhisMed1. Sequencing of the final vector construct revealed no PCR-induced mutations compared with the sequence reported in the database. The pET6xhisMed1 plasmid was used to transform BL21-DE3 cells. Expression of Med1 was induced by addition of isopropyl-β-d-thiogalactopyranoside, and Med1 was purified to >75% homogeneity from the soluble fraction by affinity chromatography on Ni-Agarose (Qiagen, Chatsworth, CA). Approximately 300 μg of purified Med1 was excised from a preparative SDS/PAGE gel and was used to immunize rabbits (Agri Sera AB).
β-Galactosidase Assays.
For the galactose induction experiment in Fig. 2, yeast cells were grown to an OD600 of 0.5 in a synthetic medium containing 3% raffinose. The cells were harvested at this point and were incubated on ice for at least 30 minutes. Galactose then was added to a final concentration of 2%, and the cells were returned to a 30°C shaking incubator. Samples were removed at indicated time points and were frozen at −70°C. The β-galactosidase assays were performed essentially as described (25).
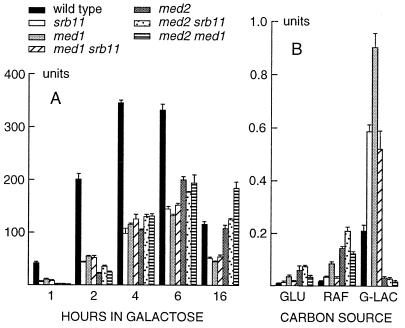
Figure 2.
Effect of med1, med2, and srb11 disruptions on GAL1 expression. (A) Single and double disrupted yeast strains as well as a wild-type control were grown to mid-log phase in the presence of 3% raffinose. Galactose then was added to a final concentration of 2%. The level of GAL1 expression was monitored at indicated timepoints after induction by using a GAL1-lacZ reporter gene as described (25). (B) GAL1-lacZ expression in the same strains when grown in 2% glucose (repressing conditions), 3% raffinose, or 3% glycerol-lactate (nonrepressing, noninducing conditions). The strains used were W303–1A, H707, H713, H715, H905, H909, and H922 (Table 1).
Protein Purification.
Purification of Med1 for amino acid sequencing was done exactly as described (4). Fractionation of whole cell extracts from wild-type and med1 cells was performed essentially as described (7) except for the for the fourth column, where we used a UNO Q-1 (Bio-Rad) column instead of a Mono Q HR5/5 column. The strains were grown in yeast extract/peptone/dextrose medium containing 1% yeast extract (Merck), 2% Bacto-Peptone (Difco), and 2% glucose.
RESULTS
Identification and Cloning of the MED1 cDNA and Gene.
Holo-RNA polymerase II was purified from S. cerevisiae as described (3). The constituent polypeptides were resolved by SDS/PAGE, were transferred to a poly(vinylidiene difluoride) membrane, and were stained with Ponceau S. The protein band migrating at 70 kDa (Med1) was excised and subjected to tryptic digestion in situ (28). Four high-confidence peptide sequences, YVETL (peptide I), LVLASNFDNFDYFNQRDGEHEK (peptide II), ESNYTDLIWFPEDFISP (peptide III), and DVSSKPSKPES (peptide IV), were obtained from separate tryptic fragments. A search of the GenBank database by using the blast program (36) revealed that all four peptides were derived from ORF YPR070w on chromosome XVI, encoding a protein with a predicted size of 64 kDa that lacks previously known function. A further analysis of the predicted Med1 sequence revealed no obvious features or motifs except for a high content of acidic residues resulting in a net charge of −32.56 at pH 7.
Loss of MED1 Causes a Complex Phenotype Affecting RNA Polymerase II-Dependent Gene Expression.
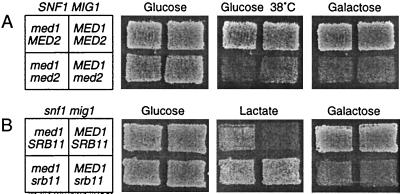
One-step disruptions of the MED1 gene were made in W303–1A congenic yeast cells. Tetrad analysis showed that med1 cells are viable but that the spores are smaller in size than wild-type spores, indicating a moderate growth defect. Because we found evidence that Med1 and Med2 may interact physically (see below), we also disrupted the MED2 gene in both wild-type and med1 cells. We found that a MED2 disruption causes a more severe phenotype than a MED1 disruption, with a pronounced growth defect, particularly on gluconeogenic carbon sources. As noted (7), the med2 cells are unable to grow on galactose. Of interest, we further found that med2 cells are temperature-sensitive at 38°C (Fig. 1A) and also cold sensitive at 16°C. However, the latter phenotype is only seen in leu2 strains such as W303–1A, which already have a reduced growth at 16°C. The med1 med2 double disrupted cells have a phenotype similar to that of med2 cells, though the growth defect is slightly more pronounced, particularly at 38°C.
Figure 1.
Growth of wild-type and mutant yeast strains under different conditions. The strains were grown on a yeast extract/peptone/dextrose plate and then were replicated to different synthetic media, as shown in the figure. The relevant genotype of each strain is shown to the left. (A) Effects of med1 and med2 disruptions in an SNF1 MIG1 wild-type background. The strains used were W303–1A, H707, H905, and H922 (Table 1). (B) Effects of med1 and srb11 disruptions in a snf1 mig1 mutant background. The strains used were H661, H668, H749, and H750 (Table 1).
To study the effects of the MED1 deletion on transcription, plasmids carrying different promoters fused to the lacZ reporter gene were transformed into the med1 strain. Expression was monitored on X-gal indicator plates containing different carbon sources. Promoters tested included GAL1, SUC2, FBP1, PCK1, CYC1, MIG1, and CAT8. We found that the MED1 deletion causes a complex phenotype similar to mutations in SRB10 and SRB11. Thus, expression was reduced below wild-type levels under conditions at which a given promoter normally is expressed. This was particularly evident for the GAL1 promoter on galactose, but a clear effect also was seen for the FBP1 promoter on acetate and for the MIG1 and CAT8 promoters on nonglucose carbon sources. On the other hand, we also found that several glucose-repressed promoters had an increased expression on glucose in the med1-disrupted strain, indicating a partial loss of repression.
Effects of med1 and med2 on the GAL1 Promoter.
The effects on the GAL1 promoter were studied in more detail by using β-galactosidase assays on liquid cultures (Fig. 2). We also included the med2 strain and an srb11 strain, as well as double mutant strains for med1, med2, and srb11. We found that GAL1 induction is severely affected in both the med1 and med2 strains, but with different kinetics. The med1 strain behaves exactly like the srb11 strain. Both strains express GAL1 at a significantly lower level (3- to 5-fold) than the wild-type strain but with a similar kinetics (Fig. 2A). GAL1 expression peaks at 4–6 h after induction and then declines. In contrast, the med2 strain differs from the wild-type also in the kinetics of induction, which is significantly delayed. Thus, expression in the med2 strain is 30× lower than in the wild-type 1 h after induction but reaches 60% of the wild-type level after 6 h and 92% after 16 h. At this point, GAL1 expression in the med2 strains is therefore twice as high as in the med1 and srb11 strains. This is surprising in view of the gal− phenotype of med2 cells. The med1 srb11 double mutant did not differ significantly from the med1 and srb11 single mutants. This is consistent with previous findings that there is little or no synergism between mutations in SRB8, SRB10, and SRB11, all of which have identical phenotypes (20, 29). The med1 med2 and med2 srb11 double mutants, finally, behave like the med2 single mutant, with a significantly delayed kinetics of induction.
We proceeded to study GAL1 expression on repressing (2% glucose) and noninducing (3% raffinose or 3% glycerol-lactate) carbon sources in which the GAL1 promoter normally is silent. All three mutations cause an increased expression under these conditions (Fig. 2B). This effect is most pronounced in the med2 strain (7-fold on glucose and 8-fold on raffinose). On glycerol-lactate, however, there was no detectable expression in the med2 strain, which hardly grew on this carbon source. The med1 disruption causes a 4-fold increase in all three cases, and the srb11 disruption causes a 2-fold increase. The med1 srb11 double mutant resembles the srb11 mutant, with a 2-fold effect, whereas the med1 med2 and med2 srb11 double mutants most closely resemble the med2 mutant (Fig. 2B). We conclude that Med1, Med2, and Srb11 all contribute to keeping the GAL1 promoter silent when it should not be expressed and that this effect is seen also in the absence of glucose repression (on glycerol-lactate). However, the levels of expression in the mutant strains are still very low, ≈1,000-fold less than in fully induced cells (2% galactose).
MED1 Is a Suppressor of snf1.
Several holopolymerase subunits were isolated in genetic screens for mutations that can suppress loss of the Snf1 protein kinase (20, 29, 30). Cells that lack Snf1 have a complex phenotype including inability to grow on all carbon sources except glucose, inability to accumulate storage carbohydrates, temperature sensitivity, sensitivity to nitrogen starvation, and a generally poor growth (31). Some of these phenotypes, such as the inability to grow on galactose and raffinose, are suppressed by a deletion of the MIG1 gene, but snf1 mig1 cells are still unable to grow on gluconeogenic carbon sources (32). However, such growth is possible after additional mutations in SRB8, SRB10, or SRB11 (20), which also permit snf1 MIG1 cells to grow on raffinose.
We therefore tested the effect of disrupting MED1 in both snf1 and snf1 mig1 strains. As a control, we included disruptions of SRB11. We found that the med1 disruption partially suppresses the snf1 phenotype, though to a somewhat lesser extent than the srb11 disruption. Thus, snf1 mig1 med1 cells can grow on gluconeogenic carbon sources, but the effect is less pronounced than in snf1 mig1 srb11 cells (Fig. 1B). Similarly, the med1 disruption permits growth of snf1 MIG1 cells on raffinose, but not as clearly as the srb11 disruption (data not shown). Other snf1 phenotypes are not significantly suppressed by either the med1 or srb11 disruption. We conclude that the med1 disruption strongly resembles the srb11 disruption in its ability to suppress some snf1 phenotypes. One clear difference between the srb11 and med1 disruptions is that snf1 mig1 srb11 cells have a reduced growth on galactose, an effect that is not seen in snf1 mig1 med1 cells. Finally, we tested the effect of a MED2 disruption in the snf1 background. We found that med2 snf1 double disrupted cells have a general growth defect that is much more severe than either of the med2 or snf1 cells. It was therefore not possible to determine whether the med2 disruption modifies any of the snf1 phenotypes.
LexA-Med1 Is a Strong Activator that Is Negatively Controlled by Cyclin C.
Several subunits of the RNA polymerase II holoenzyme can activate transcription when fused to a heterologous DNA-binding domain. Such activation may reflect the in vivo function of a given protein as a co-activator, but it also can be a more general consequence of its ability to recruit RNA polymerase II to the target promoter through interactions with other subunits (33). We therefore fused the entire Med1 protein (including an N-terminal His6 tag) to the lexA DNA-binding domain. The resulting construct was expressed from a single-copy centromeric plasmid and was tested for its ability to activate transcription from the lex operator. A lexA-VP16 fusion was used as a positive control, and a lexA-bicoid fusion (inactive in yeast) was used as a negative control. As shown in Table 2 (experiments 1–3), we found that the lexA-Med1 fusion fails to activate transcription in the wild-type strain.
Table 2.
Ability of lexA fusion proteins to activate transcription
| Fusion protein | Wild-type | med1 | srb11 | med2 | Vector |
|---|---|---|---|---|---|
| lexA-Bicoid | 0.8 (0.1) | 0.9 (0.0) | 0.9 (0.0) | 0.9 (0.0) | CEN6 |
| lexA-Med1 (exp 1) | 1.5 (0.0) | 4.3 (0.1) | 432 (5.5) | 4.4 (0.3) | CEN6 |
| lexA-Med1 (exp 2) | 1.9 (0.1) | 3.7 (0.1) | 359 (4.5) | 3.1 (0.1) | CEN6 |
| lexA-Bicoid | 1.9 (0.5) | 1.3 (0.1) | 0.9 (0.1) | 1.8 (0.3) | 2 micron |
| lexA-Med1 | 526 (130) | 763 (122) | 441 (49) | 32 (3.5) | 2 micron |
| lexA-VP16 | 2030 (113) | 2477 (21) | 2360 (41) | 1628 (416) | CEN6 |
| lexA-Med12–40-VP16 | 1998 (142) | 2451 (43) | 1644 (102) | 2174 (35) | CEN6 |
Plasmids expressing different lexA fusion proteins (see Materials and Methods) were transformed into wild-type (W303-1A), med1 (H707), srb1 (H713), and med2 (H905) yeast strains together with the pSH18-34 reporter, which has several lexA binding sites in front of a CYC1-lacZ fusion gene. The values shown are β-galactosidase units with standard errors in parentheses. All cells were grown in synthetic 8% glucose media.
We proceeded to test the lexA-Med1 fusion in med1, med2, and srb11 strains. Surprisingly, we found that it is a strong activator in the srb11 strain, in which the level of expression reaches 432 units (Table 2). This is well above the maximal level observed for the fully induced and derepressed GAL1 promoter (Fig. 2A) but still 6-fold below the activity of lexA-VP16 (Table 2). In contrast, lexA-Med1 showed very low levels of activation in the med1 and med2 strains. None of the three mutations had any significant effect on either the positive control (lexA-VP16) or the negative control (lexA-bicoid). Finally, we tested a construct in which lexA is fused to the His6 epitope tag, followed by residues 2–40 of Med1 and then the VP16 activating domain, to rule out that the observed effect is mediated by the His6 tag. As shown in Table 2, this construct behaved like the lexA-VP16 fusion protein. Therefore, the effect does not seem to involve the His6 tag or the N-terminal 40 residues of Med1.
A possible explanation for the observed effect would be if loss of Srb11 significantly increases the amount of lexA-Med1 protein. To check for this, total protein was analyzed in a Western blot with the Med1 antiserum. As a control, we also used an antiserum that reacts with tubulin (34). We found that loss of Srb11 does increase the amount of lexA-Med1, but only 3-fold, i.e. much less than the 300-fold effect on lexA-Med1 activity. Loss of Med2 also has a 3-fold effect on the amount of lexA-Med1, but, in this case, the increase in lexA-Med1 activity is only 3-fold. Neither disruption has a significant effect on the amounts of wild-type Med1 or tubulin. We proceeded to overexpress lexA-Med1 from a 2-μm plasmid. We found that lexA-Med1 under these conditions is a strong activator (300- to 800-fold) also in wild-type and med1 cells (Table 2). In med2 cells, its activity is reduced 16-fold as compared with wild-type cells. This indicates that an increase in lexA-Med1 protein can cause a significant increase in its activity and also suggests that Med2 to some extent is required for lexA-Med1 function.
Med1 Is Important for Stability of the Mediator Complex.
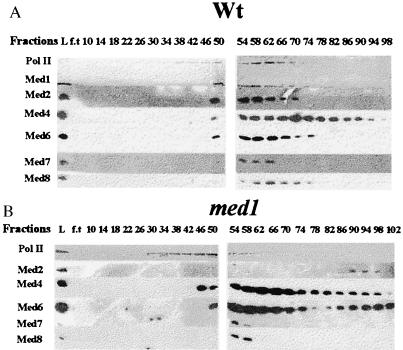
Holopolymerase was purified from both the med1 strain and the isogenic wild-type W303–1A by using the previously described procedure (3). Immunoblotting with antibodies specific for Med1 and for the recently described mediator subunits Med2, Med4/5, Med6, Med7, and Med8 showed that all subunits (except Med1 in the med1 cells) were present in the 550 mM potassium acetate DEAE-Sephacel fraction from both strains (data not shown). In the third purification step on hydroxyapatite (HAP), most mediator subunits and the second peak of the polymerase coeluted at ≈90 mM potassium phosphate in the wild-type strain (Fig. 3A). The two peaks of polymerase represent the core and holo-RNA polymerase, respectively, as reported (3). Two distinct changes were seen in the med1 strain (Fig. 3B). First, the Med2 protein is absent from the holoenzyme complex and instead elutes at a much later position in the med1 strain. Second, the Med6 subunit elutes in two separate peaks. Although most of Med6 coelutes with the holopolymerase, a significant fraction is also found in approximately the same position as the Med2 protein.
Figure 3.
Elution profiles of RNA polymerase II and mediator subunits during chromatography on hydroxyapatite. Proteins from the wild-type W303–1A (A) and med1 H707 (B) strains eluting in the 550 mM potassium acetate fraction from the DEAE-sephacel columns were applied to 10 ml HAP columns and were developed with a 100 ml-gradient of 0.01–0.2 M potassium phosphate. Fractions eluting from the columns were analyzed on a 10% SDS/PAGE gel and were immunoblotted with antibodies specific for different proteins indicated to the left. “L” represents proteins loaded to the HAP column, and “f.t” represents proteins present in the flow-through.
Fractions 50–74 from the HAP columns, which correspond to the holo-polymerase complex, were pooled and further purified on a UNO Q-1-column. In both strains, most of the mediator subunits also copurified on this column, but they were clearly separated from the major peak of RNA polymerase II that eluted at a higher concentration of potassium acetate (data not shown). This is similar to what has been reported when purifying the mediator complex from strains that are not protease deficient (7). Of interest, one mediator subunit, Med6, coeluted with the core polymerase rather than with the other mediator subunits in both med1 and wild-type strains. This suggests that the Med6 protein is more strongly associated with the core polymerase than the other mediator subunits and also that the interaction between Med6 and the other subunits might be particularly sensitive to proteolysis.
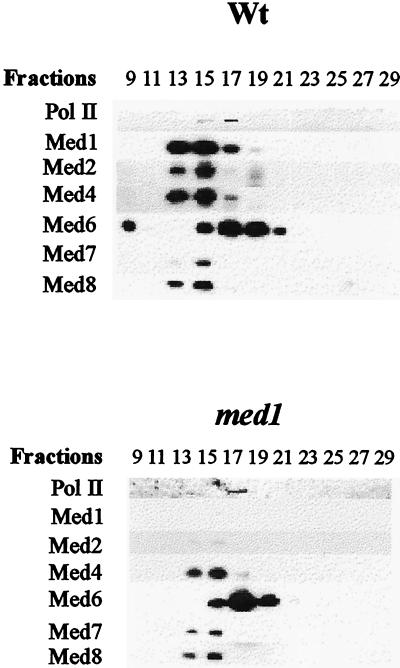
To verify these results, one fraction from each UNO Q-1 column containing both the mediator subunits and the core polymerase was fractionated further by gel filtration on a Bio-Sil SEC 400 column. Similar to the elution profile on the UNO Q-1 column, the mediator subunits split in two peaks on Bio-Sil SEC 400: one larger complex comprising Med1 (absent in the med1 strain), Med2 (also absent in med1 strain), Med4, Med7, and Med8 and one smaller complex containing Med6 and the core polymerase (Fig. 4). The molecular masses of the two complexes, as estimated from the elution profile of marker proteins on the same column, were 1.100 and 650 kDa, respectively. The fact that Med6, with a predicted molecular mass of 33 kDa, elutes as a 650-kDa protein complex confirms that the cofractionation of the core polymerase and Med6 on the UNO Q-1 column was not fortuitous.
Figure 4.
Dissociation of Med6 and core polymerase from the mediator complex. Shown is an immunoblot of Bio-Sil 400 SEC fractions from wild-type (W303–1A) and med1 (H707) yeast cells. One fraction from each UNO Q-1 column containing both the mediator subunits and the core polymerase was analyzed by gel filtration on a Bio-Sil 400 SEC column. The fractions from the gradient were separated on a 10% SDS/PAGE gel and were immunoblotted with antibodies directed against individual proteins as indicated to the left.
DISCUSSION
The yeast mediator complex was discovered during biochemical fractionations aimed at identifying factors required for regulated transcription. Several subunits of the mediator complex (Gal11, Sin4, Rgr1, Rox3, Pgd1, and Srb2, -4, -5, -6, and -7) soon were identified as proteins encoded by genes already known to be involved in RNA polymerase II-dependent transcription. In contrast, amino acid sequencing of the remaining subunits (Med1, -2, -4/5, -6, -7, and -8) showed that they are encoded by genes without previously known function (6, 7).
Our analysis of the med1 disrupted cells revealed that they have a phenotype that strongly resembles that of mutations in SRB10 and SRB11. Like these mutations, a disruption of MED1 causes both a partial defect in GAL gene induction and an increased expression under repressing or noninduced conditions in which the GAL genes normally are silent (Fig. 2). It is notable that disrupting MED1 or SRB11 causes a similar increase in expression on glucose, raffinose, and glycerol-lactate (4-fold for MED1 and 2-fold for SRB11). This suggests that these genes are not directly involved in glucose repression but, rather, may have a more general role in keeping nonexpressed genes silent. We further found that the med1 disruption suppresses some phenotypes associated with a loss of the Snf1 protein kinase, again similar to disruptions of SRB10 and SRB11. The only qualitative difference is that snf1 mig1 med1 cells, unlike snf1 mig1 srb11 cells, can grow on galactose. However, it is conceivable that this also reflects a quantitative difference and that GAL gene expression in the snf1 mig1 med1 strain is just above the level required for growth on galactose.
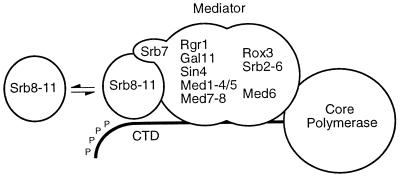
Given the pronounced similarities in the phenotypes, it seems likely that Med1 somehow is functionally connected to Srb10 and Srb11. Together with Srb8 and Srb9, Srb10 and Srb11 originally were identified in genetic screens for mutations that relieve transcriptional repression but also in a screen for mutations that can suppress the effect of a C-terminal truncation in the large subunit of RNA polymerase II (12, 20, 22, 29). Srb8–11 are present within the holopolymerase complex when purified according to one procedure (11), but they are not found in the holopolymerase preparation used in our work (7). Given these differences, it has been suggested that Srb8–11 form a separate subcomplex within the holopolymerase, which is more or less loosely associated with the mediator complex (17). Recent evidence suggests that Srb10 inhibits transcription by phosphorylating the CTD tail of RNA polymerase II, thus causing dissociation of the preinitiation complex (16). The human mediator subunit hSrb7 has been reported to associate with hSrb10 and hSrb11 (35), possibly indicating that it mediates the interaction between Srb8–11 and the mediator complex (Fig. 5). It is conceivable that Med1 also could be involved in this interaction, which would explain the phenotypic similarities between med1, srb10, and srb11 mutations. However, because the effect of a med1 disruption in some instances is weaker than that of a srb10 or srb11 disruption, proper function of the latter two proteins cannot be completely dependent on Med1.
Figure 5.
Mediator and associated protein complexes within the yeast RNA polymerase II holoenzyme.
Further evidence linking Med1 to the Srb10/11 complex comes from our finding that a lexA-Med1 fusion, which lacks activity in wild-type cells, is a strong activator of transcription in srb11 cells. It should be noted that there is a 3-fold increase in lexA-Med1 protein in srb11 cells, but we think this effect alone is unlikely to account for the >300-fold increase in activity. Our basis for this conclusion is that we find a similar 3-fold increase in lexA-Med1 protein in the med2 strain but only a 2- to 3-fold effect on activation. Furthermore, the lexA-VP16 activity is not increased in the srb11 cells, although the fusion protein is expressed from the same promoter as the lexA-Med1. Rather, we favor the possibility that Srb11 acts directly to inhibit the activity of lexA-Med1 in wild-type cells. One possible mechanism would be if Srb11 (and, presumably, also the associated Srb8, -9, and -10 proteins) bind directly to Med1, thereby blocking its ability to activate transcription. Such physical interaction is consistent with our finding that overexpression of lexA-Med1 can bypass the Srb11-dependent inhibition of its activity in wild-type cells (Table 2). However, because the Srb10/11 complex is a kinase, it is also possible that Med1 is negatively controlled by an Srb11-dependent phosphorylation that is saturated in the lexA-Med1 overproducing strain. Purification of holopolymerase complexes from wild-type and med1-disrupted yeast cells revealed that the latter differ from the former not only in the absence of Med1 but also in the complete absence of Med2. Moreover, a significant amount of the Med6 protein coeluted with Med2 in the med1 strain. This unexpected finding suggests that one function of Med1 may be to mediate the interaction between Med2 and the other subunits of the holopolymerase complex. In this context, it is noteworthy that we found the activity of lexA-Med1, when overexpressed from a 2-μm plasmid, to depend partially on MED2 (Table 2). However, this finding should be interpreted with some caution because med2 cells containing the 2-μm lexA-Med1 plasmid grew more poorly than those containing the centromeric lexA-Med1 plasmid. The effect on Med6 may reflect a further partial dissociation of the mediator complex in the absence of Med1. In agreement with this, both med1, med2, and med6ts cells are defective in GAL gene induction (6, 7). The gal− phenotype and the general effect on growth are most pronounced in the med6ts strain and least pronounced in med1. This is consistent with our biochemical results that imply interactions between these subunits in the order Med6–Med2–Med1 in which Med6 is located most proximal to the core polymerase.
The phenotypes we found for med2 are more pronounced than those described previously (7). In addition to the reduced GAL gene induction and weak temperature-sensitive phenotype, we found that med2 cells are also cold-sensitive and show a marked growth defect on gluconeogenic carbon sources. These additional phenotypes could be caused by differences in the genetic backgrounds used. Because loss of Med2 causes a more severe phenotype than loss of Med1, we consider it unlikely that Med2 is lost from the holopolymerase in med1 cells. The fact that Med2 cofractionates with the polymerase in several steps and dissociates from it only on the HAP column also suggests that it remains associated with the holopolymerase in med1 cells, though less strongly so than in wild-type cells. Because Med2 causes a severe growth defect, it was difficult to test these cells for the phenotypes associated with loss of med1 or srb11, such as partial suppression of snf1. It is, therefore, possible that further investigations will reveal functional links between Med2 and the latter two proteins.
Table 1.
Yeast strains
| Strain | Relevant genotype |
|---|---|
| H661 | mig1-Δ1∷LEU2 snf1-Δ1∷his3∷(TRP1, GAL1-lacZ) |
| H668 | mig1-Δ1∷LEU2 snf1-Δ1∷his3∷(TRP1, GAL1-lacZ) |
| srb11-Δ1∷HIS3 | |
| H707 | med1-Δ2∷HIS3 |
| H713 | srb11-Δ1∷HIS3 |
| H715 | med1-Δ2∷LEU2 srb11-Δ1∷HIS3 |
| H749 | mig1-Δ1∷LEU2 snf1-Δ1∷his3∷(TRP1, GAL1-lacZ) |
| med1-Δ2∷URA3 | |
| H750 | mig1-Δ1∷LEU2 snf1-Δ1∷his3∷(TRP1, GAS1-lacZ) |
| med1-Δ2∷URA3 srb11-Δ1∷HIS3 | |
| H905 | med2-Δ1∷HIS3 |
| H909 | med2-Δ1∷HIS3 srb11-Δ1∷LEU2 |
| H922 | med1-Δ2∷LEU2 med2-Δ1∷HIS3 |
All strains are W303-1A congenic and therefore also carry the following genetic markers: MATa ade2-1 can1-100 his3-11, 15 leu2-3,112 trp1-1 ura3-1.
Acknowledgments
We thank C. M. Gustafsson, L. C. Myers, R. D. Kornberg, and K.-D. Entian for providing us with antibodies and plasmids. We also thank Y. Li for collaboration on purification of RNA polymerase II holoenzyme and W. S. Lane for amino acid sequence determination of Med1. This work was supported by grants to S.B. from the Swedish Natural Science Research Council, the Swedish Cancer Society, the Magn. Bergvalls Stiftelse and the Foundation for Basic Science-oriented Biotechnology Research at Umeå University and by grants to H.R. from the Swedish Natural Science Research Council.
ABBREVIATION
- CTD
C-terminal domain
- HAP
hydroxyapatite
Footnotes
Present address: Department of Plant Biology, Uppsala Genetic Center, Swedish University of Agricultural Sciences, Box 7080, S-75007 Uppsala, Sweden.
References
- 1.Kelleher R J, III, Flanagan P M, Kornberg R D. Cell. 1990;61:1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 2.Flanagan P M, Kelleher R J, III, Sayre M H, Tschochner H, Kornberg R D. Nature (London) 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 3.Kim Y J, Björklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 4.Li Y, Björklund S, Jiang Y W, Kim Y J, Lane W S, Stillman D J, Kornberg R D. Proc Natl Acad Sci USA. 1995;92:10864–10868. doi: 10.1073/pnas.92.24.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gustafsson C M, Myers L C, Li Y, Redd M J, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. J Biol Chem. 1997;272:48–50. doi: 10.1074/jbc.272.1.48. [DOI] [PubMed] [Google Scholar]
- 6.Lee Y C, Min S, Gim B S, Kim Y J. Mol Cell Biol. 1997;17:4622–4632. doi: 10.1128/mcb.17.8.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koleske A J, Young R A. Nature (London) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 9.Nonet M L, Young R A. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson C M, Koleske A J, Chao D M, Young R A. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 11.Hengartner C J, Thompson C M, Zhang J, Chao D M, Liao S M, Koleske A J, Okamura S, Young R A. Genes Dev. 1995;9:897–910. doi: 10.1101/gad.9.8.897. [DOI] [PubMed] [Google Scholar]
- 12.Liao S M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J, Young R A. Nature (London) 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 13.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 14.Myer V E, Young R A. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 15.Svejstrup J Q, Li Y, Fellows J, Gnatt A, Björklund S, Kornberg R D. Proc Natl Acad Sci USA. 1997;94:6075–6078. doi: 10.1073/pnas.94.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hengartner C J, Myer V E, Liao S-M, Wilson C J, Koh S S, Young R A. Mol Cell. 1998;2:45–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 17.Carlson M. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Gancedo J M. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlson M, Osmond B C, Neigeborn L, Botstein D. Genetics. 1984;107:19–32. doi: 10.1093/genetics/107.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balciunas D, Ronne H. Nucleic Acids Res. 1995;23:4421–4425. doi: 10.1093/nar/23.21.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowry C W, Zitomer R S. Proc Natl Acad Sci USA. 1984;81:6129–6123. doi: 10.1073/pnas.81.19.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahi M, Johnson A D. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas B J, Rothstein R J. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 24.Nehlin J O, Ronne H. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Östling J, Carlberg M, Ronne H. Mol Cell Biol. 1996;16:753–761. doi: 10.1128/mcb.16.3.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guarente L, Mason T. Cell. 1983;32:1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- 27.Kern L, deMontigny J, Jund R, Lacroute F. Gene. 1990;88:149–157. doi: 10.1016/0378-1119(90)90026-n. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez J, DeMott M, Atherton D, Mische S M. Anal Biochem. 1992;201:255–264. doi: 10.1016/0003-2697(92)90336-6. [DOI] [PubMed] [Google Scholar]
- 29.Kuchin S, Yeghiayan P, Carlson M. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song W, Treich I, Qian N, Kuchin S, Carlson M. Mol Cell Biol. 1996;16:115–120. doi: 10.1128/mcb.16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson-Jaeger S, Francois J, Gaughran J P, Tatchell K. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ronne H. Trends Genet. 1995;11:12–17. doi: 10.1016/s0168-9525(00)88980-5. [DOI] [PubMed] [Google Scholar]
- 33.Ptashne M, Gann A. Nature (London) 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 34.Kilmartin J V, Wright B, Milstein C. J Cell Biol. 1982;93:576–582. doi: 10.1083/jcb.93.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho H, Orphanides G, Sun X, Yang X-J, Ogryzko V, Lees E, Nakatani Y, Reinberg D. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]