Abstract
We report here that human monocytic/macrophage THP-1 cells express the neurokinin 1 receptor (NK-1R), and that exposure of these cells to the proinflammatory cytokine IL-1β increased the expression of the NK-1R gene at the mRNA and protein levels. Because IL-1β function involves nuclear factor κB (NF-κB) activation, these data suggest that this increase in the expression of the NK-1R gene is mediated by the NF-κB transcription factor. An earlier report noted that the promoter region of the human NK-1R gene contains a putative binding site for NF-κB [Takahashi, K., Tanaka, A., Hara, M. & Nakanishi, S. (1992) Eur. J. Biochem. 204, 1025–1033]. Here we demonstrate that this is indeed a functional NF-κB-binding site, and that NF-κB is responsible for regulating the expression of the NK-1R gene by binding to the promoter region of the NK-1R gene. To further substantiate that the observed NF-κB-dependent IL-1β induction of the human NK-1R gene is regulated via a transcriptional event through this NF-κB site on the NK-1R gene promoter, we transfected THP-1 cells with a luciferase promoter-reporter construct containing the 5′ promoter region of the human NK-1R gene. Exposure of these cells to IL-1β or overexpression of NF-κB cDNAs resulted in a significant increase in the amount of luciferase activity that was diminished greatly in cells transfected with IκBα, the NF-κB inhibitor. These results directly implicate NF-κB in the regulation of the NK-1R gene and provide a molecular mechanism for the increase in expression of the NK-1R gene in responsive cells.
The 11-aa neuropeptide substance P (1) is widely distributed throughout the central nervous system (2). Apart from its known effects in nociception, smooth-muscle contraction, exocrine and endocrine gland secretion, and connective-cell proliferation, considerable evidence implicates substance P as a neuroimmune modulator. Substance P exerts its biological effects after binding to its high-affinity G protein-coupled neurokinin 1 receptor (NK-1R) expressed in a variety of cells (2). Several studies using NK-1R antagonists (3–5) and mice genetically lacking NK-1R (6–8) indicate that substance P binding to this receptor mediates inflammatory responses in various organs via mechanisms involving nuclear factor κB (NF-κB) (9) and mitogen-activated protein kinase-dependent (10) pathways and release of proinflammatory cytokines from target cells (7, 9–12). Importantly, NK-1R expression is increased during acute and chronic enterocolitis in animals and humans (13–15) even in the presence of increased ligand concentrations (14, 16). Apart from the gastrointestinal tract, increased NK-1R expression was also found in several forms of experimental inflammation in other organs including liver (17), rat skin (18), bladder (19), and dorsal horn neurons (20). However, the molecular mechanism of NK-1R up-regulation is not known.
The NF-κB transcription factor is an important regulator of the immune and inflammatory response in mammalian cells (21–24). NF-κB is activated in a great variety of cell types, and its activation leads to the rapid induction of a wide range of genes involved in the function and development of the immune system and in inflammatory and acute responses, apoptosis, cell-cycle regulation, and development (21, 23). NF-κB is found in an inactive state in the cytoplasm bound to its inhibitor IκB (inhibitor of κB) (21). After induction by a large number of stimuli, IκB is degraded, and NF-κB is free to translocate into the nucleus. In the nucleus NF-κB binds to promoters and enhancers bearing NF-κB-binding sites, and in many instances, as illustrated by the very well characterized IFN-β enhancer, forms higher-order multiprotein complexes termed “enhanceosomes” and regulates the expression of numerous genes (21, 25–27).
Previous results from our laboratory indicate that rat macrophages express NK-1R (11). Moreover, responses to this receptor were increased when macrophages were isolated from intestinal segments previously exposed to a proinflammatory enterotoxin, Clostridium difficile toxin A (11), indicating up-regulation of this receptor during intestinal inflammation.
A putative NF-κB-binding site on the NK-1R promoter was noted earlier by Takahashi et al. (28). However, no further characterization of this site has been published. In the present study we examined the possibility that NF-κB plays a role in the up-regulation of the NK-1R gene after stimulation by proinflammatory cytokines. The results presented indicate that THP-1 monocytes express the NK-1R, and expression of NK-1R in these cells is increased after exposure to the proinflammatory cytokines IL-1β and tumor necrosis factor α (TNFα). We also present direct evidence that this increase in NK-1R gene expression is mediated by the NF-κB pathway and specifically by a transcriptional event involving the NF-κB-binding site on the promoter of the NK-1R gene. Our results provide insight to the mechanism that leads to the up-regulation of the NK-1R gene expression during inflammatory responses in vivo.
Materials and Methods
Cells and Reagents.
The human monocyte cell line THP-1 was purchased from the American Type Culture Collection and cultured in suspension by using RPMI medium 1640 (GIBCO/BRL) containing 10% FCS (Sigma), 100 units of penicillin-streptomycin (GIBCO/BRL), 50 μM 2-mercaptoethanol (GIBCO/BRL), and 2 g of sodium bicarbonate (Fisher Scientific) at a pH of 7.3. Recombinant human (rh)IL-1β and rhTNFα were purchased from R & D Systems. Tissue-culture supplies were purchased from Fisher Scientific.
Human NK-1R Antibody.
An antiserum generated against a peptide representing the last 15 aa of the human NK-1R carboxyl terminus was prepared by Immuno-Dynamics (La Jolla, CA) according to the m-maleim-idobenzoyl-N-hydroxysuccinimide-coupling method described by Kitigawa and Aikawa (29) and characterized by ELISA. Immunoprecipitation experiments using the method described by MacDonald et al. (30) showed that this antibody immunoprecipitated photoaffinity-labeled NK-1R expressed in Chinese hamster ovary cells transfected with the human NK-1R (data not shown). This antibody was used previously by us to identify NK-1Rs in human colonic mucosa by confocal microscopy (31). The specificity of this antibody had been established previously by using the carboxyl-terminal 15-aa peptide used to generate the NK-1R antiserum (31).
Immunohistochemistry of NK-1R in THP-1 Cells.
THP-1 cells at a density of 1 × 105 cells per ml were incubated in RPMI medium 1640 for 1 h at 37°C with rhIL-1β (25 ng/ml), TNFα (25 ng/ml), or medium alone (control). Cells were washed with Tris-buffered saline (TBS, DAKO, Carpinteria, CA) at 4°C, fixed in 70% ethanol at −20°C, and placed onto Superfrost/Plus (Fisher) slides. Cells then were incubated in 4% paraformaldehyde (Sigma)/TBS for 10 min at room temperature, washed three times with 1× TBS, and permeabilized with 0.2% Triton X-100/TBS (5 min at room temperature). To reduce free-aldehyde groups, slides were incubated in 0.5 mg/ml sodium borohydride (Sigma)/TBS for 10 min at room temperature. Slides then were washed four times in 1× TBS, blocked for 60 min in 3% BSA (Sigma)/TBS, and then exposed to 1:1,000 dilution of human NK-1R IgG or control rabbit IgG (30 min in TBS containing 1% BSA). Cells then were washed three times in 1× TBS and incubated (40 min at room temperature) with an FITC-labeled anti-rabbit IgG antibody (1:200, Jackson ImmunoResearch). After three washes in 1× TBS, NK-1R was visualized by using a confocal microscope as described above.
Western Blotting of the NK-1R.
THP-1 cells were plated at a density of 4 × 106 cells per ml of RPMI medium 1640 and stimulated with IL-1β (25 ng/ml) or medium alone for up to 3 h at 37°C as described (32). Briefly, cells then were washed two times in ice-cold PBS and lysed with RIPA buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) and placed on ice for 45 min. After centrifugation (15,000 × g for 5 min) the supernatants were aspirated, and protein concentration was measured by the bicinchonic-acid method (Pierce). Cell lysates containing equal amounts of protein (2 mg/ml) were incubated (2 h at 4°C) with NK-1R antibody (10 μg/mg cell lysate) plus 50 μl of protein G plus agarose (Santa Cruz Biotechnology). Beads were washed two times by centrifugation (12 × g for 20 sec) with RIPA buffer at 4°C, washed in ice-cold PBS, and then boiled for 5 min in 30 μl of sample loading buffer (62.5 mM Tris, pH 6.8/10% glycerol/2% SDS/5% 2-mercaptoethanol/0.1% bromophenol blue). Immunoprecipitated proteins were fractionated on an SDS/PAGE gel and then transferred onto a nitrocellulose membrane. Membranes were blocked overnight at 4°C in 5% skim milk in PBS (pH 7.4) with 0.05% Tween-20 and then incubated with antibodies (Santa Cruz Biotechnology). Proteins were visualized by using the ECL Western blotting detection reagents (Amersham Pharmacia). Quantification of the Western blot signal was performed by using densitometry.
Cloning of the Human NK-1R Promoter Region.
An 1,837-bp fragment (containing nucleotides −1,279 to +558) of the 5′ promoter region of the human NK-1R gene (28) was prepared by PCR amplification of human genomic DNA by using primers containing internal restriction endonuclease sites and cloned into a luciferase expression vector (pGL3-Basic, Promega) between unique KpnI and BglII sites. The oligonucleotide primers used for cloning of the NK-1R promoter region were purchased from Genosys (The Woodlands, TX). The PCR was set up with 10 μl of the genomic template, 2 μl of each primer, 10 μl of Taq Plus polymerase low-salt buffer (Promega), 8 μl of 10 mM dNTP mix, 1 μl of Taq plus polymerase, and 67 μl of sterile H2O. The reaction was set up with 30 cycles of 30 sec at 94°C, 30 sec at 57°C, 3 min at 72°C, and a final extension for 5 min at 72°C. The reaction products were separated electrophoretically on a 1.2% agarose gel. By using the QIAquick kit (Qiagen, Valencia, CA), the product was extracted from the gel and purified.
RT-PCR for NK-1R.
In an RNase-free tube we added 2–5 μg of RNA, 1 μl of random primer (0.125 μg/μl), and RNase-free water to 10 μl. We heated the tube at 70°C for 2 min, put it on ice, and then added 4 μl of RNase-free water, 5 μl of 5× reverse transcription (RT) buffer, 2 μl of 10 mM dNTPs, 2 μl of 0.1 M DTT, 1 μl of RNasin (40 units/μl, Promega), and 1 μl of Moloney murine leukemia virus-RT (200 units/μl, Invitrogen). We performed the RT reaction at 37°C for 1 h and 70°C for 15 min and then left the tube on ice for PCR. The primers used for PCR of the human NK-1R gene were primer 1 (5′-GACTCCTCTGACCGCTACCA-3′) and primer 2 (5′-GGATTTCATTTCCAGCCCCT-3′). The PCR was set up as 39.75 μl of sterile water, 5 μl of 10× PCR buffer, 1 μl of 10 mM dNTPs, 1 μl of primer 1, 1 μl of primer 2, 0.25 μl of TaqDNA polymerase (50 units/μl, Qiagen), and 2 μl of RT reaction mixture. The PCR conditions for NK-1R were initial denaturation for 5 min at 94°C followed by 3-step cycling [denaturation for 0.5 min at 94°C, annealing for 0.5 min at 58°C, and extension for 0.5 min at 72°C (45 cycles)] followed by final extension for 7 min at 72°C. Quantification of the RT-PCR bands was performed by using densitometry.
Transient Transfection of THP-1 Cells.
THP-1 cells were transfected with the NK-1R promoter-reporter plasmid by using the DEAE transfection method as described (32). Briefly, cells were put into 50 ml of Falcon (Fisher) tubes at a density of 2 × 107 cells and washed by centrifugation (12 × g for 10 min) with 1× Tris buffered saline solution (TBST, 25 mM Tris⋅Cl/137 mM NaCl/5 mM KCl/0.6 mM Na2HPO4/0.7 mM CaCl2/0.5 mM MgCl2, pH, 7.4). DEAE-dextran (160 μg/ml, Sigma) was mixed with 5 μg of NK-1R plasmid alone or cotransfected with NF-κB p65 or inhibitor of NF-κB (IκBα) plasmid (generously provided by Dimitris Thanos, Columbia University, New York) in 500 μl of 1× TBST, and the mixture was added to the cells and incubated at 37°C for 10 min. The reaction was stopped by adding 15 ml of 1× TBST and after washing two times with centrifugation. Cells were resuspended in 30 ml of complete RPMI medium 1640 and incubated at 37°C for 48–72 h. Cells were placed in RPMI medium 1640 containing 10% FCS for 15 h before stimulation with rhIL-1β (25 ng/ml) and rhTNFα (25 ng/ml) for 2 h. After stimulation, cells were washed two times with ice-cold PBS, treated with 100 μl of 1× reporter lysis buffer (Promega), and placed on a shaker for 15 min. Cells then were frozen at −70°C for 15 h, and cell lysates were thawed and centrifuged at 12 × g at 4°C for 8 min. Twenty microliters of lysates were combined with 100 μl of luciferase buffer (Promega) and quantified by using a Turner TD-20e luminometer. In the experiments where IκB was transfected, samples with empty vector without the IκB insert were also performed as negative control and showed no inhibitory effect.
Preparation of Nuclear Extracts.
Nuclear extracts were prepared from 4 × 106 THP-1 cells per condition as described (33, 34). Briefly, THP-1 cells were cultured in medium alone or medium containing 10 ng/μl rhIL-1β for 2 h. Cells were washed three times by centrifugation (1,850 × g for 10 min) with ice-cold PBS. Pellets were resuspended (10 min at 4°C) in 400 μl of buffer A (10 mM Hepes, pH 7.9/1.5 mM MgCl2/10 mM KCl/0.5 mM DTT/0.1 mM EDTA/0.2 mM PMSF). Nonidet P-40 then was added to the mixture at a final concentration of 2%, and after 2 min cells were centrifuged (16,000 × g for 10 sec), and the supernatant was removed. Nuclear extracts were obtained by incubating (20 min at 4°C) the remaining pellets in hypertonic buffer (20 mM Hepes, pH 7.9/0.4 M NaCl/1 mM EDTA/1.5 mM MgCl2/0.2 mM EDTA/25% glycerol/0.5 mM DTT/0.1 mM PMSF). Suspensions then were centrifuged (16,000 × g for 2 min) and aliquots of the nuclear extracts (supernatant) were stored at −80°C.
Electrophoretic Mobility-Shift Assay (EMSA) of NF-κB on the Promoter Region of the NK-1R.
An oligonucleotide probe containing the recognition sequences for the binding site of NF-κB on the NK-1R promoter (5′-GTGGGGGTGTTTCCTAAAA-3′) was synthesized by Operon Technologies (San Francisco). This oligonucleotide was end-labeled with [32P]dCTP by Klenow DNA polymerase (New England Biolabs). The resulting probe was purified on a Quick-Sep column (Isolab), and percent binding was calculated. EMSA experiments were performed as described (35, 36). Briefly, in the binding mixture 6 μg of nuclear proteins, 2 μl of radioactive probe (80,000–100,000 cpm), binding buffer, and water were added to a final volume of 20 μl. The binding buffer consisted of 50 mM MgCl2/340 mM KCl with 3 μg/μl poly(dI-dC) in a 5:3 ratio with a secondary buffer containing 0.1 mM EDTA, pH 8.0, 40 mM KCl, 25 mM Hepes (pH 7.6), 8% Ficoll, and 1 mM of DTT. Certain reactions also contained 100-fold excess of the specific unlabeled consensus oligonucleotide to ensure specificity in the binding reaction. The binding mixtures were incubated for 15 min in room temperature and then analyzed on nondenaturing 6% polyacrylamide gels in Tris-boric-EDTA (pH 7.4). Gels were run for ≈3 h, vacuum-dried, exposed to x-ray film (Kodak), and finally developed.
Results
Induction of NK-1R Gene Expression by IL-1β.
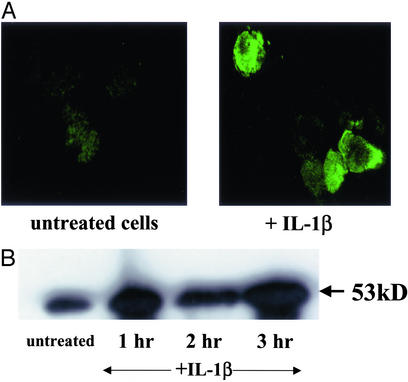
To determine whether there is a possible role for proinflammatory cytokines in the regulation of the NK-1R gene, we examined the expression patterns of the NK-1R gene products in THP-1 human monocytes. First, we performed immunohistochemistry assays in THP-1 cells and observed a great increase in the expression of the NK-1R protein localized primarily at the cell surface when comparing unstimulated cells and cells that had been stimulated with IL-1β (Fig. 1A). Similar results were obtained when we stimulated THP-1 cells with TNFα (data not shown). Next we stimulated THP-1 cells with IL-1β for various times, prepared cell extracts, subjected these cell extracts to SDS/PAGE, and performed Western blot experiments by using the NK-1R antibody. We observed an increase in the amount of NK-1R protein within 1 h (by 2-fold) of treatment with IL-1β, and this increase in protein expression persisted even after 3 h (Fig. 1B). To examine whether this increase in the amount of NK-1R protein correlates with an increase at the RNA level, we extracted total RNA from THP-1 cells that were either left untreated or treated with IL-1β for various times. We then reverse-transcribed this RNA and performed PCR by using primers for the human NK-1R gene. We observed an increase (by 2-fold) in the amount of NK-1R RNA present within 1 h of stimulation with IL-1β, and this increase persisted for up to 3 h (Fig. 2A). Levels of β-actin were measured as a control (Fig. 2A). This result directly correlates with our Western blot and immunostaining data and confirms that IL-1β induces the expression of the human NK-1R gene in human monocytes.
Figure 1.
Expression of the human NK-1R gene in THP-1 human monocytes. (A) Immunostaining of NK-1R protein in THP-1 human monocytic cells. Cells were stained with the NK-1R antibody after either no stimulation or stimulation with IL-1β (25 ng/ml × 1 h at 37°C). (Magnification ×400.) (B) Western blot of NK-1R protein in THP-1 cells. Cells were harvested either untreated or after IL-1β treatment (25 ng/ml) for the indicated time points at 37°C. Whole-cell extracts were prepared and separated by SDS/PAGE on 10% gels, and Western blots were performed by using incubation for 1 h with the NK-1R antibody. Results are representative of at least three separate experiments that gave similar results.
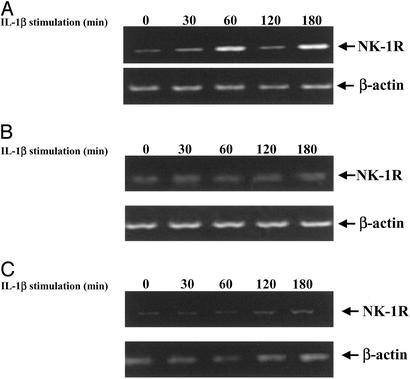
Figure 2.
RT-PCR of the NK-1R gene in IL-1β-stimulated THP-1 cells after treatment with NF-κB-specific inhibitors. (A) RT-PCR of the NK-1R and β-actin genes in THP-1 cells. Cells were either unstimulated (first lane on both Upper and Lower) or stimulated with IL-1β (10 ng/ml) for the indicated time points (in min). (B) RT-PCR of the NK-1R and β-actin genes in THP-1 cells that are overexpressing IκBα. Cells were transfected with the IκBα cDNA plasmid for 48 h and then were either left unstimulated (first lane on both Upper and Lower) or stimulated with IL-1β (10 ng/ml) for the indicated time points (in min). (C) RT-PCR of the NK-1R and β-actin genes in THP-1 cells that were incubated with 25 μM of the proteasome inhibitor MG132 for 1 h. After treatment with MG132, cells were either left unstimulated (first lane on both Upper and Lower) or stimulated with IL-1β (10 ng/ml) for the indicated time points (in min). Results are representative of at least three separate experiments that gave similar results.
Induction of NK-1R Gene Expression by IL-1β Is NF-κB-Dependent.
To examine whether the up-regulation of expression of the NK-1R gene in response to IL-1β indicates a possible involvement for NF-κB, an important regulator of the immune and inflammatory responses, we treated cells with NF-κB inhibitors and then stimulated these cells with IL-1β. Specifically, we transfected THP-1 cells with a cDNA plasmid encoding for IκBα and then treated them with IL-1β for various times. We extracted total RNA from these cells and performed RT-PCR by using the human NK-1R primers. As shown in Fig. 2B, expression of the IκBα plasmid completely abrogates the up-regulation of the human NK-1R gene we observed in Fig. 2A, indicating a possible role for this NF-κB inhibitor and the NF-κB/IκB pathway in the regulation of the NK-1R gene. Next we examined the role of another inhibitor of the NF-κB pathway, MG-132, which has the ability to inhibit signal-induced degradation of the IκB proteins by the 26S proteasome, which is a prerequisite for NF-κB activation, and we performed RT-PCR as described for Fig. 2A. As shown in Fig. 2C, treatment of cells with MG-132 for 1 h before IL-1β stimulation inhibits the up-regulation of the NK-1R gene. Taken together, our data indicate a critical role for the NF-κB pathway in the regulation of the NK-1R gene.
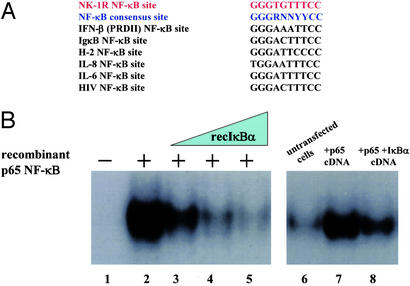
The NK-1R Gene Promoter Has a Functional NF-κB-Binding Site.
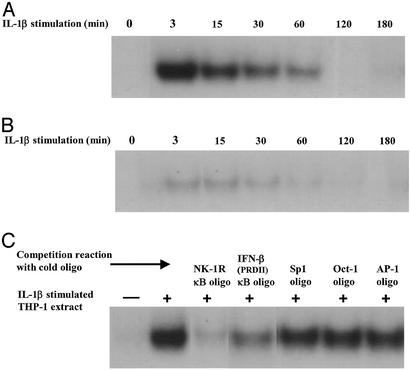
The presence of a putative NF-κB-binding site (5′-GGGTGTTTCC-3′) upstream of the transcriptional start site and the TATAA box of the human NK-1R gene has been noted (28). After sequence comparison, we observed that it closely relates to the sequence of the NF-κB consensus site and other well characterized NF-κB sites (ref. 37; Fig. 3A). To investigate whether the potential NF-κB involvement that was suggested from our results shown in Fig. 2 is due to a transcriptional event on the NK-1R promoter, we decided to examine whether this is a functional NF-κB site. We thus constructed an oligonucleotide with this putative NF-κB sequence and performed EMSAs by using radiolabeled DNA and bacterially expressed and purified recombinant p65 NF-κB and IκBα proteins. We observed that recombinant p65 NF-κB protein has the ability to bind to this putative κB site (Fig. 3B, lane 2). Moreover, increasing amounts of recombinant IκBα protein has the ability to disrupt preformed NF-κB–DNA complexes (Fig. 3B, lanes 3–5), mimicking the role of nuclear IκBα as a postinduction repressor (33). Next, we tested the ability of this putative NF-κB site to bind to cell extracts from THP-1 cells that had been transfected with mammalian expression vectors for the p65 subunit of NF-κB and its IκBα inhibitor. As shown in Fig. 3B (lane 7), this NF-κB site is indeed bound in THP-1 cell extracts where p65 is transfected. However, when the IκBα inhibitory plasmid was cotransfected along with p65, we observed a strong decrease in the amount of NF-κB–DNA binding (Fig. 3B, lane 8). Next we performed EMSAs using extracts from THP-1 cells that had been stimulated with IL-1β for various times. We were again able to observe a strong NF-κB–DNA binding in IL-1β-stimulated cells (Fig. 4A), and this binding was diminished greatly in IL-1β-stimulated cells overexpressing the inhibitory IκBα plasmid (Fig. 4B). To verify that this binding was NF-κB-specific, we added excess of unlabeled NF-κB oligonucleotide in the binding reaction of the IL-1β-stimulated cell extract (Fig. 4C). As shown in Fig. 4C, incubation with either the NK-1R NF-κB site or the PRDII NF-κB site from the IFN-β gene promoter completely abolished NF-κB binding, whereas incubation with excess cold Sp1, Oct-1, or AP-1 oligos had no effect on the ability of NF-κB proteins to bind to the NK-1R NF-κB site. These results indicate that the putative NF-κΒ site on the NK-1R promoter is in fact a true NF-κB-binding site, further suggesting that NF-κB is indeed involved in the observed up-regulation of expression of the NK-1R gene.
Figure 3.
NF-κB binds to the NK-1R gene promoter. (A) Nucleotide sequence alignment of the putative NF-κB-binding site in the promoter region of the NK-1R gene, with the consensus NF-κB-binding site, and other known NF-κB-binding sites. (B) EMSA/gel-shift experiment using the NK-1R NF-κB site as a double-stranded 32P-labeled DNA probe. The reaction mixture containing the NK-1R DNA probe was incubated with histidine-tagged, bacterially expressed and purified recombinant p65 NF-κB protein alone for 15 min (lane 2) or incubated with recombinant p65 NF-κB for 15 min and then challenged with increasing amounts of histidine-tagged, bacterially expressed and purified recombinant IκBα protein for another 15 min (lanes 3–5). Also shown is the EMSA/gel-shift experiment using the NK-1R NF-κB site as a double-stranded, 32P-labeled DNA probe. The reaction mixture containing the NK-1R DNA probe was incubated with nuclear extracts of THP-1 cells that were untransfected (lane 6), transfected with the p65 NF-κB cDNA plasmid alone (lane 7), or transfected with both the p65 NF-κB and IκBα cDNA plasmids (lane 8).
Figure 4.
The NK-1R gene promoter contains a true NF-κB-binding site. (A) EMSA/gel-shift experiment using the NK-1R NF-κB site as a double-stranded, 32P-labeled DNA probe. The reaction mixture containing the NK-1R DNA probe was incubated with THP-1 nuclear extracts of cells that were either untreated (first lane) or treated with IL-1β (10 ng/ml) for the indicated time points (in min). (B) EMSA/gel-shift experiment using the NK-1R NF-κB site as a double-stranded, 32P-labeled DNA probe. The reaction mixture containing the NK-1R DNA probe was incubated with THP-1 nuclear extracts of cells that were transfected with the IκBα plasmid that were either untreated (first lane) or treated with IL-1β for the indicated time points (in min). (C) EMSA/gel-shift experiment using the NK-1R NF-κB site as a double-stranded, 32P-labeled DNA probe. The reaction mixture containing the NK-1R DNA probe was incubated for 15 min with THP-1 nuclear extracts of cells that were either untreated or treated with IL-1β for 5 min and then competed off with excess of cold oligonucleotides for NK-1R NF-κB-, IFN-β NF-κB-, Oct-1-, Sp1-, and AP-1-binding sites for 15 min. The first lane contains just the DNA probe and the reaction mixture without any cell extract. Results are representative of at least three separate experiments that gave similar results.
The NK-1R Gene Promoter Is Inducible by IL-1β and NF-κB.
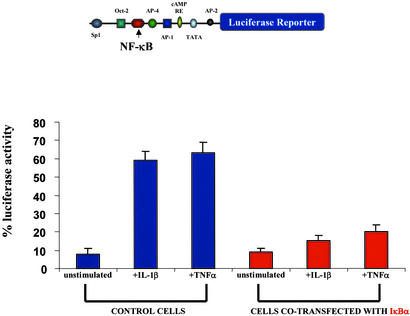
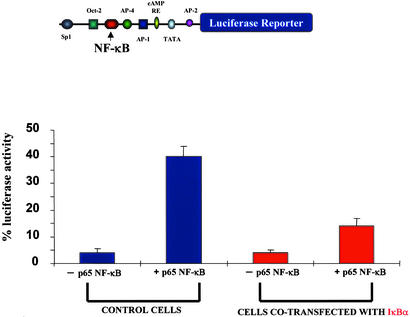
We have shown thus far that the NK-1R gene is up-regulated by IL-1β, this up-regulation is NF-κB-dependent, and the NK-1R gene promoter contains a true binding site for the NF-κB transcription factor. To examine whether the observed NF-κB-dependent IL-1β induction of the human NK-1R gene is regulated via a transcriptional event through the NF-κB site on the NK-1R gene promoter, we constructed a promoter-reporter construct containing this NF-κB site. We fused a region of the gene containing the 5′ promoter region of the human NK-1R gene to the luciferase reporter gene. We transfected this promoter-reporter construct in THP-1 cells and performed luciferase assays in unstimulated cells and cells that were stimulated with IL-1β. As shown in Fig. 5, treatment of cells with IL-1β (10 ng/ml) or TNFα (10 ng/ml) resulted in a significant increase in the amount of luciferase activity when compared with unstimulated cells. However, cells that had been transfected with the IκBα plasmid exhibited a decreased amount of stimulation by IL-1β and TNFα, indicating a role for IκBα in the induction of the NK-1R gene. To verify that this IL-1β-mediated increase in luciferase activity is indeed mediated by members of the Rel family of proteins, we transfected THP-1 cells with the reporter-promoter construct and overexpressed a plasmid encoding for the p65 subunit of NF-κB, which is ubiquitously expressed and known to possess great transcriptional activation potential (35), and we observed a large amount of transcriptional activation (Fig. 6). Next, we cotransfected THP-1 cells with the p65 NF-κB cDNA along with a plasmid expressing the IκBα inhibitor (Fig. 6), and we observed a large decrease in luciferase activity in cells overexpressing IκΒα. We therefore have shown that the NK-1R gene is up-regulated by the proinflammatory cytokine IL-1β and that this up-regulation is mediated at the transcriptional level by NF-κB transcription factor.
Figure 5.
The NK-1R gene promoter region is inducible by IL-1β and TNFα. Results from cotransfection experiments of the luciferase NK-1R promoter–reporter construct in THP-1 cells are shown. THP-1 cells were transfected with the luciferase NK-1R promoter-reporter construct for 48 h. Cells then were left unstimulated or stimulated with IL-β (10 ng/ml) or TNFα (10 ng/ml). Some samples were also cotransfected with the IκBα cDNA alone or cotransfected with the IκBα cDNA and stimulated with IL-β (10 ng/ml) or TNFα (10 ng/ml). Cells were harvested 2 days posttransfection, and luciferase activity was measured. The luciferase NK-1R promoter-reporter construct used in these experiments is shown (Upper).
Figure 6.
The NK-1R gene promoter is activated by NF-κB. Results from cotransfection experiments of the NK-1R promoter-reporter construct in THP-1 cells are shown. THP-1 cells were transfected with the luciferase NK-1R promoter–reporter construct for 48 h. Cells also were either transfected with empty PCDNA3 vector or cotransfected with the NF-κB p65 cDNA alone, with the IκBα cDNA alone, or with both the NF-κB p65 and IκBα cDNA plasmids. Cells were harvested 2 days posttransfection, and luciferase activity was measured. The luciferase NK-1R promoter–reporter construct used in these experiments is shown (Upper).
Discussion
We report here that the expression of the NK-1R in human monocytic THP-1 cells is up-regulated at the mRNA and protein levels by IL-1β, a proinflammatory cytokine known to activate the NF-κB pathway (refs. 21 and 23; Fig. 1). Consistent with the notion of NF-κB involvement in IL-1β-related signaling pathways (21, 23), we also demonstrate that expression of the NK-1R gene after IL-1β exposure is diminished greatly after either overexpression of the IκBα plasmid or treatment of cells with the NF-κB pathway and proteasome inhibitor, MG132. (ref. 23; Fig. 2). A previous study had reported on a putative NF-κB-binding site (ref. 28; Fig. 3A). Our results indicate that this putative NF-κB-binding site is indeed bound by NF-κB proteins and is a functional NF-κB site. Furthermore, using a promoter-reporter construct approach we show that the NK-1R gene is up-regulated by the proinflammatory cytokine IL-1β and that this up-regulation is mediated at the transcriptional level by NF-κB.
The results reported here may be relevant to the pathophysiology of NK-1R-dependent inflammation. For example, NK-1R levels are elevated in the intestinal mucosa early during the course of C. difficile toxin A-mediated enteritis in rats (14), and administration of NK-1R antagonists inhibits the inflammatory effects of toxin A in rat ileum (3, 11). Interestingly, toxin A can directly stimulate release of IL-1β from macrophages (38, 39). Moreover, IL-1β levels were elevated in patients with severe, C. difficile toxin-induced pseudomembranous colitis (40), a clinical condition also associated with increased intestinal NK-1R expression (41). Similarly, patients with inflammatory bowel disease have increased intracolonic release of IL-1β (42) and a substantial increase of NK-1R expression (41). Our results demonstrating that IL-1β can directly stimulate increased expression of NK-1R in THP-1 cells via NF-κB activation provide a molecular mechanism that may play a role in increased NK-1R gene and protein expression during intestinal inflammation and possibly during other inflammatory conditions in animals and humans.
An important issue that is raised by the results of this study is whether the mode of activation of the NK-1R gene after IL-1β stimulation is similar to the up-regulation of other genes involved in the inflammatory response that are regulated by the NF-κB pathway. Specifically, the stimulation by IL-1β that leads to the up-regulation of the NK-1R gene by activating the NF-κB pathway may also lead to the convergence and activation of other signaling pathways that work cooperatively to activate the NK-1R gene. For instance, treatment of cells with IL-1β leads to activation of both the NF-κB and Erk1/2 mitogen-activated protein kinase pathways known to direct the expression of the IL-8 gene by the coordinate function of the NF-κB and AP-1 transcription factors (43). The possible involvement of the mitogen-activated protein kinase-related pathway is supported further by the presence of AP-1-binding sites on the NK-1R promoter region. Indeed, examination of the 5′ region of the NK-1R gene promoter indicates the presence of putative binding sites for other transcription factors such as AP-1, Sp1, and Oct-2 (28), which have been known to be involved in the regulation of numerous inflammatory genes. Studies on the role of these transcription factors on the NK-1R promoter region will be central to our understanding of the regulation of the NK-1R gene. Furthermore, the potential for the cooperative binding of these transcription factors and NF-κB binding and its synergistic effect in the activation of transcription will be of great importance to further elucidate the mode of regulation of this gene.
Acknowledgments
We thank Dr. Dimitris Thanos of Columbia University for invaluable guidance and advice and for providing the NF-κB and IκB cDNAs used in this study. This work was supported by National Institutes of Health Grant DK 47343 (to C.P.) and a Research Fellowship Award from the Crohn's and Colitis Foundation of America (to S.S.).
Abbreviations
- NK-1
neurokinin 1
- NK-1R
NK-1 receptor
- IκB
inhibitor of κB
- TNFα
tumor necrosis factor α
- rh
recombinant human
- RT
reverse transcription
- IκBα
inhibitor of NF-κB
- EMSA
electrophoretic mobility-shift assay
References
- 1.Chang M M, Leeman S E. J Biol Chem. 1970;245:4784–4790. [PubMed] [Google Scholar]
- 2.Erspamer V, Melchiorri P, Broccardo M, Erspamer G F, Falaschi P, Improota G, Negri L, Renda T. Peptides (Tarrytown, NY) 1981;2:7–16. doi: 10.1016/0196-9781(81)90003-6. [DOI] [PubMed] [Google Scholar]
- 3.Pothoulakis C, Castagliuolo I, LaMont J T, Jaffer A, O'Keane J C, Snider R M, Leeman S E. Proc Natl Acad Sci USA. 1994;91:947–951. doi: 10.1073/pnas.91.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stucchi A F, Shofer S, Leeman S, Materne O, Beer E, McClung J, Shebani K, Moore F, O'Brien M, Becker J M. Am J Physiol. 2000;279:G1298–G1306. doi: 10.1152/ajpgi.2000.279.6.G1298. [DOI] [PubMed] [Google Scholar]
- 5.Hammond T G, Saban R, Bost K L, Harris H W, Jr, Kaysen J H, Goda F O, Wang X C, Lewis F C, Navar G L, Campbell W C, et al. Am J Physiol. 2000;278:F440–F451. doi: 10.1152/ajprenal.2000.278.3.F440. [DOI] [PubMed] [Google Scholar]
- 6.Bozic C R, Lu B, Hopken U E, Gerard C, Gerard N P. Science. 1996;273:1722–1725. doi: 10.1126/science.273.5282.1722. [DOI] [PubMed] [Google Scholar]
- 7.Castagliuolo I, Riegler M, Pasha A, Nikulasson S, Lu B, Gerard C, Gerard N P, Pothoulakis C. J Clin Invest. 1998;101:1547–1550. doi: 10.1172/JCI2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhatia M, Saluja A K, Hofbauer B, Frossard J L, Lee H S, Castagliuolo I, Wang C C, Gerard N, Pothoulakis C, Steer M L. Proc Natl Acad Sci USA. 1998;95:4760–4765. doi: 10.1073/pnas.95.8.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieb K, Fiebich B L, Berger M, Bauer J, Schulze-Osthoff K. J Immunol. 1997;159:4952–4958. [PubMed] [Google Scholar]
- 10.Fiebich B L, Schleicher S, Butcher R D, Craig A, Lieb K. J Immunol. 2000;165:5606–5611. doi: 10.4049/jimmunol.165.10.5606. [DOI] [PubMed] [Google Scholar]
- 11.Castagliuolo I, Keates A C, Qiu B, Kelly C P, Nikulasson S, Leeman S E, Pothoulakis C. Proc Natl Acad Sci USA. 1997;94:4788–4793. doi: 10.1073/pnas.94.9.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lotz M, Vaughan J H, Carson D A. Science. 1988;241:1218–1221. doi: 10.1126/science.2457950. [DOI] [PubMed] [Google Scholar]
- 13.Mantyh C R, Gates T S, Zimmerman R P, Welton M L, Passaro E P, Jr, Vigna S R, Maggio J E, Kruger L, Mantyh P W. Proc Natl Acad Sci USA. 1988;85:3235–3239. doi: 10.1073/pnas.85.9.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pothoulakis C, Castagliuolo I, Leeman S E, Wang C C, Li H, Hoffman B J, Mezey E. Am J Physiol. 1998;275:G68–G75. doi: 10.1152/ajpgi.1998.275.1.G68. [DOI] [PubMed] [Google Scholar]
- 15.Renzi D, Pellegrini B, Tonelli F, Surrenti C, Calabro A. Am J Pathol. 2000;157:1511–1522. doi: 10.1016/S0002-9440(10)64789-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldin E, Karmeli F, Selinger Z, Rachmilewitz D. Dig Dis Sci. 1989;34:754–757. doi: 10.1007/BF01540348. [DOI] [PubMed] [Google Scholar]
- 17.Cook G A, Elliott D, Metwali A, Blum A M, Sandor M, Lynch R, Weinstock J V. J Immunol. 1994;152:1830–1835. [PubMed] [Google Scholar]
- 18.Carlton S M, Coggeshall R E. Neurosci Lett. 2002;326:29–32. doi: 10.1016/s0304-3940(02)00299-9. [DOI] [PubMed] [Google Scholar]
- 19.Ishigooka M, Zermann D H, Doggweiler R, Schmidt R A, Hashimoto T, Nakada T. Pain. 2001;93:43–50. doi: 10.1016/S0304-3959(01)00288-3. [DOI] [PubMed] [Google Scholar]
- 20.Schafer M K, Nohr D, Krause J E, Weihe E. NeuroReport. 1993;4:1007–1010. doi: 10.1097/00001756-199308000-00003. [DOI] [PubMed] [Google Scholar]
- 21.Thanos D, Maniatis T. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 22.Baldwin A S., Jr Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh S, Karin M. Cell. 2002;109,Suppl.:S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 24.Silverman N, Maniatis T. Genes Dev. 2001;15:2321–2342. doi: 10.1101/gad.909001. [DOI] [PubMed] [Google Scholar]
- 25.Merika M, Thanos D. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 26.Munshi N, Yie Y, Merika M, Senger K, Lomvardas S, Agalioti T, Thanos D. Cold Spring Harbor Symp Quant Biol. 1999;64:149–159. doi: 10.1101/sqb.1999.64.149. [DOI] [PubMed] [Google Scholar]
- 27.Lomvardas S, Thanos D. Mol Cell. 2002;9:209–211. doi: 10.1016/s1097-2765(02)00463-x. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi K, Tanaka A, Hara M, Nakanishi S. Eur J Biochem. 1992;204:1025–1033. doi: 10.1111/j.1432-1033.1992.tb16724.x. [DOI] [PubMed] [Google Scholar]
- 29.Kitagawa T, Aikawa T. J Biochem (Tokyo) 1976;79:233–236. doi: 10.1093/oxfordjournals.jbchem.a131053. [DOI] [PubMed] [Google Scholar]
- 30.MacDonald D, Silberman S C, Lowe J A, III, Drozda S E, Leeman S E, Boyd N D. Mol Pharmacol. 1996;49:808–813. [PubMed] [Google Scholar]
- 31.Riegler M, Castagliuolo I, So P T, Lotz M, Wang C, Wlk M, Sogukoglu T, Cosentini E, Bischof G, Hamilton G, et al. Am J Physiol. 1999;276:G1473–G1483. doi: 10.1152/ajpgi.1999.276.6.G1473. [DOI] [PubMed] [Google Scholar]
- 32.Warny M, Keates A C, Keates S, Castagliuolo I, Zacks J K, Aboudola S, Qamar A, Pothoulakis C, LaMont J T, Kelly C P. J Clin Invest. 2000;105:1147–1156. doi: 10.1172/JCI7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran K, Merika M, Thanos D. Mol Cell Biol. 1997;17:5386–5399. doi: 10.1128/mcb.17.9.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simeonidis S, Liang S, Chen G, Thanos D. Proc Natl Acad Sci USA. 1997;94:14372–14377. doi: 10.1073/pnas.94.26.14372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merika M, Williams A J, Chen G, Collins T, Thanos D. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 36.Simeonidis S, Stauber D, Chen G, Hendrickson W A, Thanos D. Proc Natl Acad Sci USA. 1999;96:49–54. doi: 10.1073/pnas.96.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thanos D, Maniatis T. Mol Cell Biol. 1995;15:152–164. doi: 10.1128/mcb.15.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flegel W A, Muller F, Daubener W, Fischer H G, Hadding U, Northoff H. Infect Immun. 1991;59:3659–3666. doi: 10.1128/iai.59.10.3659-3666.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rocha M F, Maia M E, Bezerra L R, Lyerly D M, Guerrant R L, Ribeiro R A, Lima A A. Infect Immun. 1997;65:2740–2746. doi: 10.1128/iai.65.7.2740-2746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steiner T S, Flores C A, Pizarro T T, Guerrant R L. Clin Diagn Lab Immunol. 1997;4:719–722. doi: 10.1128/cdli.4.6.719-722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mantyh C R, Maggio J E, Mantyh P W, Vigna S R, Pappas T N. Dig Dis Sci. 1996;41:614–620. doi: 10.1007/BF02282350. [DOI] [PubMed] [Google Scholar]
- 42.Casellas F, Papo M, Guarner F, Antolin M, Segura R M, Armengol J R, Malagelada J R. Clin Sci. 1995;89:521–526. doi: 10.1042/cs0890521. [DOI] [PubMed] [Google Scholar]
- 43.Zhao D, Keates A C, Kuhnt-Moore S, Moyer M P, Kelly C P, Pothoulakis C. J Biol Chem. 2001;276:44464–44471. doi: 10.1074/jbc.M104942200. [DOI] [PubMed] [Google Scholar]