Abstract
We have identified the human papillomavirus (HPV) DNA replication initiation protein E1 as a tight-binding substrate of cyclin E/cyclin-dependent kinase (Cdk) complexes by using expression cloning. E1, a DNA helicase, collaborates with the HPV E2 protein in ori-dependent replication. E1 formed complexes with cyclin E in insect and mammalian cells, independent of Cdks and E2. Additional cyclins, including A-, B-, and F-type (but not D-type), interacted with the E1/E2 complex, and A- and E-type cyclin kinases were capable of phosphorylating E1 and E2 in vitro. Association with cyclins and efficient phosphorylation of E1 required the presence of a cyclin interaction motif (the RXL motif). E1 lacking the RXL motif displayed defects in E2-dependent HPV ori replication in vivo. Consistent with a role for Cdk-mediated phosphorylation in E1 function, an E1 protein lacking all four candidate Cdk phosphorylation sites still associated with E2 and cyclin E but was impaired in HPV replication in vitro and in vivo. Our data reveal a link between cyclin/Cdk function and activation of HPV DNA replication through targeting of Cdk complexes to the E1 replication-initiation protein and suggest a functional role for E1 phosphorylation by Cdks. The use of cyclin-binding RXL motifs is now emerging as a major mechanism by which cyclins are targeted to key substrates.
Cell cycle transitions are coordinated in large part through the action of a family of cyclin-dependent kinases (Cdks), enzymes composed of a catalytic Cdk subunit, and a regulatory cyclin subunit (1). Much of what we know concerning Cdk action in the G1/S transition comes from analysis of the Rb/E2F pathway, which has led to the identification of Rb, p107, p130, E2F-1, and DP-1 as Cdk substrates (1, 2). Phosphorylation of these proteins typically is linked with formation of tight complexes between the kinase and the substrate, most frequently through a motif in the substrate (the RXL motif) that interacts with the cyclin box. The RXL motif forms the basis for interaction of the p21 family of Cdk inhibitors with cyclins (3–6) and is important for Cdk-mediated phosphorylation of p107 (6, 7), E2F-1 (6, 8, 9), and Rb (10). Cyclin E/Cdk2 is a key regulator of S-phase initiation. Removal of cyclin E/Cdk2 activity from Xenopus egg replication systems (reviewed in ref. 11) or mammalian cells (12) blocks S-phase entry, and in Xenopus, this blockade can be overcome by addition of cyclin E/Cdk2. Conversely, ectopic expression of cyclin E can initiate replication independent of Rb inactivation in mammalian cells and in Drosophila (ref. 13, and reviewed in ref. 2). However, targets of this kinase in the preinitiation complex are unknown. We and others (14) have taken advantage of the tight association of cyclins with their substrates to identify cyclin E/Cdk-binding proteins and substrates via expression cloning techniques. Here we report the identification of human papillomavirus (HPV) replication-initiation protein E1 as a tight-binding substrate of cyclin/Cdk complexes.
Papillomavirus origin-dependent replication requires the viral E1 and E2 proteins as well as host replication proteins (15, 16). E2 is the primary ori recognition protein; it interacts with E1, thereby increasing the specificity of the E1–ori interaction (17–22). E2 also functions in preinitiation complex assembly but is not required during the elongation phase (23). In contrast, E1, the initiation helicase, interacts with Hsp40 (24) and DNA polymerase α (25–27) and is required throughout elongation (23). We show that HPV E1 binds cyclin/Cdk complexes via an RXL motif and that E1 interaction with and phosphorylation by cyclin/Cdk complexes is required for efficient E2- and ori-dependent replication in vivo. The identification of functional interactions between Cdks and replication proteins from HPV provides a vehicle to investigate the role of cyclin/Cdk complexes in initiation of DNA replication.
MATERIALS AND METHODS
Expression Cloning.
Immobilized Glutathione S-Transferase (GST)–cyclin E/Cdk complexes (30 μg) (28) were radiolabeled by autophosphorylation using 0.3 mCi of [γ-32P]ATP and eluted in 40 mM glutathione (GSH), 0.1 M Tris⋅HCl (pH 8), 0.1 M NaCl (∼5 × 107 cpm/μg). Complexes were used to probe a λGEX5-HA HeLa cell cDNA library (1.5 × 104 plaques per 150 mm plate) (29). Filters were blocked with 5% dry milk and rinsed in binding buffer (20 mM Tris⋅HCl (pH7.5)/50 mM NaCl/10 mM MgCl2/1 mM glycerol 2-phosphate/1 mM DTT/0.2 mM phenylmethylsulfonyl fluoride/0.1% Tween 20) before incubation with the probe (1−2 × 105 cpm/ml, 23°C, 1–3 h). Washed filters were subjected to autoradiography. Phage were plaque purified, and cDNAs were sequenced after recovery from λ DNA (29).
Protein Interactions and Kinase Assays.
Baculoviruses for HPV-11 E2, Glu-Glu (EE)-tagged E1, and cyclin/Cdks as well as plasmids for expression of E2, E1, cyclin E, and Cdk2 in mammalian cells were from previous studies (13, 18–20). Mutations in E1 were introduced by using a GeneEditor kit (Promega). Viruses expressing EE-E1ARA and EE-E1ΔCdk were generated by using Fastbac (Gibco/BRL). Proteins produced in insect cells (28) were purified by using either 2 μg of anti-EE antibody (Babco, Richmond, CA) bound to protein A-Sepharose or by using GSH–Sepharose. Washed complexes were subjected to SDS/PAGE and immunoblotting by using the indicated antibodies or were used for kinase assays (28). COS-1 cell transfection was performed by using FuGene6-mediated lipofection (Boehringer Mannheim). Cell lysates were made at 48 h and used for immunoprecipitation with anti-Cdk2 (M2), anti-cyclin E (C-19) (Santa Cruz Biotechnology), or anti-E1 (18). Immune complexes were immunoblotted with the indicated antibodies.
Replication Assays.
HPV–ori-dependent replication assays were performed by using 293 cells 48 h after transfection with 5 μg of pMT2-E2, 0.5 μg of HPV-11 ori plasmid, and E1 expression plasmids (18, 20). Southern blots to detect replicated ori plasmids were quantified on a PhosphorImager (Molecular Dynamics). Cell-free HPV replication assays (19, 23) employed HPV-11 ori plasmid (40 ng), E2 protein (8 ng), and EE-E1 proteins from insect cells (19) or Escherichia coli (J.-S. Liu, S.-R. Kuo, T.R. Broker, and L.T.C., unpublished results) and 293 cell extracts (100 μg).
RESULTS
HPV E1 Is Identified as a Cyclin E-Interacting Protein by Expression Cloning.
To identify cyclin E/Cdk-interacting proteins, we screened a HeLa cell cDNA library with radiolabeled cyclin E/Cdk2 and cyclin E/Cdk3 complexes. Both of these complexes have been shown to activate replication when injected into quiescent human fibroblasts (30), suggesting that they can interact with relevant substrates. From 1 × 106 plaques screened with cyclin E/Cdk3, 156 positive plaques were obtained. Thirty-six putative clones were obtained from 0.25 × 106 plaques screened with cyclin E/Cdk2. Sequence analysis of 152 cDNAs revealed several known Cdk-interacting proteins, including Cks1 (5 isolates), NPAT (2 isolates) (14), and PRC1 (9 isolates) (29), as well as several other genes. The most abundant gene identified (44 isolates) was the E1 protein from HPV-18. Although HeLa cells contain integrated HPV-18 DNA (31), expression of E1 has not been reported. Here, we report a functional and biochemical analysis of the interaction of E1 with cyclins. In subsequent experiments, we used HPV-11 E1 and E2 proteins because these have been characterized in great detail (18–20, 23, 24).
E1 and E1/E2 Associate with Cyclin E Independent of Cdk2.
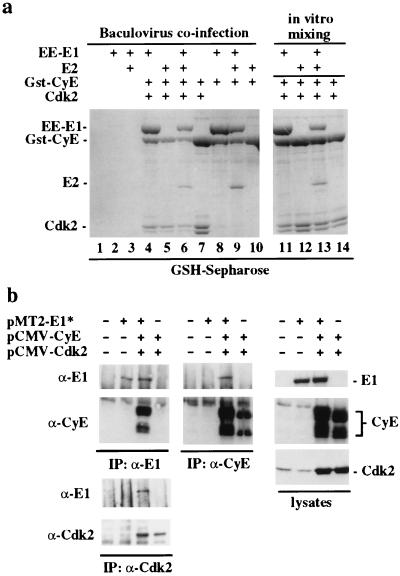
To verify the interaction of cyclin E with E1 and to examine whether the E1/E2 interaction is influenced by cyclin E binding, various combinations of viruses expressing GST–cyclin E, Cdk2, EE-E1, and E2 were coinfected into insect cells and cyclin E-containing complexes analyzed by using SDS/PAGE (Fig. 1a). Cyclin E associated with E1 and the E1/E2 complex independent of Cdk2 (Fig. 1a, lanes 4, 6, 8, 9) but did not bind E2 directly (Fig. 1a, lane 10). In control experiments, neither E1 or E2 associated with GST alone (data not shown), GST-p21CIP1 (Fig. 2a), or GSH–Sepharose in the absence of cyclin E (Fig. 1a, lanes 2 and 3). Analogous complexes could also be assembled by mixing the same lysates containing E1 alone or E1/E2 with lysates containing GST–cyclin E (Fig. 1a, lanes 11–14) or with in vitro translation products (data not shown), suggesting that the interaction can occur in vitro. Thus, the E1/cyclin E interaction appears to be direct, whereas the E2/cyclinE interaction is mediated by E1. Association of E1 with cyclin E/Cdk2 complexes also was observed in COS-1 cells transiently expressing E1, cyclin E, and Cdk2 (Fig. 1b). Anti-E1 antibodies precipitated E1 complexes containing cyclin E (Fig. 1b Left) whereas anti-cyclin E and anti-Cdk2 antibodies precipitated E1 (Fig. 1b Center). As observed previously (12), cyclin E is expressed as two major forms at 50 and 40 kDa (Fig. 1b). Thus, cyclin E/Cdk complexes can associate with E1 in multiple settings.
Figure 1.
Cyclin E interacts with HPV E1. (a) Cdk-independent interaction of E1 and E1/E2 complexes with cyclin E in insect cells. Cells were infected with the indicated viruses and complexes were analyzed by SDS/PAGE and Coomassie staining after purification using GSH–Sepharose (lanes 1–10). Proteins were confirmed by immunoblotting (data not shown). In lanes 11–14, lysates from cells expressing E1, E2, or E1/E2 were mixed with cyclin E-containing lysates before purification. (b) Association of cyclin E/Cdk2 and E1 in COS-1 cells. Lysates from cells transfected with the indicated plasmids (Right) were subjected to immunoprecipitation and immunoblotting by using the indicated antibodies (Left and Center). Quantities of plasmids were: pMT2-E1*, 10 μg; pCMV-cyclin E, 1 μg; pCMV-Cdk2, 1 μg plus empty pMT2 to maintain DNA levels.
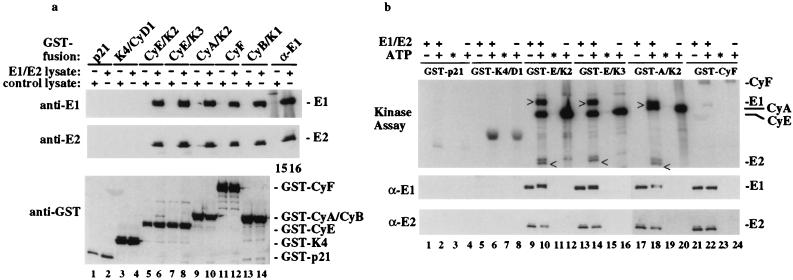
Figure 2.
Multiple cyclins associate with and phosphorylate E1 and E2 in vitro. (a) Purified cyclin/Cdk complexes or GST–p21 as a negative control were immobilized on GSH–Sepharose beads and used in binding reactions with lysates containing EE–E1/E2 complexes or uninfected lysates as indicated. Complexes were immunoblotted with antibodies against EE–E1, E2, or GST (lanes 1–14). An anti-E1 immune complex (lanes 15 and 16) served to control for the EE–E1/E2 interaction. (b) EE–E1/E2/cyclin complexes prepared as described in a were used in kinase assays. For assays containing kinase and E1, mixtures were fractionated into EE–E1 bound complexes (lanes 2, 6, 10, 14, 18, and 20) and reaction supernatants (∗; lanes 3, 7, 11, 15, 19, and 21) before SDS/PAGE and immunoblotting. Kinase activity was visualized by autoradiography of the filter and proteins identified by immunoblotting with anti-E1 and -E2 antibodies. The positions of phosphorylated EE–E1(>) and E2 (<) are indicated.
Multiple Cyclin/Cdk Complexes Associate with and Phosphorylate E1/E2 Complexes.
To examine the specificity of the E1–cyclin interaction, in vitro binding reactions were performed by incubating lysates containing E1/E2 complexes with various cyclin/Cdk complexes immobilized on GSH–Sepharose. Similar quantities of proteins were used as assessed with Western blotting, (Fig. 2a Lower) or Coomassie staining (data not shown). Human A-, B-, E-, and F-type cyclins efficiently associated with E1/E2 complexes (Fig. 2a, lanes 5–14), whereas neither GST–p21CIP1 nor GST–Cdk4/cyclin D1 did (Fig. 2a, lanes 1–4). The ratio of E1 to E2 in cyclin complexes is comparable to that found with anti-E1 immune complexes from these lysates (Fig. 2a, lanes 15 and 16), indicating that association of cyclin with E1 does not disrupt the interaction with E2. The interaction of E1/E2 with cyclin F, whose kinase partner has not been identified, was Cdk-independent.
E1 contains four conserved candidate Cdk-phosphorylation sites at residues S89, S93, S107, and T468 (Fig. 3a), and E2 also contains several candidate Cdk sites. To examine whether E1 or E2 are Cdk substrates, complexes were assembled as described in Fig. 2a and after incubation with [γ-32P]ATP, products were analyzed by using SDS/PAGE and autoradiography. E1 and E2 are both efficiently phosphorylated by E- and A-type Cdk complexes (Fig. 2b, lanes 10, 14, and 18). About 50% of E1 and E2 are phosphorylated to more slowly migrating forms by E- and A-type Cdk complexes (Fig. 2b, lanes 9 and 10, 13 and 14, and 17 and 18) and neither was released from the kinase after phosphorylation (Fig. 2b, lanes 11, 15, 19). As expected, cyclins also were phosphorylated in these reactions, independent of E1 (Fig. 2b, lanes 10, 12, 14, 16, 18, and 20). Low levels of a cyclin F-associated kinase activity copurify with cyclin F in insect cells, and in this setting, E1 was weakly phosphorylated (Fig. 2b, lane 22). Consistent with the finding that cyclin D1/Cdk4 does not associate with E1, purified cyclin D1/GST–Cdk4 was unable to phosphorylate E1 in the absence of a prebinding step but readily phosphorylated Rb in parallel assays and is inhibited by p21 (data not shown), suggesting that its inability to associate with E1 (Fig. 2a) does not reflect gross misfolding or an effect of the placement of the GST tag. Although cyclin B/Cdc2 associated with E1/E2 complexes, it phosphorylated E1 only weakly compared with cyclin E/Cdk2 (data not shown). These data indicate that a subset of cyclin/Cdk complexes can associate with and phosphorylate E1 and its associated E2 protein.
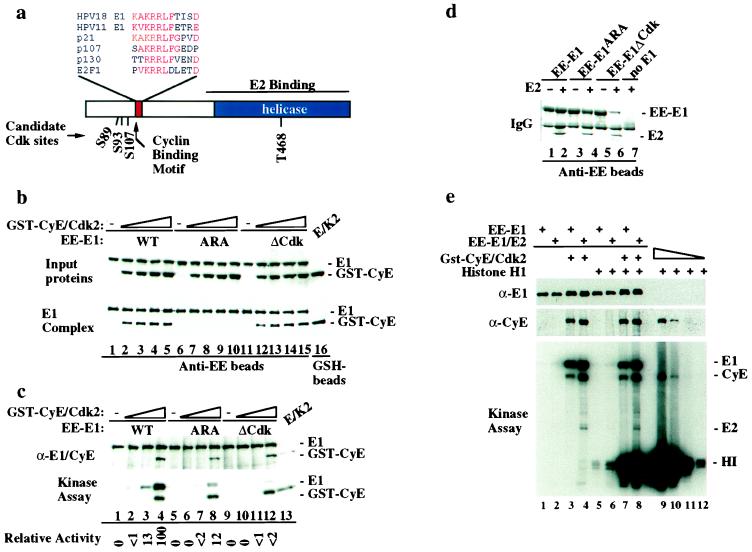
Figure 3.
An RXL motif in E1 is required for association with and phosphorylation by cyclin E/Cdk2. (a) Domains and positions of candidate Cdk phosphorylation sites in HPV E1 are shown. (b) Mutations in the RXL motif of E1 abolish association with cyclin E in vitro. Equal quantities (0.8 μg) of EE–E1 (lanes 1–5), EE–E1ARA (lanes 6–10), and EE–E1ΔCdk (lanes 11–15) were used for binding with increasing quantities of insect-cell lysates with or without cyclin E/Cdk2 and bound proteins detected by immunoblotting. To control for levels of input proteins, immunoblots of reaction mixtures prior to bead washing were performed (Top). (c) EE–E1 (lanes 1–4), EE–E1ARA (lanes 5–8), and EE–E1ΔCdk (lanes 9–12) immobilized on anti-EE beads were incubated with purified cyclin E/Cdk2 (1, 10, and 100 nM, respectively) and [γ-32P]ATP (see Materials and Methods). Products were separated by using SDS/PAGE and transferred to nitrocellulose before immunoblotting (Top) and autoradiography (Bottom). (d) E1ARA and E1ΔCdk interact with E2. Anti-EE immune complexes of insect-cell lysates derived from the indicated infections were separated by using SDS/PAGE and visualized with Coomassie blue. (e) Immobilized EE–E1 or EE–E1/E2 complexes were assembled with cyclin E/Cdk2 in vitro and the excess kinase removed before assay in the presence (lanes 5–8) or absence (lanes 1–4) of histone H1. As a control, serial dilutions of cyclin E/Cdk2 (1–1,000 ng) were used in histone H1 kinase assays (lanes 9–12).
An RXL Motif in E1 Mediates Interaction with and Phosphorylation by Cyclin/Cdk Complexes.
We found that E1 proteins contain a conserved candidate RXL motif located in the N terminus (residues 121–131) reminiscent of that found in p21CIP1 and other Cdk-interacting proteins (Fig. 3a). To examine whether this motif functioned in cyclin recognition, we mutated both R124 and L126 to alanine to generate E1ARA and tested whether immobilized EE–E1ARA, like its wild-type counterpart, could retrieve GST–cyclin E from a lysate. As shown in Fig. 3b, mutation of the RXL motif abolished the interaction with cyclin E (compare lanes 1–5 with lanes 6–10) but had no effect on association with E2 (Fig. 3d, lanes 2 and 4). We also prepared a mutation, E1ΔCdk, in which all four candidate Cdk2 phosphorylations sites were replaced by alanine (S89A-S93A-S107A-T468A). EE–E1ΔCdk stably associated with both cyclin E (Fig. 3b, lanes 12–15) and E2 (Fig. 3d, lane 6).
We then tested whether E1ARA could be phosphorylated by cyclin E/Cdk2. EE–E1 or EE–E1ARA proteins were incubated with purified cyclin E/Cdk2 and [32P]ATP (Fig. 3c). As expected, E1 was efficiently phosphorylated (Fig. 3c, lanes 1–4). In contrast, E1ARA was poorly phosphorylated (Fig. 3c, lanes 5–8). At the highest level of kinase used, the extent of phosphorylation was 12% that found with E1, whereas the extent of cyclin E autophosphorylation was similar. As expected, EE–E1ΔCdk was not phosphorylated by cyclin E/Cdk2 (Fig. 3c, lanes 9–12). These data indicate that efficient phosphorylation of E1 by cyclin E/Cdk2 requires physical interaction through the RXL motif whereas association of E1 with E2 requires neither the RXL motif or Cdk-dependent phosphorylation.
We next asked whether cyclin E/Cdk2 bound to E1 or E1/E2 could phosphorylate the exogenous substrate histone H1 (Fig. 3e). Histone H1 was readily phosphorylated by cyclin E/Cdk2 bound to E1 (lane 7) or E1/E2 (lane 8) complexes (≈30% activity compares with free cyclin E/Cdk2 measured in parallel, lanes 9–12), and as expected, E1, E2, and cyclin E became phosphorylated regardless of the presence of histone H1 (Fig. 4, lanes 3, 4, 7, 8). This result is in stark contrast to the situation with p107, which is an obligate substrate when bound to cyclin A/Cdk2 (7). Although the E1/kinase complex can phosphorylate histone H1 and E2, it would not be expected to efficiently phosphorylate substrates such as p107 or Rb, which like E1, depend on an interaction with cyclin through their RXL-motifs for phosphorylation (6, 7).
Figure 4.
Efficient HPV replication requires an intact cyclin-binding motif and Cdk-phosphorylation sites in E1. (a and b) HPV replication assays were performed as described (18, 20). Low molecular weight DNA was analyzed by Southern blotting using labeled origin plasmid as a probe to detect the presence of DpnI-resistant replication products. This probe contains sequences that also hybridize with E1 and E2 plasmids. Blots were quantified by PhosphorImager analysis. E1* reflects the use of pMT2-E1* based plasmids, which express higher levels of protein and replication activity. (c) Levels of E1*, E1*ARA, and E1*ΔCdk proteins were determined by immunoblotting with anti-E1 antibodies. (d) Cell-free replication was conducted as described in Material and Methods and products detected by incorporation of [α-32P]dCTP.
The Cyclin-Binding Motif in E1 Is Required for Efficient HPV Replication in Vivo.
We used an established transient assay that measures replication of an HPV-11 ori-containing plasmid to test whether the E1–cyclin interaction contributes to replication. Replication of this plasmid depends on coexpression of E1 and E2 and produces replication products that cannot be digested with the methylation-dependent restriction enzyme DpnI (18). With a given amount of E2 expression plasmid, the extent of replication of this ori plasmid (as determined by DpnI-resistant products) depends on the level of E1 protein. We introduced the ARA mutation into HPV-11 E1 and a more efficiently expressed, modified HPV-11 E1* (20), both under control of an adenovirus major late promoter (pMT2-E1 and pMT2-E1*, respectively). HPV-11 E1* derives its 5′-untranslated sequence and two amino acids at the amino terminus from HPV-16 E1. It is translated more efficiently than the HPV-11 E1 transcript and exhibits a 10- to 20-fold higher replication activity (ref. 20; Fig. 4; and data not shown).
With low levels of expression, the replication activity of E1ARA was reduced by 80% when compared with its wild-type counterpart (Fig. 4a, compare lane 2 with 4). The activity of E1ARA was reduced to 20% of that of wild-type E1 in the context of the high expressing pMT2-E1* constructs (Fig. 4b). The absence of E1 and E2 plasmids in samples treated with DpnI provided a control for complete digestion of the transfected plasmids. The levels of E1 proteins were comparable as determined by immunoblotting of cell lysates (Fig. 4c). Thus, efficient replication of HPV ori plasmids requires an intact cyclin interaction motif in E1. Interestingly, E1 underwent partial conversion to a more slowly migrating form reminiscent of that generated on phosphorylation by Cdks in vitro (Fig. 2) whereas E1ARA did not undergo this shift (Fig. 4c).
Cdk-Mediated E1 Phosphorylation Is Important for HPV Replication in Vitro and in Vivo.
To examine whether the replication defects in E1ARA reflect inefficient phosphorylation by Cdks, we examined the replication activity of E1ΔCdk and variants thereof. When expressed at low levels, the transient replication activity of E1ΔCdk was abolished (Fig. 4a), consistent with a requirement for E1 phosphorylation in HPV replication in vivo. The triple mutant S89A;S93A;S107A and the single mutant T468A also displayed undetectable activity, whereas the S107A mutant displayed 15% activity (Fig. 4a). When expressed at higher levels, E1*ΔCdk displayed 10% activity (Fig. 4b) compared with E1* expressed at similar protein levels (Fig. 4c). Consistent with the absence of phosphorylation, E1*ΔCdk did not undergo a shift to a more slowly migrating form (Fig. 4c). Recombinant EE-E1ΔCdk also displayed reduced activity using an E1- and E2-dependent cell-free replication assay (Fig. 4d). As described previously (19), plasmid replication generated fast-migrating Form I topoisomers and Form II open circles, as well as slow migrating θ-form replication intermediates. The activity of E1ΔCdk was ≈15% of the wild-type protein (Fig. 4d, compare lanes 1–4 to lanes 5, 6, 8, and 9), and this level of activity was not significantly increased at a higher concentration of the phosphorylation-deficient E1 protein (Fig. 4d, lanes 7 and 10). Taken together, our data indicate that the interaction of cyclin/Cdk complexes with E1 is critical for efficient replication and that one role, but perhaps not the only role, for cyclin/Cdk complexes in HPV replication is phosphorylation of E1.
DISCUSSION
An understanding of the mechanisms by which Cdks regulate DNA replication requires the identification of critical substrates involved in this process. In this study, HPV-18 E1 was the most frequently recovered cyclin E-interacting protein from a HeLa cell cDNA library. This interaction extends to the well characterized replication-competent HPV-11 E1, which interacts with A-, B-, E-, and F-type cyclins and is phosphorylated by Cdk2 and Cdk3 in vitro (Fig. 2). E1 functions together with the ori-binding protein E2 to establish the preinitiation complex (15, 16). Cyclin E can also bind the E1/E2 complex and in this context can phosphorylate E2 (Fig. 2). Mutations in the cyclin-binding RXL motif in E1 not only reduced binding to and phosphorylation by cyclin kinases but also significantly reduced E2-dependent HPV ori replication in vivo, suggesting a role for cyclin kinase-mediated E1 phosphorylation in HPV replication. Indeed, E1-containing mutations in all four Cdk consensus sites was still capable of binding E2 and cyclin E but was not phosphorylated and displayed greatly reduced replication activity in vitro and in vivo (Figs. 3 and 4). After completion of this work, Cueille et al. (32) reported that efficient E2-independent replication of a BPV ori plasmid by BPV E1 in crude Xenopus lysates correlates with the presence of cyclin E/Cdk2 and that E1 interacts with Xenopus cyclin E/Cdk2. However, the significance of this interaction for replication was not assessed.
How do cyclin/Cdk complexes and E1 or E2 phosphorylation regulate HPV replication? Given the available data (Fig. 2), it is unlikely that the formation of E1/E2 complexes is regulated by Cdks. It is conceivable that E1 and/or E2 phosphorylation facilitates assembly of higher-order E1/E2 complexes that recognize origins or facilitates their association with DNA polymerase α/primase (25–27) or replication protein A (33). Alternatively, phosphorylation may regulate E1’s helicase activity. In this regard, mutation of the single candidate Cdk phosphorylation site in the helicase domain (T468) greatly reduces the replication activity of E1 in vivo (Fig. 4b). The finding that cyclin E/Cdk2/E1 complexes retain the ability to phosphorylate bound E2 and exogenous histone H1 (Figs. 2 and 3d) leaves open the possibility that E1 physically couples Cdks to other replication components such as RPA and polymerase α that are known to be phosphorylated by Cdks and interact with E1/E2 complexes (34, 35). Although phosphorylation is important for maximal replication efficiency, it is not essential and can be overcome to a limited extent (≈10%) when E1ΔCdk is highly expressed. We note, however, that 5- to 10-fold decrease in replication efficiency would have major ramifications on the establishment phase of virus infection and subsequent DNA amplification for progeny production.
Because CIP/KIP proteins also use RXL motifs to bind cyclins, the interaction of E1 and CIP/KIP proteins with cyclins would be mutually exclusive. Thus, the use of this targeting motif by E1 ensures that its associated cyclin kinase would retain activity toward replication components, thereby facilitating HPV replication. Conversely, high levels of CIP/KIP proteins may be expected to compete with E1 for binding to cyclin E and preclude efficient viral DNA amplification. Cell-free HPV-11 ori replication is inhibited by the addition of p21CIP1 (J. S. Liu, S. R. Kuo, T. R. Broker, and L.T.C., unpublished data), reminiscent of that observed with SV40 replication (36). A reciprocal relationship also has been observed in benign HPV lesions in that high copy number of viral DNA and abundant viral RNA occurs in cells distinct from those in which p21CIP1 is induced in response to HPV infection (37).
Subversion of host cell functions has emerged as a major theme in viral life cycles. Viral genes can function to activate Cdks either by inducing their expression or by blocking the action of CKIs while simultaneously inducing a replication-competent state. Because of the critical roles of cyclin/Cdk complexes in chromosomal DNA replication, it is not surprising that viruses have also integrated the action of these enzymes into their replication pathway. The replication function of SV40 T antigen also requires specific phosphorylation by Cdks (38), suggesting that this may be a general feature of DNA viruses. The use of in vitro HPV DNA replication systems will facilitate a determination of the step(s) at which cyclin/Cdk function is employed in the HPV replication process.
Acknowledgments
We thank T. Hunter for cDNA libraries and W. Kaelin, T. Hunter, S. Elledge, E. Harlow, and J. Zhao for helpful discussions. J.W.H. was supported by Grants GM-54137 and AG-11085 from the National Institutes of Health and by the Welch Foundation. L.T.C. was supported by National Institutes of Health Grant CA36200 and by the Cystic Fibrosis Foundation.
ABBREVIATIONS
- Cdk
cyclin-dependent kinase
- HPV
human papillomavirus
- GST
glutathione S-transferase
- EE
Glu–Glu epitope
- GSH
glutathione
References
- 1.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 2.Dyson N. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 3.Russo A A, Jeffrey P D, Patten A K, Massague J, Pavletich N. Nature (London) 1996;382:325–331. doi: 10.1038/382325a0. [DOI] [PubMed] [Google Scholar]
- 4.Vlach J, Hennecke S, Amati B. EMBO J. 1997;16:5334–5344. doi: 10.1093/emboj/16.17.5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J, Saha P, Kornbluth S, Dynlacht B D, Dutta A. Mol Cell Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams P D, Sellers W R, Sharma S K, Wu A D, Nalin C M, Kaelin W G., Jr Mol Cell Biol. 1996;16:6623–6633. doi: 10.1128/mcb.16.12.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu L, Harlow E, Dynlacht B D. Genes Dev. 1995;9:1740–1752. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]
- 8.Krek W, Ewen M, Shirodkar S, Arany Z, Kaelin W G, Livingston D. Cell. 1994;78:161–172. doi: 10.1016/0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 9.Peeper D S, Parker L L, Ewen M E, Toebes M, Hall F L, Xu M, Zantema A, van der Eb A J, Piwnica-Worms H. EMBO J. 1993;12:1947–1954. doi: 10.1002/j.1460-2075.1993.tb05844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams, P. D., Li, X., Sellers, W. R., Baker, K. B., Leng, X., Harper, J. W., Taya, Y. & Kaelin, W. G., Jr. (1998) Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 11.Chevalier S, Blow J J. Curr Opin Cell Biol. 1996;8:815–821. doi: 10.1016/s0955-0674(96)80082-2. [DOI] [PubMed] [Google Scholar]
- 12.Ohtsubo M, Theodoras A M, Schumacher J, Roberts J M, Pagano M. Mol Cell Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leng X, Connell-Crowley L, Goodrich D, Harper J W. Curr Biol. 1997;7:709–712. doi: 10.1016/s0960-9822(06)00301-0. [DOI] [PubMed] [Google Scholar]
- 14.Zhao J, Dynlacht B, Imai T, Hori T, Harlow E. Genes Dev. 1998;12:456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chow L T, Broker T R. Intervirology. 1994;37:150–158. doi: 10.1159/000150373. [DOI] [PubMed] [Google Scholar]
- 16.Stenlund A. In: DNA Replication in Eukaryotic Cells. DePamphilis M L, editor. Vol. 23. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 679–697. [Google Scholar]
- 17.Yang L, Li R, Mohr I J, Clark R, Botchan M R. Nature (London) 1991;353:628–632. doi: 10.1038/353628a0. [DOI] [PubMed] [Google Scholar]
- 18.Chiang C M, Ustav M, Stenlund A, Ho T F, Broker T R, Chow L T. Proc Natl Acad Sci USA. 1992;89:5799–5803. doi: 10.1073/pnas.89.13.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo S-R, Liu J-S, Broker T R, Chow L T. J Biol Chem. 1994;269:24058–24065. [PubMed] [Google Scholar]
- 20.Zou N, Liu J-S, Kuo S-R, Broker T R, Chow L T. J Virol. 1998;72:3436–3441. doi: 10.1128/jvi.72.4.3436-3441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller F, Seo Y S, Hurwitz J. J Biol Chem. 1994;269:17086–17094. [PubMed] [Google Scholar]
- 22.Sedman J, Stenlund A. EMBO J. 1995;14:6218–6228. doi: 10.1002/j.1460-2075.1995.tb00312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J-S, Kuo S-R, Broker T R, Chow L T. J Biol Chem. 1995;270:27283–27291. doi: 10.1074/jbc.270.45.27283. [DOI] [PubMed] [Google Scholar]
- 24.Liu J S, Kuo S R, Makhov A M, Cyr D M, Griffith G D, Broker T R, Chow L T. J Biol Chem. 1998;273:30704–30712. doi: 10.1074/jbc.273.46.30704. [DOI] [PubMed] [Google Scholar]
- 25.Park P, Copeland W, Yang L, Wang T, Botchan M R, Mohr I J. Proc Natl Acad Sci USA. 1994;91:8700–8704. doi: 10.1073/pnas.91.18.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masterson P J, Stanley M A, Lewis A P, Romanos M A. J Virol. 1998;72:7407–7419. doi: 10.1128/jvi.72.9.7407-7419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conger, K. L., Liu, J.-S., Kuo, S.-R., Chow, L. T. & Wong, S.-F (1999) J. Biol. Chem., in press. [DOI] [PubMed]
- 28.Connell-Crowley L, Harper J W, Goodrich D W. Mol Biol Cell. 1997;8:287–301. doi: 10.1091/mbc.8.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang, W., Jimenez, G., Wells, N. J., Wahl, G. M., Hunter, T & Fukunaga, R. (1998) Mol. Cell, in press. [DOI] [PubMed]
- 30.Connell-Crowley L, Elledge S J, Harper J W. Curr Biol. 1998;8:65–68. doi: 10.1016/s0960-9822(98)70021-1. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz E, Freese U K, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. Nature (London) 1985;314:111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 32.Cueille N, Nougarede R, Mechali F, Phillipe M, Bonne-Andrea C. J Virol. 1998;72:7255–7262. doi: 10.1128/jvi.72.9.7255-7262.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li R, Botchan M R. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 34.Wold M S. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 35.Voitenleitner C, Fanning E, Nasheuer H P. Oncogene. 1997;14:1611–1615. doi: 10.1038/sj.onc.1200975. [DOI] [PubMed] [Google Scholar]
- 36.Flores-Rozas H, Kelman Z, Dean F B, Pan Z Q, Harper J W, Elledge S J, O’Donnell M, Hurwitz J. Proc Natl Acad Sci USA. 1994;91:8655–8659. doi: 10.1073/pnas.91.18.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt-Grimminger D C, Wu X, Jian Y, Broker T R, Chow L T. Am J Pathol. 1998;152:1015–1024. [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Bhattacharyya S, Prives C. J Virol. 1997;71:6479–6485. doi: 10.1128/jvi.71.9.6479-6485.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]