Abstract
Understanding the relationship between the structural and functional organization of the human brain is one of the most important goals of neuroscience. Individual variability in brain structure means that it is essential to obtain this information from the same subject. To date, this has been almost impossible. Even though noninvasive functional imaging techniques such as functional MRI (fMRI) are now commonplace, there is no complementary noninvasive structural technique. We present an in vivo method of examining the detailed neuroanatomy of any individual, which can then be correlated with that individual's own functional results. This method utilizes high-resolution structural MRI to identify distinct cortical regions based on cortical lamination structure. We demonstrate that the observed MR lamination patterns relate to myeloarchitecture through a correlation of histology with MRI. In vivo high-resolution MRI studies identify striate cortex, as well as visual area V5, in four individuals, as defined by using fMRI. The anatomical identification of a cortical area (V5/MT) outside of striate cortex is a significant advance, proving it possible to identify extra-striate cortical areas and demonstrating that in vivo structural mapping of the human cerebral cortex is possible.
Positron emission tomography and functional MRI (fMRI) have revealed specific activations in perceptual, cognitive, and motor tasks. Interpretation depends crucially on knowledge of brain anatomy. The key issue, and the focus of this article, is how to define functionally activated regions anatomically.
Knowledge of human brain structure comes from cyto-, myelo-, and chemoarchitectural analysis of postmortem material. If all functional imaging subjects were so studied, we could exactly relate functional results with individual cerebral anatomy. Because this level of study is not practical, the systematic correlation of structure and function, crucially important in neuroscience, has been impossible in normal living humans.
Researchers have related human findings with functional and anatomical detail of presumed comparable areas in the monkey brain (1). Despite excellent homology between human and monkey brain for some regions, there is much uncertainty over others, e.g., whether human and monkey inferior parietal lobules correspond.
Correlating human functional and structural neuroanatomy through detailed MR images was pioneered 10 years ago in relating findings to obvious features such as size, shape, and landmarks (2, 3). Traditionally, atlases and standardized coordinates are used for gross localization and to compare individuals, but such approaches are problematic. The commonly used atlas, by Talairach and Tournoux (4), gives coordinates based on the gross morphology of a poorly representative single brain that has never undergone cyto- or myeloarchitectonic analysis. Cortical areas are crudely estimated by simply projecting Brodmann's cytoarchitectonic map (5). However, brain topography varies among individuals. The motion-sensitive area, human V5, has a range of 27 mm in location (2), whereas the size, shape, and topographical relations of Brodmann's areas 17 and 18 vary highly between individuals (6, 7). One solution has been probabilistic atlases of easily identified or histologically distinct regions, but these only assist in the interpretation of functional results (7, 8).
Accurate localization of individual results can only come from their unique neuroanatomy. Our primary goal is the in vivo definition of anatomically distinct regions to correlate with individual functional results. Armed with detailed information, we may then address important questions: what is the underlying structural organization of human functional areas? Is this organization preserved across individuals? Can we predict functions of structurally defined regions about which we have no functional information?
Flechsig (9) related cerebral myelination patterns, presumed function early in life, and features of cortical folding. His most heavily myelinated fields (1–20, primary areas functioning early in life) closely relate to overlying sulci formed earlier in embryonic life. Early myelination seems to correlate with early function and may direct early, more constant sulcal patterns. In 1993, Watson et al. (2) described a strong structure-function relationship for human V5, which correlates precisely with Flechsig's Field 16 and the little human histology available (10, 11). Other Flechsig areas in the 1–20 group have structural surface folding correlates, many with important functions, e.g., striate and primary sensorimotor cortex. The functions of other areas, e.g., a dorsal field near the occipito-parietal junction, are unclear.
The work on in vivo recognition of cortical features is limited to the striate cortex (12, 13).** MRI acquisition sequences are commonly used to differentiate gray and white matter; however, the work of Clark et al. (13) was significant in showing that lamination patterns within the gray matter observed on MR images of striate cortex directly correlated with its distinctive myelination pattern. Additional work is now required to extend these observations into other cortical areas and to validate the observations by using objective observer-independent techniques.
We have developed an in vivo method to identify distinct cortical regions based on lamination structure by using high-resolution MRI. We demonstrate our method with the in vivo identification of visual area V5 in four individuals as defined in fMRI and confirm earlier demonstrations of V1.
Methods
We carried out two concurrent investigations, a correlation study of structural MRI and histology and an in vivo structure–function correlation MRI study.
Postmortem Histology and Structural MRI.
The entire brain of a normal person and a portion of striate cortex (35 × 30 × 3 mm3) from another were obtained through a Brain Donor Program (see Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org). The whole brain was formalin fixed for 2 weeks, and a left inferior lateral block was sectioned and MR imaged to determine an optimal smaller block (10 × 25 × 30 mm3) containing putative V5/MT.
The striate and putative V5/MT blocks were imaged at high resolution in a transverse orientation in a Bruker BIOSPEC 47/30 scanner, a horizontal 4.7 T magnet with 30-cm bore. T2-weighted multislice images were acquired. [Number of slices, 12 and 24, respectively; slice thickness, 0.6 mm (no gap); field of view (FOV) 30 × 35 mm2; matrix, 512 × 512; in-plane resolution, 59 × 68 μm2; echo time (TE), 66 ms; repetition time (TR), 4 s; number of excitations (NEX), 4; acquisition time, 69 min and 200 min, respectively.]
The blocks were cryoprotected and serially sectioned at 50 μm on a freezing microtome. Series of sections at 500 μm intervals were stained with cresyl violet for cytoarchitecture and black-gold for myeloarchitecture (14). Mounted sections were washed in 1% acetic acid (3 × 15 min), heated to 60°C in 0.1 M PBS for 30 min and stained with black-gold at 60°C for ≈90 min until fine fibers of the molecular layer appeared. Staining was intensified in 0.2% potassium tetrachloroaureate for 10 min at 60°C; sections were rinsed in distilled water and fixed in 3% sodium thiosulphate before dehydrating in alcohols, clearing, and coverslipping.
In Vivo Structure–Function Correlation MRI Study of Visual Area V5.
Six healthy, normal subjects (5 males; mean age, 36; range, 22–46) were scanned to identify area V5 by using fMRI. Each subject's V5 was located by using fMRI results and sulcal patterns on preliminary MRI. Each subject was scanned with high-resolution structural MRI to examine detailed neuroanatomy in these regions. All subjects gave informed consent in accordance with the Declaration of Helsinki, with ethics approval from the Howard Florey Institute and the University of Sydney.
Structural MRI.
Eight coronally oriented high-resolution surface coil T1-weighted images were acquired successively (15) on a 1.5 T scanner (Signa Echospeed, General Electric). Twin flexible surface coils (FOV, 10 cm) were used and placed directly over the regions identified preliminarily, with protective cushions and custom-made foam moulds. Aliasing artifact (superimposed ghost images of slices from the opposite end) was compensated for by setting a wide sagittal FOV. [Three-dimensional (3D) fast spoil gradient (FSPGR) sequence; 120 slices, slice thickness: five subjects, 0.5 mm; one subject, 0.7 mm (no gap); FOV, 16 cm2; matrix, 288 × 288; in-plane resolution, 0.556 × 0.556 mm2; TE, 3.5 ms; TR, 15.9 ms; inversion time, 350 ms; NEX, 1; FA of 25°.] A T1-weighted image was similarly acquired by using a volume coil, except with twice the slice thickness, and used for grayscale normalization. Raw images were transferred via a DICOM client program (Digital Jacket, Hewlett–Packard), composed into contiguous volumes, and saved in Analyze format (Biomedical Imaging Resource, Mayo Foundation, Rochester, MN).
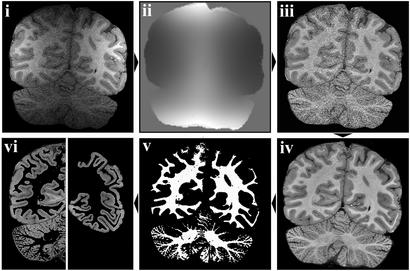
Images were manually segmented to remove nonbrain tissue by using MEDX V.3.4.1 (Sensor Systems, Sterling, VA) (Fig. 1i). Surface coil signal attenuation was corrected by gray scale normalization (16). Optimal normalization was achieved with a priori maps of gray, white matter, and cerebrospinal fluid (CSF) generated from the volume coil T1-weighted image, after first applying a nonlinear noise reduction algorithm (17). Normalization maps were estimated for each surface coil image (Fig. 1ii) and used to generate a series of grayscale-normalized surface coil (GS-SC) T1-weighted images (Fig. 1iii).
Figure 1.
Structural image processing. (i) Deskulled surface coil (SC) image. (ii) Normalization maps for SC image. (iii) Intensity-restored SC image. (iv) Mean SC image. (v) Segmented white matter mask for mean SC image. (vi) Segmented (Left) and manually edited (Right) gray matter from mean SC image.
The eight GS-SC T1-weighted images were realigned by using a linear registration tool (18), resampled at half the acquired voxel dimensions, and averaged by using a softmean function to create a single, high-resolution average GC-SC T1-weighted image (in-plane resolution, 0.28 × 0.28 mm; slice thickness, 0.25/0.35 mm; Fig. 1iv).
A segmented white matter probability map was generated (16) and manually edited as necessary (Fig. 1v). A mask of the white matter image was used to generate a gray matter image (Fig. 1vi Left), which was manually edited to remove cerebellum, CSF in deep sulci, and medial cortex unsuitable for cortical lamination analysis (Fig. 1vi Right).
A two-dimensional (2D) method of performing simple cortical lamination analysis was developed based on the analysis of intensity line profiles through gray matter. A distance transform was used to calculate a set of nonintersecting line segments through the cortical sheet, taking only the shortest segments where lines shared a common end point. A polynomial function was fitted to each segment intensity profile, and the zero points of the first derivative, corresponding to stationary points in the intensity profile, were determined. The number and position of stationary points were used to identify and compare discrete cortical regions possessing similar patterns of lamination (for example, see Fig. 4iv).
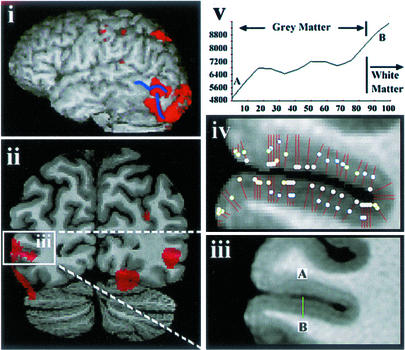
Figure 4.
In vivo functional MR and structural MR results from one subject (left hemisphere). (i) Left lateral view of surface rendering of subjects' brain with overlaid functional results of V5 study (red). Blue sulcal markings identify the ascending limb of the inferior temporal sulcus and lateral occipital sulcus. (ii) Coronal section through the functionally defined V5 region (red) overlaid on the subject's average T1 image with V5 indicated (white box). (iii) Inset from ii Lower showing the high-resolution average T1 for this region (slice thickness, 0.35 mm; in-plane resolution, 0.28 × 0.28 mm2). (iv) Cortical lamination analysis with positions of intensity line profiles (red lines) and maxima in the intensity line profiles (white dots) indicated. (v) Intensity line profile through the green line AB of (iii) showing two intensity maxima.
fMRI.
The six subjects underwent fMRI while observing a moving checkerboard, and V5 was identified (2). Subjects saw a black and white checkerboard subtending 13° horizontally and vertically, moving randomly in the four cardinal directions and diagonally at a speed of 9°/s. A Macintosh PowerBook G3 laptop using PSYSCOPE V.1.2.2 (19) generated stimuli, projected on a 1.2 × 1.0-m ground-glass screen 2.2 m from the subjects' eyes. Subjects lay supine and observed the stimuli in a mirror. The motion condition was presented in 6-s blocks randomly interleaved with 6-s blocks of the stationary condition. Subjects maintained central fixation throughout each 96-s session; eight sessions were acquired.
A whole-brain sagittal T1 anatomical scan (3D FSPGR: TR, 11.8 ms; TE, 2.4 ms; FA, 25°; NEX, 1; acquisition matrix, 256 × 256; FOV, 23 cm2; 124 slices of thickness 1.2 mm) and whole-brain axial T2 anatomical scan (2D fast spin echo sequence: TR, 5120 ms; TE, 97ms; FA, 90°; NEX, 2; acquisition matrix, 384 × 384; FOV, 27 cm2; 15 slices of thickness 7 mm) were acquired. Echoplanar imaging (EPI) was performed with 32 whole brain volumes acquired per scanning session (TR/TE = 3000/60; FA, 30o; NEX, 1; matrix 64 × 64 × 15; FOV, 27 cm2; slice thickness 7 mm) with slice orientation as for the T2 anatomical scan. Two initial EPI scans were discarded to allow for T1 saturation effects.
Each subject's functional images were spatially normalized to the whole brain T1-weighted image. A mean EPI volume was calculated for the first session, and the other session mean EPI volumes were aligned by using a linear registration tool (18). The aligned mean EPI volumes were averaged to create a mean volume for the whole study. Each subject's whole brain T2-weighted anatomical image was aligned to the whole brain T1-weighted image, and then the mean study EPI volume was aligned to this. Transformation matrices were concatenated and applied to each individual EPI volume, with the resulting EPI volumes having voxel dimensions of 1.2 × 0.9 × 0.9 mm3. A categorical comparison of the motion condition with the stationary condition was determined with SPM99 (20, 21) using a box-car response convolved with the hemodynamic response but no temporal derivative. The statistical t-maps were transformed to standardized Z-scores, thresholded at an uncorrected probability of P < 0.00001 (individual subject analyses), and visualized on each subject's anatomical T1 image. V5 was located to compare with the structural MR results.
To compare the functional and structural results, each subject's whole brain T1-weighted image was aligned to a low-resolution copy of the average GS-SC T1-weighted image. This transformation was concatenated with the previous transformations, reapplied to each EPI volume to produce volumes in registration with their average GS-SC T1-weighted image with voxel dimensions of 1.1 × 1.2 × 1.0/1.4 mm3. The EPI data were reanalyzed with SPM99 and visualized on the subject's average GS-SC T1-weighted image.
Results
Postmortem.
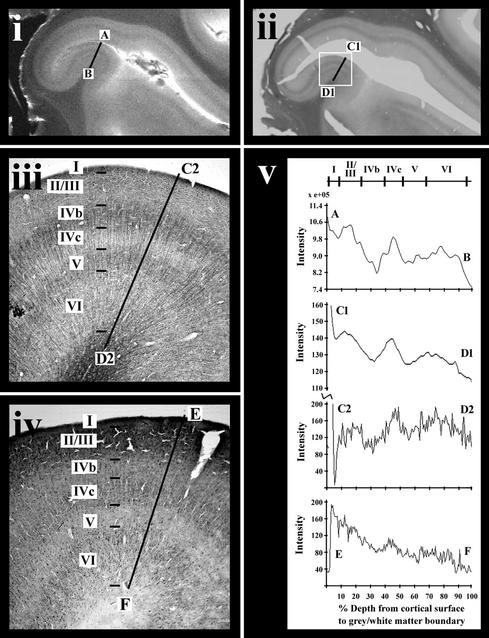
The postmortem striate brain section (Fig. 2) shows, as expected, distinctive myelarchitecture consisting of dark horizontally oriented bands within layers IV (stria of Gennari) and V (internal band of Baillarger) (Fig. 2 ii and iii; see Table 1 for summary of cyto- and myeloarchitecture). Note that the inset region (Fig. 2 ii and iii) from the top of a gyrus differs in structure from the remainder of the visible striate cortex in layer thickness, with a more prominent internal band of Baillarger. The MR image (Fig. 2i) and the corresponding myelin-stained section (Fig. 2 ii and iii) show remarkable correspondence. Both the stria of Gennari (layer IVb) and the internal band of Baillarger (layer V) are apparent in the MRI as darker bands. There is also evidence of a faint outer dark band corresponding to the fine horizontally arranged fibers in layer I apparent in the myelin-stained section (Fig. 2 iii and v). Equivalent intensity line profiles through each figure also indicate that the MR intensity line profile (Fig. 2v AB) is most similar to the low-magnification myelin stain intensity line profile (Fig. 2v C1–D1), with intensity reductions corresponding to layers I, IVb and V. Further quantitative analysis is required to ascertain the exact correspondence between the MR image signal and the histological stains.
Figure 2.
Postmortem MRI and histology of section from striate cortex. (i) T2-weighted MRI (slice thickness, 0.60 mm; in-plane resolution, 59 × 68 μm2). (ii) Low-magnification image of myelin-stained section corresponding to the MRI in i. (iii) High-magnification inset of myeloarchitecture from white box in ii. (iv) High-magnification cell density stain of adjacent section to iii. (v) Intensity line profiles through equivalent regions of each image measured through the annotated line segments in i–iv. The intensity line profile for the cell density stain (iv) has been inverted for ease of comparison with the other profiles. The horizontal scale on these profiles is expressed as a percentage of the cortical thickness measured from the cortical surface (refer to Table 1 for more detail).
Table 1.
Cyto- and myeloarchitectonic characteristics of striate cortex (see Fig. 2)
| Layer | Cytoarchitecture | Myeloarchitecture |
|---|---|---|
| I | Outer meshed layer of fine horizontally oriented fibers | |
| II | Radial arrangement of cells | Sparse fine radial fibers on a sparse mesh of fine fibers |
| III | Medium and fine radial fibers on a dense fine fiber mesh | |
| IV | Medium radially arranged fiber bundles extending from layer VI | |
| a: Not discernible from II and III | ||
| b: Nonpyramidal cells; reduced density compared with II/III | Dense fiber mesh with prominent horizontal orientation (stria of Gennari) | |
| c: Radially arranged nonpyramidal cells with high cell density | Sparser mesh without apparent orientation | |
| V | Reduced cell density compared with IV; increased numbers of small pyramidal cells and single large pyramidal cells | Dense fine fiber mesh with prominent horizontal orientation (internal band of Baillarger) |
| VI | Radially arranged pyramidal cells | Medium radially arranged fiber bundles extending into layer IV; fiber mesh without prominent orientation |
The myeloarchitecture of the postmortem putative V5 brain section is also distinctive with dark, horizontally oriented bands in layers I, IV, and V (the latter two correspond to the lines of Baillarger) and bundles of radial fibers traversing layers VI to IV (Fig. 3 ii and v; see Table 2 for summary of the cyto- and myeloarchitecture). The MR image (Fig. 3i) shows a dark band adjacent to the white matter and a second dark band approximately in the middle of the cortex, corresponding to the lines of Baillarger observable in the myelin-stained sections.
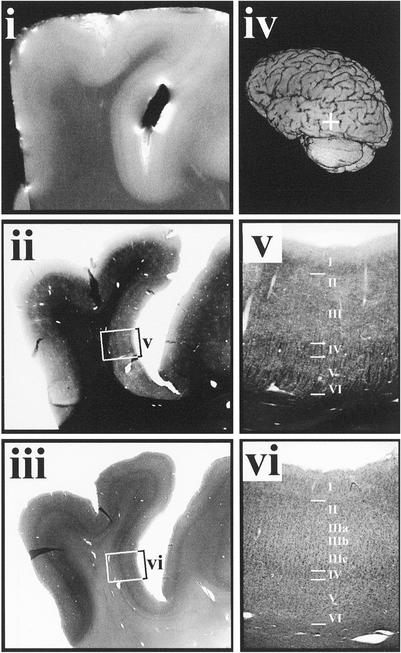
Figure 3.
Postmortem MRI and histology of section from putative V5 region. (i) T2-weighted MRI (slice thickness, 0.60 mm; in-plane resolution, 59 × 68 μm2). (ii) Low-magnification image of myelin-stained section corresponding to the MR image in i. (iii) Low-magnification image of adjacent cell density-stained section. (iv) Left posterior oblique-rendered image of the postmortem brain with region of interest marked by a white crosshair. (v) High magnification of myelarchitecture from white box in ii. (vi) High-magnification cell density image of cell density stain from white box in iii (refer to Table 2 for more detail).
Table 2.
Cyto- and myeloarchitectonic characteristics of cortex in putative human V5/MT (see Fig. 3)
| Layer | Cytoarchitecture | Myeloarchitecture |
|---|---|---|
| I | Sublayer of horizontally oriented fine fiber mesh | |
| II | Medium cell packing density | Sparse fine fiber mesh with no prominent orientation |
| III | IIIa: Medium cell packing density IIIb: Low cell packing density; small radially arranged pyramidal cells; IIIc: Large prominent pyramidal cells | Sparse fine fiber mesh with prominent vertical and horizontal clustering |
| IV | Granular layer with increased cell packing density compared with II | Medium radially arranged fiber bundles; dense mesh of horizontally oriented fine fibers (external band of Baillarger) |
| V | Medium sized pyramidal cells with similar cell packing density to III. The boundary between layers V and VI not defined | Medium radially arranged fiber bundles; sublayer with horizontally oriented fine fibers (internal band of Baillarger) |
| VI | Medium sized pyramidal cells with similar cell packing density to III. The boundary between layers V and VI not defined | Medium radially arranged fiber bundles |
In Vivo.
Two subjects were removed from the structural analysis because of motion during scans (see Supporting Text). The results for one of the remaining four subjects are shown in detail in Fig. 4. The functionally defined V5 region (Fig. 4i, red) lies at the intersection of the ascending limb of the inferior temporal sulcus and the lateral occipital sulcus (Fig. 4i, blue; ref. 2), with other areas of activation visible corresponding to lower order visual areas, most likely V2/V3. A high degree of spatial correspondence is evident between the extent of the functional V5 results, (Fig. 4ii) and the delineation of the putative structural V5 region (Fig. 4 iii and iv). In particular, intensity line profile analysis through this sulcus (Fig. 4iv) delineates the lower bank (two dots per line segment) from the immediately adjacent upper bank (one dot per line segment), with the functional results (Fig. 4ii) indicating V5 is located in the lower bank. A thicker light band is identifiable in the lower half of the cortex of the putative V5 region, and another light band is visible near the cortical surface.
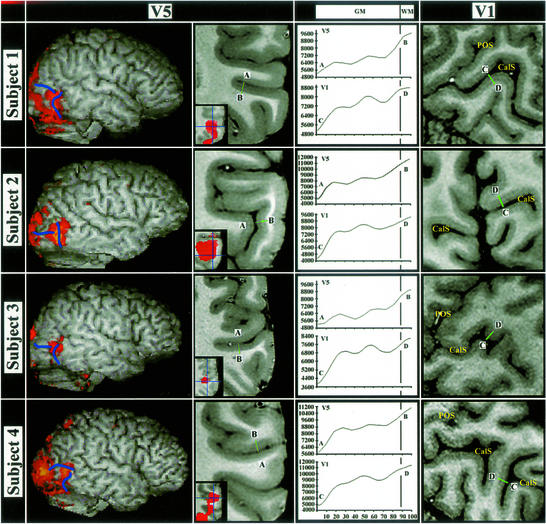
In vivo structural MRI and fMRI results for the V1 and V5 regions are shown for all four subjects' right hemispheres in Fig. 5. As in Fig. 4, there is a high degree of spatial correspondence between the extent of the functional and putative structural V5 regions (the latter again characterized by two light bands). The V1 images, with lower signal-to-noise due to lateral positioning of the surface coils, clearly show the stria of Gennari as a thick central light band. Note the difference in number, intensity, amplitude, and position of maxima in the intensity line profiles for V1 and V5.
Figure 5.
In vivo V5 structural MRI and fMRI results and in vivo V1 structural MR results in four subjects [slice thickness, 0.35 mm (subject 2)/0.25 mm (subjects 1, 3, and 4); in-plane resolution, 0.28 × 0.28 mm2]. The first column shows surface-rendered images of the right hemispheres with overlaid fMRI V5 results (red) and sulcal landmarks (blue). The second column shows coronal sections from the high-resolution average T1 image through these functionally defined V5 regions. Inset are the overlaid fMRI results for each subject (red). Green lines (A–B) indicate the putative structural V5 region, with intensity line profiles shown in the column headed GM|WM. The column at far right shows sagittal or coronal sections through each subject's right hemisphere striate cortex. The calcarine sulcus (CalS) and parieto-occipital sulcus (POS) are indicated. Intensity line profiles for V1 (green lines C–D) are shown in the column headed GM|WM.
Discussion
The MR signal is a weighted summation of cellular properties, background features, and patterns of fiber connections in the cerebral cortex. Most of this signal is derived from myelin, as evidenced by the remarkable degree of similarity between T2-weighted images of postmortem material and corresponding myelin-stained sections (Figs. 2 and 3). Moreover, we have demonstrated that intensity line profiles through the cortex can be used to characterize and delineate cortical areas, not only from the surrounding cortex (Fig. 4), but also from cortical areas located distantly (e.g., see Fig. 5 V1 and V5).
We have demonstrated human striate or primary visual cortex, reproducing earlier results of Clark et al. (13). Barbier et al. (12) recently reproduced the work of Clark et al. at 3 T field strength (resolution 350 × 350 × 600 μm3) by using a 3D acquisition protocol and with the aid of surface coils. Our work using a 3D protocol and phased array of surface coils includes a postprocessing protocol for grayscale normalization, alignment, averaging, segmentation, and quantitative analysis of the images. Importantly, this method enables combination of images from multiple studies in different sessions by using different coil placements.** Our achieved resolution (280 × 280 × 250 μm3) has enabled identification of a cortical area outside of striate cortex, area V5/MT.
V5/MT was an ideal cortical area to examine because it is easy to activate, has reliable sulci landmarks, and correlates well with Flechsig's Field 16 (2, 22). We identified functionally in vivo V5 in all of the eight hemispheres tested and, in six of these, corresponding cortex with identifiable and similar MRI line profiles through the cortical layers (Figs. 4 and 5; Supporting Text). We believe this approach will be successful in localizing other cortical areas, initially guided by Flechsig's first 20 fields. In the present study, we (as have others) observed activation foci other than V5/MT in the occipital and parietal regions. Some of these seem to correlate with structural cortical features identified in our structural study. One of these was dorsal, in the parietal cortex, where interestingly, Flechsig's Field 17 lies. Other areas had distinctive cortical line profiles in the structural MR images without corresponding functional activity, either with activation too weak to identify or because these areas were not involved in this paradigm.
Good histological information exists about V5/MT in the nonhuman primate. There are fundamental similarities in the location, architecture, function, and topographic organization of V5/MT across many nonhuman primate species (23). In owl monkey, MT is a small region in the caudal middle temporal sulcus, characterized by heavy myelination consisting of distinct outer and inner bands of Baillarger (24). In the macaque, the homologous region is found in the superior temporal sulcus and is characterized by heavy myelination concentrated in layers IV and V and often extending into layer VI (23). In the macaque, the central visual field representation of V1 and V2 project to MT, whereas the peripheral field projects to an area medial to MT (called MTp), which is characterized by much lighter myelination (25). Other visual areas, some with similar roles and structure, surround MT/MTp in the macaque (25).
In humans, V5 is located more posteriorly, in the inferior temporal sulcus, with previous myeloarchitectonic evidence (10, 11), suggesting that V5 is also heavily myelinated, particularly in the lower layers. Our in vivo results are consistent with this previous evidence, with a thick light band identified in the lower portion of the gray matter underlying the functionally activated V5 region (Figs. 4 and 5).
Recent evidence suggests that human V5/MT is surrounded by other regions with similar, but more specialized, motion sensitive roles (26) which may, as the myeloarchitecture of the macaque suggests, have similar architecture. The functional stimulus used here may have activated several of these regions, or, as fMRI records changes in blood oxygenation, the functional activation may have spilled into other regions. An important issue is to what extent do activation foci correspond to architectonic fields. The structural area we have identified may actually be part, or all, of the human V5/MT complex. Nevertheless, in all of the subjects examined, the V5 functional activation, using a consistent threshold across all subjects, closely follows the structural MR lamination patterns in both location and extent. In the majority of our subjects, there is a clear border between the putative structural V5 region and the immediately adjacent dorsal region (e.g., Fig. 4iv), whereas the border ventrally is less well defined and often appears as a gradual transition between regions.
Localization of putative V5/MT in our postmortem material was achieved by using sulcal landmarks. Although we are cautious in drawing conclusions because there are no functional data on this subject, the myelin stain of this region shows heavy myelination in the lower layers, as expected of V5/MT, which correlates extremely well with lamination patterns evident in the corresponding MR image.
Although not as sensitive as histological analysis, high-resolution structural MRI techniques sensitive to myelin concentration have the advantage of being noninvasive and applicable in vivo and may still allow the identification of many cortical areas. Recent extensions to the technique have provided preliminary evidence that quantitative parcellation of the cortex will be possible by using high-resolution structural MR images. Even areas of cortex with the same structural MRI characteristics but very different functions may still be distinguished, based on differences in location. If they are found in similar locations, this observation may be a clue to functional similarity.
We have shown that different areas of living human cortex can be distinguished on structural grounds by noninvasive MR techniques. Moreover, we have shown a direct correlation between the myeloarchitectonic characteristics of the human cerebral cortex, the MR image characteristics, both postmortem and in vivo, and the results of a functional activation study. The identification of a cortical area (V5/MT) outside of striate cortex is a significant advance and opens up both the possibility of in vivo structural mapping and true structure–function correlation in the human cerebral cortex.
Supplementary Material
Acknowledgments
We thank Katherine Podzebenko for assistance with the fMRI study and John Williams for his assistance with the initial postmortem MRI studies. This work was supported by the Australian National Health and Medical Research Council and the Brain Foundation.
Abbreviations
- fMRI
functional MRI
- GS-SC
grayscale normalized surface coil
- FOV
field of view
- EPI
echoplanar imaging
- TE
echo time
- TR
repetition time
- NEX
number of excitations
Footnotes
Walters, N. B., Jenkinson, M., Kean, M., Watson, J. D. & Egan, G., Proceedings of the Ninth International Conference on Neural Information Processing, November 18–22, 2002, Singapore, Vol. 1, pp. 193–196.
References
- 1.Felleman D J, Van Essen D C. Cereb Cortex. 1991;1:1–47. doi: 10.1093/cercor/1.1.1-a. [DOI] [PubMed] [Google Scholar]
- 2.Watson J D G, Myers R, Frackowiak R S J, Hajnal J V, Woods R P, Mazziotta J C, Shipp S, Zeki S. Cereb Cortex. 1993;3:79–94. doi: 10.1093/cercor/3.2.79. [DOI] [PubMed] [Google Scholar]
- 3.Paus T, Petrides M, Evans A C, Meyer E. J Neurophysiol. 1993;70:453–469. doi: 10.1152/jn.1993.70.2.453. [DOI] [PubMed] [Google Scholar]
- 4.Talairach J, Tournoux P. Co-Planar Stereotaxic Atlas of the Human Brain. Stuttgart: Thieme; 1988. [Google Scholar]
- 5.Brodmann K. Vergleichende Lokalisationslehre der Groβhirnrinde. Leipzig: Barth; 1909. [Google Scholar]
- 6.Stensaas S S, Eddington D K, Dobelle W H. J Neurosurg. 1974;40:747–755. doi: 10.3171/jns.1974.40.6.0747. [DOI] [PubMed] [Google Scholar]
- 7.Amunts K, Malikovic A, Mohlberg H, Schormann T, Zilles K. NeuroImage. 2000;11:66–84. doi: 10.1006/nimg.1999.0516. [DOI] [PubMed] [Google Scholar]
- 8.Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, et al. Philos Trans R Soc London B. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flechsig P. Anatomie des Menschlichen Gehirns und Rückenmarks. Leipzig: Thieme; 1920. [Google Scholar]
- 10.Clarke S, Miklossy J. J Comp Neurol. 1990;298:188–214. doi: 10.1002/cne.902980205. [DOI] [PubMed] [Google Scholar]
- 11.Tootell R B, Taylor J B. Cereb Cortex. 1995;5:39–55. doi: 10.1093/cercor/5.1.39. [DOI] [PubMed] [Google Scholar]
- 12.Barbier E L, Marret S, Danek A, Vortmeyer A, van Gelderen P, Duyn J, Bandettini P, Grafman J, Koretsky A P. Magn Reson Med. 2002;48:735–738. doi: 10.1002/mrm.10255. [DOI] [PubMed] [Google Scholar]
- 13.Clark V P, Courchesne E, Grafe M. Cereb Cortex. 1992;2:417–424. doi: 10.1093/cercor/2.5.417. [DOI] [PubMed] [Google Scholar]
- 14.Schmued L, Slikker W., Jr Brain Res. 1999;837:289–297. doi: 10.1016/s0006-8993(99)01624-8. [DOI] [PubMed] [Google Scholar]
- 15.Holmes C J, Hoge R, Collins L, Woods R, Toga A W, Evans A C. J Comput Assist Tomogr. 1998;22:324–333. doi: 10.1097/00004728-199803000-00032. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Brady M, Smith S. IEEE Trans Med Imaging. 2001;20:45–57. doi: 10.1109/42.906424. [DOI] [PubMed] [Google Scholar]
- 17.Smith S, Brady J. Int J Comp Vis. 1997;23:45–78. [Google Scholar]
- 18.Jenkinson M, Smith S M. Med Image Anal. 2001;5:143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- 19.Cohen J D, MacWhinney B, Flatt M, Provost J. Behav Res Methods Instrum Comput. 1993;25:257–271. [Google Scholar]
- 20.Friston K J, Holmes A P, Worsley K J, Poline J-P, Frith C D, Frackowiak R S J. Hum Brain Mapp. 1995;2:189–210. [Google Scholar]
- 21.Friston K J, Holmes A P, Worsley K J. NeuroImage. 1999;10:1–5. doi: 10.1006/nimg.1999.0439. [DOI] [PubMed] [Google Scholar]
- 22.Dumoulin S O, Bittar R G, Kabani N J, Baker C L, Jr, Le Goualher G, Pike B, Evans A. Cereb Cortex. 2000;10:454–463. doi: 10.1093/cercor/10.5.454. [DOI] [PubMed] [Google Scholar]
- 23.Van Essen D C, Maunsell J H R, Bixby J L. J Comp Neurol. 1981;199:293–326. doi: 10.1002/cne.901990302. [DOI] [PubMed] [Google Scholar]
- 24.Allman J M, Kaas J H. Brain Res. 1971;31:85–105. doi: 10.1016/0006-8993(71)90635-4. [DOI] [PubMed] [Google Scholar]
- 25.Desimone R, Ungerleider L G. J Comp Neurol. 1986;248:164–189. doi: 10.1002/cne.902480203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dukelow S P, DeSouza J F X, Culham J C, Van Den Berg A V, Menon R S, Vilis T. J Neurophysiol. 2001;86:1991–2000. doi: 10.1152/jn.2001.86.4.1991. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.