Abstract
Matriptase is a type II transmembrane serine protease expressed in most human epithelia, where it is coexpressed with its cognate transmembrane inhibitor, hepatocyte growth factor activator inhibitor (HAI)-1. Activation of the matriptase zymogen requires sequential N-terminal cleavage, activation site autocleavage, and transient association with HAI-1. Matriptase has an essential physiological role in profilaggrin processing, corneocyte maturation, and lipid matrix formation associated with terminal differentiation of the oral epithelium and the epidermis, and is also critical for hair follicle growth. Matriptase and HAI expression are frequently dysregulated in human cancer, and matriptase expression that is unopposed by HAI-1 potently promotes carcinogenesis and metastatic dissemination in animal models.
DISCOVERY OF MATRIPTASE
Matriptase (MT-SP1, TADG-15, epithin, ST14) was first described in 1993 as a new gelatinolytic activity in cultured breast cancer cells (1). It was molecularly cloned by four different groups at the turn of the millennium (2–5) and was shown to be the sixth member of the then-emerging family of type II transmembrane serine proteases (TTSPs), which today comprises 17 members in humans and 20 in mice (6–8). Two close cousins of matriptase, matriptase-2 and matriptase-3, were identified recently (7,9,10). Orthologs of matriptase are present in all nine vertebrate genomes examined to date (human, chimpanzee, dog, mouse, rat, chicken, zebrafish, spotted green pufferfish, and tiger pufferfish), which suggests a conserved evolutionary function (R. Szabo and T. Bugge, unpublished data). This review summarizes the progress over the past half-dozen years in unraveling the biochemistry, physiology, and pathology of this complex and fascinating cell surface serine protease.
THE COMPLICATED LIFE CYCLE OF MATRIPTASE: INHIBITOR-ASSISTED AUTOACTIVATION, INHIBITION, AND SHEDDING
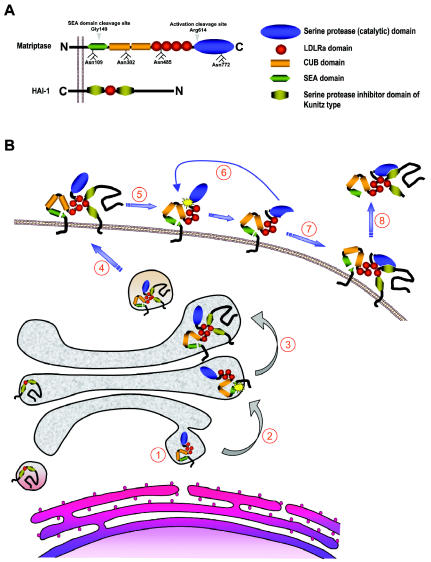
Matriptase is a 80- to 90-kDa cell surface glycoprotein with a complex modular structure that is common to all matriptases (Figure 1A). It lacks a classical signal peptide, and the N-terminal signal anchor, which is not removed during synthesis, functions as a single-span transmembrane domain that orients the protease in the plasma membrane as a type II integral membrane protein with a cytoplasmic N-terminus and an extracellular C-terminus (11). The function of the intracellular domain of matriptase (residues 1–54) is presently unknown, although it is plausible to speculate that it binds cytosolic proteins to regulate enzymatic activity and cellular distribution of the protease. Indeed, a recent report by Kim et al. (12) suggests that the direct interaction between matriptase and the actin-binding protein filamin anchors the protease to the actin cytoskeleton. This would be consistent with the observation that the exposure of immortalized human mammary epithelial cells to sphingosine-1-phosphate (see below) leads to the rapid translocation of matriptase to the cell surface and a subsequent activation in an actin cytoskeleton remodeling–dependent manner (13). The extracellular stem region of matriptase consists of a single SEA (residues 86–201), 2 CUB (residues 214–334 and 340–447), and 4 LDLRA (residues 452–486, 487–523, 524–561, and 566–604) domains. These noncatalytic modules appear to play an essential role in the cellular localization, activation, inhibition, and, likely, the substrate specificity of matriptase. The C-terminal serine protease domain (residues 614–855) is structurally highly similar to that of the other TTSPs and other members of the S1 clan of trypsin-like serine proteases (6).
Figure 1.
Structure of matriptase and HAI-1 and proposed life cycle of matriptase. (A) Domain structures of matriptase and HAI-1. The N-terminal cleavage site within the SEA domain and the activation cleavage site within the serine protease domain of matriptase are indicated by arrowheads. Also shown are the four N-glycosylation sites. (B) (1) Matriptase is synthesized on the rough endoplasmic reticulum and is oriented in the membrane as a single-span type II transmembrane protein by an N-terminal signal anchor. (2) The SEA domain of matriptase undergoes an endoproteolytic cleavage after Gly149 within the endoplasmic reticulum or Golgi apparatus. (3) SEA domain–cleaved matriptase associates with HAI-1, (4) which facilitates transport of the protease to the plasma membrane, (5) where it is activated by autocatalytic cleavage after Arg614 within the highly conserved activation cleavage site R-VVGG. (6) Activated matriptase is rapidly inhibited by HAI-1 (7), and (8) the matriptase–HAI-1 complex is shed from the plasma membrane.
Matriptase is synthesized as an inactive, single-chain zymogen. The activation of the matriptase zymogen is extraordinarily complex, unique among all serine proteases studied to date, and incompletely understood. Matriptase activation requires two sequential endoproteolytic cleavages and involves the transient interaction with its cognate inhibitor, hepatocyte growth factor activator inhibitor (HAI)-1 (Figure 1B). The mature single-chain proenzyme is first cleaved after Gly149 located in a conserved GSVIA motif in the N-terminal SEA domain by an unknown proteolytic activity or possibly by nonenzymatic hydrolysis of the peptide bond (12,14). Although this severs the covalent link to the signal anchor, matriptase remains tightly attached to the cell surface, possibly via noncovalent interactions within the cleaved SEA domain. SEA-domain cleavage appears to take place already in the secretory pathway, as only the N-terminally cleaved form of the enzyme is present on the surface of cells (14). Subsequent to and dependent on SEA domain cleavage, matriptase next is converted into its active conformation by proteolytic cleavage after Arg614 within the highly conserved activation cleavage site R-VVGG in the serine protease domain. The activation site cleavage appears to require the proteolytic activity of matriptase, as mutations in any of the residues of the catalytic triad render matriptase unable to undergo activation site cleavage. This observation led to a proposed transactivation mechanism in which a weak intrinsic proteolytic activity of SEA domain–cleaved matriptase zymogen activates neighboring SEA domain–cleaved matriptase molecules (15).
The proteolytic autoactivation of matriptase appears to be controlled by the stem region, posttranslational modifications, and the cellular localization of the protease. Inactivating mutations in the Ca2+-binding motifs of any or all of the four LDLRA domains prevents the activation of matriptase. Interestingly, however, the complete deletion of all four LDLRA domains allows efficient activation of the enzyme, suggesting an autoinhibitory role of the LDLRA modules that may prevent premature activation of matriptase in the absence of appropriate activation stimuli (15). Glycosylation also appears to be critical to activation. Matriptase contains four functional N-glycosylation sites, Asn109, 302, 485, and 772. Whereas the inactivation of Asn109 and Asn485 had no effect on the activation of matriptase, glycosylation of the first CUB domain (Asn302) and the catalytic domain (Asn772) was required for zymogen activation in cultured breast cancer cells (15).
The specific mechanisms that trigger the activation of matriptase are incompletely understood. In a pioneering study, Benaud et al. (16) showed that matriptase translocates to the cell surface and is activated within minutes after exposure of breast cancer cells to sphingosine-1-phosphate, a serum-derived lipid that signals through specific G-protein–coupled receptors. Interestingly, this activation process was shown to require actin cytoskeleton remodeling (17). Other molecules shown to induce matriptase activation linked to spatial redistribution include suramin and androgens in prostate cancer cells (18). The ability of the matriptase zymogen to become activated and presented on the cell surface may depend on the direct physical interaction with its cognate Kunitz-type serine protease inhibitor HAI-1 (Figure 1B). Thus, in the absence of HAI-1, matriptase accumulates in the Golgi compartment (19). Interestingly, a point mutation in the single LDLRA domain or the Kunitz domain 1 of HAI-1 prevented both cell surface translocation and activation of matriptase (15,19). In contrast to the wild-type enzyme, catalytically inactive matriptase mutants were readily deposited on the cell surface even in the absence of HAI-1 (19).
The inhibition of activated matriptase by HAI-1 was first documented by the identification of matriptase/HAI-1 complexes in human milk and conditioned medium of cultured mammary epithelial cells and in a number of cancer cell lines (20). Recently, the functional relevance of HAI-1 inhibition of matriptase was confirmed in a transgenic mouse model in which matriptase-induced skin tumorigenesis was completely prevented by the overexpression of HAI-1 in the same tissue (21) (see below). Kunitz domain 1, but not Kunitz domain 2, of HAI-1 is responsible for the inhibition of matriptase (15). Interestingly, the interaction also requires a functional LDLRA domain of HAI-1 (22). The N-terminal cleavage of the SEA domain makes matriptase susceptible to the shedding of its extracellular part. Indeed, the original isolation of matriptase from human milk suggests that a significant shedding of the protease takes place from the cell surface in vivo. Data obtained from cell culture systems indicate that most of matriptase released from cells is in a two-chain form, and, in fact, the deletion of the fourth LDLRA domain or the activation cleavage site, which stabilizes the zymogen form, prevents cell surface shedding, without interfering with N-terminal processing (23). The shedding of matriptase also appears to require the presence of HAI-1, as only HAI-1–complexed, but never HAI-1–free, active matriptase is detected in milk or conditioned medium (24,25). The specific molecular events that lead to matriptase/HAI-1 shedding as well as the ultimate fate of the complex are unknown.
PHYSIOLOGICAL FUNCTIONS OF MATRIPTASE
Matriptase is a strictly epithelial protease with a fairly widespread, but not ubiquitous, expression in human and mouse tissues. Expression has been documented in epidermis, cornea, salivary gland, oral and nasal cavities, thyroid, thymus, esophagus, trachea, bronchioles, alveoli, stomach, pancreas, gallbladder, duodenum, small intestine, colon, rectum, kidney, adrenals, urinary bladder, ureter, seminal vesicles, epididymis, prostate, ovaries, uterus, and vagina (26,27, K. List and T. Bugge, unpublished data). Mutations in the matriptase gene have not been identified to date in humans or other animal species. However, a gene-targeting study in mice has revealed an essential role of the membrane protease in oral epithelium, epidermis, hair follicles, and thymic epithelium (27–29). Matriptase-deficient mice develop to term but uniformly die within 48 hours of birth as a consequence of seriously compromised epidermal barrier function, which leads to rapid and fatal dehydration. Detailed analysis of this barrier defect uncovered an essential role of matriptase in the formation of the two physical structures that form the epidermal barrier: the stratum corneum lipid matrix and the cornified envelope of corneocytes.
Interestingly, at the molecular level, matriptase deficiency completely abrogates the proteolytic processing of the polyprotein profilaggrin into filaggrin monomer units and an N-terminal filaggrin S-100 regulatory protein, the latter of which translocates to the nucleus to promote terminal epidermal differentiation. As partial loss-of-function mutations in both the mouse and human profilaggrin genes are associated with epidermal barrier defects, this matriptase-dependent profilaggrin processing pathway may define one key step in the initiation of terminal epidermal differentiation and acquisition of epidermal barrier function (30,31). In this regard, it should be noted that deletion of the SPINK-5 gene, which encodes a Kazal-type serine protease inhibitor that is coexpressed with matriptase in the epidermis, leads to accelerated profilaggrin processing and premature differentiation of human and mouse skin (32,33). Loss of matriptase also impairs the growth of hair follicles and prevents vibrissae eruption owing to absence of formation of vibrissal hair canals. Furthermore, matriptase dramatically increases apoptosis of immature thymocytes in the thymus, leading to thymocyte depletion.
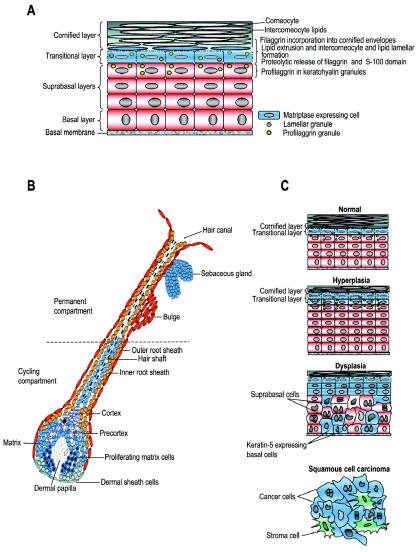
Detailed localization studies, using enzymatic gene trapping combined with immunohistochemical and ultrastructural analysis, have revealed a close association between matriptase expression and function (Figure 2A,B). In the epidermis, keratinized oral epithelium, and thymic epithelium, matriptase is exclusively expressed in postmitotic transitional-layer keratinocytes in the process of undergoing terminal differentiation. In all three tissues, matriptase colocalizes with profilaggrin, frequently displaying close proximity to profilaggrin-containing granules, as revealed by ultrastructural analysis (27). Likewise, in accordance with the important function of matriptase in hair follicle growth, matriptase is specifically expressed in growth phase (anagen) hair follicles and is located in undifferentiated and rapidly proliferating hair matrix cells, precortex, and cortex cells of the cycling portion of the hair follicle (27). The exploration of potential physiological functions of matriptase in nonepidermal tissues is complicated by the perinatal mortality of matriptase-deficient mice and awaits the identification of spontaneous matriptase mutations in humans or other animal species, or the generation of conditional matriptase knockout mice.
Figure 2.
Expression and function of matriptase in normal epidermal structures and pathogenic misexpression during squamous cell carcinogenesis. (A) In the epidermis, matriptase (blue) is strictly confined to the transitional cells undergoing terminal differentiation into dead corneocytes (squames) and the first layer of corneocytes, where it regulates the proteolytic release of filaggrin monomers and filaggrin S-100 domain from profilaggrin to facilitate cornified envelope formation and epidermal lipid extrusion. (B) In early anagen and anagen hair follicles, matriptase (blue) is expressed in proliferating hair matrix cells, precortex cells, cortex cells, and the hair shaft, where it promotes hair growth and hair eruption. Expression of matriptase is also observed in sebocytes of the hair follicle sebaceous gland. (C) In multistage epidermal squamous cell carcinogenesis, matriptase (blue) becomes expressed in rapidly proliferating keratin-5–positive basal keratinocytes during the progression of epidermal lesions from hyperplasia to dysplasia, and the protease is widely expressed in the tumor cells of malignant lesions.
MATRIPTASE IN EPITHELIAL CARCINOGENESIS
Much of the attention paid to matriptase by the biomedical research community is spurred by the consistent expression of the protease in human epithelial tumors. Thus, matriptase is expressed in carcinomas of the head and neck, mesothelium, breast, ovary, cervix, prostate, lung, and gastrointestinal tract, as well as in cell lines derived from these tumors. Matriptase does not appear to be expressed in tumors of mesenchymal origin, suggesting a specific function of the protease in epithelial carcinogenesis (34–38, K. List and T. Bugge, unpublished data). Multiple studies have assessed the level of expression of matriptase during malignant progression and the potential value of matriptase as a prognostic marker in various human cancers. The findings from these studies, which are summarized in part in Table 1, do not paint a consistent picture. In prostate and cervical cancer, matriptase mRNA and protein are upregulated in cancerous lesions compared with normal tissue, and there is a positive correlation between matriptase expression and histopathological grade of the tumor (25,35,39). In contrast, in the gastrointestinal tract, a significant downregulation of matriptase mRNA compared with normal tissue, as well as a decrease of matriptase mRNA levels with increasing tumor grade, has been reported (36). Matriptase is expressed at very low levels in the normal ovary, becomes highly expressed in early-stage ovarian carcinoma, and is then downregulated in advanced-stage tumors (38). In breast cancer, one study concluded that matriptase mRNA was not significantly increased in tumors compared with normal breast tissue (40), whereas another study reported that high matriptase expression is predictive of poor survival as assessed by immunohistochemical detection of matriptase (41).
Table 1.
Matriptase expression in human cancer
| Study | Sample No. | Method | Expression | Correlation with Grade or Stage | Correlation with Survival |
|---|---|---|---|---|---|
| Breast cancer | |||||
| Kang et al., 2003 (41) | 320 | IHC | 229 (72%) samples low expressing 91 (28%) samples high expressing |
N/A | High matriptase expression predictive of poor survival |
| Parr et al., 2004 (40) | 120 | qRT-PCR | Matriptase was not significantly increased in tumors compared with normal breast tissue | No correlation | No correlation |
| Cervical cancer | |||||
| Santin et al., 2003 (39) | 19 | IHC/RT-PCR | 0/8 (0%) in normal squamous epithelia 6/6 (100%) in SCC 2/5 (40%) in adenocarcinomas |
N/A | N/A |
| Lee et al., 2005 (25) | 89 | IHC/LCM/RT-PCR | 0/10 (0%) in normal squamous epithelia 4/19 (21%) in LSIL 12/20 (60%) in HSIL 38/40 (95%) in invasive SSC |
Frequency of matriptase detection increased with histopathological grade | N/A |
| Ovarian cancer | |||||
| Tanimoto et al., 2005 (38) | 96 | IHC/sqRT-PCR | 0/7 (0%) of normal ovarian tissue 50/89 (56%) of ovarian carcinomas |
Matriptase more frequently expressed in stage 1 tumors compared with advanced stage tumors | Positive correlation between matriptase expression and overall survival |
| Oberst et al., 2002 (43) | 54 | IHC | 39/54 (72%) of ovarian carcinomas expressed matriptase | Matriptase more frequently expressed in stage 1 tumors compared with advanced stage tumors | N/A |
| Prostate cancer | |||||
| Riddick et al., 2005 (35) | 67 | qRT-PCR | Significant increase (2–3 fold) of matriptase expression in malignant vs. nonmalignant samples | Significant positive correlation with Gleason score | N/A |
| Gastrointestinal cancer | |||||
| Zeng et al., 2005 (36) | 77 | qRT-PCR | Significant downregulation of matriptase in gastric and colorectal cancer compared with adjacent normal tissue | No significant correlation with stage or lymph node metastasis status | N/A |
IHC, immunohistochemistry; LCM, laser capture microscopy; RT-PCR, reverse-transcriptase polymerase chain reaction; qRT-PCR, quantitative RT-PCR; sqRT-PCR, semiquantitative RT-PCR; LSIL, low-grade squamous intraepithelial lesions; HSIL, high-grade squamous intraepithelial lesions.
Although variations in cancer-associated matriptase expression between different human cancers may reflect, in part, differences in quantitation methodology, tissue sampling, tissue composition, and tumor staging, it is likely that potential diverging roles of matriptase in different epithelia—under both normal physiological conditions and tumor progression—also contribute to the complexity of interpreting the relationship between matriptase expression and human cancer. Furthermore, whereas expression studies give valuable information regarding matriptase localization and total levels of matriptase mRNA or protein, the activity level and regulation of matriptase are not measured. As for most extracellular proteases, the specific detection of matriptase activity in vivo and in situ is an important, but as yet unmet, challenge (42).
Interestingly, studies of tumor and cancer cell extracts, where matriptase can be detected in an active free form or in a high-affinity complex with HAI-1, suggest that a larger proportion of total matriptase exists in its inhibitor-free form in cancer cells, compared with nontumorigenic epithelial cells (2,20,34). Matriptase and HAI-1 are coexpressed with remarkable consistency in normal epithelium, suggesting that the proteolytic activity of matriptase is strictly regulated (34). It therefore has been proposed that an imbalance in the matriptase–HAI-1 ratio, rather than absolute matriptase levels, may be indicative of a cancer-associated dysregulation of matriptase-mediated proteolysis. In support of this proposition, an increased matriptase–HAI-1 mRNA ratio that correlated with tumor grade has been reported in studies of gastrointestinal (36) and ovarian cancer, where advanced-stage tumors that expressed matriptase protein were more likely to do so in the absence of HAI-1 (43).
A recent animal study detailing the pattern of matriptase expression in epidermis during chemically induced multistage carcinogenesis yielded valuable insights into the expression of the membrane protease during carcinogenesis (27). The study revealed that matriptase was expressed in all stages of carcinogenesis at comparable levels, but underwent a dramatic spatial redistribution during the transition of epidermal lesions from hyperplasia to dysplasia (Figure 2C). Thus, whereas matriptase expression in normal and hyperplastic epidermis is narrowly confined to highly differentiated, nonproliferating keratinocytes with no potential for malignant transformation, dysplastic and malignant lesions presented expression of matriptase in a much broader subset of keratinocytes. This subset included proliferating, keratin-5–expressing basal keratinocytes with high self-renewal capacity, which include epidermal stem cells believed to be the primary target cells in epithelial carcinogenesis. That this carcinogen-induced spatial dysregulation of matriptase indeed may be functionally relevant to epithelial carcinogenesis has gained strong support from a transgenic mouse study showing that expression of matriptase even at modest levels in keratin-5–positive keratinocytes of the skin sufficed to both cause spontaneous epithelial malignancies and dramatically potentiate the effect of carcinogen exposure (21).
Several studies have addressed the role of matriptase in later stages of carcinogenesis by inhibiting the protease in established tumor cell lines from prostate, colon, and ovarian cancer using small interfering RNAs, antisense matriptase oligodeoxyribonucleotides, or synthetic active-site matriptase inhibitors. In all cases, matriptase inhibition did not affect cancer cell proliferation in vitro or in xenografted tumors, whereas tumor cell invasion was impaired (44–46). How matriptase promotes malignant progression in these model systems is unknown. Dysregulated matriptase activity may directly affect the cell environment by altered processing of extracellular components and cell-matrix adhesion proteins. Matriptase may also act through the activation or inactivation of downstream effector molecules, including growth factors and receptors, chemokines, and protease zymogens. In this respect, it is noteworthy that most of the candidate substrates for matriptase are implicated in malignant progression, including protease-activated receptor-2 (11), prohepatocyte growth factor activator/scatter factor (11,47), receptor-bound prourokinase plasminogen activator (11,47), and the src-associated transmembrane protein SIMA135/CDCP1 (48).
PERSPECTIVES
Twenty-first-century bioinformatics, proteomics, and mouse genetics have provided rapid insight into the biochemistry, physiology, and pathology of matriptase, and an epithelial transmembrane serine protease with a fascinating biochemistry and biology has been unveiled. The unprecedentedly complex posttranslational regulation of matriptase defines a new paradigm for serine protease zymogen activation and inhibition. Gene inactivation of matriptase provided the first demonstration of a cell surface serine proteolytic activity essential to oral epithelial and epidermal barrier formation, hair growth, and thymic epithelial function. Matriptase was found to be expressed in a curiously high proportion of human epithelial tumors and to promote malignant progression in a multitude of animal models. As quickly as knowledge about matriptase is gained, however, pertinent new questions arise. As a conclusion to this review, we list just a few of these questions.
What transcriptional and posttranscriptional regulatory networks govern the intricate regulation of matriptase expression, activation, and inhibition? Studies of matriptase regulation and dysregulation in cells, tissues, and animals are well-deserving, given the interesting biology and unique expression and regulation of the membrane serine protease.
Is dysregulation of matriptase causally involved in the genesis or progression of cancer in humans? In other words, is the potent tumor-promoting potential of matriptase that is so convincingly demonstrated in xenograft and transgenic animal models unleashed during human carcinogenesis? The prognostic significance of matriptase and HAI-1 expression in several human carcinomas suggests that this could be so. A final verdict may come from future clinical cancer trials using specific matriptase inhibitors.
What are the overall physiological functions of matriptase beyond the epidermis? The evolutionarily conserved and intricate expression pattern of matriptase in nonepidermal tissues certainly would suggest a generalized function of the protease in epithelial biology. Tissue-specific matriptase gene ablation to overcome the neonatal lethality would be excellently suited to provide answers to this question.
Which substrates are cleaved by matriptase to promote epidermal differentiation, hair growth, thymocyte survival, and malignant transformation?
Do matriptase-2 and -3 have the same molecular functions as matriptase in tissues where matriptase is not expressed, or did the three members of the matriptase family evolve to perform independent molecular tasks?
Future studies will provide answers to these and many other questions.
ACKNOWLEDGMENTS
We thank Drs. Robert Angerer, Silvio Gutkind, and Mary Jo Danton for critically reviewing this manuscript. This work was supported by the NIH Intramural program and by a grant from the Department of Defense (DAMD-17-02-1-0693) to T.H.B.
Footnotes
Online address: http://molmed.org
REFERENCES
- 1.Shi YE, et al. Identification and characterization of a novel matrix-degrading protease from hormone-dependent human breast cancer cells. Cancer Res. 1993;53:1409–15. [PubMed] [Google Scholar]
- 2.Lin CY, Anders J, Johnson M, Sang QA, Dickson RB. Molecular cloning of cDNA for matriptase, a matrix-degrading serine protease with trypsin-like activity. J Biol Chem. 1999;274:18231–6. doi: 10.1074/jbc.274.26.18231. [DOI] [PubMed] [Google Scholar]
- 3.Tanimoto H, et al. Ovarian tumor cells express a transmembrane serine protease: a potential candidate for early diagnosis and therapeutic intervention. Tumour Biol. 2001;22:104–14. doi: 10.1159/000050604. [DOI] [PubMed] [Google Scholar]
- 4.Kim MG, et al. Cloning and chromosomal mapping of a gene isolated from thymic stromal cells encoding a new mouse type II membrane serine protease, epithin, containing four LDL receptor modules and two CUB domains. Immunogenetics. 1999;49:420–8. doi: 10.1007/s002510050515. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi T, Shuman MA, Craik CS. Reverse biochemistry: use of macromolecular protease inhibitors to dissect complex biological processes and identify a membrane-type serine protease in epithelial cancer and normal tissue. Proc Natl Acad Sci USA. 1999;96:11054–61. doi: 10.1073/pnas.96.20.11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Netzel-Arnett S, et al. Membrane anchored serine proteases: a rapidly expanding group of cell surface proteolytic enzymes with potential roles in cancer. Cancer Metastasis Rev. 2003;22:237–58. doi: 10.1023/a:1023003616848. [DOI] [PubMed] [Google Scholar]
- 7.Szabo R, Netzel-Arnett S, Hobson JP, Antalis TM, Bugge TH. Matriptase-3 is a novel phylogenetically preserved membrane-anchored serine protease with broad serpin reactivity. Biochem J. 2005;390:231–42. doi: 10.1042/BJ20050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hobson JP, et al. Mouse DESC1 is located within a cluster of seven DESC1-like genes and encodes a type II transmembrane serine protease that forms serpin inhibitory complexes. J Biol Chem. 2004;279:46981–94. doi: 10.1074/jbc.M403299200. [DOI] [PubMed] [Google Scholar]
- 9.Hooper JD, Campagnolo L, Goodarzi G, Truong TN, Stuhlmann H, Quigley JP. Mouse matriptase-2: identification, characterization and comparative mRNA expression analysis with mouse hepsin in adult and embryonic tissues. Biochem J. 2003;373:689–702. doi: 10.1042/BJ20030390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velasco G, Cal S, Quesada V, Sanchez LM, Lopez-Otin C. Matriptase-2, a membrane-bound mosaic serine proteinase predominantly expressed in human liver and showing degrading activity against extracellular matrix proteins. J Biol Chem. 2002;277:37637–46. doi: 10.1074/jbc.M203007200. [DOI] [PubMed] [Google Scholar]
- 11.Takeuchi T, et al. Cellular localization of membrane-type serine protease 1 and identification of protease-activated receptor-2 and single-chain urokinase-type plasminogen activator as substrates. J Biol Chem. 2000;275:26333–42. doi: 10.1074/jbc.M002941200. [DOI] [PubMed] [Google Scholar]
- 12.Kim C, et al. Filamin is essential for shedding of the transmembrane serine protease, epithin. EMBO Rep. 2005;6:1045–51. doi: 10.1038/sj.embor.7400534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee MS, Kiyomiya K, Benaud C, Dickson RB, Lin CY. Simultaneous activation and hepatocyte growth factor activator inhibitor 1-mediated inhibition of matriptase induced at activation foci in human mammary epithelial cells. Am J Physiol Cell Physiol. 2005;288:C932–41. doi: 10.1152/ajpcell.00497.2004. [DOI] [PubMed] [Google Scholar]
- 14.Cho EG, et al. N-terminal processing is essential for release of epithin, a mouse type II membrane serine protease. J Biol Chem. 2001;276:44581–9. doi: 10.1074/jbc.M107059200. [DOI] [PubMed] [Google Scholar]
- 15.Oberst MD, Williams CA, Dickson RB, Johnson MD, Lin CY. The activation of matriptase requires its noncatalytic domains, serine protease domain, and its cognate inhibitor. J Biol Chem. 2003;278:26773–9. doi: 10.1074/jbc.M304282200. [DOI] [PubMed] [Google Scholar]
- 16.Benaud C, et al. Sphingosine 1-phosphate, present in serum-derived lipoproteins, activates matriptase. J Biol Chem. 2002;277:10539–46. doi: 10.1074/jbc.M109064200. [DOI] [PubMed] [Google Scholar]
- 17.Hung RJ, et al. Assembly of adherens junctions is required for sphingosine 1-phosphate-induced matriptase accumulation and activation at mammary epithelial cell-cell contacts. Am J Physiol Cell Physiol. 2004;286:C1159–69. doi: 10.1152/ajpcell.00400.2003. [DOI] [PubMed] [Google Scholar]
- 18.Kiyomiya KI et al. (2006) Matriptase activation and subsequent shedding with HAI-1 is induced by steroid sex hormones in human prostate cancer cells, but not in breast cancer cells. Am. J. Physiol. Cell Physiol. Feb 8 [Epub ahead of print]. [DOI] [PubMed]
- 19.Oberst MD, et al. HAI-1 regulates activation and expression of matriptase, a membrane-bound serine protease. Am J Physiol Cell Physiol. 2005;289:C462–70. doi: 10.1152/ajpcell.00076.2005. [DOI] [PubMed] [Google Scholar]
- 20.Benaud CM, et al. Deregulated activation of matriptase in breast cancer cells. Clin Exp Metastasis. 2002;19:639–49. doi: 10.1023/a:1020985632550. [DOI] [PubMed] [Google Scholar]
- 21.List K, et al. Deregulated matriptase causes ras-independent multistage carcinogenesis and promotes ras-mediated malignant transformation. Genes Dev. 2005;19:1934–50. doi: 10.1101/gad.1300705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirchhofer D, et al. Tissue expression, protease specificity, and Kunitz domain functions of hepatocyte growth factor activator inhibitor-1B (HAI-1B), a new splice variant of HAI-1. J Biol Chem. 2003;278:36341–9. doi: 10.1074/jbc.M304643200. [DOI] [PubMed] [Google Scholar]
- 23.Cho EG, Schwartz RH, Kim MG. Shedding of membrane epithin is blocked without LDLRA4 and its protease activation site. Biochem Biophys Res Commun. 2005;327:328–34. doi: 10.1016/j.bbrc.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Lin CY, Anders J, Johnson M, Dickson RB. Purification and characterization of a complex containing matriptase and a Kunitz-type serine protease inhibitor from human milk. J Biol Chem. 1999;274:18237–42. doi: 10.1074/jbc.274.26.18237. [DOI] [PubMed] [Google Scholar]
- 25.Lee JW, et al. Increased expression of matriptase is associated with histopathologic grades of cervical neoplasia. Hum Pathol. 2005;36:626–33. doi: 10.1016/j.humpath.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Oberst MD, et al. Characterization of matriptase expression in normal human tissues. J Histochem Cytochem. 2003;51:1017–25. doi: 10.1177/002215540305100805. [DOI] [PubMed] [Google Scholar]
- 27.List K, Szabo R, Molinolo A, Nielsen BS, Bugge TH. Delineation of matriptase protein expression by enzymatic gene trapping suggests diverging roles in barrier function, hair formation, and squamous cell carcinogenesis. Am J Pathol. 2006;168:1513–25. doi: 10.2353/ajpath.2006.051071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.List K, et al. Matriptase/MT-SP1 is required for postnatal survival, epidermal barrier function, hair follicle development, and thymic homeostasis. Oncogene. 2002;21:3765–79. doi: 10.1038/sj.onc.1205502. [DOI] [PubMed] [Google Scholar]
- 29.List K, et al. Loss of proteolytically processed filaggrin caused by epidermal deletion of matriptase/MT-SP1. J Cell Biol. 2003;163:901–10. doi: 10.1083/jcb.200304161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith FJ, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006;38:337–42. doi: 10.1038/ng1743. [DOI] [PubMed] [Google Scholar]
- 31.Presland RB, et al. Loss of normal profilaggrin and filaggrin in flaky tail (ft/ft) mice: an animal model for the filaggrin-deficient skin disease ichthyosis vulgaris. J Invest Dermatol. 2000;115:1072–81. doi: 10.1046/j.1523-1747.2000.00178.x. [DOI] [PubMed] [Google Scholar]
- 32.Chavanas S, et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat Genet. 2000;25:141–2. doi: 10.1038/75977. [DOI] [PubMed] [Google Scholar]
- 33.Descargues P, et al. Spink5-deficient mice mimic Netherton syndrome through degradation of desmoglein 1 by epidermal protease hyperactivity. Nat Genet. 2005;37:56–65. doi: 10.1038/ng1493. [DOI] [PubMed] [Google Scholar]
- 34.Oberst M, et al. Matriptase and HAI-1 are expressed by normal and malignant epithelial cells in vitro and in vivo. Am J Pathol. 2001;158:1301–11. doi: 10.1016/S0002-9440(10)64081-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riddick AC, et al. Identification of degradome components associated with prostate cancer progression by expression analysis of human prostatic tissues. Br J Cancer. 2005;92:2171–80. doi: 10.1038/sj.bjc.6602630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zeng L, Cao J, Zhang X. Expression of serine protease SNC19/matriptase and its inhibitor hepatocyte growth factor activator inhibitor type 1 in normal and malignant tissues of gastrointestinal tract. World J Gastroenterol. 2005;11:6202–7. doi: 10.3748/wjg.v11.i39.6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoang CD, et al. Gene expression profiling identifies matriptase overexpression in malignant mesothelioma. Chest. 2004;125:1843–52. doi: 10.1378/chest.125.5.1843. [DOI] [PubMed] [Google Scholar]
- 38.Tanimoto H, et al. Transmembrane serine protease TADG-15 (ST14/matriptase/MT-SP1): expression and prognostic value in ovarian cancer. Br J Cancer. 2005;92:278–83. doi: 10.1038/sj.bjc.6602320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santin AD, et al. The novel serine protease tumor-associated differentially expressed gene-15 (matriptase/MT-SP1) is highly overexpressed in cervical carcinoma. Cancer. 2003;98:1898–904. doi: 10.1002/cncr.11753. [DOI] [PubMed] [Google Scholar]
- 40.Parr C, Watkins G, Mansel RE, Jiang WG. The hepatocyte growth factor regulatory factors in human breast cancer. Clin Cancer Res. 2004;10:202–11. doi: 10.1158/1078-0432.ccr-0553-3. [DOI] [PubMed] [Google Scholar]
- 41.Kang JY, et al. Tissue microarray analysis of hepatocyte growth factor/Met pathway components reveals a role for Met, matriptase, and hepatocyte growth factor activator inhibitor 1 in the progression of node-negative breast cancer. Cancer Res. 2003;63:1101–5. [PubMed] [Google Scholar]
- 42.Sloane BF, et al. Functional imaging of tumor proteolysis. Ann Rev Pharmacol Toxicol. 2006;46:301–15. doi: 10.1146/annurev.pharmtox.45.120403.095853. [DOI] [PubMed] [Google Scholar]
- 43.Oberst MD, et al. Expression of the serine protease matriptase and its inhibitor HAI-1 in epithelial ovarian cancer: correlation with clinical outcome and tumor clinicopathological parameters. Clin Cancer Res. 2002;8:1101–7. [PubMed] [Google Scholar]
- 44.Forbs D, et al. In vitro inhibition of matriptase prevents invasive growth of cell lines of prostate and colon carcinoma. Int J Oncol. 2005;27:1061–70. [PubMed] [Google Scholar]
- 45.Suzuki M, et al. Inhibition of tumor invasion by genomic down-regulation of matriptase through suppression of activation of receptor-bound prourokinase. J Biol Chem. 2004;279:14899–908. doi: 10.1074/jbc.M313130200. [DOI] [PubMed] [Google Scholar]
- 46.Galkin AV, et al. CVS-3983, a selective matriptase inhibitor, suppresses the growth of androgen independent prostate tumor xenografts. Prostate. 2004;61:228. doi: 10.1002/pros.20094. [DOI] [PubMed] [Google Scholar]
- 47.Lee SL, Dickson RB, Lin CY. Activation of hepatocyte growth factor and urokinase/plasminogen activator by matriptase, an epithelial membrane serine protease. J Biol Chem. 2000;275:36720–5. doi: 10.1074/jbc.M007802200. [DOI] [PubMed] [Google Scholar]
- 48.Bhatt AS, Erdjument-Bromage H, Tempst P, Craik CS, Moasser MM. Adhesion signaling by a novel mitotic substrate of src kinases. Oncogene. 2005;24:5333–43. doi: 10.1038/sj.onc.1208582. [DOI] [PMC free article] [PubMed] [Google Scholar]