Abstract
The centromere of eukaryotic chromosomes is essential for the faithful segregation and inheritance of genetic information. In the majority of eukaryotic species, centromeres are associated with highly repetitive DNA, and as a consequence, the boundary for a functional centromere is difficult to define. In this study, we demonstrate that the centers of rice centromeres are occupied by a 155-bp satellite repeat, CentO, and a centromere-specific retrotransposon, CRR. The CentO satellite is located within the chromosomal regions to which the spindle fibers attach. CentO is quantitatively variable among the 12 rice centromeres, ranging from 65 kb to 2 Mb, and is interrupted irregularly by CRR elements. The break points of 14 rice centromere misdivision events were mapped to the middle of the CentO arrays, suggesting that the CentO satellite is located within the functional domain of rice centromeres. Our results demonstrate that the CentO satellite may be a key DNA element for rice centromere function.
INTRODUCTION
The centromere is the most characteristic landmark of chromosomes in higher eukaryotic species and appears cytologically as a distinct primary constriction on condensed metaphase chromosomes. The centromere is responsible for sister chromatid cohesion and is the site for kinetochore assembly and spindle fiber attachment, allowing for the faithful pairing and segregation of sister chromatids during cell division. Determining the precise DNA boundary of a centromere has proven to be a difficult task. In the majority of eukaryotic species, centromeres are embedded within megabases of highly repetitive DNA that cannot be sequenced precisely by even the most tenacious genome-sequencing projects (Arabidopsis Genome Initiative, 2000; International Human Genome Sequencing Consortium, 2001).
The budding yeast Saccharomyces cerevisiae is the most striking exception to the rule of DNA sequence complexity in eukaryotic centromeres. Each of the “point” centromeres in S. cerevisiae consists of only ∼125 bp of unique sequence (Clarke, 1990, 1998). By contrast, the centromeres of most other model eukaryotic species, including fission yeast (Schizosaccharomyces pombe), Drosophila melanogaster, and human, encompass long tracts of highly repetitive DNA sequences (for review, see Henikoff et al., 2001). Satellite repeats often are the major DNA component of centromeres in higher eukaryotic species (Csink and Henikoff, 1998). The best-characterized centromeric satellite DNA is the α satellite located in human centromeres.
The α satellite DNA is composed of arrays of an ∼171-bp monomer repeat. The amount of the α satellite DNA in human centromeres varies from ∼250 kb to >4 Mb (Wevrick and Willard, 1989; Oakey and Tyler-Smith, 1990). Human artificial chromosomes have been assembled successfully using either synthetic or cloned α satellite DNA as the centromere component (Harrington et al., 1997; Ikeno et al., 1998; Henning et al., 1999), suggesting that a long stretch of α satellite DNA can act as a functional human centromere.
The centromeres of the model plant Arabidopsis have been defined genetically (Copenhaver et al., 1999) and contain various types of repetitive DNA elements, including retroelements, transposons, and telomere-like repeats (Richards et al., 1991; Thompson et al., 1996; Brandes et al., 1997; Haupt et al., 2001). The most abundant DNA element within functional Arabidopsis centromeres is the pAL1 repeat, a 180-bp satellite repeat family that constitutes 3% of the Arabidopsis genome (Martinez-Zapater et al., 1986; Maluszynska and Heslop-Harrison, 1991; Round et al., 1997). Kumekawa et al. (2001) recently demonstrated that the amount of the pAL1 repeat in Arabidopsis centromeres was underestimated previously and that each Arabidopsis centromere may contain several megabases of this satellite repeat.
The centromeres of supernumerary B chromosomes of maize have been well studied. A repetitive element specific to the B centromere was isolated (Alfenito and Birchler, 1993). A functional role was proposed for this repeat, in part because of its strong homology with the knob repeat of maize. The maize knobs are cytologically distinct heterochromatin structures on meiotic pachytene chromosomes and can function as neocentromeres in certain genetic backgrounds. Thus, the B centromere repeat may contribute to centromere function through a mechanism similar to that of the knob repeat. Kaszas and Birchler (1996) demonstrated that the B centromere repeat is present in significantly rearranged B centromeres. Transmission studies and molecular characterization of rearranged B centromeres eventually may define the critical component of a functional B centromere (Kaszas and Birchler, 1998).
Several centromeric repetitive DNA elements have been reported in rice (Dong et al., 1998; Nonomura and Kurata, 1999). Sequence analysis revealed that most of these centromeric DNA elements are derived from a Ty3/gypsy-class retrotransposon family that is specific to the centromeric regions of grass chromosomes (Miller et al., 1998a; Presting et al., 1998; Langdon et al., 2000). However, the 155-bp satellite repeat CentO, previously named RCS2 (Dong et al., 1998), is unique to rice and is located exclusively in rice centromeres.
In this study, we conducted high-resolution cytological mapping of the CentO satellite repeat in the rice genome. The distribution and organization of the CentO satellite, together with the centromere-specific retrotransposon CRR, in the 12 rice centromeres are illustrated using a combination of cytological and molecular methods. Quantification of the CentO repeat in normal and telocentric rice chromosomes revealed that the break points of centromere misdivisions always are located within the CentO loci, suggesting that the CentO satellite is a key component of functional rice centromeres.
RESULTS
The CentO Satellite Is Located in Cytologically Defined Rice Centromeres
The CentO satellite was identified originally in the clone pRCS2, which contains four tandemly arranged monomers with units between 154 and 165 bp and maps to the centromeric regions of somatic metaphase chromosomes of rice (Dong et al., 1998). Because of the low mapping resolution using somatic metaphase chromosomes, it is not known if the CentO satellite is associated with the primary constrictions. However, meiotic pachytene chromosomes of rice are >10 times the length of somatic metaphase chromosomes and provide superior resolution for fluorescence in situ hybridization (FISH) mapping (Cheng et al., 2001b).
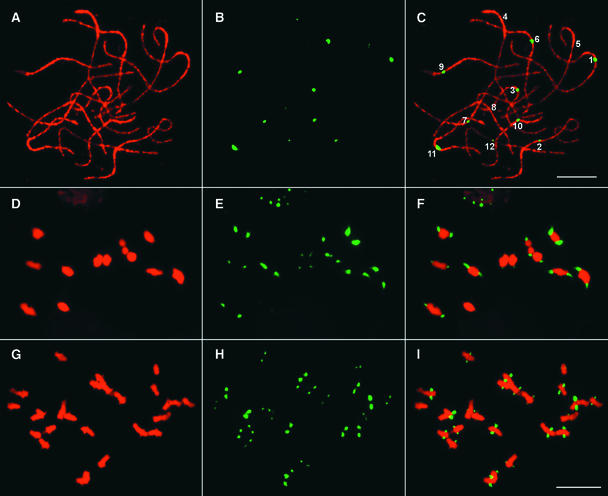
Pachytene FISH analysis using pRCS2 revealed a single CentO locus on each of the 12 rice chromosomes (Figures 1A to 1C). The pRCS2 FISH signals clearly were associated with the primary constriction in pachytene cells, in which the morphology of the chromosomes was well preserved after the FISH procedure. Noncentromeric signals were not observed on pachytene chromosomes, even under conditions of low hybridization stringency (30% formamide at 37°C), indicating that the CentO satellite is highly specific to the centromeres. On meiotic metaphase I chromosomes, the FISH signals are located consistently on the tips (the most poleward positions) of the bivalent chromosomes (Figures 1D to 1F), suggesting that the chromosomal regions containing the CentO satellite are associated with the kinetochore protein complex.
Figure 1.
Centromeric Localization of the CentO Satellite in Rice.
(A) to (C) A single CentO locus is detected on each of the 12 meiotic pachytene chromosomes prepared from var Nipponbare rice.
(A) A complete pollen mother cell at the pachytene stage.
(B) FISH signals derived from the CentO probe pRCS2. Note that the sizes and intensities of the FISH signals are significantly different among the 12 centromeres.
(C) A merged image of the pachytene chromosomes and the FISH signals. The individual pachytene chromosomes are identified based on morphology. Bar = 10 μm.
(D) to (F) Locations of the CentO satellite on meiotic metaphase I chromosomes prepared from var Wuyujing 8 rice.
(D) A complete pollen mother cell at metaphase I.
(E) FISH signals derived from the CentO probe pRCS2.
(F) A merged image of the bivalent chromosomes and the FISH signals. Note that the signals are located on the stretched terminal regions on every bivalent chromosome.
(G) to (I) Locations of the CentO satellite on mitotic metaphase chromosomes prepared from var Nipponbare rice.
(G) A complete somatic cell at metaphase.
(H) FISH signals derived from the CentO probe pRCS2.
(I) A merged image of the chromosomes and the FISH signals. Note that the signals are located on the stretched chromosomal regions. Bar = 10 μm.
We also observed a unique FISH hybridization pattern in some mitotic metaphase cells. The centromeric regions of the chromosomes in these cells were stretched out (Figure 1G), possibly as a result of the attachment of spindle fibers and the mechanical stretching of the spindle fibers imposed by the crushing in the cytological preparation. Interestingly, the FISH signals derived from the CentO satellite always colocalized with the stretched centromeric regions (Figures 1H and 1I). This unique FISH signal pattern indicates that the CentO satellite is located within the chromosomal regions that are the sites of kinetochore formation and spindle fiber attachment.
Complex Composition of the CentO Satellite
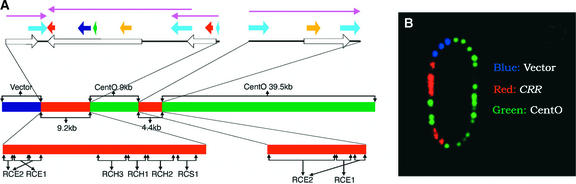
We previously identified a rice BAC clone (17p22) that is derived from a centromeric region of Oryza sativa subsp indica var IR-BB21. Several repetitive DNA elements, including the CentO satellite sequence, were isolated from this BAC (Dong et al., 1998). To obtain more sequence information from 17p22, we conducted 10-fold redundant shotgun sequencing of an M13 library prepared from 17p22. A rigorous assembly process (see Methods) coupled with fiber-FISH analysis resulted in a single contig of 62 kb (Figures 2A and 2B).
Figure 2.
Structure of Rice Centromeric BAC 17p22.
(A) The middle diagram shows that the insert of BAC 17p22 contains two blocks of the CentO satellite that are 9 and 39.5 kb, respectively, and that are separated by two CRR-related DNA sequences. The top diagram shows the structure of the two CRR-related fragments. The 9.2-kb fragment contains three truncated or rearranged CRR elements. The 4.4-kb fragment contains a single CRR element with two LTRs. Each CRR element is represented by a purple arrow. Open arrows indicate open reading frames; light blue arrows indicate LTRs; the dark blue arrow indicates the reverse transcriptase domain; red arrows indicate the integrase domain; the green arrow indicates the protease domain; yellow arrows indicate the gag domain. The bottom diagram shows that the two CRR-related DNA fragments are covered by six plasmid clones: pRCE1, pRCE2, pRCH1, pRCH2, pRCH3, and pRCS1. These six plasmid clones were used as the CRR probe for FISH analysis.
(B) A fiber-FISH image of a single 17p22 molecule visualized by three probes: BAC vector (blue), CRR probe (red), and CentO probe pRCS2 (green). The sequence assembly of BAC 17p22 in (A) is confirmed by the fiber-FISH result.
The 62-kb insert of 17p22 consists of two uninterrupted CentO arrays of 9 and 39.5 kb that are separated by a single retrotransposon insertion and flanked on one side by a complex arrangement of additional retrotransposon sequences (Figure 2A). All CentO monomers are arranged in the same orientation, but their organization and polymorphism are surprisingly complex. There are two distinct subfamilies, with consensus lengths of 155 bp (252 copies) and 164 bp (61 copies), which differ principally as a result of a single 10-bp insertion in the larger family (Figure 3). All 164-bp monomers are flanked on both sides by the 155-bp monomers.
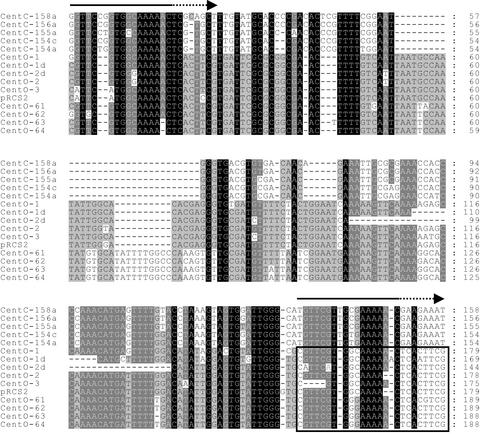
Figure 3.
Sequence Similarity between Rice CentO and Maize CentC Centromeric Satellite Repeats.
Five CentC monomers (top) are compared with CentO sequences from the 17p22 or pRCS2 clones. CentC contains an internal deletion and a degenerate duplication (CentC numbering is as in the database entry; the degenerate duplications are marked by arrows above). CentC sequences shown are from single, complete monomers, whereas CentO sequences represent single monomers followed by part (∼24 bp) of the adjacent downstream monomer (boxed). The CentO variants displayed include two partial deletions and four members of the 164-bp subfamily. The region removed in the two partial deletions has the maximum predicted potential for bending within the CentO consensus. Sequence conservation between the displayed repeats is indicated by background shading (black represents 100% conservation, dark gray represents at least 80%, and light gray represents at least 60%).
Within each subfamily, there are many variants, consisting mainly of single base changes and/or single base insertions/deletions relative to the consensus. There are a limited number of common polymorphisms, although all monomers are >90% identical to the relevant consensus. Few bases are conserved absolutely, and there are no simple universal consensus motifs present. Two variants have internal deletions (11 and 35 bp), and the deletions are centered on the same consensus position, yet otherwise these variants appear unrelated by polymorphism at other positions (Figure 3). The majority of variants occur only once, but some (including the 35-bp deletion) are repeated moderately (6 to 14 copies). Repeated variants generally are dispersed widely throughout the clone, and there are no simple tandem arrays of identical units.
Sequence Similarity between the CentO Satellite and the Maize Centromeric Repeat CentC
An initial search in GenBank using several monomers of the CentO satellite revealed partial sequence similarity with a 156-bp satellite repeat, CentC, located in maize centromeres (Ananiev et al., 1998). Alignment of the two satellite families confirms that they are most likely descended from a common ancestor (Figure 3). Relative to the published CentC orientation (Ananiev et al., 1998), CentC is highly divergent from CentO in the central region, where a deletion of 22 or 32 bp is present compared with the 155- and 164-bp subfamilies, respectively. However, CentO and CentC retain strong similarities at the 5′ and 3′ ends.
The central CentC deletion appears to have been balanced by a degenerate duplication of the terminal 25 bp (shown in Figure 3 as a 3′ extension relative to CentO), such that the length of both satellite families remains similar, suggesting that this may have functional importance. Interestingly, the conserved 3′ region is predicted to have significant potential to direct bending, based on the criteria of the bend.it server (http://www2.icgeb.trieste.it/~dna/bend_it.html; see Methods), whereas both deletions found in the CentO variants have occurred at this point, suggesting that it may be susceptible to breakage. Therefore, it is likely that structural constraints play a role in the pattern of sequence conservation observed between CentC and CentO.
Quantification of the CentO Satellite in Individual Rice Centromeres
The sizes and intensities of the FISH signals derived from the CentO satellite differ significantly on individual rice chromosomes (Figures 1B, 1E, and 1H), suggesting that the copy number of the CentO repeat is highly variable among the 12 rice centromeres. We quantified the amount of the CentO satellite in each of the 12 rice centromeres in O. sativa subsp japonica var Nipponbare and O. sativa subsp indica var Zhongxian 3037 (Table 1). The fluorescence intensities of the FISH signals derived from the CentO satellite were measured on rice pachytene chromosomes using IPLab software.
Table 1.
Distribution of the CentO Satellite among Different Centromeres in Two Rice Varieties
| Nipponbare (japonica rice)
|
Zhongxian 3037 (indica rice)
|
|||
|---|---|---|---|---|
| Centromere | Relative Intensity of the FISH Signala |
CentO Contentb (kb) |
Relative Intensity of the FISH Signala |
CentO Contentb (kb) |
| 1 | 22.04 ± 2.46 | 1,418 ± 158.3 | 23.45 ± 1.35 | 1,898 ± 109 |
| 2 | 11.14 ± 1.06 | 716.6 ± 68.19 | 11.17 ± 0.72 | 904.1 ± 58.28 |
| 3 | 2.83 ± 0.78 | 182.1 ± 50.18 | 2.27 ± 0.20 | 183.7 ± 16.19 |
| 4 | 1.68 ± 0.70 | 108.1 ± 45.03 | 1.22 ± 0.16 | 98.75 ± 12.95 |
| 5 | 2.40 ± 0.56 | 154.4 ± 36.02 | 7.40 ± 0.63 | 599.0 ± 50.99 |
| 6 | 12.69 ± 2.46 | 816.3 ± 158.3 | 1.93 ± 0.21 | 156.2 ± 17.00 |
| 7 | 4.96 ± 0.56 | 319.1 ± 36.02 | 1.44 ± 0.11 | 116.6 ± 8.903 |
| 8 | 1 | 64.33 ± 25.73c | 1 | 80.94 ± 11.33c |
| 9 | 9.57 ± 1.14 | 615.6 ± 73.33 | 2.57 ± 0.30 | 208.0 ± 24.28 |
| 10 | 7.32 ± 1.64 | 470.9 ± 105.5 | 1.59 ± 0.25 | 128.7 ± 20.24 |
| 11 | 29.48 ± 3.21 | 1,896 ± 206.5 | 5.19 ± 0.62 | 420.1 ± 50.18 |
| 12 | 2.43 ± 0.59 | 156.3 ± 37.95 | 2.90 ± 0.36 | 234.7 ± 29.14 |
Distribution values are based on 10 measurements for each data point.
The intensity of the FISH signal in centromere 8 was calibrated as 1, and the intensities of the signals in the other centromeres were converted into relative values.
The error for the CentO content in each centromere was calculated by multiplying the error of the relative intensity of the FISH signals by 64.33 or 80.94 kb (the average length of CentO-8). The error for the CentO-8 was calculated based on fiber-FISH data.
The physical sizes of the CentO array in centromere 8 were measured on 10 and 6 genomic DNA fibers of Nipponbare and Zhongxian 3037, respectively (see Figure 4A). The length of the fiber-FISH signals was converted into kilobases by 3.18 kb/μm (unpublished fiber-FISH calibration parameter in rice by Z. Cheng and J. Jiang). The sizes of the CentO loci in other centromeres were calculated based on the values of relative fluorescence intensity.
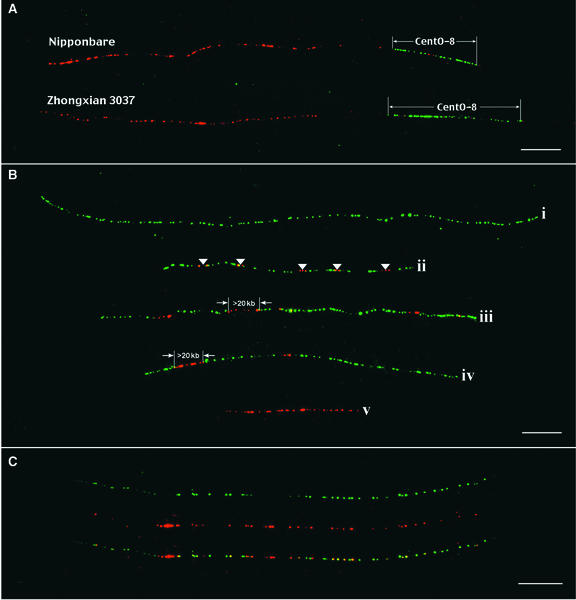
The 10 best cells in which all 12 pachytene bivalents were identified unambiguously were selected for measurements. The intensity of the FISH signal derived from centromere 8 was calibrated as 1, and the relative intensities of the signals derived from other centromeres were calculated and are summarized in Table 1. A BAC clone, RC8-1, was found to be closely linked to the CentO locus of chromosome 8. Using RC8-1 as a reference marker, we were able to determine the length of the CentO locus of chromosome 8 (CentO-8) using fiber-FISH (Figure 4A). The sizes of the CentO-8 in Nipponbare and Zhongxian 3037 were measured as 64 and 81 kb, respectively, by fiber-FISH analysis (Figure 4A, Table 1). The amount of the CentO satellite in other centromeres was calculated based on relative fluorescence intensities compared with centromere 8 (Table 1).
Figure 4.
Fiber-FISH Analysis of Rice Centromeric DNA.
(A) BAC clone RC8-1 (red) is located ∼50 kb away from the CentO locus in rice chromosome 8 (CentO-8). Fiber-FISH signals derived from CentO-8 (green) can be identified unambiguously and measured using RC8-1 as a reference marker. The top signal is from var Nipponbare rice; the bottom signal is from var Zhongxian 3037 rice. Note that the red signals within the CentO-8 loci were derived from the CRR sequences contained in BAC RC8-1. Bar = 10 μm.
(B) Representative fiber-FISH signals demonstrate highly variable densities and nesting of the CRR elements (red signals) within CentO arrays (green signals). An ∼400-kb CentO signal (fiber i) contains no CRR insertion. Fiber ii (∼200 kb) shows five insertions (arrowheads). Long CRR signals (>20 kb) inserted into CentO satellite DNA are detected within fibers iii and iv. A CRR signal independent of CentO satellite DNA (fiber v) contains ∼100-kb CRR-related sequences. The long CRR signals most likely are derived from nested CRR elements. Bar = 10 μm.
(C) A fiber-FISH signal consists of interspersed signals from CRR and CentO. The top fiber signal (green) is from CentO; the middle fiber signal (red) is from CRR; and the bottom fiber signal is a merged image. Bar = 10 μm.
We also identified DNA markers that are closely linked to the CentO locus on chromosome 11 (CentO-11). Using a strategy similar to that demonstrated in Figure 4A, we measured the sizes of the CentO-11 in Nipponbare and Zhongxian 3037 to be 1.64 and 0.64 Mb, respectively, based on five fiber-FISH signals from each rice variety (data not shown). These measurements are similar to the estimated sizes of CentO-11 (1.90 and 0.42 Mb) based on relative fluorescence intensities (Table 1). These results demonstrated an appreciable accuracy of the quantification method based on measurements of FISH signal intensities.
The total amounts of the CentO satellite are ∼7 and 5 Mb in Nipponbare and Zhongxian 3037, respectively, and account for 1.6 and 1.2% of the 430-Mb rice genome (Arumuganathan and Earle, 1991). The centromeres of chromosomes 1, 2, 3, 4, 8, and 12 in the two rice varieties, which represent two different subspecies, show similar amounts of the CentO satellite. But the centromeres of the remaining chromosomes in the two varieties differ significantly in their CentO contents. For example, centromere 6 in Nipponbare and Zhongxian 3037 contains ∼820 and 160 kb of the CentO satellite, representing a fivefold difference.
The Break Points of Rice Centromere Misdivisions Are Located in the Middle of the CentO Arrays
Telocentric chromosomes and isochromosomes are derived from misdivisions of the centromeres. Mapping the break points of centromere misdivisions is an important approach in defining the functional centromeres (Kaszas and Birchler, 1996, 1998; Yu and Dawe, 2000; Zhang et al., 2001). We have developed numerous rice telotrisomic stocks, which contain 24 normal chromosomes and one telocentric chromosome, in Zhongxian 3037 (Cheng et al., 2001c). To discover whether misdivision break points of the telocentric rice chromosomes are located within the CentO arrays, we conducted quantitative FISH mapping using 14 different telotrisomic stocks involving all 12 rice chromosomes (Table 2).
Table 2.
The Content of the CentO Satellite in Telocentric Rice Chromosomes
| Telotrisomic Stock |
CentO Content in the Telocentric Chromosome (%) |
|---|---|
| 2n+•1S | 60.97 ± 5.36 |
| 2n+•1L | 65.01 ± 7.48 |
| 2n+•2S | 57.80 ± 3.29 |
| 2n+•3L | 47.81 ± 4.41 |
| 2n+•4S | 56.39 ± 3.10 |
| 2n+•5L | 70.91 ± 4.30 |
| 2n+•6S | 84.41 ± 4.83 |
| 2n+•7S | 57.09 ± 3.82 |
| 2n+•8L | 77.47 ± 4.82 |
| 2n+•9S | 52.17 ± 7.44 |
| 2n+•10S | 67.57 ± 3.31 |
| 2n+•11L | 66.87 ± 4.21 |
| 2n+•12S | 69.65 ± 4.50 |
| 2n+•12L | 76.25 ± 4.67 |
CentO content data are based on five measurements for each data point and are presented as the percentage of the corresponding normal chromosome.
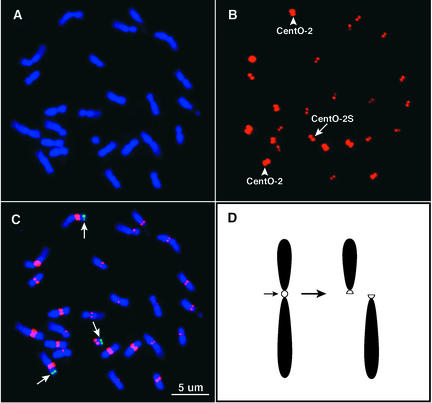
Somatic metaphase chromosomes prepared from the telotrisomic stocks were hybridized to the CentO satellite probe pRCS2. Chromosome arm–specific BAC clones (Cheng et al., 2001a) also were included in FISH analysis to unambiguously detect the telocentric chromosome and its corresponding normal chromosome (Figures 5A to 5C). The relative fluorescence intensities of the telocentric chromosomes and the two normal chromosomes were measured using IPLab software (Table 2). Quantitative FISH results demonstrated that the telocentric chromosomes always contain a lower amount of the CentO satellite than their respective normal centromeres in all 14 telotrisomic stocks. The centromeres of telocentric chromosomes contain an average of 65% (ranging from 48 to 84%) of the CentO satellite compared with the normal centromeres. These results demonstrate that the break points of all of the centromere misdivisions are located within the CentO arrays.
Figure 5.
Quantification of the CentO Satellite in the Centromere of Telocentric Chromosome 2S.
(A) A complete prometaphase cell of telotrisomic 2S.
(B) FISH signals derived from the CentO satellite. The arrow indicates the signal on telocentric chromosome 2S. Arrowheads point to the signals on normal chromosome 2.
(C) Merged signals of chromosomes and FISH signals derived from the CentO satellite (red) and BAC clone a0095P12 (green) specific to the short arm of rice chromosome 2 (Cheng et al., 2001a). Note that the FISH signal derived from CentO in the telocentric chromosome is significantly weaker and smaller than those in normal chromosome 2.
(D) Diagram of centromere misdivision. A division in the middle of the centromere results in two functional centromeres associated with the two telocentric chromosomes.
Distribution and Organization of the Centromere-Specific Retrotransposon in the Rice Genome
We previously demonstrated that the centromeres of grass species, including rice, contain a Ty3/gypsy-class retrotransposon family (Miller et al., 1998a; Langdon et al., 2000). This centromere-specific family in rice is referred to as CRR (Centromeric Retrotransposon of Rice). The insert of BAC 17p22 contains two regions, 4.4 and 9.2 kb, respectively, that are derived from the CRR family (Figure 2A).
The 4.4-kb region represents an insertion of a nonautonomous element into the CentO satellite array. This insertion has occurred relatively recently, as judged by the lack of divergence of its long terminal repeat (LTR) sequences. As with other members of the nonautonomous subfamily of CRR elements, an open reading frame that has homology with the gag gene of full-length elements is fused to a reading frame of unknown function (Langdon et al., 2000). No canonical enzymatic motifs are present, but elements that share a similar structure to this 4.4-kb region are found in numerous rice DNA sequence entries in GenBank, consistent with their dispersal by recent retrotransposition. The 5-bp duplication “signature” observed at the junctions of retrotransposed elements also is found at the terminus of the 4.4-kb CRR element.
The 9.2-kb region is derived from three CRR elements, each of which is truncated differently (Figure 2A). The most 5′ element is a nonautonomous element whose upstream sequences have been lost in cloning that has inserted into the integrase region of a full-length element. The 5′ LTR of the full-length element is recombined exactly with the 3′ LTR of a second full-length element, generating two tandem overlapping elements. The second full-length element has lost almost all upstream sequences, apparently as a consequence of a deletion event that extended into the CentO array. This deletion may have occurred during propagation of the BAC clone, because we found recently that many BAC clones containing tandem repeats are not stable in Escherichia coli (Song et al., 2001).
To investigate the distribution of CRR elements in the rice genome, we screened a Nipponbare BAC library (http://www.genome.clemson.edu/orders/Product.html) using the CentO satellite pRCS2 probe and the pRCS1 probe derived from the integrase domain of CRR (Miller et al., 1998a). Probes pRCS2 and pRCS1 hybridized to 569 (1.50%) and 712 (1.87%) of the 38,016 clones, respectively. A total of 157 BACs hybridized to both probes. We expect that the CRR elements in these 157 BACs are either located adjacent to or inserted in the middle of the CentO arrays.
All of the CentO- and CRR-positive clones were used to search the BAC fingerprint database developed by Clemson University (http://www.genome.clemson.edu/projects/rice/fpc/). Among the 569 clones associated with the CentO repeat, 267 clones are singletons and the other 302 clones are distributed in 26 different contigs. Among the 712 clones associated with CRR, 604 clones are distributed on 70 contigs and 98 clones are singletons. These results suggest that the CentO satellite is organized into long arrays with few disruptions by other sequences, whereas the CRR elements are dispersed in a wider range of chromosomal regions than the CentO satellite. In addition, many of the BACs containing the CentO satellite may not produce distinct restriction patterns; thus, they appear as singletons.
To gain a cytological view of the distribution of the CRR elements, we conducted FISH analysis by pooling the six plasmid clones derived from different parts of the CRR element as a single probe (Figure 2A; referred to as CRR probe hereafter). FISH signals derived from the CRR probe were highly enriched in the centromeric regions on both somatic metaphase and meiotic pachytene chromosomes (Figure 6A). However, cross-hybridization of the CRR probe to other chromosomal regions was observed. The noncentromeric signals were not consistent among different cells and generally were much weaker than the signals detected in the centromeres. We could not determine whether such signals were derived from noncentromeric members of the CRR family or from cross-hybridization with other retrotransposon sequences. Based on the size and intensity of the FISH signals, the copy number of the CRR element varied significantly among the 12 rice centromeres. The CRR elements were distributed in a much wider range of the centromeric regions than the CentO satellite (Figures 6B to 6E), which is consistent the BAC library screening results.
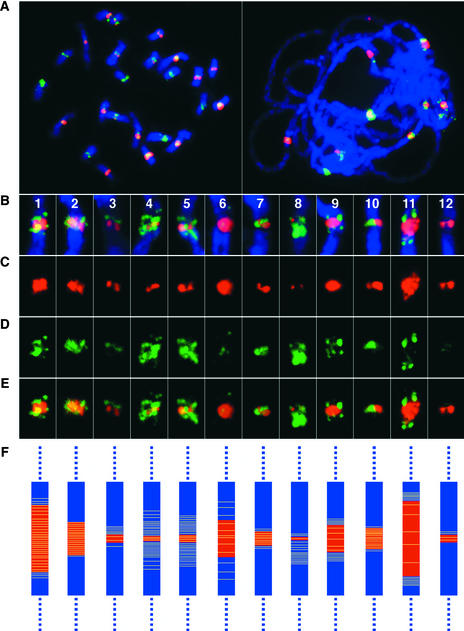
Figure 6.
Distribution and Organization of the CentO Satellite and the Centromere-Specific Retrotransposon CRR in Rice Chromosomes.
(A) Localization of the CentO satellite (red) and the CRR elements (green) on mitotic prometaphase chromosomes (left) and meiotic pachytene chromosomes (right).
(B) to (E) Images of individual rice centromeres of pachytene chromosomes hybridized with the CentO satellite (red) and the CRR probe (green) (B). The composite images are separated digitally into images of FISH signals derived from the CentO satellite (C), images of FISH signals derived from the CRR probe (D), and merged images of FISH signals from both probes (E).
(F) Diagrams depicting the distribution of the CentO satellite (red) and CRR elements (yellow) in individual rice centromeres. The sizes of the red bars represent the relative amount of the CentO satellite (see Table 1). The density of the yellow lines represents the relative density of the CRR elements in different centromeres.
The FISH signals derived from the CRR probe were not distributed uniformly in the majority of the centromeres. In several centromeres, the signals from the CRR probe were clearly enriched on both sides of the CentO locus (Figures 6C to 6E). In some chromosomes, the CRR signal on one side of the centromere was significantly stronger than the signal on the other side of the centromere. The FISH results also indicated that the majority of the CRR elements flank the CentO arrays rather than insert into the CentO arrays.
The CRR and CentO probes were used for FISH analysis on extended DNA fibers prepared from Nipponbare. Insertions of the CRR elements into the CentO arrays were visualized (Figure 4B). The density of the CRR elements inserted in the CentO arrays varied significantly among different fiber- FISH signals (Figure 4B). We observed CentO arrays as long as 500 kb without disruptions by CRR elements (Figure 4B). Another interesting observation is the nesting of the CRR elements in some centromeric regions. Most of the fiber-FISH signals derived from the CRR probe consisted of two to four fluorescent spots, representing ∼1 to 3 μm or 3 to 9 kb. However, contiguous CRR signals as long as 50 μm were observed. Such signals most likely were derived from nesting of multiple CRR elements (Figure 4B). The 9.2-kb CRR-related region in BAC 17p22 (Figure 2A) may represent one such nesting case. Although in the majority of the fiber-FISH images, the CRR signals appear to occupy separate domains from the CentO signals, images with significant interspersing of the CRR and CentO signals were observed (Figure 4C).
DISCUSSION
DNA sequences associated with centromeric regions have been reported in numerous plant species (Alfenito and Birchler, 1993; Harrison and Heslop-Harrison, 1995; Aragon-Alcaide et al., 1996; Jiang et al., 1996; Brandes et al., 1997; Miller et al., 1998b; Nagaki et al., 1998; Francki, 2001; Gindullis et al., 2001; Hudakova et al., 2001; Page et al., 2001; Saunders and Houben, 2001). However, large-scale sequencing and organization studies of centromeric DNA have been documented in only a few plant species. The most abundant DNA element in Arabidopsis centromeres is the 180-bp satellite repeat pAL1. The pAL1 arrays may be interrupted by the 106B repeat, a diverged copy of the LTR of the Athila retrotransposon (Fransz et al., 2000), but complete Athila elements, the most dominant retrotransposon family in Arabidopsis, are highly enriched in the pericentromeric region rather than in functional centromeres (Fransz et al., 2000). Satellite repeats and retrotransposons also are the major component of centromeric DNA in barley (Hudakova et al., 2001) and Beta species (Gindullis et al., 2001).
In this study, we demonstrate that the centers of rice centromeres are occupied by the CentO satellite repeat sequence. Cytological mapping revealed that the CentO satellite is located within the chromosomal regions at which the kinetochore is formed and the spindle fibers are attached (Figures 1D to 1F and 1G to 1I). A Ty3/gypsy-class retrotransposon family, CRR, is colocalized with the CentO satellite within rice centromeres. CRR is highly specific to the centromeres, and its homologous sequences have been found in the centromeric regions of all grass chromosomes (Ananiev et al., 1998; Miller et al., 1998a; Presting et al., 1998; Langdon et al., 2000).
Despite its centromeric specificity, the copy number and density of CRR elements within and outside of the CentO arrays are highly variable among the 12 rice centromeres. In addition, a majority of the CRR elements flank the CentO arrays rather than insert into the CentO satellite sequences. Frequent and drastic nesting of the CRR element was detected (Figure 4B). It will be interesting to determine if this retrotransposon has any direct or indirect role in rice centromere function. Other retroelements that are not specific to the centromeres have been identified in rice centromeric regions (Nonomura and Kurata, 2001).
The mechanism of centromere function is an intriguing puzzle for biologists. The functional role of the centromere in cell division, including both meiosis and mitosis, is highly conserved in all eukaryotic species. Several proteins involved in centromeric function have been found to be conserved in highly divergent eukaryotic species, including S. cerevisiae, Caenorhabditis elegans, D. melanogaster, human, and plants (for reviews, see Dobie et al., 1999; Yu et al., 2000). However, the centromeric DNA sequences are extremely variable even among closely related species.
Satellite repeats often are the major component of complex eukaryotic centromeres (Csink and Henikoff, 1998). The centromeric satellite repeats generally have a similar monomer length, ranging from 150 to 180 bp (for review, see Henikoff et al., 2001), which is close to the range of nucleosomal unit length. Henikoff et al. (2001) recently proposed that adaptive modifications of the centromeric histone 3 in response to centromeric satellite expansion may be the driving force of centromere evolution.
Although satellite repeats are the most common feature of eukaryotic centromeres and have been proposed to be the key centromeric DNA component in many species, their true functional role is difficult to verify. Human α satellite is the only centromeric satellite whose function has been demonstrated by artificial chromosome assays (Harrington et al., 1997; Ikeno et al., 1998; Henning et al., 1999; Schueler et al., 2001), and its in vivo association with centromeric histone H3 also has been demonstrated by immunoprecipitation (Vafa and Sullivan, 1997).
We have obtained several lines of evidence that indicate that the CentO satellite is the key component of functional rice centromeres. First, the CentO satellite is cytologically located at the tip of the bivalent chromosomes at metaphase I of meiosis, indicating that the CentO satellite is located within the chromosomal regions at which the kinetochore forms and the spindle fibers attach. Second, sequence comparison revealed that the CentO satellite repeats share significant similarity with the maize centromeric satellite repeat CentC. Both the localization of these two satellites and the features that appear to have been conserved during their divergence since a common origin support a functional role for these sequences. Third, and most importantly, we have demonstrated that the break points of all 14 centromere misdivision events are located in the middle of the CentO loci, suggesting that the CentO satellite occupies the center of functional rice centromeres.
The human and Arabidopsis genomes have been sequenced using a clone-by-clone approach (Arabidopsis Genome Initiative, 2000; International Human Genome Sequencing Consortium, 2001). However, the centromeres of these chromosomes were all left as “gaps” in the sequencing data. Thus, even the most rigorous clone-by-clone sequencing approach has not yielded data on the complete DNA sequences of a centromere from either of these two species. There are arguments about whether sequencing of centromeres is necessary in genome projects, because it is known that the centromeres contain mainly known satellite repeats. However, in humans, only part of the α satellite arrays in the centromeres is involved in kinetochore assembly, because antibodies to kinetochore proteins localize to only a portion of the α satellite DNA (Warburton et al., 1997) and not all α satellite arrays can form artificial chromosomes (Ikeno et al., 1998). Characterization of the functional centromere of human X chromosome by Schueler et al. (2001) demonstrated that a DNA sequencing effort, together with functional assays, represents a powerful approach not only to define functional centromeres but also to reveal the plasticity and evolution of eukaryotic centromeres. Schueler et al. (2001) showed that only a subset of the α satellite repeats is responsible for the function of the X chromosome.
Rice provides an excellent model for functional and evolutionary studies of centromeres. Several rice centromeres contain only a limited amount of satellite repeat compared with human and Arabidopsis centromeres. It should be technically feasible to construct BAC contigs that span the entire centromeres of such chromosomes. The BAC contigs would provide an unprecedented resource for centromere sequencing.
It has been demonstrated in several eukaryotic species that a centromeric motif would have to be reiterated over hundreds of kilobases to achieve the minimum size of fully functional centromeres. In humans, a minimum of 100 kb of α satellite DNA is required to maintain the centromere function of minichromosomes derived from the Y chromosome (Yang et al., 2000). The centromere of a Drosophila minichromosome is contained within a 420-kb region of centric repetitive DNA. Deletion of DNA in this region leads to a progressive reduction of transmission (Sun et al., 1997). A similar minichromosome-based study in maize also demonstrated that 500 kb of centromeric repeat seems to be the minimum for fully functional B centromeres (Kaszas and Birchler, 1998). Therefore, it is surprising that some rice centromeres contain <100 kb of centromeric satellite repeat. It would be of great interest to determine if 60 to 80 kb of CentO is sufficient to act as a functional centromere in rice or if the DNA sequences flanking the CentO satellite are involved in centromere function.
METHODS
Materials
Rice (Oryza sativa subsp japonica var Nipponbare and Wuyujing 8 and O. sativa subsp indica var Zhongxian 3037) was used for cytological studies. The telotrisomic stocks used in this study were developed from Zhongxian 3037 (Cheng et al., 2001c). All BAC clones used for fluorescence in situ hybridization (FISH) analysis were selected from a BAC library developed from Nipponbare by Clemson University (http://www.genome.clemson.edu/orders/Product.html). The rice chromosome-specific BAC markers and rice centromeric DNA probes, including pRCS1, pRCS2, pRCH1, pRCH2, pRCH3, pRCE1, and pRCE2, were described previously (Dong et al., 1998; Cheng et al., 2001a).
FISH and Fiber-FISH
The preparation of somatic metaphase chromosomes and meiotic pachytene chromosomes was as described (Cheng et al., 2001a). The FISH procedure applied to both mitotic and meiotic preparations was essentially the same as the published protocols (Jiang et al., 1995). DNA probes were labeled with biotin-dUTP, digoxigenin-dUTP, or fluorescein isothiocyanate–dUTP (Boehringer Mannheim). Chromosomes were counterstained with propidium iodide or 4′,6-diamidino-2-phenylindole in Vectashield antifade solution (Vector Laboratories, Burlingame, CA). FISH procedures on genomic DNA fibers and BAC molecules were as described previously (Jackson et al., 1998, 1999). Hybridization signals in two-color fiber-FISH were detected with a three-layer antibody detection system as described (Jackson et al., 1998). Signal detection in three-color fiber-FISH was according to Hsieh et al. (2000).
Cytological Measurements and Analysis
All images were captured digitally using a SenSys charge-coupled device camera (Roper Scientific, Tucson, AZ) attached to an Olympus BX60 epifluorescence microscope (Tokyo, Japan). The charge-coupled device camera was controlled using IPLab Spectrum version 3.1 software (Signal Analytics, Vienna, VA) on a Macintosh computer. Gray-scale images were captured for each color channel and then merged. Measurements were made on the digital images of the FISH signals within IPLab Spectrum software. For quantitative FISH analysis, the intensity of 5 to 10 signals from the same chromosome was measured, and the numerical data were analyzed in Microsoft Excel 98 using the data analysis package. Only the mitotic metaphase cells or meiotic pachytene cells in which all 12 chromosomes or all targeted chromosomes could be identified unambiguously were selected for quantitative FISH analysis.
DNA Sequencing and Analysis
An M13 library was constructed from nebulized, size-fractionated DNA prepared from BAC 17p22. DNA templates were purified from random library clones, and sequences were collected using dye-terminator-labeled fluorescent cycle sequencing Prism reagents and ABI377 automated sequencers (Applied Biosystems, Foster City, CA). Sequences were assembled into contigs with the Seqman II program (DNASTAR, Madison, WI), and clones were selected for sequencing from the opposite end to fill coverage, resolve ambiguities, and close gaps (Burland et al., 1993). Final coverage was ∼10-fold.
Unique sequences (vector and nonsatellite) were readily assembled as described previously (Blattner et al., 1997). The assembly of highly repetitive sequences presented a challenge. Because the highly repetitive satellite sequences led to misassembly under the usual conditions, they were reassembled under higher stringency and using clone coverage information (dual-end sequence pairs derived from the same clone of ∼1 to 2.5 kb). Each initial clonal region was microedited to resolve ambiguities and then used as a longer sequence for another round of assembly. This rigorous recursive assembly process was repeated until all collected data were added to the project.
Sequence alignments were refined manually and displayed using GeneDoc (http://www.psc.edu/biomed/genedoc). DNA bending predictions were made using the bend.it server (http://www2.icgeb.trieste.it/~dna/bend_it.html). This server predicts DNA curvature from DNA sequences, calculated as a vector sum of dinucleotide geometries (roll, tilt, and twist angles) using the BEND algorithm of Godsell and Dickerson and expressed as degrees per helical turn (Munteanu et al., 1998).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Number
The GenBank accession number for BAC clone 17p22 is AY101510.
Acknowledgments
This research was supported by a grant from the Consortium for Plant Biotechnology Research, Novartis Seeds, and Dow AgroSciences, by Department of Energy Grant DE-FG02-01ER15266, and by funds from the Graduate School of the University of Wisconsin-Madison to J.J. This work also was partially supported by Chinese Project 973 Grant G1999011601 to Z.C. and M.G. Funding support to C.R.B. included U.S. Department of Agriculture Grant 99-35317-8275, National Science Foundation Grant DBI998282, and Department of Energy Grants DE-FG02-99ER20357 and DE-FG01-01ER15265.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003079.
References
- Alfenito, M.R., and Birchler, J.A. (1993). Molecular characterization of a maize B chromosome centric sequence. Genetics 135, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev, E.V., Phillips, R.L., and Rines, H.W. (1998). Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95, 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Aragon-Alcaide, L., Miller, T., Schwarzacher, T., Reader, S., and Moore, G. (1996). A cereal centromeric sequence. Chromosoma 105, 261–268. [DOI] [PubMed] [Google Scholar]
- Arumuganathan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218. [Google Scholar]
- Blattner, F.R., et al. (1997). The complete sequence of Escherichia coli K-12. Science 277, 1453–1474. [DOI] [PubMed] [Google Scholar]
- Brandes, A., Thompson, H., Dean, C., and Heslop-Harrison, J.S. (1997). Multiple repetitive DNA sequences in the paracentric regions of Arabidopsis thaliana L. Chromosome Res. 5, 238–246. [DOI] [PubMed] [Google Scholar]
- Burland, V., Daniels, D.L., Plunkett, G., III, and Blattner, F.R. (1993). Genome sequencing on both strands: The Janus strategy. Nucleic Acids Res. 21, 3385–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z.K., Buell, C.R., Wing, R.A., Gu, M.H., and Jiang, J. (2001. a). Toward a cytological characterization of the rice genome. Genome Res. 11, 2133–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z.K., Presting, G.G., Buell, C.R., Wing, R.A., and Jiang, J. (2001. b). High resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157, 1749–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z.K., Yan, H., Yu, H., Tang, S., Jiang, J., Gu, M.H., and Zhu, L. (2001. c). Development and applications of a complete set of rice telotrisomics. Genetics 157, 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, L. (1990). Centromeres of budding and fission yeasts. Trends Genet. 6, 150–154. [DOI] [PubMed] [Google Scholar]
- Clarke, L. (1998). Centromeres: Proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Curr. Opin. Genet. Dev. 8, 212–218. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G.P., et al. (1999). Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286, 2468–2474. [DOI] [PubMed] [Google Scholar]
- Csink, A.K., and Henikoff, S. (1998). Something from nothing: The evolution and utility of satellite repeats. Trends Genet. 14, 200–204. [DOI] [PubMed] [Google Scholar]
- Dobie, K.W., Hari, K.L., Maggert, K.A., and Karpen, G.H. (1999). Centromere proteins and chromosome inheritance: A complex affair. Curr. Opin. Genet. Dev. 9, 206–217. [DOI] [PubMed] [Google Scholar]
- Dong, F., Miller, J.T., Jackson, S.A., Wang, G.-L., Ronald, P.C., and Jiang, J. (1998). Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc. Natl. Acad. Sci. USA 95, 8135–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francki, M.G. (2001). Identification of Bilby, a diverged centromeric Ty1-copia retrotransposon family from cereal rye (Secale cereale L.). Genome 44, 266–274. [DOI] [PubMed] [Google Scholar]
- Fransz, P.F., Armstrong, A., de Jong, J.H., Parnell, L.D., van Drunen, C., Dean, C., Zabel, P., Bisseling, T., and Jones, G.H. (2000). Integrated cytogenetic map of chromosome arm 4S of A. thaliana: Structural organization of heterochromatic knob and centromere region. Cell 100, 367–376. [DOI] [PubMed] [Google Scholar]
- Gindullis, F., Desel, C., Galasso, I., and Schmidt, T. (2001). The large-scale organization of the centromeric region in Beta species. Genome Res. 11, 253–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, J.J., Bokkelen, G.V., Mays, R.W., Gustashaw, K., and Willard, H.F. (1997). Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nat. Genet. 15, 345–355. [DOI] [PubMed] [Google Scholar]
- Harrison, G.E., and Heslop-Harrison, J.S. (1995). Centromeric repetitive DNA in the genus Brassica. Theor. Appl. Genet. 90, 157–165. [DOI] [PubMed] [Google Scholar]
- Haupt, W., Fischer, T.C., Winderl, S., Fransz, P., and Torres-Ruiz, R.A. (2001). The CENTROMERE1 (CEN1) region of Arabidopsis thaliana: Architecture and functional impact of chromatin. Plant J. 27, 285–296. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., and Malik, H.S. (2001). The centromere complex: Stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- Henning, K.A., Novotny, E.A., Compton, S.T., Guan, X.-Y., Liu, P.P., and Ashlock, M.A. (1999). Human artificial chromosomes generated by modification of a yeast artificial chromosome containing both human alpha satellite and single-copy DNA sequences. Proc. Natl. Acad. Sci. USA 96, 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, H.B., Wang, M., Lersch, R.A., Kim, U.-J., and Weier, H.-U.G. (2000). Rational design of landmark probes for quantitative DNA fiber mapping (ODFM). Nucleic Acids Res. 28, e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudakova, S., Michalek, W., Presting, G.G., ten Hoopen, R., dos Santos, K., Jasencakova, Z., and Schubert, I. (2001). Sequence organization of barley centromeres. Nucleic Acids Res. 24, 5029–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeno, M., Grimes, B., Okazaki, T., Nakano, M., Saitoh, K., Hoshino, H., McGill, N.I., Cooke, H., and Masumoto, H. (1998). Construction of YAC-based mammalian artificial chromosomes. Nat. Biotechnol. 16, 431–439. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- Jackson, S.A., Dong, F., and Jiang, J. (1999). Digital mapping of bacterial artificial chromosomes by fluorescence in situ hybridization. Plant J. 17, 581–587. [DOI] [PubMed] [Google Scholar]
- Jackson, S.A., Wang, M.L., Goodman, H.M., and Jiang, J. (1998). Application of Fiber-FISH in genome analysis of Arabidopsis thaliana. Genome 41, 566–572. [PubMed] [Google Scholar]
- Jiang, J., Gill, B.S., Wang, G.-L., Ronald, P.C., and Ward, D.C. (1995). Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc. Natl. Acad. Sci. USA 92, 4487–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, J., Nasuda, S., Dong, F., Scherrer, C.W., Woo, S.S., Wing, R.A., Gill, B.S., and Ward, D.C. (1996). A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc. Natl. Acad. Sci. USA 93, 14210–14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaszas, E., and Birchler, J.A. (1996). Misdivision analysis of centromere structure in maize. EMBO J. 15, 5246–5255. [PMC free article] [PubMed] [Google Scholar]
- Kaszas, E., and Birchler, J.A. (1998). Meiotic transmission rates correlate with physical features of rearranged centromeres in maize. Genetics 150, 1683–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumekawa, N., Hosouchi, T., Tsuruoka, H., and Kotani, H. (2001). The size and sequence organization of the centromeric region of Arabidopsis thaliana chromosome 4. DNA Res. 8, 285–290. [DOI] [PubMed] [Google Scholar]
- Langdon, T., Seago, C., Mende, M., Leggett, M., Thomas, H., Forster, J.W., Thomas, H., Jones, R.N., and Jenkins, G. (2000). Retrotransposon evolution in diverse plant genomes. Genetics 156, 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maluszynska, J., and Heslop-Harrison, J.S. (1991). Localization of tandemly repeated DNA sequences in Arabidopsis thaliana. Plant J. 1, 159–166. [Google Scholar]
- Martinez-Zapater, J.M., Estelle, M.A., and Somerville, C.R. (1986). A high repeated DNA sequence in Arabidopsis thaliana. Mol. Gen. Genet. 204, 417–423. [Google Scholar]
- Miller, J.T., Dong, F., Jackson, S.A., Song, J., and Jiang, J. (1998. a). Retrotransposon-related DNA sequences in the centromeres of grass chromosomes. Genetics 150, 1615–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J.T., Jackson, S.A., Nasuda, S., Gill, B.S., Wing, R.A., and Jiang, J. (1998. b). Cloning and characterization of a centromere-specific repetitive DNA element from Sorghum bicolor. Theor. Appl. Genet. 96, 832–839. [Google Scholar]
- Munteanu, M.G., Vlahovicek, K., Parthasaraty, S., Simon, I., and Pongor, S. (1998). Rod models of DNA: Sequence-dependent anisotropic elastic modelling of local bending phenomena. Trends Biochem. Sci. 23, 341–346. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Tsujimoto, H., and Sasakuma, T. (1998). A novel repetitive sequence of sugar cane, SCEN family, locating on centromeric regions. Chromosome Res. 6, 295–302. [DOI] [PubMed] [Google Scholar]
- Nonomura, K.I., and Kurata, N. (1999). Organization of the 1.9-kb repeat unit RCE1 in the centromeric region of rice chromosomes. Mol. Gen. Genet. 261, 1–10. [DOI] [PubMed] [Google Scholar]
- Nonomura, K.I., and Kurata, N. (2001). The centromere composition of multiple repetitive sequences on rice chromosome 5. Chromosoma 110, 284–291. [DOI] [PubMed] [Google Scholar]
- Oakey, R., and Tyler-Smith, C. (1990). Y chromosome DNA haplotyping suggests that most European and Asian men are descended from one of two males. Genomics 7, 325–330. [DOI] [PubMed] [Google Scholar]
- Page, B.T., Wanous, M.K., and Birchler, J.A. (2001). Characterization of a maize chromosome 4 centromeric sequence: Evidence for an evolutionary relationship with the B chromosome centromere. Genetics 159, 291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presting, G.G., Malysheva, L., Fuchs, J., and Schubert, I. (1998). A Ty3/gypsy retrotransposon-like sequence localizes to the centromeric regions of cereal chromosomes. Plant J. 16, 721–728. [DOI] [PubMed] [Google Scholar]
- Richards, E.J., Goodman, H.M., and Ausubel, F.M. (1991). The centromere region of Arabidopsis thaliana chromosome 1 contains telomere-similar sequences. Nucleic Acids Res. 19, 3351–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round, E.K., Flowers, S.K., and Richards, E.J. (1997). Arabidopsis thaliana centromere regions: Genetic map positions and repetitive DNA structure. Genome Res. 7, 1045–1053. [DOI] [PubMed] [Google Scholar]
- Saunders, V.A., and Houben, A. (2001). The pericentromeric heterochromatin of the grass Zingeria biebersteiniana (2n=4) is composed of Zbcen1-type tandem repeats that are intermingled with accumulated dispersedly organized sequences. Genome 44, 955–961. [DOI] [PubMed] [Google Scholar]
- Schueler, M.G., Higgins, A.W., Rudd, M.K., Gustashaw, K., and Willard, H.F. (2001). Genomic and genetic definition of a functional human centromere. Science 294, 109–115. [DOI] [PubMed] [Google Scholar]
- Song, J., Dong, F., Lilly, J.W., Stupar, R.M., and Jiang, J. (2001). Instability of bacterial artificial chromosome (BAC) clones containing tandemly repeated DNA sequences. Genome 44, 463–469. [PubMed] [Google Scholar]
- Sun, X., Wahlstrom, J., and Karpen, G.H. (1997). Molecular structure of a functional Drosophila centromere. Cell 91, 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, H.L., Schmidt, R., and Dean, C. (1996). Identification and distribution of seven classes of middle-repetitive DNA in the Arabidopsis thaliana genome. Nucleic Acids Res. 24, 3017–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa, O., and Sullivan, K.F. (1997). Chromatin containing CENP-A and satellite DNA is a major component of the inner kinetochore plate. Curr. Biol. 7, 897–900. [DOI] [PubMed] [Google Scholar]
- Warburton, P.E., Cooke, C.A., Bourassa, S., Vafa, O., Sullivan, B.A., Stetten, G., Gimelli, G., Warburton, D., Tyler-Smith, C., Sullivan, K.F., Poirier, G.G., and Earnshaw, W.C. (1997). Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 7, 901–904. [DOI] [PubMed] [Google Scholar]
- Wevrick, R., and Willard, H.F. (1989). Long-range organization of tandem arrays of a satellite DNA at the centromeres of human chromosomes: High frequency array-length polymorphism and meiotic stability. Proc. Natl. Acad. Sci. USA 86, 9394–9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J.W., Pendon, C., Yang, J., Haywood, N., Chand, A., and Brown, W.R.A. (2000). Human minichromosomes with minimal centromeres. Hum. Mol. Genet. 9, 1891–1902. [DOI] [PubMed] [Google Scholar]
- Yu, H.-G., and Dawe, R.K. (2000). Functional redundancy in the maize meiotic kinetochore. J. Cell Biol. 151, 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H.-G., Hiatt, E.N., and Dawe, R.K. (2000). The plant kinetochore. Trends Plant Sci. 5, 543–547. [DOI] [PubMed] [Google Scholar]
- Zhang, P., Friebe, B., Lukaszewski, A.J., and Gill, B.S. (2001). The centromere structure in Robertsonian wheat-rye translocation chromosomes indicates that centric breakage-fusion can occur at different positions within the primary constriction. Chromosoma 110, 335–344. [DOI] [PubMed] [Google Scholar]