Abstract
A convenient in vitro chemical ligation strategy has been developed that allows folded recombinant proteins to be joined together. This strategy permits segmental, selective isotopic labeling of the product. The src homology type 3 and 2 domains (SH3 and SH2) of Abelson protein tyrosine kinase, which constitute the regulatory apparatus of the protein, were individually prepared in reactive forms that can be ligated together under normal protein-folding conditions to form a normal peptide bond at the ligation junction. This strategy was used to prepare NMR sample quantities of the Abelson protein tyrosine kinase-SH(32) domain pair, in which only one of the domains was labeled with 15N. Mass spectrometry and NMR analyses were used to confirm the structure of the ligated protein, which was also shown to have appropriate ligand-binding properties. The ability to prepare recombinant proteins with selectively labeled segments having a single-site mutation, by using a combination of expression of fusion proteins and chemical ligation in vitro, will increase the size limits for protein structural determination in solution with NMR methods. In vitro chemical ligation of expressed protein domains will also provide a combinatorial approach to the synthesis of linked protein domains.
Keywords: segmental labeling, expressed protein ligation, structural biology, Src homology domains
Many large cellular and extracellular proteins are composed of independently folded protein modules with distinct biochemical properties. Specific recombinations of these modules provide the overall functional character of the complete protein in vivo (1, 2). Consequently, there is some interest in understanding the structural and functional interplay that occurs between such domains in the context of the multidomain protein. Experimentally, this can be achieved by manipulating the spatial and/or functional organization of the domains by using standard recombinant DNA techniques. An alternative protein-engineering strategy would involve the in vitro assembly of multidomain proteins from individual “off-the-shelf” protein domains. Advantages of the latter strategy include the ability to prepare a large number of chimeric proteins from a small number of premade building blocks, the ability to prepare fused proteins that are cytotoxic from individually expressed domains that are not, the potential incorporation of nonnatural residues in an efficient combination of in vivo and chemosynthetic approaches, and the labeling of one segment of a protein for structural or biochemical investigation. In this paper we demonstrate that ligation of native expressed domains can be accomplished and that segmental labeling, especially valuable for NMR, can be achieved.
A practical size limit for protein structural determination by NMR spectroscopy arises because of the length of the protein, which is a function of the number of residues, n (3). This limit is attributable to the loss of resolution, proportional to n, occurring because signals with longer correlation times exhibit increased line widths and because of the increased number of signals that have similar chemical shifts.
Isotopic labeling can be used for the selection of coupled nuclei pairs, the perturbation of relaxation of complex or isochronous spin systems, and the observation of low sensitivity nuclei (specifically 13C and 15N). The application of this labeling to proteins is well exploited (for examples, see refs. 4 and 5). Although early examples of highly tailored isotopic syntheses of peptides by chemical means (for example, see ref. 6) were useful, that approach was subsumed by the more general ability to uniformly label proteins by overexpression in isotopically substituted media. However, labeling a segment of a protein remains an important goal generally and especially in connection with the study of multidomain or modular proteins (for examples, see refs. 7 and 8). Labeling a segment permits the direct assignment of chemical shifts in that segment, because of reduced spectral complexity. Moreover, in cases in which the subdomains are individually folded, segmental labeling permits the structural determination of the independent segment and possible comparison of the structure in isolated and multidomain forms. Segmental labeling also permits simplified observation of the individual subdomain for spin relaxation, residual dipolar coupling analysis (9), or study of ligand binding by chemical shift perturbation/structure-activity-relationship (SAR)-by-NMR (10).
In principle, selectively labeled proteins can be obtained by joining labeled and unlabeled recombinant proteins together in vitro. Along these lines, Yamazaki et al. (11) recently exploited trans-protein splicing (12–14) to generate a segmentally labeled protein for NMR analysis. By using a genetically dissected protein-splicing system, this group was able to join together labeled and unlabeled peptides derived from the α subunit of Escherichia coli RNA polymerase (11). Although elegant, this strategy resulted in the insertion of five unwanted amino acids at the splice junction and required a chemical denaturation step. These features, along with the moderate yields often associated with the trans-splicing process (12) may reduce the general applicability of this approach.
Previously, we described a protein-engineering approach, expressed protein ligation, that allows synthetic peptides to be chemically ligated to the C terminus of recombinant proteins (15, 16). Key to this process is the generation of a recombinant protein α-thioester derivative that can react with an N-terminal cysteine residue in the peptide to form a normal peptide bond. Reactive recombinant protein α-thioesters can be prepared by exploiting the natural process, protein splicing, a posttranslational event known to involve thioester intermediates (17). It is possible to chemically intercept the splicing process with a suitable thiol by appending the recombinant fragment in question to the N terminus of a genetically modified protein-splicing element (an intein), thereby generating the corresponding recombinant protein α-thioester (15, 16). Suitable protein expression vectors are commercially available that allow recombinant proteins to be expressed as an N-terminal intein–CBD fusion, in which the CBD is a chitin-binding domain affinity handle (17). After affinity purification on chitin beads, the immobilized fusion protein is exposed to an aqueous solution containing the synthetic peptide and a catalytic amount of thiophenol at pH 7.0. Under these conditions, near quantitative ligation of the peptide to the protein is observed (15, 16). Expressed protein ligation has thus far been used only to generate semisynthetic proteins (15, 16, 18); however, the approach could, in principle, be adapted to allow two recombinant, folded proteins to be ligated together. Such an extension permits segmental isotopic labeling of multidomain proteins for use in multidimensional NMR analysis, as well as other uses of combinatorial chemistry with protein domains.
MATERIALS AND METHODS
Analytical.
Electrospray mass spectrometry (ESMS) was performed on a Perkin–Elmer-Sciex (Thornhill, ON, Canada) API-100 mass spectrometer. Predicted masses were calculated by using macbiomass (S. Verumi and T. Lee, City of Hope, Duarte, CA). Analytical HPLC was performed on a Hewlett–Packard 1100 series instrument. Preparative HPLC was performed on a Waters DeltaPrep 4000 system. Linear gradients of 0.1% aqueous trifluoroacetic acid (solvent A) versus 90% CH3CN plus aqueous 0.1% trifluoroacetic acid (solvent B) were used for all runs.
Cloning and Expression of Abl-[C121]SH2.
Suitable SH2 constructs were generated from a pGEX2T vector containing the human Abl-SH(32)-coding sequence (19). Two restriction sites, NcoI and XmaI, were introduced on either side of the linker region between SH3 and the SH2 domains by using PCR mutagenesis. After the plasmid was treated with NcoI and XmaI and alkaline phosphatase, a double-stranded 5′-phosphorylated DNA cassette (comprising synthetic oligonucleotides 5′-CCG GTC ATC GAA GGT CGT TGC CTG GAG AAA CAT TCC TGG TAT-3′ and 5′-C ATG ATA CCA GGA ATG TTT CTC CAG GCA ACG ACC TTC GAT GA-3′) was inserted into the pGEX2T plasmid. This oligonucleotide creates an insertion of a factor Xa cleavage site (IEGR-C) and a S121 → C point mutation in the coding sequence. DNA sequencing was used to confirm the presence of the insertion and mutation. The glutathione S-transferase-Abl-SH3-IEGRC-SH2 fusion protein was expressed in E. coli DH5-α cells grown in M9 medium with 15N-ammonium chloride. Mid-log phase cells were induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside for 4 h at 37°C and harvested by centrifugation. Cells were resuspended in 4.3 mM sodium phosphate, 137 mM NaCl, 2.7 mM KCl, and 1.4 mM potassium phosphate, pH 7.2, which contained 100 mM EDTA, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1% (vol/vol) Triton X-100, and 1% (wt/vol) aprotinin, and then lysed by using sonication. The soluble fraction was then passed over glutathione agarose beads, which were then washed with 137 mM NaCl, 8 mM sodium phosphate, 2.7 mM KCl, and 1.4 mM potassium phosphate, pH 7.2, which contained 100 mM EDTA. Abl-SH3-IEGRC-SH2 was cleaved from the glutathione beads with thrombin (20). After thrombin cleavage, Abl-SH3-IEGRC-SH2 was exchanged with factor Xa reaction buffer (1 mM CaC12/100 mM NaC1/50 mM Tris⋅HCl, pH 7.8, with 0.01% NaN3). About 200 units of factor Xa (Pharmacia) were used to cleave 15 mg of Abl-SH3-IEGRC-SH2 in 4 ml of a reaction buffer at room temperature for 20 h. The resulting Abl-[C121]SH2 was purified by fast protein liquid chromatography with a Superdex-75 gel filtration column (Pharmacia) and 137 mM NaCl, 4.3 mM sodium phosphate, 2.7 mM KCl, and 1.4 mM potassium phosphate, pH 7.2, with 2 mM EDTA and 0.1 mM sodium azide as the eluent. The purified protein was concentrated to 0.5 mM with a Centricon 3 concentrator (Amicon). Purity and characterization were confirmed by analytical HPLC and ESMS: observed = 11,997.8 ± 1.4 Da; expected (average isotope composition) = 11,998.2 Da.
Cloning and Expression of Abl-SH3-Intein-CBD.
The DNA encoding the Abl-SH3 domain (residues L65–V119) was isolated by PCR from a cloned Abl-SH(32) gene (pGEX2T, ref. 19) with the oligonucleotide primers Abl 1 (5′-GGA TCC CCT GGT CAT ATG CTT TTT GTG GCA CTC TAT GAT TTT GTG-3′) and Abl 2 (5′-ATG TTT CTC CAG GCT GTT AAC GGG GGT GAT GTA GTT GCT TGG-3′). The PCR-amplified SH3 domain was purified and digested simultaneously with NdeI and HpaI and then recloned into the NdeI–SmaI-treated plasmid pTYB2 (New England Biolabs). The resulting plasmid, pTYB2Abl-SH3, expresses the Abl-SH3 domain fused via a single G residue to the intein-CBD from an isopropyl-1-thio-β-d-galactopyranoside-inducible T7 promoter. The pTYB2Abl-SH3 plasmid was shown to be free of mutations in the Abl-SH3-coding region by DNA sequencing. E. coli BL21 cells transformed with pTYB2Abl-SH3 were grown to mid-log phase in Luria–Bertani medium and induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside at 37°C for 5 h. No protein was detected by SDS/PAGE in the soluble fraction of the cell lysate under these conditions. Expression conditions were modified by inducing mid-log phase cells with 0.1 mM isopropyl-1-thio-β-d-galactopyranoside at room temperature for 2 h to yield protein in the soluble fraction. After centrifugation, cells were resuspended in 60 ml of lysis buffer (25 mM Hepes, pH 8.0/0.1 mM EDTA/250 mM NaCl/5% glycerol/1.0 mM phenylmethylsulfonyl fluoride) and lysed with a French press. The lysate was clarified first by low speed centrifugation and further clarified by ultracentrifugation. The clarified lysate (≈45 ml) was loaded onto a 15-ml chitin column preequilibrated in column buffer (20 mM Hepes, pH 7.0/250 mM NaCl/1 mM EDTA/0.1% Triton X-100), and the column was extensively washed with the same buffer and then stored at 4°C until further use. The column loading was determined by treating 100 μl of beads overnight with a buffer containing 0.2 M phosphate, pH 7.2, 0.2 M NaCl, and 100 mM DTT. After the beads were washed extensively with 1:1 acetonitrile:water, the amount of cleaved Abl-[G120]SH3 in solution was quantified by analytical HPLC through comparison to an Abl-SH3 standard of known concentration. This analysis indicated a loading of ≈0.35 mg/ml Abl-[G120]SH3. Results of ESMS of the cleavage product were: observed = 6,259.4 ± 0.5 Da; expected (average isotope composition) = 6,260.0 Da.
Peptide Synthesis.
A model peptide NH2-CGRGRGRK[fluorescein]-CONH2 was chemically synthesized on a methylbenzhydrylamine resin with in situ neutralization/2-[1H-benzotriazolyl]-1,1,3,3-tetramethyluronium hexafluorophosphate activation protocols for t-butyloxycarbonyl solid phase peptide synthesis (21). Orthogonal protection of the ɛ-amino group of the C-terminal K residue with fluorenylmethoxycarbonyl allowed solid phase attachment of fluorescein (activated as a succinimide ester) before the final cleavage step. The peptide was purified by reverse-phase HPLC and characterized by ESMS: observed mass = 1,245.9 ± 0.5 Da; expected (average isotope composition) = 1,246.5 Da.
Model Ligation Reactions.
Typically, 100 μl of chitin beads were equilibrated with a buffer containing 0.2 M phosphate and 0.2 M NaCl at pH 7.2. To these beads was added a solution of synthetic peptide (1 mg/ml) in the above buffer (100 μl) along with 1.5% (vol/vol) thiophenol. The suspension was then gently agitated at room temperature overnight, the supernatant was removed, and the beads were washed with 1:1 acetonitrile:water. The combined supernatant and washes were then analyzed by analytical HPLC and ESMS, which indicated the presence of the ligation product in excellent (>90%) yield: observed mass = 7,488.0 ± 1.5 Da; expected (average isotope composition) = 7,488.5 Da.
Preparation of Abl-[G120]SH3-Ethyl-α-thioester.
The chitin column, loaded and washed as described above, was equilibrated and suspended in 0.2 M phosphate, pH 6.0, and 0.2 M NaCl buffer to which 3% (vol/vol) ethanethiol was then added. This suspension was agitated overnight, the supernatant was removed, and the beads were washed several times with 1:1 acetonitrile:water. All washes were combined with the supernatant and purified by preparative reverse-phase HPLC by using a Vydac (Hesperia, CA) C18 column. The purity and composition of the resulting Abl-[G120]SH3-ethyl-α-thioester were confirmed by analytical HPLC and ESMS: observed mass = 6,305.4 ± 1.5 Da; expected (average isotope composition) = 6,304.2 Da.
Preparation of Abl-[G120C121][SH2-15N]SH(32).
Purified Abl-[G120]SH3-ethyl-α-thioester (2 mg) and purified 15N-labeled Abl-[C121]SH2 (8 mg) were reacted in 1.5 ml of 0.2 M phosphate, pH 7.2, and 0.2 M NaCl buffer containing both thiophenol and benzyl mercaptan each at final concentrations of 1.5% (vol/vol). After ≈90 h of reaction, the desired ligation product was purified by preparative HPLC and characterized by ESMS: observed mass = 18,240.1 ± 5.4 Da; expected (average isotope composition) = 18,240.2 Da. The lyophilized ligated product (≈2.5 mg) was then dissolved in 200 μl of 6 M Gdn⋅HCl (where Gdn is guanidine), 0.2 M phosphate, pH 7.2, and 0.2 M NaCl buffer and refolded by rapid dilution (10-fold) into 0.2 M phosphate, pH 7.2, and 0.2 M NaCl buffer. Note that the SH3 domain was also prepared with 15N labeling: observed mass = 6,376.8 ± 0.5 Da; expected (average isotope composition) = 6,378.0 Da. This material could be ligated to [C121]SH2, resulting in analytical quantities of [G120C121][SH3-15N]SH(32): observed mass = 18,163.3 ± 6.0 Da; expected (average isotope composition) = 18,156.2 Da.
NMR Measurements on Abl-[SH2-15N]SH(32).
Protein samples were exchanged in 200 mM NaCl, 4.3 mM sodium phosphate, 2.7 mM KCl, and 1.4 mM potassium phosphate, pH 7.2, which contained 8% (vol/vol) D2O, 2 mM [2H12]EDTA, 0.02% (wt/vol) NaN3, and either 2 or 10 mM DTT-2H10 for wild-type [U-15N]SH(32) and [SH2-15N]SH(32), respectively. The final concentration of the ligated sample was 0.2 mM and that of the wild-type sample was 0.8 mM. 1H-15N heteronuclear single-quantum correlation spectroscopy was performed at 35°C on a DMX-500 NMR spectrometer (Bruker, Billerica, MA) with a 5-mm probe (Nalorac Cryogenics, Martinez, CA). The spectral widths were 14 ppm for the 1H axis and 33 ppm for the 15N axis. The spectra were processed by using XWINNMR (Bruker). The resulting resolution in the final spectra was 1.75 Hz in the proton dimension and 3.2 Hz in the 15N dimension.
Fluorescence Binding Assay.
The equilibrium dissociation constants of the protein constructs for the consolidated ligand were determined by using the previously described fluorescence-based titration assay (22). The binding constant for the segment labeled construct was 300 (±100) nM. Experiments were performed on a Fluorolog-3 (Spex Industries, Metuchen, NJ) spectrophotometer fitted with a Neslab Instruments (Portsmouth, NH) temperature control unit.
RESULTS AND DISCUSSION
In this report we describe the development of procedures that allow two folded recombinant protein domains to be efficiently linked together by in vitro chemical ligation. This strategy has been used to prepare NMR quantities of the Abl regulatory apparatus, Abl-SH(32), in which only one domain was uniformly labeled with 15N.
The cellular signaling protein, c-Abl, is one of the few nonreceptor protein tyrosine kinases directly linked to human malignancies (23). The kinase activity of c-Abl is tightly controlled in vivo and is thought to be partly regulated by specific interactions of its SH3 and SH2 domains with other cytoplasmic and nuclear proteins (19, 24). The three-dimensional structures of the Abl-SH3 and Abl-SH2 domains have been studied in solution by NMR methods, both individually (20, 25) and together in the context of the domain pair (20). This level of structural characterization combined with the importance of these regulatory domains in c-Abl function suggested the Abl-SH(32) domain pair as an excellent target system for segmental labeling studies.
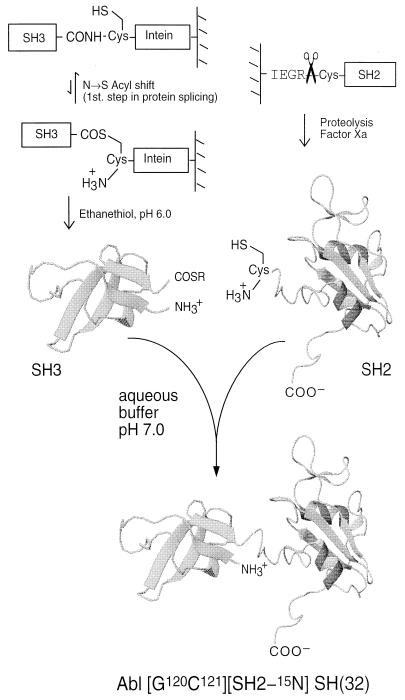
As illustrated in Fig. 1, our in vitro chemical ligation strategy called for the generation of a recombinant Abl-SH3 domain activated at its C terminus as an α-thioester and a recombinant Abl-SH2 domain containing an N-terminal cysteine residue. These two, folded protein domains should, when combined under physiological conditions, chemoselectively react via the well established native chemical ligation reaction (26, 28) to form an amide linkage at the ligation junction. The location of the ligation site was chosen to be within the short linker region that connects the two domains and involved mutation of the wild-type residues N120 and S121 to G and C, respectively. The S → C mutation was required to facilitate the ligation reaction, whereas the N → G mutation was expected to improve the kinetics of ligation.¶ Residue numbering is referenced to the complete Abl protein; the C121 mutation is then the N terminus of the Abl-SH2 domain. Previous studies had indicated this linker region to be relatively flexible (20), and it was anticipated that the mutations would lead to minimal significant structural perturbations.
Figure 1.
In vitro chemical ligation of folded recombinant proteins is illustrated by the preparation of Abl-SH(32). The Abl-SH3 domain is generated as an ethyl α-thioester derivative from the corresponding intein fusion protein, and the Abl-SH2 domain is generated with a cysteine at the N terminus via a factor Xa proteolysis strategy. Note that the linkage between the SH3 domain and the fused intein protein-splicing domain is naturally in equilibrium between an amide and a thioester (15–17). Exposure of this fusion protein to ethanethiol at pH 6.0 results in the formation of an ethyl α-thioester derivative of the SH3 domain. Combining these SH3 and SH2 protein derivatives under conditions that maintain them as folded results in a chemoselective ligation reaction and the generation of a normal peptide bond at the ligation junction (26). The sequence of the final ligation product is m{65}LFVALYDFVASGDNTLSITKGEKLRVLGYNHNGEWAEAQTKNGQGWVPSNYITPVGCLEKHSWYHGPVSRNAAEYLLSSGINGSFLVRESESSPGQRSISLRYEGRVYHYRINTASDGKLYVSSESRFNTLAELVHHHSTVADGLITTLHYPAPKR{220}gihrd. Lowercase letters indicate nongene residues from the expression systems used. This construct uses a C101 → S mutation internal to the SH3, which had previously been inserted to improve stability for NMR experiments. This is also in the “wild-type” sequence. Note that native chemical ligation reactions can be performed in the presence of multiple internal cysteine residues in either of the reacting segments (27); only the N-terminal cysteine participates in the ligation reaction.
The Abl-SH3 sequence (residues L65–V119) was subcloned into the commercially available pTYB2 expression vector, which allowed the generation of an Abl-SH3-intein-CBD fusion protein. After soluble expression in E. coli, the desired fusion protein was affinity purified on chitin beads. A small aliquot of the loaded beads was treated overnight with DTT, and the reaction supernatant was analyzed by reverse-phase HPLC and ESMS. This indicated that the expected Abl-SH3 construct was present in >90% homogeneity (data not shown) and that approximately 0.35 mg of the Abl-SH3 domain was immobilized per ml of chitin beads.
Initial attempts to generate the [C121]SH2 construct involved cyanogen bromide cleavage of a glutathione S-transferase-Abl-SH(32) fusion containing a unique M-C unit at the appropriate position within the interdomain linker.‖ This synthetic strategy was unsuccessful because of irreversible oxidation of the C residue to cysteic acid during the chemical cleavage step; the resulting Cys(O3H)-Abl-SH2 analog could not participate in subsequent chemical ligation reactions. An alternative approach was therefore used that applied the factor Xa cleavage strategy previously described by Verdine and coworkers (29). With this approach a glutathione S-transferase-Abl-SH(32) fusion protein was generated that contained an -I-E-G-R-C- motif inserted within the linker region connecting the Abl-SH3 and Abl-SH2 domains, before L122. Proteolysis of this fusion protein with factor Xa afforded the desired [C121]SH2 construct in good yield. A similar strategy was also used to prepare uniformly 15N-labeled [C121]SH2 (see Materials and Methods).
In preliminary ligation studies, we investigated whether a short synthetic peptide, NH2-CGRGRGRK[fluorescein]-CONH2, could be reacted with the immobilized Abl-SH3-intein-CBD fusion protein. Consistent with previously published examples (15, 16), nearly quantitative ligation of the synthetic peptide to the recombinant Abl-SH3 domain was observed, as indicated by reverse-phase HPLC, ESMS, and fluorescence spectroscopy (data not shown). These studies thus established that expressed protein ligation reactions could be performed on the folded Abl-SH3 domain.
Initial attempts to ligate [C121]SH2 to the immobilized SH3/thioester domain produced no detectable product formation. These studies used approximately equimolar amounts of the two reactants, requiring ≈2 ml of beads (≈700 μg of SH3 α-thioester) for every mg of [C121]SH2 used. The net effect of performing this reaction directly from the chitin beads was, therefore, to dilute greatly the [C121]SH2 domain (<50 μM), leading to a kinetically unfavorable reaction.** Note that this kinetic problem was not encountered with the model ligation described above because the synthetic peptide was present in large molar excess and millimolar concentration. However, emulating these pseudo-first-order conditions for the [C121]SH2 ligation was impractical because of the large amounts of the protein required (e.g., ≈100 mg of [C121]SH2 would be required for a preparative scale 10-ml reaction).
An alternative and more efficient synthetic approach was developed that overcame the kinetic problems associated with the immobilized Abl-SH3-intein-CBD fusion protein. This approach generates a soluble, stable α-thioester derivative of Abl-SH3 that can be easily purified and stored but whose reactivity can be modulated through transthioesterification during the ligation reaction. Previous studies have shown that alkyl α-thioester derivatives of synthetic peptides are relatively unreactive as acyl donors (33, 34). We found that overnight exposure of the chitin beads to ethanethiol at pH 6.0 led to the generation of an ethyl α-thioester derivative of the Abl-SH3 domain (Fig. 1). This transthioesterification/cleavage reaction was found to be remarkably clean as indicated by HPLC/ESMS analyses of the reaction supernatant and SDS/PAGE analysis of the residual immobilized protein on the chitin beads. The Abl-SH3 ethyl α-thioester derivative was easily purified by HPLC (gel filtration or dialysis could also be used provided the pH is kept at 6.0 or below) and could be stored as a lyophilized powder for several months.
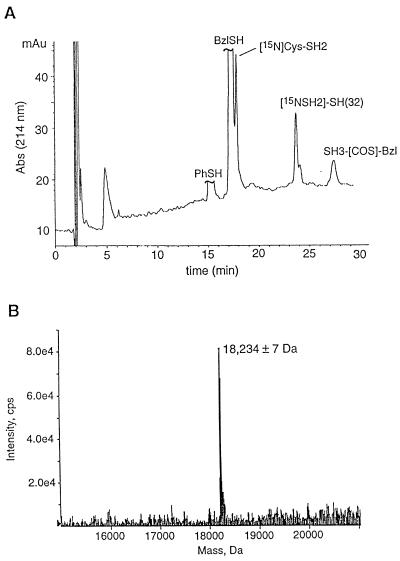
The [G120]SH3 ethyl α-thioester derivative and [C121][U-15N]SH2 domain were combined in phosphate buffer at pH 7.2, conditions under which the two protein domains are known to adopt stable tertiary folds (20, 25). To our knowledge this is the first time that the chemical ligation of two folded proteins has been attempted.‡‡ Three steps were thus taken to ensure efficient reaction, namely: the two domains were kept at a moderately high concentration (≈0.5 mM); one of the reactants, [C121]SH2, was added in molar excess; and the cofactors, thiophenol and benzyl mercaptan, were each included in the reaction medium. These cofactors are known to catalyze native chemical ligation reactions through in situ transthioesterification (30). The progress of the ligation reaction was monitored by using a combination of analytical HPLC and ESMS, which indicated that the reaction had gone to ≈70% completion after 4 days (Fig. 2A). At this point, the ligation product, Abl-[G120C121][SH2-15N]SH(32), was purified by preparative HPLC, and its covalent structure was characterized by ESMS (Fig. 2B).
Figure 2.
Chemical ligation of Abl-[G120]SH3 to Abl-[C121][U-15N]SH2. (A) Analytical reverse-phase HPLC profile of the crude ligation mixture after a 90-h reaction. A linear gradient of 32–46% of solvent B over 30 min was used. ESMS was used to identify the various components in the mixture, which are labeled accordingly. Note that the Abl-SH3 domain is converted to the more reactive benzyl- and phenyl-α-thioester derivatives in situ. (B) Electrospray mass spectrum (mass reconstruction) of the purified product, Abl-[G120C121][SH2-15N]SH(32); expected mass (average isotope composition) = 18,240.2 Da.
Preliminary studies had indicated that HPLC-purified recombinant Abl-SH(32) could be lyophilized and then refolded by rapid dilution from a 6 M Gdn⋅HCl-containing buffer into phosphate buffer at pH 7.2. Under these conditions, no protein precipitation was observed, and NMR analysis indicated that the sample had adopted a native fold (data not shown). A similar strategy was therefore used to prepare the complete [SH2-15N]SH(32) construct for functional and structural analysis. The binding affinity of Abl-[G120C121][SH2-15N]SH(32) for the consolidated ligand,†† NH2-PVpYENVG6>(PPAYPPPPVPKconh2), which binds both the SH3 and the SH2 domains simultaneously (22), was studied by a fluorescence-based titration assay. This revealed the equilibration dissociation constant for binding to the ligand, 300 nM, was essentially that previously reported for the Abl-SH(32) construct, 249 nM (22). This affinity is characteristic of the dual domain construct.
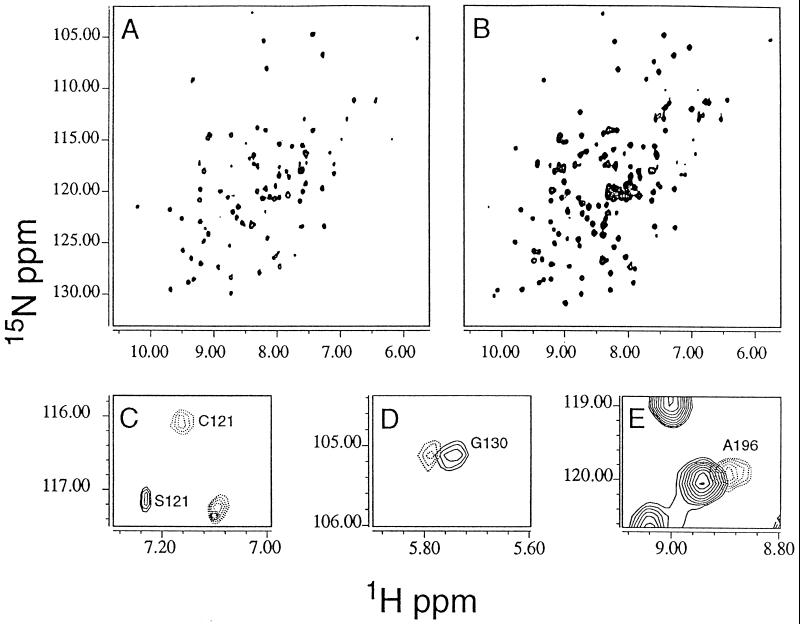
The purified ligation product was stable under NMR sample conditions. In Fig. 3A, the 1H{15N} heteronuclear single-quantum correlation spectroscopy map of the [G120C121][SH2-15N]SH(32) may be compared with the [U-15N]SH(32); these spectra are essentially fingerprints of the folded proteins. All peaks in the heteronuclear single-quantum correlation spectroscopy map of [G120C121][SH2-15N]SH(32) almost exactly coincide with those of [U-15N]SH(32) and are in agreement with the previous assignments by analogy (20) and from triple resonance data (R.X., unpublished results). There are no extraneous peaks. These NMR data are highly indicative that the structures are very similar and that the ligation reaction did not affect folding. At the ligation site, chemical shift changes are expected and observed for the NS → GC double mutation. The 15N-labeled amide of C121, assigned by analogy and difference, indicates the expected standard amide bond formation after the ligation reaction. The G120 is not labeled. The new spectra permit identification of the amide for E123, previously only ambiguously identified because of low intensity, and overlap with an SH3 amide resonance. Some subtle, but experimentally significant, shifts are observed for G130 and A196 (Fig. 3 D and E). From the expected contacts (20, 25) and observed flexibility of the linker (20, 36), these two residues are believed to be spatially close to the ligation site, where minor effects of the N → G and S → C mutations might be expected for changes in the side chain environment. The small magnitude of these chemical shift perturbations (<0.06 ppm, 1H; <0.1 ppm 15N, excluding the S121 →C mutation) further support the conclusions that the [G120C121][SH2-15N]SH(32) is topologically very similar to the wild type.
Figure 3.
1H{15N} NMR spectra at 500 MHz of Abl-[G120C121][SH2-15N]SH(32) (A) and wild-type Abl-SH(32) (B) with uniform 15N labeling. The peaks in A are the SH2-associated subset of those in B. (C–E) The peaks showing detectable chemical shift changes away from their position in the wild type are illustrated. (C) S121 in the wild type is mutated to C121 in the segment-labeled material. (C–E) The wild type subspectrum is shown in solid lines, and the segment-labeled protein is shown in dashed lines. Residue G130 shows a small 1H chemical shift (D), as does A196 (E). Both of these residues are spatially close to the junction between SH3 and SH2 and presumably are slightly structurally perturbed.
CONCLUSIONS
Significantly larger protein systems can be studied with new methods for spectral observation and structure determination by NMR (9, 37). The approach of segment labeling makes possible assignment and high resolution structural determination of large proteins without requiring the natural spectral simplification that occurs because of molecular symmetry. For example, it would seem practical to obtain highly resolved fragment spectra for about 100 residues of an 800-residue protein (≈110 kDa), comparable to those reported for the highly symmetric 7,8-dihydroneoptrin aldolase, a homooctomer (5). The effects of “context” of the surrounding domains on a segmentally labeled domain can now be practically studied by appropriate mutation and chemical ligation. Fragment labeling also permits segmental determination of dynamic properties, residual dipolar couplings (9), and SAR-by-NMR (10). Unlike the previously described trans-splicing approach (11), the chemical ligation strategy presented here can be extended to allow three recombinant protein segments to be regioselectively linked together; the feasibility of such an approach was recently demonstrated in a model synthetic peptide system (33). In principle, this important extension would allow internal domains of a protein to be isotopically labeled for NMR analysis. Other applications of our approach include the incorporation of selenomethionyl-labeled domains into a larger protein, facilitating structure determination of proteins by using multi-wavelength anomalous dispersion x-ray experiments for phasing (38), the incorporation of 2H-labeled segments for neutron scattering or diffraction, and the incorporation of highly magnetically anisotropic domains to provide additional orientation for NMR dipolar coupling measurements (39).
Acknowledgments
We gratefully acknowledge the cooperation of, and discussion with, the members of the Muir and Cowburn laboratories. We are indebted to Prof. B. J. Mayer for helpful suggestions concerning cloning and constructs. This work was supported by Grant R29-GM55843-01 (to T.W.M.), Grant R01-GM47021 (to D.C.), and Grant F32-AI-09537 (to R.X.) from the National Institutes of Health; a grant from the Pew Charitable Trusts (to T.W.M); and a grant from the National Leukemia Research Association (to T.W.M.).
ABBREVIATIONS
- ESMS
electrospray mass spectrometry
- CBD
chitin-binding domain
- SH2
src homology type 2 domain
- Abl
human Abelson protein tyrosine kinase
- SAR
structure-activity-relationship
Footnotes
A Commentary on this article begins on page 332.
Recent studies indicate that the majority of naturally occurring amino acids (with the exception of I, V, E, D, N, Q, and P) can be tolerated at the N-terminal side of the ligation junction without dramatically altering ligation yield/kinetics (Hackeng, P. M., Griffin, J. H. & Dawson, P. E., presented at the Twelfth Symposium of the Protein Society, San Diego, July 25–29, 1998). Thus, in future applications only a single amino acid mutation (i.e., X → C) may be necessary for expressed protein ligation.
The M-C unit was introduced into the linker region connecting the Abl-SH3 and Abl-SH2 domains by cassette mutagenesis by using an NcoI and XmaI restriction strategy. This resulted in N120 → M and S121 → C mutations in the Abl-SH(32) construct. The Abl-SH(32) sequence does not contain any endogenous M residues.
It is well established that efficient chemical ligation reactions require high concentrations (near mM) of both reactants (26–28, 30–33).
Although chemical denaturants were not present in the example here, such agents can be added if required and do not interfere with native ligation chemistry (26–28, 30–33, 35).
> denotes that the C-terminal glycyl residue is linked to the Nɛ of lysyl in the second peptide segment.
References
- 1.Jaenicke R. Biochemistry. 1991;30:3147–3161. doi: 10.1021/bi00227a001. [DOI] [PubMed] [Google Scholar]
- 2.Bork P, Schultz J, Ponting C P. Trends Biochem Sci. 1997;22:296–298. doi: 10.1016/s0968-0004(97)01084-0. [DOI] [PubMed] [Google Scholar]
- 3.Wuthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 4.Clore, G. M. & Gronenborn, A. M. (1997) Nat. Struct. Biol. 4, Suppl., 849–853. [PubMed]
- 5.Wuthrich, K. (1998) Nat. Struct. Biol. 5, Suppl., 492–495. [DOI] [PubMed]
- 6.Cowburn D, Live D H, Fischman A J, Agosta W C. J Am Chem Soc. 1983;105:7435–7442. [Google Scholar]
- 7.Campbell, I. D. & Downing, A. K. (1998) Nat. Struct. Biol. 5, Suppl., 496–499. [DOI] [PubMed]
- 8.Kuriyan J, Cowburn D. Annu Rev Biophys Biomol Struct. 1997;26:259–288. doi: 10.1146/annurev.biophys.26.1.259. [DOI] [PubMed] [Google Scholar]
- 9.Tjandra N, Bax A. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 10.Shuker S B, Hajduk P J, Meadows R P, Fesik S W. Science. 1996;274:1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 11.Yamazaki T, Otomo T, Oda N, Kyogoku Y, Uegaki K, Ito N. J Am Chem Soc. 1998;120:5591–5592. [Google Scholar]
- 12.Southworth M W, Adam E, Panne D, Bayer R, Kautz R, Perler F. EMBO J. 1998;17:918–926. doi: 10.1093/emboj/17.4.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shingledecker K, Jiang S Q, Paulus H. Gene. 1998;207:187–195. doi: 10.1016/s0378-1119(97)00624-0. [DOI] [PubMed] [Google Scholar]
- 14.Mills K V, Lew B M, Jiang S, Paulus H. Proc Natl Acad Sci USA. 1998;95:3543–3548. doi: 10.1073/pnas.95.7.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Severinov K, Muir T W. J Biol Chem. 1998;273:16205–16209. doi: 10.1074/jbc.273.26.16205. [DOI] [PubMed] [Google Scholar]
- 16.Muir T W, Sondhi D, Cole P A. Proc Natl Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chong S, Mersha F B, Comb D G, Scott M E, Landry D, Vence L M, Perler F B, Benner J, Kucera R B, Hirvonen C A, et al. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 18.Evans T C, Jr, Benner J, Xu M-Q. Protein Sci. 1998;7:2256–2264. doi: 10.1002/pro.5560071103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayer B J, Baltimore D. Mol Cell Biol. 1994;14:2883–2894. doi: 10.1128/mcb.14.5.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosser Y Q, Zheng J, Overduin M, Mayer B J, Cowburn D. Structure. 1995;3:1075–1086. doi: 10.1016/s0969-2126(01)00243-x. [DOI] [PubMed] [Google Scholar]
- 21.Alewood P, Alewood D, Miranda L, Love S, Meutermans W, Wilson D. Methods Enzymol. 1997;289:14–29. doi: 10.1016/s0076-6879(97)89041-6. [DOI] [PubMed] [Google Scholar]
- 22.Cowburn D, Zheng J, Xu Q, Barany G. J Biol Chem. 1995;270:26738–26741. doi: 10.1074/jbc.270.45.26738. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg N, Witte O N. Adv Virus Res. 1988;35:39–81. doi: 10.1016/s0065-3527(08)60708-3. [DOI] [PubMed] [Google Scholar]
- 24.Muller A J, Pendergast A M, Havlik M H, Puil L, Pawson T, Witte O N. Mol Cell Biol. 1992;12:5087–5093. doi: 10.1128/mcb.12.11.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Overduin M, Rios C, Mayer B, Baltimore D, Cowburn D. Cell. 1992;70:697–704. doi: 10.1016/0092-8674(92)90437-h. [DOI] [PubMed] [Google Scholar]
- 26.Dawson P E, Muir T W, Clark-Lewis I, Kent S B H. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 27.Hackeng T M, Mounier C M, Bon C, Dawson P E, Griffin J H, Kent S B H. Proc Natl Acad Sci USA. 1997;94:7845–7850. doi: 10.1073/pnas.94.15.7845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muir T W, Dawson P E, Kent S B H. Methods Enzymol. 1997;289:266–298. doi: 10.1016/s0076-6879(97)89052-0. [DOI] [PubMed] [Google Scholar]
- 29.Erlanson D A, Chytil M, Verdine G L. Chem Biol. 1996;3:981–991. doi: 10.1016/s1074-5521(96)90165-9. [DOI] [PubMed] [Google Scholar]
- 30.Dawson P E, Churchill M J, Ghadiri M R, Kent S B H. J Am Chem Soc. 1997;119:4325–4329. [Google Scholar]
- 31.Canne L, Bark S, Kent S B H. J Am Chem Soc. 1996;118:5891–5896. [Google Scholar]
- 32.Lu W, Qasim M A, Laskowski M, Kent S B H. Biochemistry. 1997;36:673–679. doi: 10.1021/bi9625612. [DOI] [PubMed] [Google Scholar]
- 33.Camarero J A, Cotton G J, Adeva A, Muir T W. J Pept Res. 1998;51:303–316. doi: 10.1111/j.1399-3011.1998.tb00428.x. [DOI] [PubMed] [Google Scholar]
- 34.Hojo H, Aimoto S. Bull Chem Soc Jpn. 1991;64:111–117. [Google Scholar]
- 35.Camarero J P, Pavel J, Muir T W. Angew Chem Int Ed Engl. 1998;37:345–348. doi: 10.1002/(SICI)1521-3773(19980216)37:3<347::AID-ANIE347>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 36.Nam H J, Haser W G, Roberts T M, Frederick C A. Structure. 1996;4:1105–1114. doi: 10.1016/s0969-2126(96)00116-5. [DOI] [PubMed] [Google Scholar]
- 37.Pervushin K, Riek R, Wider G, Wuthrich K. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hendrickson W A. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- 39.Prestegard, J. H. (1998) Nat. Struct. Biol. 5, Suppl., 517–522. [DOI] [PubMed]