Abstract
The POLARIS (PLS) gene of Arabidopsis was identified as a promoter trap transgenic line, showing β-glucuronidase fusion gene expression predominantly in the embryonic and seedling root, with low expression in aerial parts. Cloning of the PLS locus revealed that the promoter trap T-DNA had inserted into a short open reading frame (ORF). Rapid amplification of cDNA ends PCR, RNA gel blot analysis, and RNase protection assays showed that the PLS ORF is located within a short (∼500 nucleotides) auxin-inducible transcript and encodes a predicted polypeptide of 36 amino acid residues. pls mutants exhibit a short-root phenotype and reduced vascularization of leaves. pls roots are hyperresponsive to exogenous cytokinins and show increased expression of the cytokinin-inducible gene ARR5/IBC6 compared with the wild type. pls seedlings also are less responsive to the growth-inhibitory effects of exogenous auxin and show reduced expression of the auxin-inducible gene IAA1 compared with the wild type. The PLS peptide-encoding region of the cDNA partially complements the pls mutation and requires the PLS ORF ATG for activity, demonstrating the functionality of the peptide-encoding ORF. Ectopic expression of the PLS ORF reduces root growth inhibition by exogenous cytokinins and increases leaf vascularization. We propose that PLS is required for correct auxin-cytokinin homeostasis to modulate root growth and leaf vascular patterning.
INTRODUCTION
Hormone signaling systems coordinate plant growth and development through a range of complex interactions. It is not clear how interactions are coordinated between different classes of the classic hormones (auxin, cytokinin, ethylene, gibberellin, and abscisic acid) and between other signal molecules such as brassinosteroids (Altmann, 1999) or peptides (Lindsey et al., 2002). However, significant progress has been achieved in understanding the molecular basis of plant signaling systems through extensive genetic screens using Arabidopsis. A number of studies have identified diverse components in the signaling pathways of auxin (Leyser and Berleth, 1999), ethylene (McGrath and Ecker, 1998; Woeste and Kieber, 1998), gibberellins (Fridborg et al., 1999), abscisic acid (Finkelstein and Lynch, 2000), and cytokinins (Kakimoto, 1998; Vogel et al., 1998; Riou-Khamlichi et al., 1999; Hwang and Sheen, 2001; Inoue et al., 2001).
One experimental system, which has attracted much attention recently as a model in which to study hormonal interactions in development, is the Arabidopsis root (Dolan et al., 1993), which is readily amenable to mutational screens. For example, genes such as MONOPTEROS (Hardtke and Berleth, 1998) and BODENLOS (Hamann et al., 1999) are required for normal root development and encode components of the auxin signaling pathway. The MONOPTEROS gene, which encodes an auxin response factor transcription factor, also is required for correct patterning of vascular tissues in the cotyledon and leaf (Hardtke and Berleth, 1998). The aux1 mutant was identified on the basis of a defective root gravitropic response and encodes a putative component of the auxin influx carrier system (Bennett et al., 1996).
The EIR1/AGR1/AtPIN2 gene also is required for a normal gravitropic response and for ethylene sensitivity and encodes a likely component of the auxin efflux complex (Chen et al., 1998; Luschnig et al., 1998; Muller et al., 1998). The axr1 mutant is resistant to both auxin and ethylene and exhibits altered root branching (Lincoln et al., 1990). This and other mutants, such as axr3 and monopteros, also exhibit abnormal auxin distribution (Sabatini et al., 1999).
However, we still have an incomplete notion of the molecular mechanisms of root formation and growth control and of how hormones interact to elicit the diverse developmental pathways found in plants. For example, cytokinins can act as important regulators of cell division and appear to be synthesized in root tips, although exogenous cytokinins can inhibit root growth (Sossountov et al., 1988; Riou-Khamlichi et al., 1999; Chiappetta et al., 2001). Cytokinins can act antagonistically with auxin in some developmental contexts, such as in lateral root development, but they also can act synergistically, as in ethylene biosynthesis (Vogel et al., 1998).
Key components of the cytokinin signaling pathway have been identified on the basis of resistance to the growth-inhibitory effects of exogenous cytokinins (Brandstatter and Kieber, 1998), including the identification of a predicted cytokinin receptor (Inoue et al., 2001). Nevertheless, the mechanisms by which hormonal interactions occur remain unclear.
To gain new insight into the control of root growth and development, we screened a population of Arabidopsis promoter trap transgenic lines for genes expressed in, and required for, root development. Promoter trapping can facilitate expression analysis of genes by the characterization of in vivo fusion gene expression, the investigation of tagged gene function by mutant analysis, and the cloning of tagged genes (Lindsey et al., 1998).
Previously, we identified a promoter trap transgenic line that has β-glucuronidase (GUS) fusion activity detectable from the heart stage of embryogenesis, in the basal region of the embryo, and in the seedling root tip (Topping et al., 1994; Topping and Lindsey, 1997). Here, we describe the cloning and characterization of the tagged gene, designated POLARIS (PLS). It encodes a short transcript and a predicted small polypeptide. It is required for correct responses to cytokinins and auxins, for correct cell expansion in the root, and for vascular patterning in the leaf.
RESULTS
pls Is Defective in Root Growth and Leaf Vascularization
A screen of GUS-expressing promoter trap lines of Arabidopsis identified the line AtEM101. It contains a single-copy T-DNA and exhibits GUS fusion activity predominantly in the embryonic root from the heart stage and in the seedling primary and lateral root tips (Topping et al., 1994) (Figures 1A and 1B).
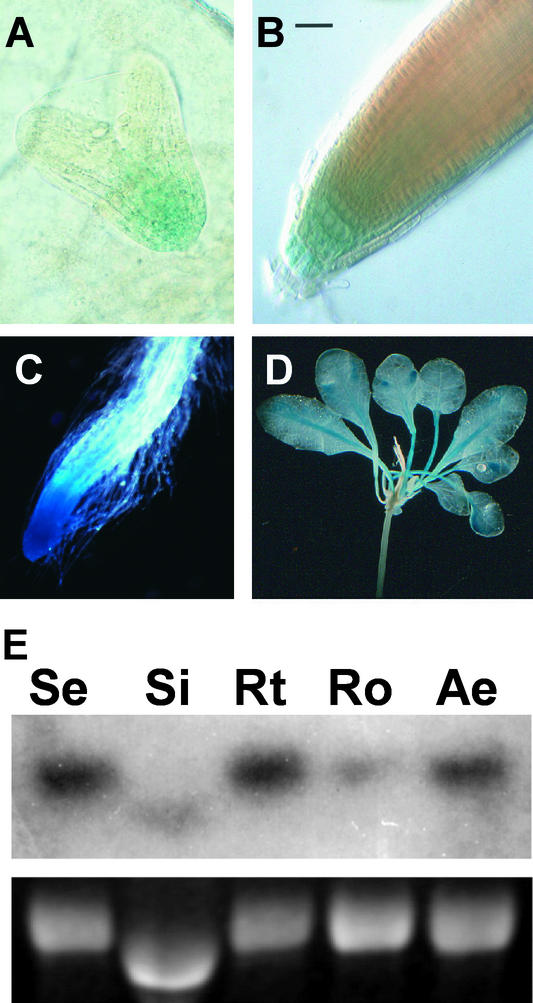
Figure 1.
GUS Expression in the PLS-GUS Promoter Trap Line AtEM101.
(A) Heart-stage embryo, with 5 h of GUS staining, showing GUS activity in the basal region. Magnification ×100.
(B) Seven-day-old seedling root tip, with 5 min of GUS staining, showing activity in columella and, weakly, in young vascular cells. Bar = 50 μM.
(C) Seven-day-old seedling root tip, with 1 h of GUS staining, showing activity throughout the root tip. Magnification ×7.
(D) Aerial parts of a 12-day-old seedling, with 8 h of GUS staining, showing activity in leaf vascular tissues. Magnification ×5.
(E) RNA gel blot analysis showing PLS-GUS fusion transcript levels in the pls mutant using 10 μg of RNA from 7-day-old total seedlings (Se), siliques (Si), root tips (Rt), roots with tips removed (Ro), and aerial parts (Ae). The top gels shows a GUS fusion transcript, and the bottom gels shows ethidium bromide–stained 25S rRNA. Note that silique RNA runs more rapidly through gels than that from other tissues.
A time course of histochemical staining of the root of AtEM101 revealed that, after a short period (5 min) of incubation in 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, GUS activity was detectable only in the columella initials and lateral root cap (Figure 1B), similar to the pattern observed for the auxin-regulated DR5::GUS gene fusion (Sabatini et al., 1999). Longer incubation periods in 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (>1 h) revealed GUS activity more widely in the root tip, notably in the meristematic region (Figure 1C). Overnight staining revealed GUS activity predominantly in both the primary and lateral root tips, but lower levels of activity also were detectable in aerial parts of the seedling. In leaves, GUS activity was detected principally in vascular tissues of the lamina and petiole (Figure 1D).
To investigate further the transcriptional regulation of the PLS-GUS fusion gene, RNA gel blot analysis was performed using total RNA from 7-day-old seedlings and from siliques of 21-day-old plants. The results are presented in Figure 1E. A gusA probe identified an ∼3-kb transcript, which is ∼300 nucleotides larger than the predicted gusA mRNA plus T-DNA left border region, indicating a transcriptional fusion. The relative abundance of the fusion transcript in different regions of the root, and in different organs, was consistent with the histochemical staining pattern of GUS activity. In particular, root tips showed greater transcript abundance compared with the older, proximal region of the root.
To determine whether there is a defective seedling phenotype associated with the T-DNA insertion, AtEM101 seedlings homozygous for the T-DNA were backcrossed to the wild type to reveal segregating mutants. Seedlings of selfed heterozygotes were grown on vertical agar plates for 14 days in the light, and three classes of seedlings were identified. Approximately 25% (43 of 186) were GUS negative and exhibited a relatively long root, comparable to that of wild-type seedlings (mean length, 75 ± 4 mm [n = 10]); 25% (41 of 186) were GUS positive and exhibited a significantly shorter root than the wild type (mean length, 38 ± 2 mm [n = 10]; i.e., ∼51% at 14 days after germination) (Figure 2A). These seedlings were homozygous for the T-DNA insertion, as demonstrated by 100% of the progeny showing this GUS-positive short-root phenotype on selfing.
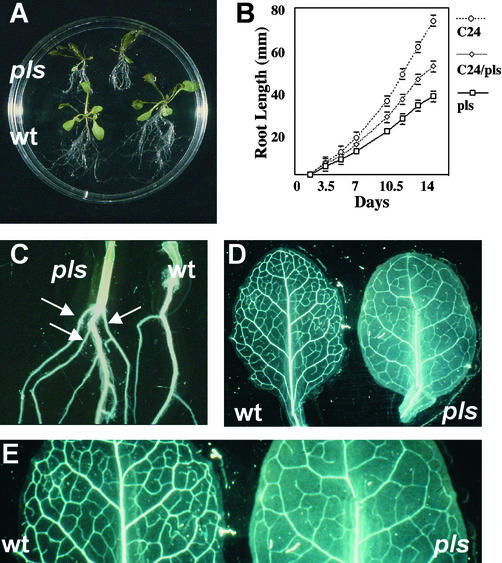
Figure 2.
pls Mutant Phenotype.
(A) pls (top) and wild-type (wt; bottom) seedlings, 14 days after germination, showing the short-root phenotype of the pls mutant.
(B) Primary root growth of wild-type C24, pls mutant, and C24/pls heterozygote seedlings.
(C) The hypocotyl-root junction of pls (left) and wild-type (right) seedlings, 14 days after germination, showing the increased frequency of anchor roots (arrows) in the pls mutant.
(D) Rosette leaves of wild-type (left) and pls mutant (right) seedlings, 12 days after germination, showing the decreased frequency of higher order vascular strands in the pls mutant.
(E) Enlargement of leaves shown in (D) showing detail of the reduced vascularization of the pls mutant.
The third class, which comprised ∼50% of the seedlings (102 of 186), was GUS positive, although with reduced GUS activity, as determined by histochemical staining compared with the previous class. It exhibited an intermediate rate of root growth (mean length, 53 ± 3 mm; i.e., ∼71% the length of wild-type roots at 14 days after germination). These seedlings were heterozygous for the T-DNA insertional mutation, as shown by segregation of the GUS-associated root phenotype on selfing. The mutant was designated pls.
To investigate further the root phenotype of the pls mutant and heterozygote, seedlings were germinated and grown in the light on vertical agar plates and primary root length was measured over a time course. The results presented in Figure 2B show that the rate of increase in root length of seedlings homozygous for the T-DNA insertion was reduced compared with that in the wild type, such that by day 14 after germination, the length of the mutant primary root was ∼50% of that of the wild type. The rate of elongation of the primary roots of seedlings heterozygous for the T-DNA insertion was intermediate between those of wild-type and homozygous seedlings, indicating that the pls mutation is semidominant. Additionally, pls mutants typically produced more anchor roots at the root-hypocotyl junction (three or four) than did wild-type seedlings (usually one; Figure 2C).
Microscopic analysis of cleared pls roots (day 7 after germination) revealed that the cells of the root meristem and the cortex of the primary root were shorter and more radially expanded than in the wild type. Cortical cells in the maturation zone were a mean length of 123.2 ± 1.2 μm (n = 68) for the wild type and 98.8 ± 0.9 μm (n = 68) for pls. The pls root was wider than the wild-type root, both across the region of the quiescent center (wild type, 69.0 ± 1.5 μm [n = 10]; pls, 74.8 ± 1.7 μm [n = 10]) and 100 μm farther back from the root tip (wild type, 86.8 ± 1.0 μm [n = 10]; pls, 96.5 ± 1.9 μm [n = 10]). The reduction in axial cell elongation in the root would only partially account for the pls short-root phenotype, indicating that the mutant meristem cells divide less frequently than wild-type cells.
Aerial parts of seedlings and mature plants were not obviously abnormal in their morphology. However, microscopic analysis of the rosette leaves of 12-day-old plants showed that the extent of vascularization of the rosette leaves was reduced compared with that in the wild type, with fewer higher order veins arising from the major strands (Figure 2D). Plants hemizygous for the pls mutation showed levels of venation intermediate between those in homozygous mutants and the wild type (data not shown).
Together, these data suggest that the T-DNA insertion had caused a semidominant mutation affecting root growth and architecture, and vascular patterning in leaves, under the conditions studied.
PLS Encodes a Short Transcript
To investigate the molecular basis of the mutation, part of the PLS locus (166 bp of T-DNA flanking sequence) was cloned by inverse PCR (Topping et al., 1994) and used as a molecular probe to identify a corresponding Arabidopsis genomic clone. A 3.6-kb EcoRI genomic subclone, hybridizing to the inverse PCR fragment, and another 1.7 kb of contiguous genomic region, were sequenced. The T-DNA had inserted in a small open reading frame (ORF) of 108 bp in a 755-bp region between two larger genes (Figure 3). There was no rearrangement of the adjacent genomic DNA. One gene, 450 bp upstream of the T-DNA, encodes a predicted protein of 92 amino acid residues of unknown identity (designated GENE X). The second gene is BRI1, which locates PLS to the bottom of chromosome 4 (Li and Chory, 1997).
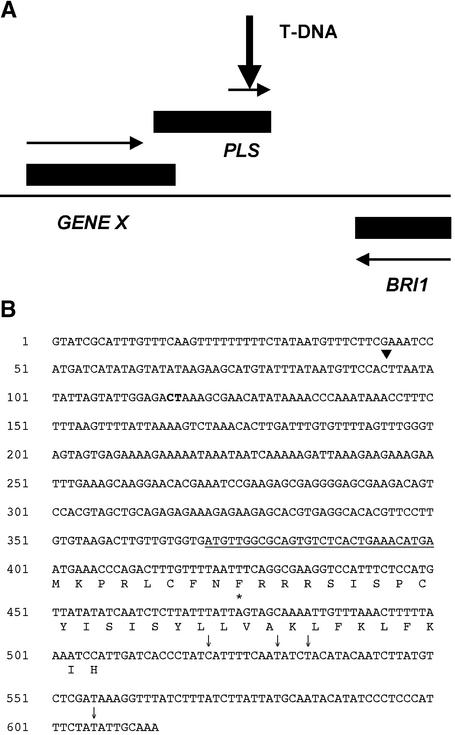
Figure 3.
PLS Encodes a Small Transcript.
(A) PLS locus. Closed rectangles indicate transcripts, and arrows indicate ORFs (for GENE X and PLS). The T-DNA insertion site is indicated.
(B) The PLS gene sequence shows transcription start site 2 (inverted arrowhead), the GENE X polyadenylation site (boldface), the 9–amino acid uORF (underlined), the 36–amino acid PLS ORF (amino acids shown), and the PLS polyadenylation sites (arrows). The asterisk marks the position of the T-DNA insertion site (in the 25th codon; L).
It was suspected at first that the ORF into which the promoter trap had inserted (designated the PLS ORF) was an exon of one of the adjacent two genes. However, it was found to be part of a separately transcribed gene. 3′ rapid amplification of cDNA ends (RACE) of GENE X demonstrated that its transcription terminated with a polyadenylation sequence 356 bp upstream of the T-DNA insertion site (data not shown). RNA gel blot analysis of 7-day-old AtEM101 seedlings, probed with the 32P-labeled 3′ RACE product and a gusA gene probe, revealed two distinct transcripts: the GENE X transcript (∼0.5 kb) and the GUS fusion transcript (∼3 kb; data not shown). Furthermore, BRI1 is located on the nontranscribed strand (i.e., in the wrong orientation to activate the T-DNA promoter trap) and is ∼400 bp downstream of the T-DNA insertion site.
5′ RACE PCR from the GUS fusion transcript was used to clone part of the PLS cDNA, and this sequence was used to clone the remainder by 3′ and 5′ RACE PCR from the wild-type plant. RNA gel blot analysis, using PLS cDNA as a probe, revealed the presence of a single low-abundance transcript of ∼500 nucleotides, representing the predicted PLS transcript (Figure 4A). The transcript was detectable only when gel blots of at least 1 μg of poly(A)+ RNA were probed and autoradiographed for at least 4 weeks. Cloning and sequencing of an RNA-specific PCR product corresponding to this transcript confirmed its identity. DNA gel blot analysis revealed a single hybridizing band, indicating a single copy per haploid genome (data not shown).
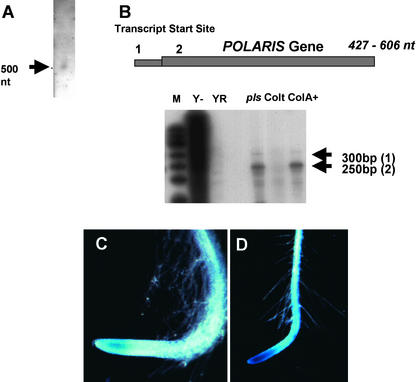
Figure 4.
PLS Expression.
(A) RNA gel blot [5 μg of poly(A)+ RNA] showing the ∼500-nucleotide (nt) PLS transcript.
(B) RNase protection assays to map the PLS transcript initiation site in wild-type and AtEM101 seedlings using a radiolabeled antisense PLS transcript. Transcript start sites (1 and 2) are shown in the bottom panel (arrows) and in diagrammatic form above. M, size markers (radiolabeled phiX174 Hinf1 DNA); Y−, yeast RNA control; YRNase, RNase-treated yeast RNA control; pls, 50 μg of total RNA from AtEM101 seedlings; Colt, 50 μg of total RNA from wild-type seedlings (Columbia); ColA+, 5 μg of poly(A)+ RNA from wild-type seedlings (Columbia).
(C) Activity of the 370-bp promoter region (pPLS::GUS expression) in the root tip of a 7-day-old transgenic seedling with 24 h of GUS staining. Magnification ×7.
(D) Activity of the 1190-bp promoter region in the root tip of a 7-day-old transgenic seedling with 24 h of GUS staining. Magnification ×5.
PLS Gene Transcription Initiates within an Upstream Gene
The site of transcript initiation was determined by 5′ RACE PCR and RNase protection assays. It was found that the transcription start site is the same for both wild-type PLS and PLS-gusA fusion transcripts (Figure 4B). Interestingly, two transcription start sites were found ∼95 bp apart. Start site 1, ∼117 bp upstream of the GENE X poly(A) site, has the sequence ATCCGTAT (the G represents the initiation site). Start site 2, which is ∼23 bp upstream of the poly(A) site, has the sequence CCACTTAATA; RACE results were unable to resolve which of the three underlined bases is likely to represent the initiation site.
The RNase protection assay results show that start site 2, which generates the shorter transcript, is used more frequently than start site 1. Start site 2 has the predicted TATA sequence TATATAA (positions −32 to −26). Start site 1 has a poorer TATA-like sequence, AATAATA (positions −35 to −29). Sequencing of 3′ RACE products revealed that the PLS transcript had a variable 3′ end, with the transcript being between 427 and 606 nucleotides long, according to the transcription start and polyadenylation sites used.
To confirm that PLS transcript initiation is driven by sequences within GENE X, transgenic lines were produced containing an upstream genomic fragment fused to the gusA reporter gene. This fragment, which comprised 370 bp upstream of the predicted transcription initiation start site 2 and lacked the ATG codon of the putative 92–amino acid GENE X ORF, was designated pPLS::GUS. T1 and T2 transformants were identified on the basis of T-DNA–mediated resistance to kanamycin. Six families of independent pPLS::GUS transgenic lines were analyzed histochemically for GUS activity in 7-day-old seedlings. All showed GUS activity in the root tip (Figure 4C). A GUS fusion with a longer promoter region, 1190 bp upstream of the predicted transcription initiation start site 2, showed stronger GUS activity than the shorter promoter (Figure 4D), with activity also detectable in the aerial parts of the seedling, as in line AtEM101 (data not shown).
The PLS Transcript Encodes a Predicted 36–Amino Acid Polypeptide
Sequencing of the PLS locus in C24 (Figure 3) showed that the T-DNA inserted into the 25th codon (Leu) of an ORF encoding a predicted 36–amino acid residue PLS polypeptide, with a predicted molecular mass of 4.6 kD and no significant homology with known proteins. The Columbia allele also was cloned and found to be identical in sequence. The T-DNA is expected to disrupt PLS function. The PLS ORF is preceded by an in-frame 27-bp ORF encoding a predicted nine–amino acid polypeptide; its TGA stop codon immediately precedes the ATG of PLS. A predicted eight–amino acid ORF overlaps with PLS 4 bp upstream of the PLS ATG.
No other ATG codons appear in the 5′ untranslated region (UTR) initiating from transcriptional start site 2. The N-terminal 24 amino acids of the PLS putative polypeptide are predicted to form two β-sheets, whereas the remaining 12 amino acids are likely to form an α-helix. Between the two β-sheets are three basic Arg residues (positions 10 to 12) that may form a turn region and a possible cleavage site. The second β-sheet contains a repeated SIS separated by four residues. The proximity of these Ser residues to the basic Arg residues of the β-turn may signify a cAMP- and cGMP-dependent protein kinase phosphorylation site.
The C-terminal α-helix also contains the repeat KLFKLFK. The Lys residues and the terminal His residue represent the only charged residues in this helical region. The three–amino acid spacing between each of the Lys residues indicates that they would all lie on the same face of the α-helix, creating an amphipathic helix with both hydrophobic and charged faces. The fact that the predicted helical region is Leu rich indicates the potential for a Leu zipper motif, suggesting the possibility for protein–protein interactions. Two of the Leu residues form a heptad (Leu residues 25 and 32), which may be significant given the small size of the polypeptide.
PLS Gene Expression Is Upregulated Rapidly by Exogenous Auxin
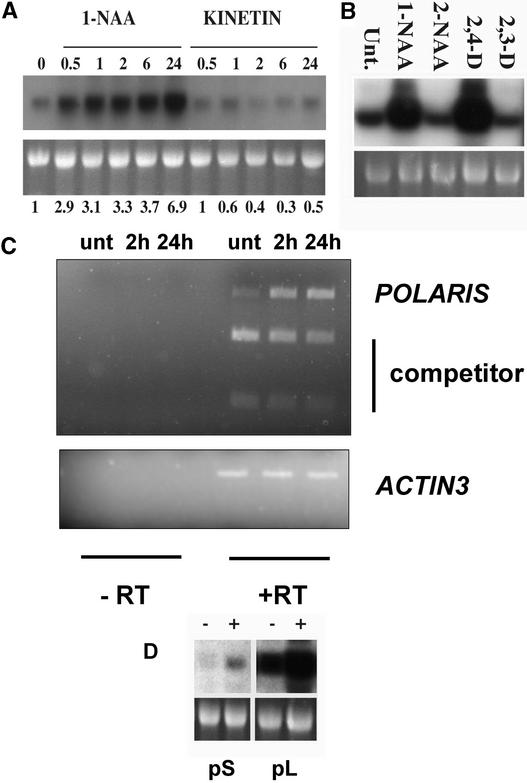
PLS-GUS activity is upregulated in response to a polarizing signal in the embryo and root, a candidate for which is auxin (Topping and Lindsey, 1997). To investigate this further, RNA gel blot analysis of the PLS-GUS transcript was used to examine the effects of exogenous auxin, cytokinin, and inactive auxin analogs on the activity of the PLS gene promoter. AtEM101 seedlings were grown for 7 days after germination on hormone-free medium and then transferred to medium containing 10 μM 1-naphthalene acetic acid (1-NAA) or 10 μM kinetin for 30 min to 24 h before RNA extraction. Total RNA was fractionated, blotted, and probed with a gusA gene fragment.
The transcript abundance increased approximately threefold within 30 min and approximately sevenfold by 24 h of treatment with auxin (Figure 5A). This effect also was seen with 2,4-D but not with the nonfunctional auxin analogs 2-NAA and 2,3-D (Figure 5B). Histochemical analysis showed that when AtEM101 seedlings were treated with high auxin concentrations (e.g., 10 μM 2,4-D), GUS activity was detectable in all root tissues and in leaves (data not shown). By contrast, treatment with kinetin led to reduced gusA transcript abundance, to a level of ∼40% of that in the untreated seedlings, within 2 h (Figure 5A).
Figure 5.
The PLS Gene Promoter is Auxin Inducible.
(A) RNA gel blot showing PLS-GUS fusion transcript levels after treatment with auxin (10 μM 1-NAA) or cytokinin (10 μM kinetin). Seedlings were grown for 7 days on hormone-free medium and then transferred to the hormones for the times indicated. The top gel shows the PLS-GUS transcript, and the bottom gel shows ethidium bromide–stained 25S RNA. Relative transcript levels were normalized compared with untreated controls, and values are shown below. Each lane contained 10 μg of total RNA.
(B) PLS-GUS fusion transcript levels after treatment of 7-day-old AtEM101 seedlings with 10 μM active auxins (1-NAA and 2,4-D) and inactive analogs (2-NAA and 2,3-D) for 24 h. Each lane contained 20 μg of total RNA. Unt., untreated.
(C) Competitor PCR analysis showing the increase in abundance of the native PLS transcript (POLARIS) in untreated 7-day-old wild-type seedlings (unt) and after treatment of 7-day-old wild-type seedlings with 10 μM 1-NAA for 2 and 24 h. The competitor lanes show the abundance of amplicon derived from a cleaved (with ClaI) synthetic PLS cDNA fragment that was identical to wild-type PLS sequence except for an introduced ClaI site and that was introduced into the PCR as an internal standard. The ACTIN3 transcript was amplified as a control, and control amplifications lacking reverse transcriptase are shown (−RT).
(D) PLS-GUS fusion transcript levels in transgenic lines containing either the 370-bp (pS) or 1190-bp (pL) promoter fragments linked to gusA after growth on 10 μM 1-NAA for 24 h (+) or unsupplemented medium (−). Each lane contained 20 μg of total RNA.
To determine whether transcription of the native PLS gene also is regulated by auxin, competitor reverse transcription (RT)–PCR was used to determine PLS mRNA relative abundance in seedlings either treated or untreated with auxin. The results, presented in Figure 5C, showed increased PLS transcript abundance in seedlings treated with 10 μM 1-NAA for 2 or 24 h compared with untreated seedlings. The relative increase in the native PLS amplicon was clear compared with the abundance of amplicon derived from a synthetic cDNA fragment, which was identical to the PLS sequence except for an introduced ClaI site and was introduced into the PCR as an internal standard. Actin controls showed that no general increase in mRNA abundance occurred in response to auxin treatment.
The cloned PLS promoter region in pPLS:GUS also showed auxin inducibility after treatment of seedlings with 10 μM 1-NAA, as shown by GUS histochemistry and confirmed by RNA gel blot analysis (Figure 5D, pS). There was an ∼2.5-fold increase in transcript levels at 24 h after auxin treatment compared with untreated controls. Therefore, the 370-bp region within pPLS:GUS is sufficient for auxin-inducible transcription. The 1190-bp promoter::GUS also showed auxin inducibility after treatment of transgenic seedlings with 10 μM 1-NAA (Figure 5D, pL).
The pPLS promoter contains a number of TGTCTC-like putative auxin-responsive elements (AuxREs) (Ulmasov et al., 1997). Two sequences, TGTCTC and TGTCGG, occur on the opposite strand (AuxREs are functional on both DNA strands). Two overlapping elements were identified starting at position −146 (TGTCTTGTCTA) relative to transcriptional start site 1. These were just upstream of the sequence AATAAT (position −130), which is similar to the sequence AATAAG found close to the TGTCTC element in two AuxREs in the soybean GH3 promoter (Liu et al., 1994). Furthermore, at positions −85 and −61 are putative TGTCTC-like elements, with the sequence TGTTTC separated by a T-rich tract. Because conservation at positions 1 to 4 is more important than at positions 5 and 6, these elements may be nonfunctional (Ulmasov et al., 1997).
pls Mutants Show Hyperresponsiveness to Cytokinin and Reduced Responsiveness to Auxin
Reduced longitudinal cell expansion and increased radial expansion, seen in the pls mutant root, can be caused by increased accumulation or sensitivity to any of several hormones, including ethylene (Abeles et al., 1992), auxin (Ljung et al., 2001), or cytokinin (Beemster and Baskin, 2000). To investigate the sensitivity of pls root growth to exogenous hormones, homozygous mutant and wild-type seedlings were grown in the presence of different concentrations of exogenous auxin (2,4-D, 100 pM to 100 nM; 1-NAA, 100 pM to 250 nM), cytokinin (benzyladenine [BA], 100 pM to 1 μM), and the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC; 10 pM to 10 nM).
For auxin treatment, 3-day-old seedlings were transferred from hormone-free medium to plates containing auxin for another 3 days in the light. For cytokinin and ACC treatments, seedlings were grown continuously in the presence of either BA or ACC for 7 days after germination in the dark (conditions used previously to identify mutants defective in cytokinin and ethylene signaling) (Vogel et al., 1998). Changes in primary root lengths of mutant and wild-type seedlings were determined (Table 1).
Table 1.
Changes in Primary Root Lengths of pls and Wild-Type C24 Seedlings after 7 Days of Continuous Growth on Cytokinin (BA) and ACC and after Transfer at 3 Days after Germination from Hormone-Free to Auxin (2,4-D, 1-NAA)–Containing Medium for another 3 Days
| C24 (n = 10)
|
pls (n = 10)
|
|||
|---|---|---|---|---|
| Mean Root Length (mm ± se) | Percent Change from Control | Mean Root Length (mm ± se) | Percent Change from Control | |
| 1/2MS10 | 18.0 ± 0.7 | 100 | 11.6 ± 0.4 | 100 |
| BA (100 pM) | 17.9 ± 0.7 | 99.4 | 9.2 ± 0.4 | 79.3 |
| BA (1 nM) | 17.3 ± 0.8 | 96.1 | 9.4 ± 0.4 | 81.0 |
| BA (10 nM) | 16.9 ± 0.8 | 93.9 | 8.3 ± 0.4 | 71.6 |
| BA (100 nM) | 9.6 ± 0.2 | 53.3 | 4.9 ± 0.3 | 42.2 |
| BA (1μM) | 9.0 ± 0.3 | 50.0 | 3.3 ± 0.3 | 28.4 |
| ACC (10 pM) | 14.6 ± 0.8 | 81.1 | 8.9 ± 0.3 | 76.7 |
| ACC (100 pM) | 11.3 ± 0.5 | 62.8 | 7.4 ± 0.4 | 63.8 |
| ACC (1 nM) | 7.2 ± 0.2 | 40.0 | 4.8 ± 0.3 | 41.4 |
| ACC (10 nM) | 3.5 ± 0.2 | 19.4 | 1.9 ± 0.2 | 16.4 |
| 1/2MS10 | 36.0 ± 1.0 | 100 | 11.9 ± 0.7 | 100 |
| 2,4-D (100 pM) | 32.8 ± 0.6 | 91.1 | 15.1 ± 0.7 | 126.5 |
| 2,4-D (1 nM) | 30.5 ± 0.8 | 84.6 | 12.9 ± 0.6 | 108.4 |
| 2,4-D (10 nM) | 7.6 ± 0.4 | 21 | 5.5 ± 0.3 | 46 |
| 2,4-D (100 nM) | 2.3 ± 0.1 | 6.4 | 1.8 ± 0.1 | 15.1 |
| NAA (100 pM) | 33.6 ± 0.8 | 93.3 | 15.1 ± 0.5 | 126.9 |
| NAA (1 nM) | 34.5 ± 0.6 | 95.8 | 14.4 ± 0.6 | 120.6 |
| NAA (10 nM) | 30.3 ± 0.6 | 84 | 12.7 ± 0.3 | 106.3 |
| NAA (100 nM) | 11.4 ± 0.3 | 31.7 | 5.1 ± 0.2 | 42.9 |
| NAA (250 nM) | 5.6 ± 0.1 | 15.6 | 3.4 ± 0.1 | 28.2 |
The relative change in primary root length is shown as percent change compared with control seedlings grown on hormone-free medium (1/2MS10). The seedings treated with BA and ACC were grown on horizontal agar plates, whereas the seedlings treated with auxins were grown on vertical agar plates, accounting for root length differences between controls of the respective treatments.
Both cytokinin and ACC treatments resulted in reduced primary root growth in both mutant and wild-type seedlings. Interestingly, the pls roots were proportionally shorter than wild-type roots in the presence of exogenous BA over a range of concentrations. On the lowest BA concentration tested (100 pM), there was an ∼20% reduction in pls root elongation, although the wild type was unaffected. There was no difference between the relative growth inhibition of pls and wild-type roots in the presence of ACC over the concentration range tested, although significant inhibitory effects were seen in both sets of seedlings.
However, there was a significantly reduced growth-inhibitory effect of auxin on the pls root compared with the wild-type root. Indeed, at 100 pM and 1 nM 2,4-D and 1-NAA, respectively, the roots of pls mutant seedlings were significantly longer than those of seedlings grown in the absence of exogenous auxin. These results suggest that the growth of the pls mutant primary root is hyperresponsive to the inhibitory effects of exogenous cytokinin, whereas the pls mutation causes a suppression of the growth-inhibitory effects of auxin on root length and, at the lowest auxin concentrations tested, leads to root growth enhancement.
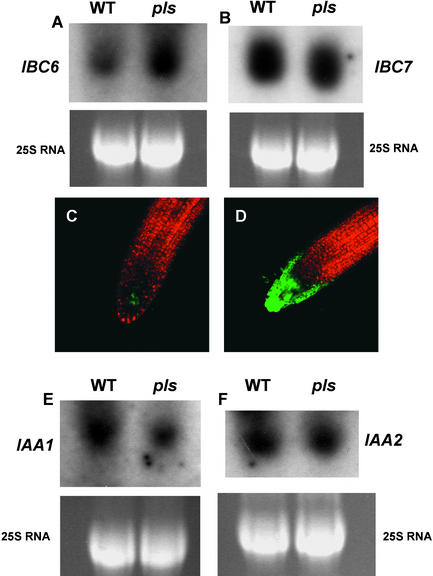
One prediction from the observation that the pls mutant exhibits increased cytokinin responsiveness would be that the expression of cytokinin-regulated genes might be increased in pls seedlings. To test this possibility, the expression levels of the IBC6/ARR5 and IBC7/ARR6 genes (Brandstatter and Kieber, 1998; D'Agostino et al., 2000), which are upregulated transcriptionally by cytokinin, were investigated in pls and wild-type seedlings in the absence of exogenous cytokinin.
Densitometric scanning of RNA gel blots showed that IBC6/ARR5 was upregulated threefold in the pls mutant, whereas IBC7 transcript was unchanged (Figures 6A and 6B). Consistent with this finding, the expression of a green fluorescent protein (GFP) gene driven by the IBC6/ARR5 gene promoter and introduced into the pls mutant background by crossing (to avoid position effects on the level of expression) was increased in the root tips of seedlings in a pls mutant background compared with expression in a wild-type background (Figures 6C and 6D). This result indicated that at least one gene, specifically regulated by cytokinins, is upregulated in the pls mutant.
Figure 6.
Gene Expression in a pls Mutant Background.
(A) and (B) RNA gel blot analysis of transcript abundances of ARR5/IBC6 (A) and ARR6/IBC7 (B) genes in wild-type (WT) and pls mutant seedlings 7 days after germination. 25S RNA is shown as a loading control. Each lane contained 10 μg of total RNA.
(C) and (D) ARR5/IBC6::GFP gene fusion activities in the root tips of transgenic wild-type (C) and pls mutant (D) seedlings 7 days after germination, viewed with the confocal microscope. Magnification ×100.
(E) and (F) RNA gel blot analysis of transcript abundances of IAA1 (E) and IAA2 (F) genes in wild-type and pls mutant seedlings 7 days after germination. 25S RNA is shown as a loading control. Each lane contained 10 μg of total RNA.
A prediction from the observation that the pls mutant showed reduced responses to exogenous auxin would be that the expression of auxin-inducible genes might be downregulated in pls seedlings compared with the wild type. To test this prediction, the expression levels of the IAA1 and IAA2 genes (Abel and Theologis, 1996), which are upregulated transcriptionally by auxin, were investigated in pls and wild-type seedlings in the absence of exogenous auxin. Densitometric scanning of RNA gel blots showed that IAA1 transcript abundance in the pls mutant was downregulated to ∼45% of the level found in wild-type seedlings, whereas the expression of IAA2 was unchanged (Figures 6E and 6F).
PLS cDNA Complements the pls Mutant Phenotype
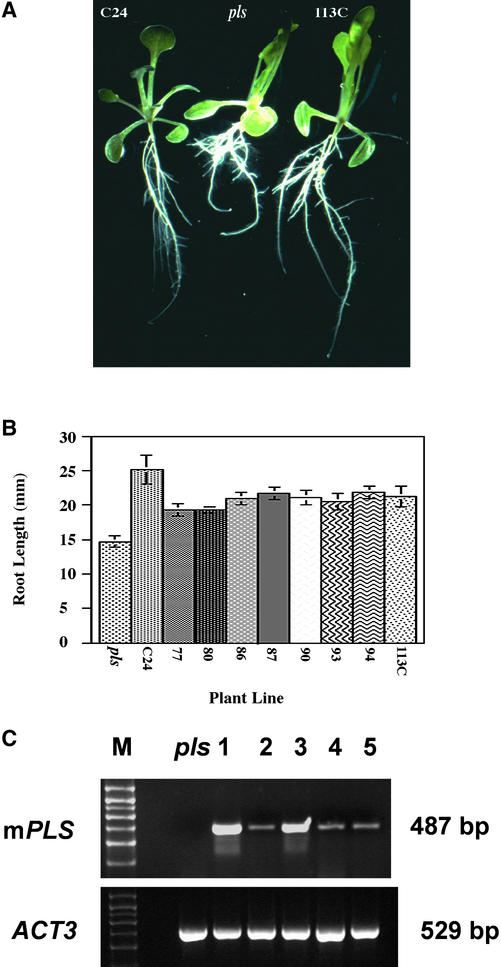
To confirm that mutation of the PLS gene is responsible for the observed short-root phenotype, the wild-type cDNA was introduced into the homozygous pls mutant under the transcriptional control of the 370-bp pPLS promoter. This promoter contained only a small region of the 3′ end of the GENE X ORF, which would not be expected to produce a functional protein. T1 transformants were selfed, and T2 and T3 seedlings representing individual families were analyzed for root phenotype after growth on hormone-free medium for 9 days. To ensure that no wild-type contaminants had been included accidentally, T2 seedlings from each potential complementing line were stained for PLS-GUS activity. All lines examined showed GUS staining in the root tips. RNA-specific PCR analysis of potentially complementing transgenic lines demonstrated that each showed expression of the wild-type PLS transcript, which was absent in the pls mutant (data not shown).
The complementation experiment showed that the short-root mutant phenotype was partially rescued by expression of the wild-type PLS allele (Figures 7A and 7B). For example, at 9 days after germination, pls mutant roots had a mean length of 14.7 ± 0.8 mm compared with 25.1 ± 2.1 mm for the wild type (n = 10). The mean root length of eight independent T3 complementing transgenic lines (Figure 7B) ranged from 19.3 ± 0.3 mm for line 77 to 21.65 ± 0.8 mm for line 87. These lines represent a mixture of homozygotes and hemizygotes for the complementing T-DNA. All eight transgenic lines had longer roots than those of the pls mutant. These data demonstrate the functionality of the PLS cDNA.
Figure 7.
Complementation of pls with the PLS ORF-Containing Partial cDNA.
(A) Fourteen-day-old seedlings of the wild type (C24), pls mutant, and complementation line 113C showing restoration of wild-type root architecture to the complemented mutant.
(B) Primary root length of pls, the wild type (C24), and eight independent complementation lines (77 to 113C) containing the partial PLS cDNA at 9 days after germination. All lines show significantly longer roots than the pls mutant.
(C) RT-PCR analysis of seedlings of the pls mutant (pls) and transgenic lines mPLS ORF 1 to 5 (lanes 1 to 5, respectively). The top gel shows amplification of the 487-bp mPLS ORF cDNA from RNA extracted from each of the five mPLS ORF transgenic lines but not from the pls mutant. The bottom gel shows amplification of the control ACT3 cDNA in pls and each mPLS ORF transgenic line. Control experiments lacking reverse transcriptase showed no amplification products (data not shown). M, molecular mass markers (kD).
To determine whether the PLS ORF is required for complementation activity, a mutant PLS cDNA was constructed in which the PLS ORF ATG codon was mutated to ATC, cloned behind the 35S promoter of Cauliflower mosaic virus, sequenced, and introduced into pls mutant plants. Mutation of the PLS ATG would prevent its recognition as a translation initiation codon. The sequence of the nine–amino acid ORF immediately upstream of the PLS ORF (Figure 3) was unchanged. Primary root length measurements were determined at 7 days after germination for seedlings that were (1) pls mutants, (2) transgenic for the wild-type PLS ORF transcript, and (3) transgenic for the mutant PLS ORF transcript.
The results, presented in Table 2, showed that although the wild-type PLS ORF transgenic seedlings had significantly longer roots than the pls mutant seedlings, all five independent lines transgenic for the mutant PLS ORF (designated mPLS ORF1 to ORF5) showed no significant difference in root length compared with the pls mutant seedling root. RT-PCR analysis confirmed that each of these lines expressed the mutant transcript, whereas the control (untransformed) pls seedlings showed no amplification product (Figure 7C). This finding strongly suggests that the functionality of the PLS gene requires a functional translation initiation codon in the PLS ORF.
Table 2.
Root Lengths of pls Seedlings, pls Transgenic Lines Containing a Wild-Type PLS ORF (PLS ORF), and Five Independent pls Transgenic Lines Containing a Mutated PLS ORF ATG Codon (mPLS ORFs) at 7 Days after Germination
| n | Mean Primary Root Length mm ± se | |
|---|---|---|
| pls | 20 | 16.6 ± 0.6 |
| PLS ORF | 20 | 20.3 ± 0.8 |
| mPLS ORF1 | 20 | 16.9 ± 0.5 |
| mPLS ORF2 | 20 | 16.4 ± 0.7 |
| mPLS ORF3 | 13 | 16.9 ± 1.1 |
| mPLS ORF4 | 21 | 17.7 ± 0.6 |
| mPLS ORF5 | 20 | 16.5 ± 0.4 |
n = number of seedlings measured per line. There was no significant difference between the pls mutant and any of the five pls transgenic lines containing the mutated PLS ORF ATG codon.
Overexpression of a Partial PLS cDNA Confers Reduced Root Growth Inhibition in the Presence of Exogenous Cytokinin and Increased Leaf Venation
The enhanced response of pls mutant roots to exogenous BA suggested that the PLS gene can negatively regulate cytokinin responses. A prediction would be that seedlings overexpressing the wild-type PLS gene would exhibit reduced responses to cytokinin.
To test this possibility, several independent transgenic lines (Columbia ecotype background) were produced that contained a partial (3′ region of the PLS) cDNA of 270 bp, which included the 9– and 36–amino acid ORFs, under the transcriptional control of the 35S promoter of Cauliflower mosaic virus. The short cDNA was used to determine whether the ORF-containing region of the transcript, rather than the entire cDNA, was sufficient for biological activity. Root length in the presence of exogenous BA was measured as an indicator of sensitivity to cytokinin.
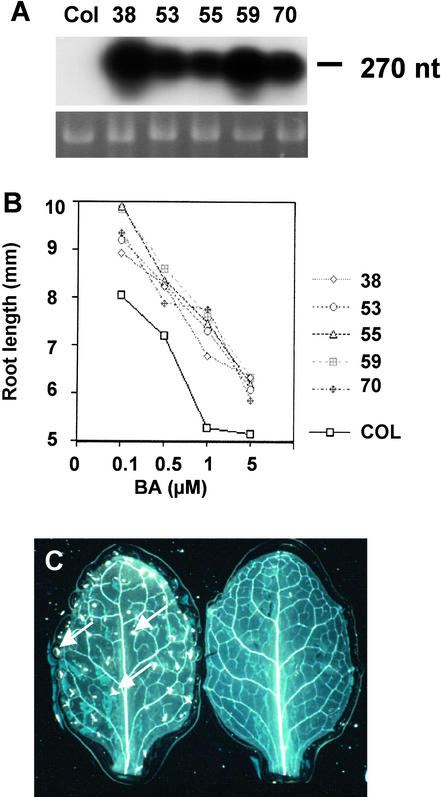
T4 seedlings of each of five independent transgenic lines overexpressed PLS transcript to relatively high levels (up to ∼100-fold) compared with the wild type (Figure 8A). Wild-type seedlings and T4 PLS overexpressers were grown for 7 days in the presence and absence of exogenous cytokinin. When grown in the absence of exogenous BA, there was no significant difference in primary root length between the control wild-type and transgenic seedlings (mean of 17.9 ± 2.4 mm [n = 20] for wild-type compared with 20.6 ± 1.9 mm [n = 100] for the overexpressers).
Figure 8.
Effects of Overexpressing a PLS ORF-Containing Partial cDNA in Wild-Type Seedlings.
(A) RNA gel blot analysis of wild-type (Columbia [Col]) and five independent transgenic overexpressers of the partial PLS cDNA (designated 38 to 70) probed with radiolabeled PLS cDNA. 25S RNA is shown as a loading control. Under the exposure conditions used, the autoradiograph does not reveal the low-abundance PLS transcript in the wild-type sample. Each lane contained 10 μg of total RNA.
(B) Primary root length of wild-type (Columbia) and five independent PLS-overexpressing lines grown for 7 days in the presence of 0.1 to 5 μM BA. Note that wild-type and overexpressing seedlings, grown in the absence of exogenous BA but under otherwise identical conditions, exhibited no significant difference in primary root length.
(C) Rosette leaves of wild-type (left) and PLS-overexpressing (right) seedlings 12 days after germination showing the increased frequency of higher order vascular strands and lack of trichomes in the overexpressers. Arrows indicate trichomes.
However, each of the PLS overexpressers exhibited significantly longer roots when grown in the presence of BA (Figure 8B). For example, on 1 μM BA, line 59 showed a mean root length of 7.6 ± 0.2 mm compared with 5.3 ± 0.3 mm for the wild type (n = 20; P = 5.4 × 10−6); that is, the root was 43% longer under these conditions. In particular, the slope of the graph (Figure 8B) suggests that the PLS overexpressers had the most reduced response to BA at between 0.5 and 1.0 μM. This finding demonstrates that the PLS gene acts to partially suppress the growth-inhibitory effects of cytokinins.
When grown for 7 days on 10 pM to 5.0 μM 1-NAA in the light, there was no significant difference in primary root length of the PLS overexpressers compared with that of wild-type seedlings (data not shown). However, a comparison of the vascular patterning of rosette leaves of wild-type lines (Columbia ecotype) and the PLS-overexpressing transgenic lines (also Columbia background) showed clearly that the overexpressers had more complex venation, with more higher order strands than the wild type (Figure 8C). It should be noted that Columbia ecotype rosette leaves had less complex venation than did leaves of C24 wild-type plants (cf. Figure 2D), so direct comparisons between different ecotypes cannot be made.
These results are consistent with the observation that pls mutant leaves had less complex venation than wild-type leaves (Figure 2D). In addition, the PLS-overexpressing lines also showed a dramatic loss of leaf trichomes compared with the Columbia wild type (Figure 8C). Although the wild-type Columbia leaves typically had ∼30 to 40 trichomes on the adaxial rosette leaf surface under the growth conditions used, the PLS overexpressers had none.
DISCUSSION
In this article, we describe the cloning of the Arabidopsis PLS gene that, when mutated, results in a short-root phenotype, altered vascular patterning in the leaf, and altered responses to exogenous cytokinins and auxins. Analysis of pls mutants, genetic complementation studies, and characterization of transgenic overexpressers strongly suggest that the gene encodes a small polypeptide that is required for correct development and cytokinin and auxin responses.
PLS Shows Unusual Organization
Eukaryotic genes typically are separated by noncoding regions containing regulatory elements. However, the PLS and GENE X transcripts overlap at their respective 5′ and 3′ UTRs. Partial complementation of the pls mutant phenotype was achieved using a genomic clone of PLS that lacked the predicted translational start of the GENE X ORF. Furthermore, analysis of the pPLS::GUS transgenic lines showed that the GENE X coding region contains sequences having promoter activity that correlates with the expression of PLS with respect to spatial pattern and auxin inducibility (Figures 3 to 5); GENE X and PLS encode transcripts of different sizes. It should be noted that this short promoter region is relatively weak (Figure 4C), accounting at least in part for the incomplete complementation, although transgene position effects also might affect the extent of complementation.
A similar gene organization has been reported for the EhMCM3 and EhPAK genes of Entamoeba histolytica (Gangopadhyay et al., 1997), which overlap and also may have two transcriptional start sites. The OTC and AUL1 genes of Arabidopsis also have a 22-bp overlap of their 3′ UTRs (Quesada et al., 1999). The CCT8 and TRP1 genes of Candida albicans overlap across the ORFs of both genes (Gerads and Ernst, 1998). Examples of gene overlap also exist in mouse and human (Speek et al., 1996; Koskimies et al., 1997).
Active RNA Versus Polypeptide?
An interesting question is whether the PLS gene encodes one or more functional polypeptides or a biologically active RNA molecule. Very few genes have been identified in plants that encode peptides that function in signaling, but PLS adds to the growing list of putative and actual plant peptide factors (reviewed in Lindsey et al., 2002). These include CLAVATA3 (Fletcher et al., 1999), the phytosulfokines (Matsubayashi et al., 1999), ENOD40 (Sousa et al., 2001), systemin (Pearce et al., 1991, 2001), the SCR proteins (Schopfer et al., 1999), and analogs of mammalian atrial natriuretic peptides (Billington et al., 1997).
We have yet to detect a PLS polypeptide by protein gel blot analysis using polyclonal antibodies to the N-terminal 18 amino acids. However, the genetic evidence demonstrates that a partial PLS cDNA that contains only the sequence encoding the three short ORFs (9, 8, and 36 amino acids) is functional and that the functionality of the cDNA requires that the PLS ORF has an ATG codon. Therefore, these data suggest that the PLS gene encodes a functional polypeptide rather than a biologically active RNA molecule.
The region of 5′ UTR between start sites 1 and 2 contains three very small ORFs of six, three, and three amino acids, respectively. Because start site 1 is used relatively infrequently and the ORFs are so small, they may have little functional significance. However, the 5′ UTR extending downstream from start site 2 contains two small ORFs of predicted nine and eight amino acids. Upstream ORFs (uORFs) are rare in eukaryote messages. The small sizes of the nine- and eight-residue polypeptides at the PLS locus suggest that they are unlikely to represent functional gene products. The position of the uORFs closer to the 5′ end of the PLS transcript means that they are likely to be identified by the ribosome before the larger PLS ORF.
Studies by Sousa et al. (2001) indicate that ORFs smaller than ∼30 amino acids may not be translated and do not inhibit the translation of a larger downstream ORF, and certainly the gusA ATG in the PLS promoter trap obviously is recognized and is functional. A number of studies in both plants (Wang and Wessler, 1998) and other eukaryotes (Kozak, 1989; Oliveira and McCarthy, 1995; Linz et al., 1997) indicate a role for uORFs in the regulation of both transcript stability and the expression of the main ORF. Therefore, the uORFs may play a role in controlling both PLS translation and also PLS transcript stability.
Polypeptide signaling molecules are common in animals and typically are cleaved from larger proproteins via such enzymes as proprotein convertases, endoproteases, and carboxypeptidases (Canaff et al., 1999). Similar processing enzymes now are known to be required for correct development in plants (Berger and Altmann, 2000; Helliwell et al., 2001; Li et al., 2001). The amino acid sequence and predicted structure of PLS suggest processing of the N terminus, with the C terminus mediating interactions with other proteins. Studies using the yeast two-hybrid system suggest that the C-terminal domain of PLS interacts with other proteins (P.M. Chilley and K. Lindsey, unpublished data). The potential for cleavage, and the possible interaction with other proteins that might produce an immunologically distinct complex, may explain why the predicted polypeptide was undetectable.
PLS Gene Expression Is Auxin Inducible
The PLS gene promoter, which contains potential TGTCTC-like AuxREs, is activated by functional auxins but not by nonfunctional analogs. This auxin inducibility is consistent with the pattern of PLS gene transcription. PLS promoter activity is strongest in the root tip, and the root tip is a site of relatively high auxin concentration or sensitivity (Sabatini et al., 1999). GUS histochemistry indicates that the greatest activity of the PLS promoter is in the columella initials, similar to that of the auxin-inducible DR5 promoter (Sabatini et al., 1999). PLS also is expressed strongly at the site of lateral root initiation (Topping and Lindsey, 1997), a process that is induced by auxin (Celenza et al., 1995; Tian and Reed, 1999). PLS also is expressed in aerial organs, albeit at lower levels than in the root. We conclude that auxin is a key component of the signaling system that regulates the spatial patterning of PLS expression.
PLS Function
Phenotypic analysis of the pls mutant and PLS overexpressers indicates that PLS is required for correct responses to exogenous cytokinins and auxins, correct root growth, and correct vascular patterning and trichome development of the rosette leaf. The PLS-overexpressing transgenic lines (in the Columbia background) were found to lack trichomes, indicating that PLS acts as a negative regulator of trichome development in this ecotype, but at present, we do not understand the mechanistic basis for this. Ecotype C24, the background in which the pls mutant was identified, has no trichomes, and no effect of the mutation was discernible. Reduced levels of PLS expression were found to limit root growth, as seen by the semidominant effect of the mutation.
However, there was not a linear relationship between PLS expression and root growth: the PLS overexpressers did not have longer roots in the presence of exogenous BA in linear proportion to the level of PLS expression. Furthermore, the PLS overexpressers did not show enhanced responsiveness to the root-inhibitory effects of exogenous 1-NAA. It is possible that the PLS gene product interacts with one or more other components to modulate root growth, which may become rate limiting at high PLS concentrations (such as in the PLS overexpressers).
Because cytokinins are synthesized in root tips and are at relatively high concentrations in roots (Sossountov et al., 1988; Chiappetta et al., 2001), we can speculate that a mechanism exists to reduce the sensitivity of cells at the root tip to cytokinins to suppress their growth-inhibitory effects. This system may involve the gene STUNTED PLANT1 (Baskin et al., 1995), and PLS also may encode a component of this mechanism.
Supporting this hypothesis are the observations that (1) the PLS gene is expressed strongly in root tips, (2) the pls mutant exhibits enhanced root growth inhibition in the presence of the cytokinin BA, (3) overexpression of the partial PLS cDNA confers reduced growth inhibition by exogenous BA, and (4) the pls mutant exhibits increased expression of the cytokinin-inducible ARR5/IBC6::GFP gene, which is expressed in the root tip (Brandstatter and Kieber, 1998). Therefore, the mutant is altered in its responses to cytokinins, so PLS might play a role in suppressing cell sensitivity to cytokinins or in cytokinin biosynthesis.
Plant development involves complex interactions between the “classic five” hormones: cytokinin, ethylene, auxin, gibberellin, and abscisic acid. In the case described here, PLS is activated transcriptionally by auxin, and the gene product appears to be required for correct responses to cytokinins and auxin. Only subsets of auxin- and cytokinin-regulated genes are altered in the pls mutant (i.e., ARR5/IBC6 but not ARR6/IBC7, and IAA1 but not IAA2), suggesting specificity of action. Furthermore, both cytokinins and auxin can induce ethylene biosynthesis (Vogel et al., 1998), and both cytokinins and auxin can inhibit root growth at high concentrations (Cary et al., 1995).
It also is possible that the observed increase in cytokinin-mediated responses in the pls mutant may be mediated by downstream ethylene effects or reduced auxin responses, because auxins and cytokinins interact and often have apparently antagonistic effects, such as in shoot branching, root branching, and vascularization (Ljung et al., 2001).
This antagonism with auxin is suggested by the enhanced responsiveness of pls to exogenous cytokinin, the reduced growth inhibition in the presence of exogenous auxins, and the upregulation of the ARR5/IBC6 gene and the downregulation of the IAA1 gene in the pls mutant. Because it is known that auxin plays an essential role in the patterning of vascular tissues (Przemeck et al., 1996; Hardtke and Berleth, 1998), possible alterations to auxin–cytokinin interactions in the pls mutant, which are attributable to increased cytokinin levels or sensitivity or reduced auxin levels or sensitivity, could account for the observed reduced vascularization in the pls leaf.
In support of this model of modified cytokinin-auxin responses in the pls mutant, we have found (P.M. Chilley, S.A. Casson, and K. Lindsey, unpublished data) that the pls mutation acts as a phenotypic suppressor of the rooty/superroot mutant, which is defective in auxin homeostasis (Boerjan et al., 1995; King et al., 1995). Furthermore, the cytokinin-insensitive cin5 mutant (Vogel et al., 1998) has a phenotype showing characteristics that contrast with those of the cytokinin-sensitive pls mutant (e.g., no triple response in the presence of exogenous cytokinins in the dark, compared with the enhanced triple response of pls in these conditions; P.M. Chilley, S.A. Casson, and K. Lindsey, unpublished data). The results presented here support the model that the PLS gene is required for correct auxin-cytokinin homeostasis to modulate root growth and leaf vascular patterning. Current experiments are aimed at further distinguishing between the effects of PLS expression on auxin, cytokinin, and ethylene interactions.
METHODS
Materials and Growth Conditions
The transgenic line AtEM101 (Arabidopsis thaliana ecotype C24) contains the promoter trap pΔgusBin19 (Topping et al., 1991, 1994). Arabidopsis seeds transgenic for the IBC6/ARR5::green fluorescent protein gene fusion were kindly provided by Joe Kieber (University of North Carolina, Chapel Hill). For in vitro growth studies, seeds were vernalized and surface-sterilized (Clarke et al., 1992) and plated on growth medium (half-strength Murashige and Skoog [1962] medium [Sigma], 1% Suc, and 0.8% agar [Difco, Detroit, MI]) at 22 ± 2°C at a PPFD of ∼150 μmol·m−2·s−1. For hormone application experiments, seeds were germinated aseptically on growth medium containing various concentrations of hormones and assayed according to the particular experiment.
Gene Expression Analysis
Tissue localization of β-glucuronidase (GUS) enzyme activity was performed as described (Topping and Lindsey, 1997). For transcript analysis, RNA was extracted using the RNeasy Plant RNA Extraction kit (Qiagen Ltd., Surrey, UK) and the PolyATract mRNA isolation system (Promega, Southampton, UK). RNA was blotted, hybridized, and probed as described (Wei et al., 1997). RNA markers were Promega G319. PLS gene sequences were used to design PCR primers for rapid amplification of cDNA ends (RACE) PCR and to generate a 299-bp sequence for RNase protection assays (RPAs).
RPAs were performed using the RPA III kit (Ambion, Austin, TX) using either 50 μg of total RNA or 5 μg of poly(A)+ RNA mixed with 4 × 105 cpm of labeled RNA probe. Reaction samples were separated on a mini denaturing polyacrylamide gel, transferred to Whatman paper, and exposed to x-ray film for 1 to 7 days without drying. RNA probes for RPAs were made using the MAXIscript T7/T3 In Vitro Transcription kit (Ambion). Templates for the transcription reaction were prepared by cloning DNA fragments into the pCR2.1-TOPO vector (Invitrogen, Groningen, The Netherlands).
For RNA-specific (RS)–PCR, the following oligonucleotide primers were used: 5′-CTTATACGGATATCCTGGCAATTCGGACTTGATAGGGTGATCAATGGA-3′ (the underlined region is complementary to the 3′ end of the PLS transcript), 5′-CTTATACGGATATCCTGG-CAATTCGGACTT-3′, and 5′-GGAGACTAAAGCGAACATATAAAACC-3′. Genes for RNA gel blot analysis were isolated by reverse transcription (RT)–PCR from RNA isolated from 7-day-old C24 seedlings. Reverse transcription was performed with an oligo(dT) primer using 15 μg of total RNA essentially as described previously (Althorpe et al., 1999).
Primers used for amplification were as follows: for ARR5/IBC6, 5′-CACGAGTCACGATCCTACTC-3′ and 5′-CAGGACATGCATGTG-TGTG-3′; for ARR6/IBC7, 5′-CATCGAGAGATTGCTTCG-3′ and 5′-CGACGACGACGTCAACAC-3′; for IAA1, 5′-CTTAAGGACACAGAGCTTCG-3′ and 5′-GATCCTTTCATGATTCTGAG-3′; and for IAA2, 5′-GAGGCAATAGAGATGGAC-3′ and 5′-GTCTAGAGCAGG-AGCGTCG-3′. For mutant (m)PLS open reading frame (ORF) RT-PCR, the primers 5′-TATCTAGACCTTTATCGAGACATAAGATTG-3′ and 5′-ATGGATCCACTTAATATATTAGTATTGG-3′ were used; and for ACT3, ACT For (5′-GATCCTAACCGAGCGTGGTTAC-3′) and ACT Rev (5′-GACCTGACTCGTCATACTCTGC-3′) were used.
mPLS ORF and ACT3 RT-PCR reaction conditions were 94°C for 2 min followed by 30 cycles of 94°C denaturation for 30 s, 60°C primer annealing for 30 s, 72°C extension for 60 s, and a final extension at 72°C for 7 min. RNA gel blot analysis was performed using 50 μg of total RNA isolated from 7-day-old wild-type and pls seedlings.
Gene Cloning and DNA Analysis
The Arabidopsis Columbia λGEM 11 genomic library (Ronald Davis, Stanford, University, Stanford, CA) was provided by Jeff Dangl (formerly of the European Commission Arabidopsis T-Project DNA Centre, Köln, Germany) and screened as described previously (Wei et al., 1997). Arabidopsis DNA was extracted using cetyl-trimethyl-ammonium bromide (Ausubel et al., 1996) purified with the Qiagen genomic-tip 100/G, as described previously (Wei et al., 1997). For 3′ and 5′ RACE-PCR cDNA synthesis, the oligo(dT) primer was 5′-CCAAGCTTCTGCAGGAGCTCTTTTTTTTTTTTTTT-3′. 5′ RACE-PCR of the PLS transcript was performed on poly(A)+ and total RNA from 7-day-old seedlings using primers 5′-CCAGGTGTTCGGCGTGGT-GTAGAGC-3′ (for the GUS fusion transcript) and 5′-GGTTTCATT-CATGTTTCAGTGAG-3′. Products were cloned as described previously (Wei et al., 1997).
3′ RACE of the PLS transcript was performed on total RNA from 7-day-old seedlings using primers 5′-CCAAGCTTCTGCAGGAGCTC-3′ (3′ RACE anchor primer) and 5′-GGAACACGAAATCCGAAGAGCGAG-3′ as follows: 94°C for 2 min followed by 35 cycles of 94°C denaturation for 30 s, 60°C primer annealing for 30 s, 72°C extension for 45 s, and a final extension at 72°C for 7 min. DNA sequencing was performed using an ABI 373 DNA sequencer (Applied Biosystems, Foster City, CA) and dye terminator labeling reactions as described previously (Wei et al., 1997). DNA was prepared for DNA gel blot analysis and PCR amplification from ∼1 g wet weight of plant tissue using the Phytopure kit (Nucleon Biosciences, Lanarkshire, UK). DNA gel blot analyses were performed as described (Wei et al., 1997).
Nucleotide and deduced protein sequences were used to search for homologies in the National Center for Biotechnology Information peptide sequence databases using the BLAST network service (Altschul et al., 1990). Protein motif homologies were determined using the ScanProsite program (http://www.expasy.ch/tools/scnpsit1.html). The putative PLS polypeptide secondary structure was modeled using the Protean program (Lasergene Navigator; DNAStar, Madison, WI).
Competitor RS-PCR
Construction of the PLS Competitor
A ClaI restriction site was introduced into PLS by PCR. This also mutated the PLS ORF ATG codon for functional analysis of the ORF in transgenic studies. Using a plasmid genomic clone of the PLS locus, two overlapping fragments of the PLS gene were produced by PCR. The first fragment of ∼300 bp was amplified with the primers BamHI Transc. For (5′-ATGGATCCACTTAATATATTAGTATTGG-3′) and ClaI Mut Rev (5′-CTGGGTTATCGATTCATGTTTCAGTGAGAC-3′; the engineered ClaI site is underlined), and the second fragment of ∼150 bp was amplified with the primers ClaI Mut For (5′-CATGAATCG-ATAACCCAGACTTTGTTTTAATTTCAG-3′) and XbaI Transc. Rev (5′-TATCTAGAGTAGATATTGAAAATGATAGG-3′).
The fragments were digested with ClaI, ligated, and used to amplify a mutant form of PLS carrying a ClaI site using the primers BamHI Transc. For and XbaI Transc. Rev. Sequencing confirmed the inclusion of a ClaI site. The PLS competitor was cloned into the pCR2.1-TOPO vector (Invitrogen) in the sense orientation with respect to the T7 RNA polymerase promoter. In vitro transcription of the PLS competitor was performed using the Maxiscript kit (Ambion) according to the manufacturer's instructions. Dilutions of the PLS competitor RNA were titrated against a fixed quantity of total RNA (10 μg), according to the method described below, to determine the dilution at which the quantity of both PLS and competitor were approximately the same after PCR amplification.
DNase Treatment of RNA
DNase treatment of RNA for PCR amplification was based on the method of Sanyal et al. (1997). To 10 μg of total RNA was added 3 μL of the PLS competitor RNA (between 1 × 105 and 1 × 106 dilution), followed by 2 μL of 5 × RT buffer (supplied with reverse transcriptase from Moloney murine leukemia virus; Promega) and 2 units of RQ1-DNase (RNase free; Promega). The volume was made up to 10 μL, and the mixture was incubated at room temperature for 15 min. DNase was inactivated by the addition of 1 μL of 25 mM EGTA, pH 8.0, and heating at 65°C for 10 min.
Reverse Transcription
For reverse transcription, 2 μL of the primer POL-RS-PCR (5′-CTTATACGGATATCCTGGCAATTCGGACTTGATAGGGTGATCAAT-GGA-3′) (10 pmol/μL) was added to the DNased RNA, heated for 10 min at 70°C, and placed on ice. To a RT master mixture (3 μL of 5 × Avian myeloblastosis virus buffer, 2 μL of deoxynucleotide triphosphate [12.5 mM], 1 μL [40 units] of RNasin, 2 μL [20 units] of reverse transcriptase from Moloney murine leukemia virus, and 6.5 μL of water) was added 6.5 μL of the RNA:primer mixture, and the reaction was incubated at 42°C for 1 h. A negative RT reaction was performed on the remaining RNA:primer mixture. cDNA was purified using the High Pure Product Purification kit (Roche Biochemicals, Mannheim, Germany) and eluted in a volume of 50 μL.
PCR
Five microliters of the eluted cDNA was used as a template for PCR amplification using the primers (10 pmol/μL) 5′TEST (5′-GGAGACTAAAGCGAACATATAAAACC-3′) and RS-PCR AD (5′-CTTATACGG-ATATCCTGGCAATTCGGACTT-3′). Hot-start PCR was used with 40 cycles of 94°C for 30 s, 63°C for 30 s, 72°C for 60 s, and a final extension of 72°C for 10 min.
Purification and Digestion of PCR Products
PCR products were purified using the High Pure Product Purification kit (Roche Biochemicals) and eluted in a volume of 100 μL. The products then were precipitated in ethanol (one-tenth volume of sodium acetate 4M, pH 6, and 2.5 volumes of ethanol) and resuspended in 10 μL of water. Digestion was performed in a total volume of 25 μL of 1 × reaction buffer and 10 units of ClaI at 37°C for 2 h. Products then were analyzed by electrophoresis on a 2% TAE gel stained with ethidium bromide.
PCR Amplification of ACTIN3 as a Loading Control
To determine that RNA loading was correct for each reaction, PCR amplification of the ACTIN3 cDNA was performed. DNase treatment of the RNA was performed as described. Reverse transcription was performed with an oligo(dT)15 primer, and the cDNA was purified as described. Five microliters of the eluted cDNA was used as a template for PCR amplification using the primers ACT For and ACT Rev (10 pmol/μL). Hot-start conditions were used with 20 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 45 s, and a final extension of 72°C for 10 min.
Gene Constructs and Plant Transformation
Promoter fragments were amplified using the Expand High Fidelity PCR system and ligated into T-tailed pBluescript SK− or pCR2.1-TOPO vector (Invitrogen), then transferred to pΔGUS (Topping et al., 1991). The promoter–GUS–nopaline synthase terminator cassette was transferred as a HindIII-EcoRI fragment into the binary vector pCIRCE (a gift from M. Bevan, John Innes Centre, Norwich, UK). For complementation analysis, a 1.15-kb DNA fragment, cognate to the PLS transcript and native promoter region, was amplified from a 3.6-kb genomic clone, introduced into the pCR2.1-TOPO vector (Invitrogen), and then transferred to the binary vector pMOG1006 (a gift from Mogen, Leiden, The Netherlands). For overexpression studies, a 270-bp partial PLS cDNA containing the 36–amino acid ORF was cloned behind the 35S promoter of Cauliflower mosaic virus in pCIRCE. All constructs were validated by sequencing. Plant transformation was performed by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens C58C3 (Dale et al., 1989).
Microscopy
Tissues were cleared and mounted for light microscopy in chloral hydrate (Topping and Lindsey, 1997) or 20% glycerol. To reveal leaf vascular tissues, leaves were cleared by incubating at 70°C for 30 min in 90% ethanol, followed by incubation at 70°C for 30 min in lactic acid:phenol:glycerol:water (1:1:1:1) and viewed under dark-field illumination (Telfer and Poethig, 1994). Photographs were taken on Ektachrome 160 tungsten-balanced film using Nikon Optiphot (Tokyo, Japan), Zeiss Axioskop (Jena, Germany), and Olympus SZH10 (Tokyo, Japan) microscopes. Confocal images were taken with a Bio-Rad Radiance 2000 microscope after counterstaining of tissues with 10 μg/mL propidium iodide. Images were processed in Adobe Photoshop 5.0 (Mountain View, CA).
Accession Number
The GenBank accession number for the predicted PLS transcript is AF285768.
Acknowledgments
We are grateful for financial support from the Biotechnology and Biological Sciences Research Council, the European Commission (FPIV contract BIO 4 CT 960217), and The Gatsby Charitable Foundation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002618.
References
- Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles, F., Morgan, P., and Saltveit, M. (1992). Ethylene in Plant Biology. (San Diego, CA: Academic Press).
- Althorpe, N.J., Chilley, P.M., Thomas, A.T., Brammar, W.J., and Wilkins, B.M. (1999). Transient transcriptional activation of the IncI1 plasmid anti-restriction gene (ardA) and SOS inhibition gene (psiB) early in conjugating recipient bacteria. Mol. Microbiol. 31, 133–142. [DOI] [PubMed] [Google Scholar]
- Altmann, T. (1999). Molecular physiology of brassinosteroids revealed by the analysis of mutants. Planta 208, 1–11. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Meyers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1996). Current Protocols in Molecular Biology. (New York: Wiley Interscience).
- Baskin, T.I., Cork, A., Williamson, R.E., and Gorst, J.R. (1995). STUNTED PLANT 1, a gene required for expansion in rapidly elongating but not dividing cells and mediating root growth responses to applied cytokinin. Plant Physiol. 107, 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster, G.T.S., and Baskin, T.I. (2000). STUNTED PLANT 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root of Arabidopsis. Plant Physiol. 124, 1718–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, M.J., Marchant, A., Green, H.G., May, S.T., Ward, S.P., Millner, P.A., Walker, A.R., Schulz, B., and Feldmann, K.A. (1996). Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 273, 948–950. [DOI] [PubMed] [Google Scholar]
- Berger, D., and Altmann, T. (2000). A subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 14, 1109–1131. [PMC free article] [PubMed] [Google Scholar]
- Billington, T., Pharmawati, M., and Gehring, C.A. (1997). Isolation and immunoaffinity purification of biologically active plant natriuretic peptide. Biochem. Biophys. Res. Commun. 235, 722–725. [DOI] [PubMed] [Google Scholar]
- Boerjan, W., Cervera, M.-T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., Caboche, M., Van Onckelen, H., Van Montagu, M., and Inzé, D. (1995). superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7, 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstatter, I., and Kieber, J.J. (1998). Two genes with similarity to bacterial response regulators are rapidly and specifically induced by cytokinin in Arabidopsis. Plant Cell 10, 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canaff, L., Bennett, H.P., and Hendy, G.N. (1999). Peptide hormone precursor processing: Getting sorted? Mol. Cell. Endocrinol. 156, 1–6. [DOI] [PubMed] [Google Scholar]
- Cary, A.J., Liu, W., and Howell, S.H. (1995). Cytokinin action is coupled to ethylene in its effects on the inhibition of root and hypocotyl elongation in Arabidopsis thaliana seedlings. Plant Physiol. 107, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza, J.L., Grisafi, P.L., and Fink, G.R. (1995). A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 9, 2131–2142. [DOI] [PubMed] [Google Scholar]
- Chen, R., Hilson, P., Sedbrook, J., Rosen, E., Caspar, T., and Masson, P.H. (1998). The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. USA 95, 15112–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappetta, A., De Witte, W., Racchi, M.L., Bitonti, M.B., Van Onckelen, H., and Innocenti, A.M. (2001). Altered cytokinin distribution in the shootless maize mutant ed*41. Aust. J. Plant Physiol. 28, 307–313. [Google Scholar]
- Clarke, M.C., Wei, W., and Lindsey, K. (1992). High frequency transformation of Arabidopsis thaliana by Agrobacterium tumefaciens. Plant Mol. Biol. Rep. 10, 178–189. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- D'Agostino, I.B., Deruere, J., and Kieber, J.J. (2000). Characterization of the response of the Arabidopsis response regulator gene family to cytokinin. Plant Physiol. 124, 1706–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale, P.J., Marks, M.S., Brown, M.M., Woolston, C.J., Gunn, H.V., Mullineaux, P.M., Lewis, D.M., Kemp, J.M., and Chen, D.F. (1989). Agroinfection of wheat: Inoculation of in vitro grown seedlings and embryos. Plant Sci. 63, 237–245. [Google Scholar]
- Dolan, L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., and Scheres, B. (1993). Cellular organisation of the Arabidopsis thaliana root. Development 119, 71–84. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher, J.C., Brand, U., Running, M.P., Simon, R., and Meyerowitz, E.M. (1999). Signalling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. [DOI] [PubMed] [Google Scholar]
- Fridborg, I., Kuusk, S., Moritz, T., and Sundberg, E. (1999). The Arabidopsis dwarf mutant shi exhibits reduced gibberellin responses conferred by overexpression of a new putative zinc finger protein. Plant Cell 11, 1019–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangopadhyay, S.S., Ray, S.S., Sinha, P., and Lohia, A. (1997). Unusual genome organisation in Entamoeba histolytica leads to two overlapping transcripts. Mol. Biochem. Parasitol. 89, 73–83. [DOI] [PubMed] [Google Scholar]
- Gerads, M., and Ernst, J.F. (1998). Overlapping coding regions and transcriptional units of two essential chromosomal genes (CCT8, TRP1) in the fungal pathogen Candida albicans. Nucleic Acids Res. 26, 5061–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann, T., Mayer, U., and Jürgens, G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development 126, 1387–1395. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell, C.A., Chin-Atkins, A.N., Wilson, I.W., Chapple, R., Dennis, E.S., and Chaudhury, A. (2001). The Arabidopsis AMP1 gene encodes a putative glutamate carboxypeptidase. Plant Cell 13, 2115–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. [DOI] [PubMed] [Google Scholar]
- Inoue, T., Higuchi, M., Hashimoto, Y., Seki, M., Kobayashi, M., Kato, T., Tabata, S., Shinozaki, K., and Kakimoto, T. (2001). Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature 409, 1060–1063. [DOI] [PubMed] [Google Scholar]
- Kakimoto, T. (1998). Cytokinin signalling. Curr. Opin. Plant Biol. 1, 399–403. [DOI] [PubMed] [Google Scholar]
- King, J.J., Stimart, D.P., Fisher, R.H., and Bleecker, A.B. (1995). A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell 7, 2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskimies, P., Spiess, A.-N., Lahti, P., Huhtaniemi, I., and Ivell, R. (1997). The mouse relaxin-like factor gene and its promoter are located within the 3′ region of the JAK3 genomic sequence. FEBS Lett. 419, 186–190. [DOI] [PubMed] [Google Scholar]
- Kozak, M. (1989). The scanning model for translation: An update. J. Cell Biol. 108, 229–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser, O., and Berleth, T. (1999). A molecular basis for auxin action. Cell Dev. Biol. 10, 131–137. [DOI] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Li, J., Lease, K.A., Tax, F.E., and Walker, J.C. (2001). BRS1, a serine carboxypeptidase, regulates BRI1 signalling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98, 5916–5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the Axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey, K., Casson, S., and Chilley, P. (2002). Peptides: New signalling molecules in plants. Trends Plant Sci. 7, 78–83. [DOI] [PubMed] [Google Scholar]
- Lindsey, K., Topping, J.F., and Wei, W. (1998). Identification of plant genes by entrapment and activation tagging. In Transgenic Plant Research, K. Lindsey, ed (Reading, PA: Harwood Academic), pp. 75–90.
- Linz, B., Koloteva, N., Vasilescu, S., and McCarthy, J.E.G. (1997). Disruption of ribosomal scanning on the 5′-untranslated region, and not restriction of translational initiation per se, modulates the stability of nonaberrant mRNAs in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 272, 9131–9140. [DOI] [PubMed] [Google Scholar]
- Liu, Z.-B., Ulmasov, T., Shi, X., Hagen, G., and Guilfoyle, T. (1994). Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung, K., Bhalerao, R.P., and Sandberg, G. (2001). Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative development. Plant J. 28, 465–474. [DOI] [PubMed] [Google Scholar]
- Luschnig, C., Gaxiola, R.A., Grisafi, P., and Fink, G.R. (1998). EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12, 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi, Y., Morita, A., Matsunaga, E., Furuya, A., Hanai, N., and Sakagami, Y. (1999). Physiological relationships between auxin, cytokinin, and a peptide growth factor, phytosulfokine-α, in stimulation of asparagus cell proliferation. Planta 207, 559–565. [Google Scholar]
- McGrath, R.B., and Ecker, J.R. (1998). Ethylene signalling in Arabidopsis: Events from the membrane to the nucleus. Plant Physiol. Biochem. 36, 103–113. [Google Scholar]
- Muller, A., Guan, C., Gälweiler, L., Tanzler, P., Huijser, P., Marchant, A., Parry, G., Bennett, M., Wisman, E., and Palme, K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17, 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Oliveira, C.C., and McCarthy, J.E.G. (1995). The relationship between eukaryotic translation and mRNA stability. J. Biol. Chem. 270, 8936–8943. [DOI] [PubMed] [Google Scholar]
- Pearce, G., Moura, D.S., Stratmann, J., and Ryan, C.A. (2001). Production of multiple plant hormones from a single polyprotein precursor. Nature 411, 817–820. [DOI] [PubMed] [Google Scholar]
- Pearce, G., Strydom, D., Johnson, S., and Ryan, C.A. (1991). A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253, 895–898. [DOI] [PubMed] [Google Scholar]
- Przemeck, G.K.H., Mattson, J., Hardtke, C.S., Sung, Z.R., and Berleth, T. (1996). Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200, 229–237. [DOI] [PubMed] [Google Scholar]
- Quesada, V., Ponce, M.R., and Micol, J.L. (1999). OTC and AUL1, two convergent and overlapping genes in the nuclear genome of Arabidopsis thaliana. FEBS Lett. 461, 101–106. [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi, C., Huntley, R., Jacqmard, A., and Murray, J.A.H. (1999). Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283, 1541–1544. [DOI] [PubMed] [Google Scholar]
- Sabatini, S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J., Benfey, P., Leyser, O., Bechtold, N., Weisbeek, P., and Scheres, B. (1999). An auxin-dependent organizer of pattern and polarity in the Arabidopsis root. Cell 99, 463–472. [DOI] [PubMed] [Google Scholar]
- Sanyal, A., O'Driscoll, S.W., Bolander, M.A., and Sarkar, G. (1997). An effective method of completely removing contaminating genomic DNA from an RNA sample to be used for PCR. Mol. Biotechnol. 8, 135–137. [DOI] [PubMed] [Google Scholar]
- Schopfer, C.R., Nasrallah, M.E., and Nasrallah, J.B. (1999). The male determinant of self-incompatibility in Brassica. Science 286, 1697–1700. [DOI] [PubMed] [Google Scholar]
- Sossountov, L., Maldiney, R., Sotta, B., Sabbagh, I., Habricot, Y., Bonnet, M., and Migniac, E. (1988). Immunocytochemical localization of cytokinins in Craigella tomato and a sideshootless mutant. Planta 175, 291–304. [DOI] [PubMed] [Google Scholar]
- Sousa, C., Johansson, C., Charon, C., Manyani, H., Sautter, C., Kondorosi, A., and Crespi, M. (2001). Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame-containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol. Cell. Biol. 21, 354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speek, M., Barry, F., and Miller, W.L. (1996). Alternate promoters and alternate splicing of human tenascin-X, a gene with 5′ and 3′ ends buried in other genes. Hum. Mol. Genet. 5, 1749–1759. [DOI] [PubMed] [Google Scholar]
- Telfer, A., and Poethig, R.S. (1994). Leaf development in Arabidopsis. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 389–401.
- Tian, Q., and Reed, J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721. [DOI] [PubMed] [Google Scholar]
- Topping, J.F., Agyeman, F., Henricot, B., and Lindsey, K. (1994). Identification of molecular markers of embryogenesis in Arabidopsis thaliana by promoter trapping. Plant J. 5, 895–903. [DOI] [PubMed] [Google Scholar]
- Topping, J.F., and Lindsey, K. (1997). Promoter trap markers differentiate structural and positional components of polar development in Arabidopsis. Plant Cell 9, 1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping, J.F., Wei, W., and Lindsey, K. (1991). Functional tagging of regulatory elements in the plant genome. Development 112, 1009–1019. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guifoyle, T.J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Vogel, J.P., Woeste, K.E., Theologis, A., and Kieber, J.J. (1998). Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. USA 95, 4766–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., and Wessler, S.R. (1998). Inefficient reinitiation is responsible for upstream open reading frame-mediated translational repression of the maize R gene. Plant Cell 10, 1733–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, W., Twell, D., and Lindsey, K. (1997). A novel nucleic acid helicase identified in Arabidopsis thaliana by promoter trapping. Plant J. 11, 1307–1314. [DOI] [PubMed] [Google Scholar]
- Woeste, K., and Kieber, J.J. (1998). The molecular basis of ethylene signalling in Arabidopsis. Philos. Trans. R. Soc. Lond. B 353, 1431–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]