Abstract
The molecular mechanisms underlying the initiation and maintenance of the embryonic pathway in plants are largely unknown. To obtain more insight into these processes, we used subtractive hybridization to identify genes that are upregulated during the in vitro induction of embryo development from immature pollen grains of Brassica napus (microspore embryogenesis). One of the genes identified, BABY BOOM (BBM), shows similarity to the AP2/ERF family of transcription factors and is expressed preferentially in developing embryos and seeds. Ectopic expression of BBM in Arabidopsis and Brassica led to the spontaneous formation of somatic embryos and cotyledon-like structures on seedlings. Ectopic BBM expression induced additional pleiotropic phenotypes, including neoplastic growth, hormone-free regeneration of explants, and alterations in leaf and flower morphology. The expression pattern of BBM in developing seeds combined with the BBM overexpression phenotype suggests a role for this gene in promoting cell proliferation and morphogenesis during embryogenesis.
INTRODUCTION
Embryogenesis is the starting point of the life cycle for both plants and animals. In plants, embryogenesis is not strictly dependent on fertilization, because many species naturally produce asexually derived embryos in the seed (apomixis) or can be induced to form embryos in tissue culture. Apomictic development is characterized by the avoidance of both meiosis and egg cell fertilization to produce an embryo that is genetically identical to the maternal parent (Asker and Jerling, 1992). Apomictic embryos may develop directly from the somatic tissue of the surrounding ovule, from parthenogenesis of the egg cell of a somatic embryo sac, or from parthenogenesis of an unreduced female gametophyte (Koltunow, 1993). Asexually derived embryos also can be induced to form in vitro from a wide range of somatic and gametophytic donor tissues (Mordhorst et al., 1997). In most cases, the addition of plant hormones or growth regulators, or the application of a stress treatment, is necessary for embryo induction. The combination of donor tissue and induction treatment determines whether the embryos develop directly from single cells or indirectly through an intermediary callus phase.
Although the initiation of zygotic, apomictic, and in vitro embryogenesis are activated by different signals and often begin from different starting tissues, it is not unreasonable to assume that all three processes converge at a very early stage on the same signaling pathway. For example, allelic variations in the genes that control the initiation of zygotic embryo development may confer differences in the temporal and/or spatial expression patterns of genes involved in parthenogenesis in apomictic species (Petrov, 1970). Likewise, the induction of in vitro embryogenesis from differentiated tissues probably arises from a transient spatiotemporal reprogramming of regulatory genes that control zygotic embryo development.
Attempts to identify these early embryo control genes have been largely disappointing; the majority of identified genes appear to control basic developmental processes, such as the establishment of meristems, polarity, and tissue patterning, that function throughout the life cycle of the plant (reviewed in Kaplan and Cooke, 1997). More recently, a number of genes have been identified that play specific roles in the induction and maintenance of embryogenesis in plants. The LEAFY COTYLEDON1 (LEC1) and LEAFY COTYLEDON 2 (LEC2) genes were identified originally as loss-of-function mutants showing defects in both embryo identity and seed maturation processes (Meinke et al., 1994; West et al., 1994).
lec1 and lec2 mutant embryos exhibit morphological characteristics that are normally expressed postembryonically, that fail to accumulate seed-specific storage products, and that, unlike wild-type seeds at maturity, are partially (lec2) or fully (lec1) desiccation intolerant. Both of these LEC genes encode seed-expressed transcription factors: LEC1 encodes a HAP3 subunit of a CBF protein (Lotan et al., 1998), whereas LEC2 encodes a B3 domain protein (Stone et al., 2001). Ectopic expression of either gene promotes somatic embryo formation on the vegetative tissues of the plant.
The conversion of postgerminative plant organs into embryo-like structures is also induced by loss-of-function mutations in another Arabidopsis gene, PICKLE (PKL). The roots of pkl seedlings express embryonic characteristics and form somatic embryos when excised and placed on minimal tissue culture medium (Ogas et al., 1997). PKL encodes a CHD chromatin remodeling factor that has been shown in animal systems to be a component of transcriptional repressor complexes (Ogas et al., 1999). The loss-of-function pkl phenotype, combined with the identification of PKL as a potential transcriptional repressor, suggest a role for PKL as a repressor of embryo gene expression programs.
A non-mutant-based approach to gain insight into the molecular events associated with the initiation of the embryonic phase of plant development involves the identification of genes that are differentially expressed during the early stages of embryo development. This is a particularly useful method for species in which mutagenesis approaches are not feasible. Unfortunately, the small size of most zygotic embryos, combined with their inaccessibility within the seed coat, makes it technically challenging to isolate sufficient material for the identification of early embryo–expressed genes.
An alternative approach is to use in vitro embryo cultures to generate large numbers of embryos at developmental stages that would normally be inaccessible in seeds. In vitro embryo cultures derived from both somatic and gametophytic tissues have been used successfully as tools to identify genes expressed during early embryo development (Wilde et al., 1988; Aleith and Richter, 1990; Reynolds and Kitto, 1992; Wurtele et al., 1993; Boutilier et al., 1994; Zarsky et al., 1995; Reynolds and Crawford, 1996; Schmidt et al., 1997; Vrinten et al., 1999). One of these genes, SOMATIC EMBRYOGENESIS RECEPTOR KINASE, was recently shown to enhance Arabidopsis somatic embryo development (Hecht et al., 2001).
We are using Brassica napus microspore embryo cultures as a model system to identify gene expression programs that are associated with the initiation of embryo development in plants. This Brassica microspore embryo culture system is based on the ability of the vegetative cell of an immature pollen grain to develop into an embryo in vitro (Keller et al., 1987; Pechan et al., 1991; Custers et al., 1994). When late uninucleate microspores and early binucleate pollen are isolated and cultured at 25°C or lower, they continue to divide and form trinucleate pollen grains. Culturing the same starting material at higher temperatures for at least 8 h induces an irreversible developmental shift from pollen to embryo development. After an initial symmetric cell division, these embryogenic cells continue to divide and form sequentially the globular, heart-, torpedo-, and cotyledon-shaped structures characteristic of zygotic embryo development.
Here, we describe a screening approach to identify genes that are differentially expressed during the switch from pollen- to microspore-derived embryo development. One of the genes we identified, BABY BOOM (BBM), encodes an AP2 domain transcription factor and is preferentially expressed in developing embryos and seeds. Our functional analyses show that BBM activates signal transduction pathways leading to the induction of embryo development from differentiated somatic cells. Therefore, BBM is likely to be a key regulator of embryo development in plants.
RESULTS
Isolation of Early Embryo-Expressed Genes
We used a subtractive hybridization approach to isolate genes expressed during the period spanning approximately the 8- to 32-cell stage of microspore-derived embryo development (preglobular). To reduce the chance of cloning genes induced specifically by the heat-stress treatment, we used RNA from microspores cultured at 25°C for 24 h followed by culture at 32.5°C for 72 h (Pechan et al., 1991; Boutilier et al., 1994) in the construction of a subtracted probe. Although these microspores have been heat stressed, they do not form embryos upon transfer to 25°C, nor do they continue dividing to form mature pollen grains. We identified six clones corresponding to five independent cDNAs that were consistently differentially expressed between embryogenic and nonembryogenic microspore cultures. Here, we present the characterization of one of these genes, BBM.
BBM Shows Similarity to the AP2/ERF Family of Transcription Factors
A single truncated BBM cDNA clone was isolated after the initial subtractive screening of the embryogenic microspore cDNA library. Subsequent isolation of full-length BBM cDNAs and genomic clones from both Brassica and Arabidopsis identified two BBM genes in Brassica (BnBBM1 and BnBBM2) and a single ortholog in Arabidopsis (AtBBM). The predicted Arabidopsis and Brassica BBM amino acid sequences are highly conserved, with the two predicted Brassica sequences showing 98% similarity to each other and 85% similarity to the Arabidopsis sequence. The positions of the intron/exon boundaries between the Arabidopsis and Brassica genomic sequences are perfectly conserved.
Search of the sequence databases indicated that the BBM translation products show similarity to the AP2/ERF family of proteins. These proteins are a plant-specific class of putative transcription factors that have been shown to regulate a wide range of developmental processes (reviewed in Riechmann and Meyerowitz, 1998). AP2/ERF proteins are characterized by the presence of a so-called AP2/ERF DNA binding domain. This domain was first identified in the APETALA2 (AP2) and ethylene-responsive element (ERE) binding (EREBP) proteins and is defined by up to 70 amino acid residues containing a conserved central core of 18 amino acids (Jofuku et al., 1994; Ohme-Takagi and Shinshi, 1995; Weigel, 1995; Okamura et al., 1997a). This conserved core is predicted to form an amphipathic α-helix that binds DNA (Ohme-Takagi and Shinshi, 1995; Stockinger et al., 1997; Zhou et al., 1997; Hao et al., 1998; Liu et al., 1998; Kagaya et al., 1999; Menke et al., 1999) and may mediate protein–protein interactions (Jofuku et al., 1994).
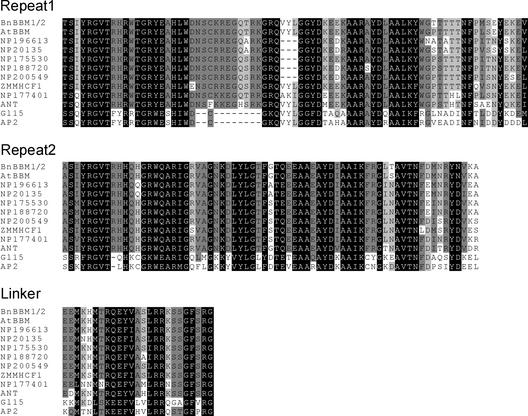
AP2/ERF proteins have been subdivided into two distinct subfamilies based on whether they contain one (ERF subfamily) or two (AP2 subfamily) DNA binding domains (Zhou et al., 1997). The BBM proteins belong to the AP2 subfamily, members of which include APETALA2 (AP2; Jofuku et al., 1994), Indeterminate spikelet1 (Ids1; Chuck et al., 1998), AINTEGUMENTA (ANT; Elliot et al., 1996; Klucher et al., 1996), Glossy15 (Gl15; Moose and Sisco, 1996), and ZMMHCF1 (Daniell et al., 1996). The Arabidopsis and Brassica BBM proteins show 98 to 99% similarity with each other in the region spanning the two AP2 domains and 60 to 89% similarity in the same region with other AP2 subfamily proteins (Figure 1).
Figure 1.
Sequence Comparison of the BBM Proteins and Related AP2 Domain–Containing Proteins.
The amino acid sequences of the first AP2/ERF domain repeat (Repeat 1), the second AP2/ERF domain repeat (Repeat 2), and the linker region lying between the two repeats (Linker) of the Brassica and Arabidopsis BBM proteins were aligned with the amino acid sequences of related AP2/ERF domain–containing proteins. The shaded boxes indicate the percentage of sequences at that position with the same amino acid identity (dark gray, 100%; medium gray, 80%; light gray, 60%). Protein names or accession numbers are indicated at the left. BnBBM1/2, Brassica BBM1 and BBM2 (the BnBBM1 and BnBBM2 protein sequences are identical in the AP2/ERF domain and linker regions); AtBBM, Arabidopsis BBM; ANT, Arabidopsis AINTEGUMENTA; ZMMHCF1, maize ZMMHCF1; GL15, maize Glossy15; AP2, Arabidopsis APETALA2. NP196613, NP20135, NP175530, NP188720, NP200549, and NP177401 are accession numbers for hypothetical Arabidopsis proteins.
The AP2 domains and linker regions of the sequences encoded by ANT, ZMMHCF1, and a number of recently identified hypothetical proteins show the highest similarity to the BBM proteins. A 10–amino acid insertion in the first AP2 DNA binding domain further distinguishes this related subgroup of proteins from two other AP2 domain–containing proteins, AP2 and Gl15 (Elliot et al., 1996). The BBM proteins do not show large stretches of sequence similarity with other AP2/ERF proteins outside of the AP2 DNA binding domain region.
BBM Genes Are Expressed Preferentially in Developing Seeds
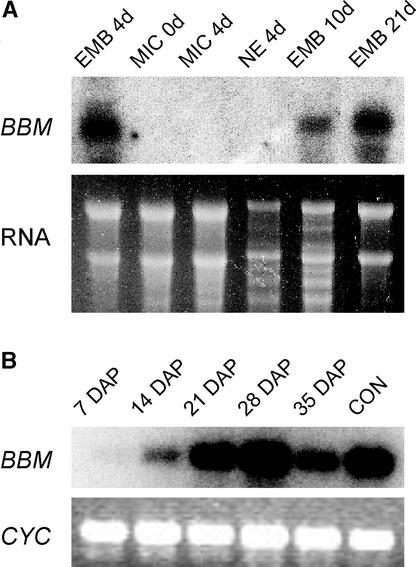
RNA gel blot analysis, reverse transcriptase–mediated (RT)–PCR, and mRNA in situ hybridization were used to determine the temporal and spatial patterns of BBM gene expression in microspore cultures and developing seeds. Figure 2A shows that BBM transcripts are present in RNA samples from 4-day-old embryogenic cultures but are not detected in the nonembryogenic cultures of the same age. BBM transcripts were detected during the subsequent globular to cotyledon stages of microspore-derived embryo development.
Figure 2.
BBM Gene Expression in Brassica.
(A) RNA gel blot analysis of BBM gene expression in Brassica microspore cultures. Total RNA was isolated from microspores at the start of culture (MIC 0d), after 4 days in culture at 32.5°C (EMB 4d), after 4 days in culture at 25°C (MIC 4d), after 1 day of culture at 25°C followed by 3 days of culture at 32.5°C (NE 4d), and from purified microspore-derived embryos at the globular (EMB 10d) and cotyledon (EMB 21d) stages of development. RNA samples (15 μg) were blotted and hybridized to a BBM1/BBM2-specific probe. Ethidium bromide staining of RNA was used to compare sample loading. EMB, embryogenic microspore culture; MIC, microspores and pollen; NE, nonembryogenic microspore culture.
(B) RT-PCR analysis of BBM transcripts in developing seeds. RNA samples for RT-PCR analysis were isolated from whole seeds collected at specific days after pollination (DAP). These developmental time points correspond approximately to the globular (7 DAP), heart/torpedo (14 DAP), early cotyledon (21 DAP), mid cotyledon (28 DAP), and mid to late cotyledon (35 DAP) stages of development. RT-PCR analysis of torpedo-stage microspore-derived embryos was included as a positive control (CON). Ethidium bromide staining of RT-PCR–amplified CyP cyclophilin cDNA (CYC) from the same reverse-transcribed RNAs was used as a control for cDNA synthesis.
BBM mRNAs are difficult to detect on gel blots containing total Brassica seed RNA; therefore, we used nonquantitative RT-PCR and hybridization to monitor BBM mRNA accumulation during seed development in this species (Figure 2B). The temporal pattern of BBM mRNA accumulation during seed development reflects the pattern observed in microspore-derived embryos: BBM mRNA was detected at the earliest time point analyzed, corresponding to the globular stage of embryo development, and continued to be expressed until late in seed development. RT-PCR analysis and gel blot hybridization of RNA from nonseed tissues revealed a barely detectable level of BBM expression in most tissues analyzed (data not shown).
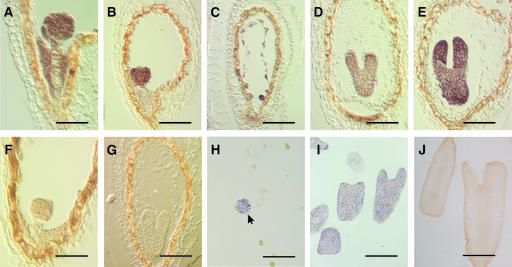
mRNA in situ hybridization was used to determine the spatial distribution of BBM mRNAs during Brassica microspore-derived embryo and Arabidopsis seed development. As shown in Figure 3, BBM expression was detected throughout the developing microspore and zygotic embryo at both early and late stages of development. In seeds, BBM expression also was observed in the free nuclear endosperm, but it decreased dramatically once endosperm cellularization began. Together, these results suggest that the BBM genes are expressed preferentially during embryo and seed development.
Figure 3.
In Situ Localization of BBM mRNA in Embryos and Seeds.
Sections of Brassica microspore-derived embryos and Arabidopsis seeds were hybridized with antisense (AtBBM, [A] to [E]; BnBBM, [H] and [I]) or sense (AtBBM, [F] and [G]; BnBBM, [J]) digoxigenin-UTP–labeled probes. The transcript hybridization signal is purple-brown.
(A) to (G) Longitudinal sections through developing Arabidopsis seeds.
(H) Globular-stage microspore embryo (arrow) and undeveloped microspores from an 8-day-old Brassica microspore embryo culture.
(I) and (J) Fourteen-day-old Brassica microspore-derived embryo culture.
Bars = 25 μm in (A) and (F), 50 μm in (B) to (E) and (G), 100 μm in (H), 350 μm in (I), and 200 μm in (J).
BBM Overexpression Indu ces Embryo Formation
The identification of the BBM proteins as putative transcription factors, combined with their preferential accumulation throughout embryo development, suggested that these proteins play a central role in regulating embryo-specific pathways. We investigated the effect of ectopically overexpressing BBM genes in Arabidopsis and Brassica by placing them under the control of two semiconstitutive promoters.
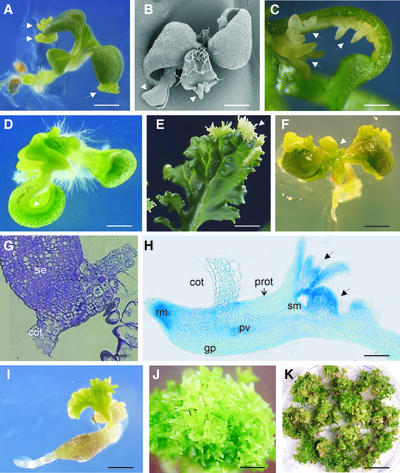
As shown in Figure 4, Arabidopsis and Brassica plants transformed with the 35S::BBM and UBI::BBM constructs spontaneously formed somatic embryos and/or cotyledon-like structures on postgermination organs. In Arabidopsis, somatic embryos developed from the margin of the blade of the cotyledon or leaf (Figures 4A and 4B), from the cotyledon or leaf petiole (Figure 4C), and from the shoot apex (Figure 4D). Similar phenotypes were observed in Brassica (Figure 4E), except that somatic embryos also formed on the hypocotyls of germinated seedlings (data not shown).
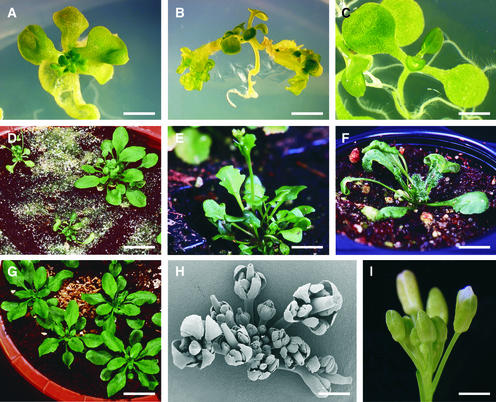
Figure 4.
Ectopic Expression of BBM Induces Somatic Embryo Formation in Arabidopsis and Brassica.
All images correspond to transgenic plants.
(A) Somatic embryos (arrows) formed on the cotyledon margins of a 35S::BBM Arabidopsis (C24) seedling.
(B) Scanning electron microscopy image of a 35S::BBM Arabidopsis (Columbia) seedling showing somatic embryos (arrows) on the cotyledons and leaves. Note the absence of trichomes on the somatic embryos.
(C) Somatic embryos (arrows) formed on the petiole and shoot apex of a UBI::BBM Arabidopsis (C24) plant.
(D) Formation of somatic embryos on the shoot apex and activation of cell division (arrow) along the cotyledon margin of a 35S::BBM Arabidopsis (C24) seedling.
(E) Somatic embryos (arrow) formed on the leaf margin of a 35S::BBM Brassica plant.
(F) Recurrent embryogenesis along the cotyledon margin and petiole of a 35S::BBM Arabidopsis (C24) seedling. The leaves of this seedling have been replaced by cotyledon-like organs (arrow).
(G) Longitudinal semithin section through the point of attachment of a single 35S::BBM (C24) somatic embryo (se) to the underlying cotyledon (cot).
(H) Longitudinal semithin section through one of the cotyledons and attached embryos of the seedling shown in (A). The somatic embryo attached to the seedling cotyledon (cot) is bipolar and contains all of the organs and tissue types seen in zygotic embryos. Secondary somatic embryos (arrows) initiate from the cotyledons of the primary somatic embryo. gp, ground parenchyma; prot, protoderm; pv, provascular tissue; rm, root meristem; sm, shoot meristem.
(I) A 35S::BBM (C24) transgenic seedling showing recurrent embryogenesis from the shoot apex and callus formation on the root/hypocotyl.
(J) Recurrent somatic embryogenesis from the shoot apex of a 35S::BBM (C24) seedling.
(K) Recurrent embryogenesis and cell proliferation in a 35S::BBM Arabidopsis (C24) seedling. The photograph shows one of 10 such dishes obtained after 3 months of biweekly subculture of a single 35S::BBM seedling on minimal tissue culture medium.
Bars = 1 mm in (A), (C), (D), (F), (I), and (J), 0.16 mm in (B), 20 mm in (E), 0.03 mm in (G), 0.2 mm in (H), and 5 mm in (K).
In both Arabidopsis and Brassica, primary somatic embryos were attached to the underlying tissue at their lateral sides (Figure 4H), at their root poles (Figure 4G), or by their cotyledons (Figure 4C). In both species, somatic embryos were formed as individual embryos or as clustered structures that also formed secondary somatic embryos or cotyledon-like structures on their surface. The formation of somatic embryos on the leaf margins generally was restricted to the first two leaves of the plant, although in one transgenic Brassica line, somatic embryos continued to develop on leaves that arose later in development (Figure 4E).
In Arabidopsis, cotyledon-like organs (i.e., embryo-like structures lacking defined shoot and root meristems) developed from the margins of seedling cotyledons and petioles, or from the shoot apex, either randomly or at the position normally occupied by leaves (Figure 4F). Analysis of semithin sections revealed that cells in either the L1 layer alone, or both the L1 and L2 layers of the cotyledons or leaves, contributed to the formation of the somatic embryos (Figures 4G and 4H).
Semithin sections of 35S::BBM Arabidopsis plants also revealed that individual BBM-derived somatic embryos are similar in organization to zygotic embryos. They generally are bipolar, consisting of (multiple) cotyledons, a shoot and root meristem, and an axis that has the typical radial arrangement of three tissue types (Figure 4H). The provascular tissue of these somatic embryos was not attached to the vascular tissues of the underlying cotyledons and leaves. As with zygotic embryos, the BBM-induced somatic embryos and cotyledon-like structures did not develop the characteristic trichomes found on the true leaves of many Arabidopsis ecotypes and Brassica cultivars (Figure 4B). Somatic embryos also expressed seed-specific molecular markers such as the 2S albumin storage protein–encoding genes (data not shown).
The postgermination fate of Arabidopsis and Brassica seedlings expressing the 35S::BBM and UBI::BBM constructs is dependent on the severity of the somatic embryo phenotype. UBI::BBM seedlings and the majority of 35S::BBM seedlings develop only a limited number of somatic embryos or cotyledon-like structures and then resume additional postgermination growth (Figure 4A). However, in a number of 35S::BBM lines, overexpression of BBM leads to a reiteration of the embryo-forming process, with the result that new embryos or cotyledons are formed continuously on the cotyledons of preexisting embryos.
Seedlings showing strong recurrent embryogenesis are severely compromised with respect to further vegetative development; the roots grow slowly and the root/hypocotyl forms callus (Figure 4I), whereas shoot outgrowth is inhibited as the embryo structures continue to divide and form balls of embryos and cotyledons (Figure 4J). Leaf-like structures and callus eventually arise from the embryo mass, although new somatic embryos are formed quickly on the leaf margins, leading to a reiteration of the process (Figure 4K).
BBM Overexpression Induces Pleiotropic Phenotypes
Additional pleiotropic effects of ectopic BBM expression on vegetative and generative development were observed at a low penetrance in both 35S::BBM and UBI::BBM Arabidopsis and Brassica transgenic seedlings. These included pinched or lobed cotyledons and leaves (Figure 5A), thickened or callused hypocotyls, formation of ectopic shoots (Figure 5B), short roots (Figure 5B), callus formation, and anthocyanin accumulation. The penetrance of these phenotypes was variable within and between independent transgenic lines.
Figure 5.
Ectopic Overexpression of BBM Induces Pleiotropic Phenotypes in Arabidopsis.
(A) UBI::BBM seedling (C24) showing lobed cotyledons and leaves, and increased shoot production at the apex.
(B) 35S::BBM seedling (C24) showing ectopic shoots on the cotyledon edge and short roots.
(C) Wild-type (C24) seedling.
(D) Homozygous, single-locus 35S::BBM line (C24) showing the class I phenotype of rounded leaves and decreased size.
(E) 35S::BBM (C24) plant with serrated leaves.
(F) 35S::BBM plant (Columbia) with rumpled leaves.
(G) Wild-type (C24) plants.
(H) 35S::BBM (C24) flowers showing alterations in floral organ length.
(I) Wild-type C24 flowers.
Bars = 1 mm in (A), 1.5 mm in (B) and (C), 10 mm in (D), 8 mm in (E) and (F), 17 mm in (G), 0.05 mm in (H), and 1.5 mm in (I).
Transgenic Arabidopsis plants ectopically expressing the 35S::BBM construct exhibited growth alterations at later stages of development, which could be divided into two major phenotypic classes. Plants in class I were dwarf to medium in size and produced rounded leaves (Figure 5D) that occasionally were wrinkled or showed an increase in serration. These plants grew very slowly, produced an excess of rosette leaves, and were slow to flower but were fully fertile. Plants in class II were wild type to medium in size, often had elongated and/or epinastic leaf petioles, and produced leaves that were severely wrinkled or that showed an increase in serration (Figures 5E and 5F). These plants often contained increased levels of anthocyanin or lacked epicuticular wax, exhibited a decrease in the length of the anthers, petals, and sepals relative to the carpels (Figure 5H), and showed decreased male and female fertility. Floral organ abnormalities such as narrow sepals and petals, greenish petals, carpels with elongate protrusions, or a wrinkled appearance at the region of the carpel adjacent to the stigma were observed. Wrinkled leaves, as well as leaves with increased lobing, were also observed in the 35S::BBM Brassica transgenic plants (Figure 4E), although the limited number of transgenic plants (six) did not permit the classification of phenotypic severity.
A number of the phenotypes described above suggest that the ectopic expression of BBM stimulates cell proliferation and subsequent differentiation. Therefore, we determined whether ectopic BBM expression was sufficient to enhance in vitro regeneration, a process that also relies on the activation of cell division and differentiation. The effect of BBM gene expression on in vitro regeneration was examined using transgenic Arabidopsis UBI::BBM plants. The weaker penetrance and expressivity of the somatic embryo phenotype in UBI::BBM transgenic plants allowed us to examine the effect of BBM gene expression on regeneration without the added complication of prolific somatic embryo production on the explant material.
Leaf and hypocotyl explants from 10-day-old seedlings of wild-type plants and seven independent transgenic lines were placed on either basal medium or media supplemented with plant growth regulators to stimulate regeneration via organogenesis. In the experiments involving plant growth regulators, explants were first placed for 3 days on callus-inducing medium (high auxin-to-cytokinin ratio) and then transferred to shoot-inducing medium (high cytokinin-to-auxin ratio) as described in Vergunst et al. (1998). Both wild-type leaf and hypocotyl explants placed on growth regulator–supplemented media produced callus on the cut end of the explant (Figures 6A and 6B), which was followed later by shoot formation (data not shown). Shoot regeneration from UBI::BBM explants placed on the same media was more vigorous and accelerated compared with that of the wild-type explants (Figure 6B).
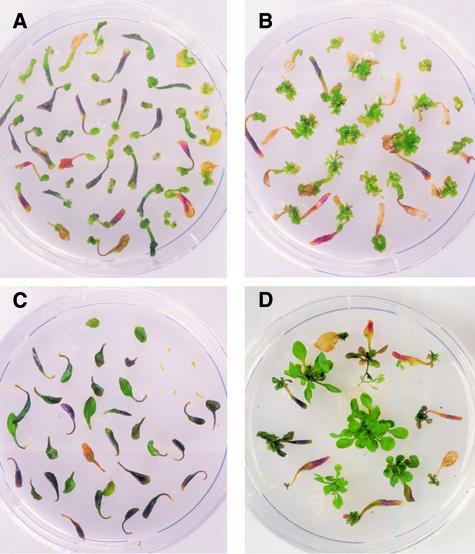
Figure 6.
Regenerative Capacity of Wild-Type and Transgenic UBI::BBM Arabidopsis Plants.
Leaf and hypocotyl explants from wild-type C24 ([A] and [C]) and transgenic UBI::BBM seedlings ([B] and [D]) were placed successively on medium containing callus-inducing growth regulators followed by shoot-inducing growth regulators ([A] and [B]) or medium lacking growth regulators ([C] and [D]). UBI::BBM explants regenerate on hormone-free medium and exhibit enhanced regeneration in the presence of growth regulators.
The effect of BBM expression on explants growing on media lacking plant growth regulators was even more pronounced. Wild-type leaf and hypocotyl explants placed on media lacking plant growth regulators routinely produced callus or regenerated roots at the cut end of the explant, although shoot formation was never observed (Figure 6C). In contrast to wild-type explants, UBI::BBM explants regenerated into complete plantlets in the absence of added plant growth regulators (Figure 6D). Organogenesis appeared to be direct, because no callus formation was observed. These results indicate that ectopic expression of BBM stimulates pathways that promote cell division and differentiation. This process is not dependent on the addition of cytokinin and auxin but can be enhanced by the presence of these growth regulators.
DISCUSSION
BBM Proteins Are Similar to the AP2/ERF Family of Transcription Factors
The proteins encoded by the Brassica and Arabidopsis BBM genes show similarity to members of the AP2/ERF family of transcription factors (Riechmann and Meyerowitz, 1998). Although exceptions exist (Wilson et al., 1996; van der Graaff et al., 2000; Banno et al., 2001), the function of the AP2/ERF proteins can be defined broadly by the number of AP2/ERF DNA binding domains they contain (Moose and Sisco, 1996). Members of the ERF subfamily, which include CBF, PTI, EREBP, ORCA, ABI4, and DREB, contain a single AP2/ERF domain, and in general, they appear to regulate processes related to abiotic and environmental stress (Ohme-Takagi and Shinshi, 1995; Stockinger et al., 1997; Zhou et al., 1997; Finkelstein et al., 1998; Liu et al., 1998; Menke et al., 1999).
By contrast, proteins of the AP2 subfamily, including AP2, ANT, Gl15, and Ids1, contain two AP2/ERF DNA binding domains and generally can be considered as developmental regulators of cell/organ identity and fate (Jofuku et al., 1994; Elliot et al., 1996; Klucher et al., 1996; Moose and Sisco, 1996; Chuck et al., 1998). The presence of two AP2/ERF DNA binding domains within the BBM proteins is consistent with a role of this subfamily of proteins in mediating developmental processes.
The AP2/ERF DNA binding domains of the BBM proteins, as well as the linker region connecting these two domains, are most similar to a subgroup of the AP2 domain subfamily that includes the Arabidopsis ANT and maize ZMMHCF1 proteins as well as a number of hypothetical proteins from Arabidopsis. ZMMHCF1 is expressed in maize postpollination endosperm. The protein has been shown to complement an l-isoaspartyl methyltransferase–deficient mutant of Escherichia coli, although the function of the ZMMHCF1 protein during plant development has not been defined (Daniell et al., 1996). The Arabidopsis ANT protein has been studied in depth using both loss- and gain-of-function mutants. Loss-of-function ant mutants show reduced cell number and organ size (Elliot et al., 1996; Klucher et al., 1996), whereas ectopic expression of ANT increases cell number and organ size and promotes spontaneous callus formation from which new organs may arise (Krizek, 1999; Mizukami and Fischer, 2000). Both the loss- and gain-of-function phenotypes suggest that ANT plays a general role in maintaining meristematic competence in plants (Mizukami and Fischer, 2000).
Both BBM and ANT, in addition to showing sequence similarity in their DNA binding domains, also appear to promote cell proliferation when overexpressed ectopically. This function is also shared by a third AP2/ERF transcription factor gene, LEAFY PETIOLE (LEP). LEP was identified originally in a T-DNA activation tagging screen for leaf developmental mutants (van der Graaff et al., 2000). LEP contains a single DNA binding domain that is most similar to those of the ERF subfamily. Upregulation of the endogenous LEP promoter by the activation tag results in conversion of the proximal petiole into an ectopic leaf blade.
The constitutive expression of LEP results in more severe phenotypes, and, similar to BBM overexpression, results in increased and ectopic cell divisions along the cotyledon/leaf margin and the hypocotyl of transgenic seedlings. The similarity in the ANT, BBM, and LEP gain-of-function phenotypes suggests that the pathways activated by these three AP2/ERF proteins may intersect at some point downstream or that these proteins activate/repress an overlapping set of target genes when expressed ectopically. Nonetheless, BBM, ANT, and LEP are clearly distinct proteins with unique functions.
BBM Regulates the Embryonic Phase of Development
The manner in which the BBM gene was isolated, its identification as a putative transcription factor, its preferential expression in seeds, and its ability to induce somatic embryo formation when expressed ectopically all combine to suggest a key role for BBM in defining the embryo phase of plant development. Similar conclusions were drawn from gain-of-function studies on two additional seed-expressed transcription factor genes, LEC1 and LEC2; it was shown that 35S::LEC1 and 35S::LEC2 constructs promote spontaneous somatic embryo formation on vegetative tissues (Lotan et al., 1998; Stone et al., 2001).
With respect to the 35S::BBM phenotype, the 35S::LEC1 phenotype is very weak, with only a few plants showing sporadic embryo development, whereas the 35S::LEC2 phenotype is comparable to that observed for 35S::BBM plants. The similarity between the BBM and LEC genes in terms of their putative function as transcription factors, their preferential expression in the seed, and the similarities in their gain-of-function phenotypes make it tempting to speculate that these genes have overlapping functions.
In the absence of a loss-of-function BBM mutant, we used RT-PCR to provide preliminary insight into the relationship between BBM and one of the LEC genes, LEC1. Our results indicate that BBM is expressed in seeds of the lec1-3 mutant (Raz et al., 2001; data not shown). This result suggests that BBM expression is not dependent per se on the presence of the LEC1 protein, although we cannot exclude the possibility that LEC1 modulates BBM expression levels. BBM may function upstream of LEC1, or BBM and LEC1 may function in separate but overlapping pathways. An overlap in function between BBM and the LEC genes might explain why lec1 and lec2 embryo development is relatively normal during the early stages of development (Meinke et al., 1994). The elucidation of the epistatic relationship between BBM and the LEC genes, as well as the identification of their respective target genes, will be important in defining the role of each protein during embryo development.
Role for BBM in Stimulating Cell Proliferation and Morphogenesis
The mechanism whereby BBM induces embryo formation is not known; however, other characteristics of the ectopic overexpression phenotype, such as callus and ectopic shoot formation, alterations in leaf morphology, and hormone-free regeneration of explants, suggest that BBM acts by stimulating cell proliferation and morphogenesis pathways.
Although ectopic BBM expression induces a number of pleiotropic phenotypes, the major phenotype is the spontaneous formation of somatic embryos, suggesting that the wild-type role of this gene is to induce and/or maintain embryo development after fertilization. In this respect, it is interesting that in both UBI::BBM and 35S::BBM overexpression lines, the somatic embryos are derived primarily from cells of the shoot apex or the marginal tissue of seedlings.
Newly formed angiosperm leaves are characterized by a marginal region, or blastozone, consisting of small, densely cytoplasmic cells (Hagemann and Gleissberg, 1996). These cells exhibit a transient period of high organogenetic capacity, giving rise to leaflets, lobes, and serrations, among other features. The shoot apex also is composed of a group of pluripotent stem cells that give rise to most of the aboveground parts of the plant during its life cycle. Thus, the cells that are most competent to respond to an ectopic BBM signal appear to be the same cells that are normally competent to divide and differentiate. Although the shoot meristem and the leaf margin of newly initiated leaves retain their organogenetic capacity for most of the plant life cycle, somatic embryo formation in BBM gain-of-function plants is generally restricted to seedlings and the first two sets of true leaves. In older plants, the competence of a cell to respond to an ectopic BBM signal is not lost but results in a different morphological response. This suggests that a relatively undifferentiated cell state is important for defining the competence to respond to BBM but that the specific developmental stage of the plant appears to determine the morphogenetic outcome of this cell proliferation.
One clue to the mechanistic action of BBM lies in the observation that neither the induction of somatic embryogenesis, nor the formation of ectopic shoots and callus, nor the ability to stimulate regeneration from explants requires the addition of plant hormones or growth regulators to the medium. In most species, organogenesis in vitro is dependent on the addition of cytokinin or a combination of cytokinin and auxin to the growth medium (Skoog and Miller, 1957). Likewise, somatic embryo development in many species, including Arabidopsis, is induced by plant growth regulators (Mordhorst et al., 1997, 1998). The ability of BBM to promote organogenesis and embryogenesis in the absence of exogenously applied growth regulators suggests that BBM may act by stimulating the production of plant hormones and/or increasing the sensitivity of the cell to these substances. Klucher et al. (1996) speculated that AP2/ERF domain proteins, being unique to plants, might have coevolved with plant-specific pathways such as hormone signal transduction.
A role for AP2/ERF domain proteins in hormone signaling pathways is not a general characteristic of this protein family; nonetheless, a large number of AP2/ERF domain genes are regulated by plant hormones at the transcriptional level and/or function in hormone signaling pathways (Ohme-Takagi and Shinshi, 1995; Büttner and Singh, 1997; Okamura et al., 1997b; Finkelstein et al., 1998; Menke et al., 1999; Gu et al., 2000; Banno et al., 2001; van der Fits and Memelink, 2001).
METHODS
Microspore Culture
Brassica napus cv Topas DH 4079 was used as the donor plant for microspore embryo cultures. The Brassica plant growth and microspore culture conditions have been described previously (Boutilier et al., 1994).
Subtractive Probe Construction and Library Screening
Poly(A)+ mRNA, isolated from Brassica microspores cultured for 4 days at 32.5°C to induce embryogenesis, was used to synthesize first-strand cDNA (Riboclone; Promega, Madison, WI). The first strand cDNA was hybridized to a fivefold excess (by weight) of poly(A)+ RNA from a heat-stressed, nonembryogenic sample that was obtained by culturing microspores for 1 day at 25°C followed by 3 days at 32.5°C.
The subtractive hybridization was performed essentially as described in Sambrook et al. (1989). The single-stranded cDNA recovered after subtraction was used as a probe for screening a cDNA library derived from embryogenic microspores (Boutilier et al., 1994). Approximately 1.5 × 105 plaque-forming units of the cDNA library were screened with (1) the subtracted probe, (2) a heat-stressed, nonembryogenic sample cDNA probe, and (3) a cDNA probe for napin seed storage protein genes (Crouch et al., 1983), which are abundant in the library (Boutilier et al., 1994).
Plaques hybridizing to the subtracted probe, but not to the nonembryogenic or napin probes, were selected for further analysis. A partial BBM1 cDNA was one of the differentially expressed cDNA clones that was isolated and subsequently characterized. The full-length Brassica BBM1 and BBM2 cDNAs were obtained by stringent screening of a Brassica globular-stage, microspore-derived embryo cDNA library (UniZAPII; Stratagene). The Arabidopsis thaliana BBM ortholog (AtBBM) was isolated by screening an Arabidopsis (ecotype C24) genomic λ phage library (λ-GEM 11; Promega) using the entire BBM1 cDNA as a probe. The BnBBM1 genomic clone was isolated from Brassica using the GenomeWalker kit (Clontech, Palo Alto, CA).
Nucleic Acid Isolation and Analysis
Total RNA was isolated as described in Ouellet et al. (1992) or using Trizol reagent (Invitrogen Life Technologies, Breda, The Netherlands). Poly(A)+ RNA isolation and RNA formaldehyde gel electrophoresis were performed as described in Sambrook et al. (1989). A cDNA fragment corresponding to nucleotides 1 to 411 of the BnBBM1 cDNA (upstream of the AP2/ERF domain) was used as a gene-specific probe on gel blots. Reverse transcriptase–mediated PCR (RT-PCR) was performed using Superscript II reverse transcriptase (Invitrogen Life Technologies), an oligo(dT) primer, and 5 μg of DNase-treated RNA.
Brassica BBM cDNAs were amplified by PCR using a gene-specific primer set (5′-ACCTTTTACCAAGAACTCGTTAGATCA-3′ and 5′-AAGTAGATGAGTTCATTGAGAGGGACA-3′) that spans the first intron of the BBM genomic sequence. BBM RT-PCR products were blotted and hybridized to the gene-specific BBM cDNA fragment. The constitutively expressed cyclophilin CyP gene (Gasser et al., 1990) was used as a cDNA synthesis control for RT-PCR experiments.
Microscopy and in Situ Hybridization
All plant material was fixed for 24 h at 4°C in 0.1 M phosphate buffer, pH 7.0, containing 4% paraformaldehyde. Samples for scanning electron microscopy were processed as described in Dornelas et al. (2000), and digital images were obtained using an Orion Framegrabber (Matrox Electronic Systems, Unterhaching, Germany). Samples for light microscopy were embedded in Technovit 7100 (Hereaus Kulzer, Wehrheim, Germany), stained with toluidine blue, and mounted in Euparal (Chroma-Gesellschaft, Kongen, Germany).
Samples for mRNA in situ hybridization were fixed as described above, except that microspore embryos were fixed initially for 4 h, embedded in 1% agarose plugs, and fixed for an additional 20 h. Tissue was dehydrated by passage through a graded ethanol series and then infiltrated with Paraplast X-tra (Oxford Labware, Oxford, UK). Twenty-micrometer sections were mounted onto Superfrost/Plus microscope slides (Fisher Scientific) and dewaxed in xylene.
Gene-specific fragments corresponding to the region of the Brassica and Arabidopsis BBM sequences lying upstream of the AP2/ERF domain were used as templates for digoxigenin-labeled RNA transcripts (Boehringer Mannheim). The Brassica BBM template is described above. The AtBBM template corresponds to a 202-bp HindIII-SspI fragment of the AtBBM genomic clone.
Prehybridization, hybridization, and high-stringency posthybridization washing steps and detection of digoxigenin-labeled probes were performed essentially as described in Jackson (1991). Staining reactions were performed for 24 h. Slides were dehydrated through a graded ethanol series and mounted in Permount (Fisher Scientific). Digital images were recorded using a charge-coupled device camera and edited using Adobe Photoshop 4 (Mountain View, CA).
Vector Construction and Transformation
The 35S::BBM construct contains the Brassica BBM1 cDNA coding and 3′ untranslated regions under the control of a double-enhanced 35S promoter and the translational enhancer of the Alfalfa mosaic virus (Datla et al., 1993) in the binary vector pBINPLUS (van Engelen et al., 1995). The UBI::BBM construct contains the BBM2 cDNA 5′ untranslated, coding, and 3′ untranslated regions under the control of a modified (lacking the first intron and ATG translational start site) sunflower UbB1 POLYUBIQUITIN promoter (Binet et al., 1991) in the binary vector pBINPLUS.
The 35S::BBM and UBI::BBM binary vectors were electroporated into Agrobacterium tumefaciens strain C58C1pMP90 and used to transform Arabidopsis ecotypes C24 and Columbia (Clough and Bent, 1998). Transgenic Brassica plants were produced by Agrobacterium-mediated transformation of haploid microspore-derived embryos. Details of the transformation protocol are available on request from J.B.M.C.
Upon request, all novel materials described in this article will be made available for academic noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for academic noncommercial research purposes.
Accession Numbers
The accession numbers for the BBM sequences described in this article are as follows: BnBBM1 cDNA (AF317904); BnBBM2 cDNA (AF317905); BnBBM1 genomic clone (AF317906); and AtBBM genomic clone (AF317907). The accession numbers for the other sequences shown in Figure 1 are as follows: Arabidopsis ANT (AAB17364); maize ZMMHCF1 (AAC49567); maize GL15 (AAC49567); and Arabidopsis AP2 (AAC13770).
Acknowledgments
We thank Claire Rouvière and Marcel van Blijderveen for help with the in vitro regeneration experiments and the maintenance of transgenic plant lines, Jindrich Novotny for assistance in constructing the globular-stage, microspore-derived embryo cDNA library, Adrie van't Hooft for art work, and Oscar Goddijn for the Arabidopsis genomic DNA library. This work was partially supported by visiting fellowships in a Canadian government laboratory to K.B. and T.O., by The Netherlands Organization for Scientific Research visiting fellowships to K.B. and V.K.S., by a Rockefeller postdoctoral fellowship to V.K.S., and by the Dutch Ministry of Agriculture, Nature Management, and Fisheries (DWK 281-392).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001941.
References
- Aleith, F., and Richter, G. (1990). Gene expression during induction of somatic embryogenesis in carrot cell suspensions. Planta 183, 17–24. [DOI] [PubMed] [Google Scholar]
- Asker, S.E., and Jerling, L. (1992). Apomixis in Plants. (Boca Raton, FL: CRC Press).
- Banno, H., Ikeda, Y., Niu, Q.-W., and Chua, N.-H. (2001). Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell 13, 2609–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binet, M.N., Lepetit, M., Weil, J.H., and Tessier, L.H. (1991). Analysis of a sunflower polyubiquitin promoter by transient expression. Plant Sci. 79, 87–94. [Google Scholar]
- Boutilier, K.A., Gines, M.J., DeMoor, J.M., Huang, B., Baszczynski, C.L., Iyer, V.N., and Miki, B.L. (1994). Expression of the BnmNAP subfamily of napin genes coincides with the induction of Brassica microspore embryogenesis. Plant Mol. Biol. 26, 1711–1723. [DOI] [PubMed] [Google Scholar]
- Büttner, M., and Singh, K.B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein, interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 94, 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck, G., Meeley, R.B., and Hake, S. (1998). The control of maize spikelet meristem fate by the APETALA2-like gene indeterminate spikelet1. Genes Dev. 12, 1145–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Crouch, M.L., Tenbarge, K.M., Simon, A.E., and Ferl, R. (1983). cDNA clones for Brassica napus seed storage proteins: Evidence from nucleotide sequence analysis that both subunits of napin are cleaved from a precursor polypeptide. J. Mol. Appl. Genet. 2, 273–283. [PubMed] [Google Scholar]
- Custers, J.B.M., Cordewener, J.H.G., Nöllen, Y., Dons, J.J.M., and Van Lookeren Campagne, M.M. (1994). Temperature controls both gametophytic and sporophytic development in microspore cultures of Brassica napus. Plant Cell Rep. 13, 267–271. [DOI] [PubMed] [Google Scholar]
- Daniell, T.J., Fordham-Skelton, A.P., Vergani, P., and Edwards, R. (1996). Isolation of a maize cDNA (accession no. Z47554) (PGR 96–013) encoding APETALA-2-like binding domains by complementation cloning of an l-isoaspartyl methyltransferase-deficient mutant of Escherichia coli. Plant Physiol. 110, 1435.
- Datla, R.S.S., Bekkaoui, F., Hammerlindl, J.K., Pilate, G., Dunstan, D.I., and Crosby, W.L. (1993). Improved high-level constitutive foreign gene expression in plants using an AMV RNA4 untranslated leader sequence. Plant Sci. 94, 139–149. [Google Scholar]
- Dornelas, M., van Lammeren, A., and Kreis, M. (2000). Arabidopsis thaliana SHAGGY-related protein kinases (AtSK11 and 12) function in perianth and gynoecium development. Plant J. 21, 419–429. [DOI] [PubMed] [Google Scholar]
- Elliot, R.C., Betzner, A.S., Huttner, E., Oakes, M.P., Tucker, W.Q.J., Gerentes, D., Perez, P., and Smyth, D.R. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., Wang, M.L., Lynch, T.J., Rao, S., and Goodman, H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 10, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser, C.S., Gunning, D.A., Budelier, K.A., and Brown, S.M. (1990). Structure and expression of cytosolic cyclophilin/peptidyl-prolyl cis-trans isomerase of higher plants and production of active tomato cyclophilin in Escherichia coli. Proc. Natl. Acad. Sci. USA 87, 9519–9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y.-Q., Yang, C., Thara, V.K., Zhou, J., and Martin, G.B. (2000). Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12, 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann, W., and Gleissberg, S. (1996). Organogenetic capacity of leaves: The significance of marginal blastozones in angiosperms. Plant Sys. Evol. 199, 121–152. [Google Scholar]
- Hao, D.Y., Ohme-Takagi, M., and Sarai, A. (1998). Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plants. J. Biol. Chem. 273, 26857–26861. [DOI] [PubMed] [Google Scholar]
- Hecht, V., Vielle-Calzada, J.P., Hartog, M.V., Schmidt, E.D., Boutilier, K., Grossniklaus, U., and de Vries, S.C. (2001). The Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASE 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 127, 803–816. [PMC free article] [PubMed] [Google Scholar]
- Jackson, D. (1991). In-situ hybridisation in plants. In Molecular Plant Pathology: A Practical Approach, S.J. Gurr, D.L. Bowles, and M.J. McPherson, eds (Oxford, UK: Oxford University Press), pp. 163–174.
- Jofuku, K.D., den Boer, B.G.W., van Montagu, M., and Okamuro, J.K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6, 1211–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya, Y., Ohmiya, K., and Hattori, T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, D.R., and Cooke, T.J. (1997). Fundamental concepts in the embryogenesis of dicotyledons: A morphological interpretation of embryo mutants. Plant Cell 9, 1903–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, W.A., Fan, Z., Pechan, P., Long, N., and Grainger, J. (1987). An efficient method for culture of isolated microspores of Brassica napus. In Proceedings of the 7th International Rapeseed Congress, Vol. I (Poznan, Poland: Panstwowe Wydawnictwo Rolnicze i Lesne), pp. 152–157.
- Klucher, K.M., Chow, H., Reiser, L., and Fischer, R.L. (1996). The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koltunow, A.M. (1993). Apomixis: Embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 5, 1425–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek, B.A. (1999). Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Dev. Genet. 25, 224–236. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi Shinozaki, K., and Shinozaki, K. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan, T., Ohto, M., Yee, K.M., West, M.A.L., Lo, R., Kwong, R.W., Yamagishi, K., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93, 1195–1205. [DOI] [PubMed] [Google Scholar]
- Meinke, D.W., Franzmann, L.H., Nickle, T.C., and Yeung, E.C. (1994). Leafy cotyledon mutants of Arabidopsis. Plant Cell 6, 1049–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke, F.L.H., Champion, A., Kijne, J.W., and Memelink, J. (1999). A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene Str interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J. 18, 4455–4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami, Y., and Fischer, R.L. (2000). Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose, S.P., and Sisco, P.H. (1996). Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev. 10, 3018–3027. [DOI] [PubMed] [Google Scholar]
- Mordhorst, A.P., Toonen, M.A.J., and de Vries, S.C. (1997). Plant embryogenesis. Crit. Rev. Plant Sci. 16, 535–576. [Google Scholar]
- Mordhorst, A.P., Voerman, K.J., Hartog, M.V., Meijer, E.A., van Went, J., Koornneef, M., and de Vries, S.C. (1998). Somatic embryogenesis in Arabidopsis thaliana is facilitated by mutations in genes repressing meristematic cell divisions. Genetics 149, 549–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas, J., Cheng, J.-C., Sung, R.Z., and Somerville, C. (1997). Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277, 91–94. [DOI] [PubMed] [Google Scholar]
- Ogas, J., Kaufmann, S., Henderson, J., and Somerville, C. (1999). PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc. Natl. Acad. Sci. USA 96, 13839–13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene responsive element. Plant Cell 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura, J.K., Caster, B., Villarroel, R., van Montagu, M., and Jofuku, K.D. (1997. a). The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 7076–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura, J.K., Szeto, W., Lotys-Prass, C., and Jofuku, K.D. (1997. b). Photo and hormonal control of meristem identity in the Arabidopsis flower mutants apetala2 and apetala1. Plant Cell 9, 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet, T., Rutledge, R.G., and Miki, B.L. (1992). Members of the acetohydroxyacid synthase multigene family of Brassica napus have divergent patterns of expression. Plant J. 2, 321–330. [PubMed] [Google Scholar]
- Pechan, P.M., Bartels, D., Brown, D.C.W., and Schell, J. (1991). Messenger-RNA and protein changes associated with induction of Brassica microspore embryogenesis. Planta 184, 161–165. [DOI] [PubMed] [Google Scholar]
- Petrov, D.F. (1970). Genetically regulated apomixis as a method of fixing heterosis and its significance in breeding. In Apomixis and Breeding, S.S. Khokhlov, ed (New Delhi: Amerind Publishing Co.), pp. 18–28.
- Raz, V., Bergervoet, J.H.W., and Koornneef, M. (2001). Sequential steps for developmental arrest in Arabidopsis seeds. Development 128, 243–252. [DOI] [PubMed] [Google Scholar]
- Reynolds, T.L., and Crawford, R.L. (1996). Changes in the abundance of an abscisic acid-responsive, early cysteine-labeled metallothionein transcript during pollen embryogenesis in bread wheat (Triticum aestivum). Plant Mol. Biol. 32, 823–829. [DOI] [PubMed] [Google Scholar]
- Reynolds, T.L., and Kitto, S.L. (1992). Identification of embryo-abundant genes that are temporally expressed during pollen embryogenesis in wheat anther cultures. Plant Physiol. 100, 1744–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., and Meyerowitz, E.M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379, 633–646. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schmidt, E.D.L., Guzzo, F., Toonen, M.A.J., and de Vries, S.C. (1997). A leucine-rich repeat containing receptor-like kinase marks somatic plant cells competent to form embryos. Development 124, 2049–2062. [DOI] [PubMed] [Google Scholar]
- Skoog, F., and Miller, C.O. (1957). Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Soc. Exp. Biol. 11, 118–131. [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L., Kwong, L.W., Yee, K.M., Pelletier, J., Lepiniec, L., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2001). LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc. Natl. Acad. Sci. USA 98, 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits, L., and Memelink, J. (2001). The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 25, 43–53. [DOI] [PubMed] [Google Scholar]
- van der Graaff, E., Den Dulk-Ras, A., Hooykaas, P.J.J., and Keller, B. (2000). Activation tagging of the LEAFY PETIOLE gene affects leaf petiole development in Arabidopsis thaliana. Development 127, 4971–4980. [DOI] [PubMed] [Google Scholar]
- van Engelen, F.A., Molthoff, J.W., Conner, A.J., Nap, J.P., Pereira, A., and Stiekema, W.J. (1995). pBINPLUS: An improved plant transformation vector based on pBIN19. Transgenic Res. 4, 288–290. [DOI] [PubMed] [Google Scholar]
- Vergunst, A.C., de Waal, E.C., and Hooykaas, P.J.J. (1998). Root transformation by Agrobacterium tumefaciens. In Arabidopsis Protocols, J.M. Martinez-Zapater and J. Salinas, eds (Totowa, NJ: Humana Press), pp. 227–244. [DOI] [PubMed]
- Vrinten, P.L., Nakamura, T., and Kasha, K.J. (1999). Characterization of cDNAs expressed in the early stages of microspore embryogenesis in barley (Hordeum vulgare) L. Plant Mol. Biol. 41, 455–463. [DOI] [PubMed] [Google Scholar]
- Weigel, D. (1995). The APETALA2 domain is related to a novel type of DNA binding domain. Plant Cell 7, 388–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, M.A.L., Yee, K.M., Danao, J., Zimmerman, J.L., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (1994). LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell 6, 1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde, H.D., Nelson, W.S., Booik, H., de Vries, S.C., and Thomas, T.L. (1988). Gene-expression programs in embryogenic and non-embryogenic carrot cultures. Planta 176, 205–211. [DOI] [PubMed] [Google Scholar]
- Wilson, K., Long, D., Swinburne, J., and Coupland, G. (1996). A Dissociation insertion causes a semidominant mutation that increases expression of TINY, an Arabidopsis gene related to APETALA2. Plant Cell 8, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtele, E.S., Wang, H., Durgerian, S., Nikolau, B.J., and Ulrich, T.H. (1993). Characterization of a gene that is expressed early in somatic embryogenesis of Daucus carota. Plant Physiol. 102, 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarsky, V., Garrido, D., Eller, N., Tupy, J., Vicente, O., Schöffl, F., and Heberle-Bors, E. (1995). The expression of a small heat shock gene is activated during induction of tobacco pollen embryogenesis by starvation. Plant Cell Environ. 18, 139–147. [Google Scholar]
- Zhou, J., Tang, X., and Martin, G.B. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 16, 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]