Abstract
We present the cloning and characterization of an Arabidopsis gene, FRD3, involved in iron homeostasis. Plants carrying any of the three alleles of frd3 constitutively express three strategy I iron deficiency responses and misexpress a number of iron deficiency–regulated genes. Mutant plants also accumulate approximately twofold excess iron, fourfold excess manganese, and twofold excess zinc in their shoots. frd3-3 was first identified as man1. The FRD3 gene is expressed at detectable levels in roots but not in shoots and is predicted to encode a membrane protein belonging to the multidrug and toxin efflux family. Other members of this family have been implicated in a variety of processes and are likely to transport small organic molecules. The phenotypes of frd3 mutant plants, which are consistent with a defect in either iron deficiency signaling or iron distribution, indicate that FRD3 is an important component of iron homeostasis in Arabidopsis.
INTRODUCTION
In soils, as in any aerobic environment, iron exists primarily in the ferric [Fe(III)] form. This poses a problem for plants that need to take in this essential nutrient, because Fe(III) is highly insoluble at neutral or basic pH. Furthermore, what little Fe(III) is in solution usually is chelated. Therefore, mechanisms that drive more iron into solution or that allow the use of chelated forms of Fe(III) are necessary to facilitate iron uptake. These mechanisms must be regulated carefully because excess iron can be toxic. The same redox properties that allow iron to serve as a critical redox cofactor also allow it to catalyze the formation of damaging oxygen radicals (Halliwell and Gutteridge, 1992).
Vascular plants can be divided into two groups based on their iron uptake responses (Römheld, 1987). Nongraminaceous plants rely on a set of iron deficiency responses termed strategy I. This strategy is based on the reduction of Fe(III) to the more soluble Fe(II) and then uptake of Fe(II) by a specific transporter (Römheld, 1987). The grasses use a chelation-based mechanism termed strategy II, which involves the release and subsequent use of low molecular mass, Fe(III)-specific chelators called siderophores. This type of mechanism also is used by many species of bacteria and fungi (Guerinot, 1994). The yeast Saccharomyces cerevisiae uses both reduction and chelation strategies (Askwith and Kaplan, 1998; Yun et al., 2000a, 2000b).
Strategy I consists of three biochemical responses—proton release, Fe(III) chelate reductase activity, and ferrous transport—upregulated in roots under conditions of iron deficiency. Recent work has identified the molecular basis for many of the strategy I responses. Fe(III) chelate reductase activity can be attributed to the product of the FRO2 (FERRIC REDUCTASE OXIDASE2) gene in Arabidopsis, which was identified by parallel mutational and sequence similarity approaches (Robinson et al., 1999). The Arabidopsis IRT1 gene (IRON-REGULATED TRANSPORTER1) also has been identified (Eide et al., 1996); its product has been shown to be the major transporter for the uptake of iron from the soil (Vert et al., 2002). As is the case for FRO2, IRT1 mRNA accumulates in the roots only under iron deficiency (Eide et al., 1996). The proton release probably is attributable to 1 of the 12 members of the AHA (ARABIDOPSIS H+-ATPase) gene family (Palmgren, 2001).
By contrast, little is known about the mechanisms that sense iron status in the plant and regulate the expression of the iron deficiency responses. To identify factors involved in iron homeostasis, we screened mutagenized Arabidopsis plants grown on sufficient iron to repress Fe(III) chelate reductase activity in wild-type plants. Our goal was to identify mutant individuals that continue to express root Fe(III) chelate reductase activity under these iron-sufficient conditions, reasoning that such plants would be likely to have defects in iron homeostasis. Defects in factors necessary for either regulation or iron accumulation and distribution could cause this phenotype. Two alleles of a single Arabidopsis locus were identified through this screen (Yi, 1995) and named frd3-1 and frd3-2 (ferric reductase defective). We demonstrated that frd3-1 constitutively expresses the iron deficiency–inducible transporter IRT1 in its roots (Eide et al., 1996).
In this work, we present further characterization of the frd3 mutant phenotype. We discovered that frd3 is allelic to the previously identified man1 mutant. This mutant, isolated originally as a manganese overaccumulator, also displays constitutive Fe(III) chelate reductase activity and overaccumulates a variety of metals in addition to manganese (Delhaize, 1996). Therefore, man1 has been renamed frd3-3. Plants that contain any of the three frd3 mutant alleles constitutively express strategy I iron deficiency responses, behaving as if they are iron deficient even though they contain higher levels of iron than the wild type. FRD3 encodes a 12–transmembrane domain protein belonging to the multidrug and toxin efflux (MATE) family of transmembrane efflux proteins. We also speculate on the role of FRD3 in iron homeostasis.
RESULTS
man1 Is Allelic to frd3
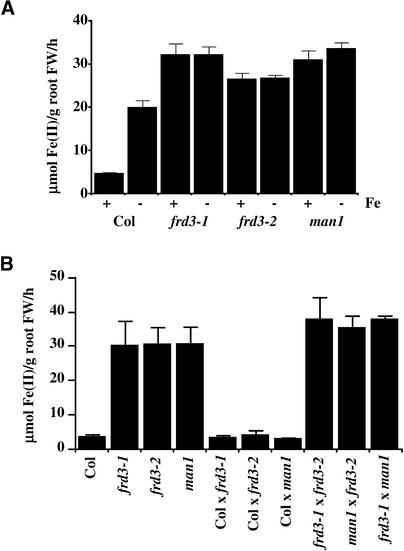
A comparison of wild-type, frd3-1, frd3-2, and man1 Fe(III) chelate reductase activities in both iron-sufficient and iron-deficient plants is shown in Figure 1A. In the wild type (ecotype Columbia), Fe(III) chelate reductase activity was induced approximately fourfold by iron deficiency. However, in all three of the mutants, Fe(III) chelate reductase activity was equivalent under iron-sufficient and iron-deficient growth conditions. In addition, cupric [Cu(II)] reductase activity also was expressed constitutively (data not shown). The Arabidopsis frd1 mutant lacks both Fe(III) chelate reductase activity and Cu(II) reductase activity (Yi and Guerinot, 1996). Both reductase activities are restored by the addition of a wild-type FRO2 gene (Robinson et al., 1999). Therefore, Cu(II) chelate reductase activity, like Fe(III) chelate reductase activity, is attributed to the FRO2 protein.
Figure 1.
Fe(III) Chelate Reductase Activity.
(A) frd3 and man1 exhibit constitutive Fe(III) chelate reductase activity. Plants grown with (+) or without (−) Fe(III) EDTA for 3 days were assayed for Fe(III) chelate reductase activity.
(B) frd3 and man1 are recessive and allelic. All plants were grown with Fe(III) EDTA for 3 days.
Data shown are means and se values of nine plants. Experiments were performed at least twice, and representative data sets are shown. FW, fresh weight.
Figure 1B shows Fe(III) chelate reductase activity of F1 progeny of frd3 mutants crossed to the wild-type parent and to each other. These plants were grown under iron-sufficient conditions to emphasize the mutant phenotype. F1 progeny from the wild type crossed to each of the mutants showed low, wild-type levels of Fe(III) chelate reductase activity, demonstrating that all three of the mutations are recessive; in fact, all three segregate as single recessive Mendelian loci (data not shown). F1 progeny from mutant-to-mutant crosses all showed high levels of reductase activity (Figure 1B), similar to the phenotypes of both parents. This finding indicates that none of the three mutants complement each other and that all of them carry mutant alleles of the same locus. Therefore, man1 has been renamed frd3-3.
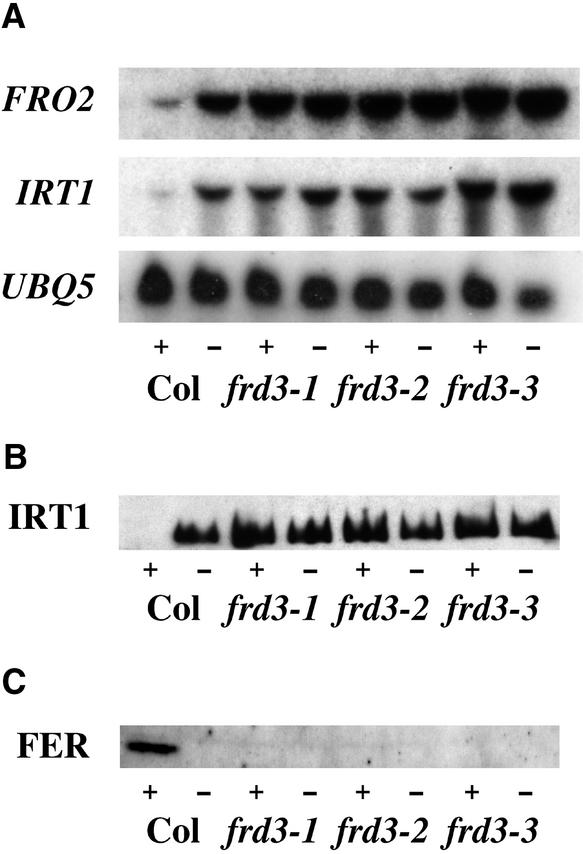
To further investigate Fe(III) chelate reductase activity, the expression of the FRO2 Fe(III) chelate reductase gene was examined by RNA gel blot hybridization. FRO2 encodes the iron deficiency–induced root Fe(III) chelate reductase (Robinson et al., 1999). Unlike the situation in wild-type plants, FRO2 was expressed constitutively in the roots of mutants homozygous for any of the three alleles of frd3 (Figure 2A). This result indicates that Fe(III) chelate reductase is misregulated at the transcriptional level in frd3 mutant plants.
Figure 2.
Expression of Iron Deficiency Responses.
(A) frd3 constitutively expresses IRT1 and FRO2. An RNA gel blot with 5 μg of total root RNA per lane that was probed with IRT1 and FRO2 is shown. RNA was extracted from plants grown with (+) or without (−) Fe(III) EDTA for 3 days. UBQ5 is shown as a loading control.
(B) frd3 accumulates IRT1 protein. An immunoblot of 10 μg of total root protein per lane that was probed with anti-IRT1 antibody is shown. Protein was extracted from roots of plants grown with (+) or without (−) Fe(III) EDTA for 3 days. A similar gel was stained with Coomassie blue to check for equal loading (data not shown).
(C) frd3 does not accumulate ferritin (FER) protein. An immunoblot of 30 μg of total shoot protein per lane that was probed with anti-ferritin antibody is shown. Protein was extracted from shoots of plants grown with (+) or without (−) Fe(III) EDTA for 3 days. A similar gel was stained with Coomassie blue to check for equal loading (data not shown).
These experiments were repeated twice, and similar results were obtained. Col, Columbia wild type.
frd3 Constitutively Expresses All Three Strategy I Responses
To determine if frd3 constitutively expresses another strategy I iron deficiency response, Fe(II) transport, the expression of the iron-regulated transporter gene IRT1 was examined. As shown in Figure 2A, the expression of IRT1 paralleled that of FRO2, with expression in the roots of frd3 mutant plants under both iron-sufficient and iron-deficient conditions. The IRT1 protein has been shown to be subject to post-transcriptional regulation (Connolly et al., 2002), so it cannot be assumed that increased mRNA levels correspond to increased levels of IRT1 protein. Therefore, IRT1 protein levels were assayed by immunoblot analysis (Figure 2B). The IRT1 protein accumulated in iron-sufficient roots of the frd3 mutant, providing additional evidence that frd3 cannot sense iron levels and is truly acting as if it is iron deficient under iron-sufficient growth conditions.
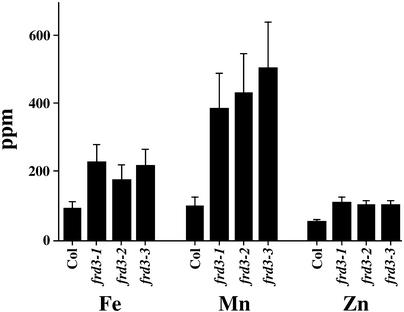
Because IRT1 was shown to transport iron, manganese, and zinc when expressed in yeast (Eide et al., 1996; Korshunova et al., 1999) and is overexpressed in frd3, shoot metal levels in the frd3 mutants were examined. As shown in Figure 3, mutants homozygous for any of the three alleles of frd3 showed two to three times higher levels of iron, two times higher levels of zinc, and three to four times higher levels of manganese in their shoots than the wild type. Copper levels were unchanged in the mutants (data not shown). frd3-3 (man1) has been shown to have higher levels of zinc and manganese but not iron in its shoots (Delhaize, 1996). This difference might be explained by the different levels of iron in the media used in the two studies.
Figure 3.
frd3 Alleles Accumulate More Fe, Mn, and Zn in Their Shoots.
Pooled samples of 2-week-old shoots from plants grown on B5 plates were subjected to elemental analysis. This experiment was repeated, and similar results were obtained. Col, Columbia wild type.
In this study, plants were grown on 100 μM ferrous sulfate, whereas Delhaize used 20 μM Fe(III) ethylenediamine di(o-hydroxyphenylacetic acid). In this study, iron levels of soil-grown plants were similar in the wild type and the frd3 mutants (data not shown), in agreement with results reported previously for soil-grown man1 (frd3-3) plants (Delhaize, 1996). Recently, seeds produced by man1 mutant plants were shown to have metal levels similar to those of seeds from wild-type plants (Lott and West, 2001); this finding is in agreement with our results (data not shown).
The iron storage protein ferritin accumulates in response to increased cellular levels of iron. Therefore, ferritin protein levels can be used as an indirect measurement of iron levels. Because ferritin synthesis has been shown to be controlled at both the transcriptional and post-transcriptional levels (Briat et al., 1999), only the levels of ferritin protein were examined by immunoblot analysis. As shown in Figure 2C, in the shoots, ferritin accumulated to detectable levels only in wild-type plants grown under iron-sufficient conditions.
In iron-deficient wild-type or mutant plants of either iron status, no ferritin protein was observed in the shoots. In plants, ferritin is localized to plastids, including chloroplasts (Briat et al., 1999). The lack of ferritin protein in frd3 mutant plants may indicate that their chloroplasts contain lower levels of iron than chloroplasts of wild-type plants. Iron-sufficient roots of wild-type and frd3 mutant plants contained approximately equal levels of ferritin, demonstrating that the mutants are capable of producing ferritin protein.
Because levels of the iron chelator nicotianamine (NA) have been shown to parallel iron levels in plant tissue (Pich et al., 2001), NA levels in the roots and shoots of wild-type and frd3-1 mutant plants grown under both iron-sufficient and iron-deficient conditions were examined by thin layer chromatography. All samples from frd3 mutant plants contained at least twofold higher amounts of NA compared with the corresponding samples from wild-type plants (data not shown).
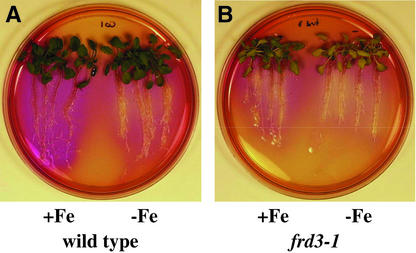
As shown with pH indicator plates in Figure 4, frd3-1 acidified the medium surrounding its roots when grown under both iron-sufficient and iron-deficient conditions. By contrast, the wild type acidified the surrounding medium only after being grown under iron-deficient conditions. This finding demonstrates that frd3 mutant plants constitutively efflux protons, another strategy I iron deficiency response. Thus, frd3 mutant plants constitutively express all three of the known strategy I iron deficiency responses.
Figure 4.
frd3-1 Exhibits Constitutive Acidification.
After being grown with (+Fe) or without (−Fe) Fe(III) EDTA for 3 days, plants were transferred to plates containing bromocresol purple for 18 h.
(A) Wild type (ecotype Columbia).
(B) frd3-1.
Iron-deficient wild type and iron-sufficient and iron-deficient frd3-1 reduced the pH of the medium to <5.2, as indicated by the yellow color. Iron-sufficient wild type caused the pH to increase to >7.0, as indicated by the purple color.
Cloning of FRD3 Using a Map-Based Approach
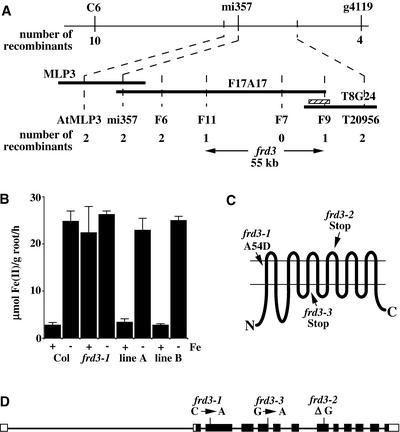
To identify the molecular basis of the frd3 phenotype, frd3-1 was crossed to Landsberg erecta and mapped using cleaved amplified polymorphic sequence markers (Konieczny and Ausubel, 1993). frd3 mapped to the top of chromosome 3, in agreement with published mapping data for man1 (Delhaize, 1996). Approximately 820 homozygous mutant F2 progeny from the interecotype cross were examined to refine the map position to a 55-kb interval, as shown in Figure 5A. This interval was covered completely by a single BAC, F17A17, that was sequenced as part of the Arabidopsis Genome Initiative (2000). Predicted open reading frames in this region were sequenced from one or more of the frd3 alleles in a search for differences between the mutant and wild-type sequences. Nonsynonymous single-base-pair alterations were found in all three alleles in one open reading frame in this region.
Figure 5.
Positional Cloning and Structure of the FRD3 Gene.
(A) The region of chromosome 3 containing FRD3. The chromosome is depicted by the uppermost horizontal line with the flanking markers C6 and g4119. Below that are three BACs from the Arabidopsis Genome Initiative (2000) minimal tiling path: MLP3, F17A17, and T8G24. Markers (see Methods), the number of recombinant chromosomes from the 1640 examined, and the final 55-kb interval containing FRD3 are shown below. The striped bar indicates the segment of genomic DNA used to complement frd3-1.
(B) Complementation of frd3-1. An 11-kb segment of genomic DNA, when expressed in frd3-1, restores the repression of Fe(III) chelate reductase activity in plants grown for 3 days in the presence of Fe(III) EDTA. Values shown are means of nine individual plants, and error bars depict se. Col, Columbia wild type.
(C) Predicted topology of the FRD3 protein. The 12 transmembrane domains, as predicted by HMMTOP, and the location and nature of the mutations carried by the three mutant alleles are shown.
(D) Intron/exon structure of FRD3. The narrow lines depict intron sequences, and the boxes depict exon sequences. The closed boxes correspond to the open reading frame, and the open boxes correspond to the 5′ and 3′ untranslated regions. Line lengths are approximately to scale.
Expression of wild-type genomic DNA containing only this open reading frame (striped box in Figure 5A) in frd3-1 complemented the chlorotic phenotype and restored the iron deficiency inducibility of Fe(III) chelate reductase activity (Figure 5B). This finding proves that the gene containing the mutations responsible for the frd3 phenotypes has been identified. FRD3 is predicted to encode an integral membrane protein of 526 amino acids.
The computer topology prediction program HMMTOP (Tusnady and Simon, 2001) predicts 12 transmembrane domains, as diagrammed in Figure 5C, with the N and C termini on the cytoplasmic side of the membrane. FRD3 is predicted to localize to the plasma membrane according to PSORT (Nakai and Kanehisa, 1992) and TargetP (Emanuelsson et al., 2000). Although mitochondrial, chloroplastic, and secretory system targeting sequences can be predicted with 85% accuracy, predictions of the final location of proteins within the secretory system are significantly less accurate (Emanuelsson et al., 2000).
The FRD3 gene matched an EST sequence; the corresponding cDNA clone was obtained from the Kazusa DNA Research Institute (Kisarazu, Japan) and sequenced completely. The cDNA sequence has been deposited in GenBank. A string of A's at the 3′ end of the sequence and 5′ rapid amplification of cDNA ends confirmed that this clone was full length. The transcriptional start site is 117 bp upstream of the ATG. The cDNA sequence is consistent with the protein sequence predicted by the Arabidopsis Genome Initiative (2000).
Comparison of the FRD3 genomic and cDNA sequences revealed that the FRD3 gene has 13 exons and 12 introns, as diagrammed in Figure 5D. It is notable that the first intron is in the 5′ untranslated region and is almost 2.6 kb in length; this is much larger than the ∼170-bp average for Arabidopsis introns (Arabidopsis Genome Initiative, 2000). Long introns in other Arabidopsis genes have been shown to play important roles in the regulation of gene expression (Jeon et al., 2000).
Figure 5D also indicates the single nucleotide sequence changes in the three frd3 mutant alleles. frd3-1 has a C-to-A transversion. In the protein, this causes the substitution of Asp for Ala at position 54 in the first transmembrane domain (Figure 5C). frd3-2 has a deletion of a single G in the eighth exon, causing a frameshift and the addition of seven novel amino acids followed by a premature stop codon. Thus, frd3-2 codes for approximately two-thirds of the wild-type protein.
frd3-3 has a G-to-A transition in the first nucleotide of the fifth intron. Because this G is part of the required GT in the splice donor site, this change is predicted to lead to the retention of the intron. Sequence data from a frd3-3 reverse transcriptase–mediated (RT)–PCR product confirms that this intron is retained (data not shown). The retention of this intron shifts the reading frame at a point approximately halfway through the protein, leading to the addition of two novel amino acids followed by a premature stop codon.
FRD3 Expression
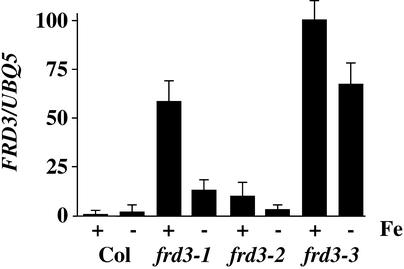
The expression of FRD3 in the roots of the wild type and the three frd3 mutants is shown in Figure 6. No expression was detected in the shoots of the wild type or any of the mutant plants by RNA gel blot hybridization or by RT-PCR (data not shown). Although RT-PCR is a highly sensitive method for the detection of mRNAs (Foley et al., 1993), we cannot exclude low-level expression of FRD3 in the shoots, especially at levels corresponding to one to two mRNAs per cell, or in a small subset of cells of the shoot. In wild-type roots, FRD3 was expressed under both iron-sufficient and iron-deficient conditions. After normalization to the control gene UBQ5, FRD3 mRNA levels were approximately twofold higher under iron deficiency in the wild type.
Figure 6.
FRD3 Expression Levels Depend on Iron Status and Genotype.
The RNA gel blot used in Figure 3 was reprobed with FRD3. The expression level of FRD3 was normalized to UBQ5 and is shown in arbitrary units. Results are presented as a graph to emphasize the very large differences in expression levels. Values shown are means of three replicate experiments, and error bars indicate se. Col, Columbia wild type.
It is interesting that plants homozygous for any of the three mutant alleles had FRD3 mRNA levels considerably higher than the wild type, and these were higher under iron-sufficient than under iron-deficient conditions. The difference in FRD3 mRNA levels under iron sufficiency varied from ∼10-fold higher than the wild type in frd3-2 to almost 100-fold higher in frd3-3. This finding implies that FRD3 itself is regulated by a process influenced by its gene product. Because FRD3 is predicted to be an integral membrane protein, this is probably an indirect effect.
The MATE Gene Family
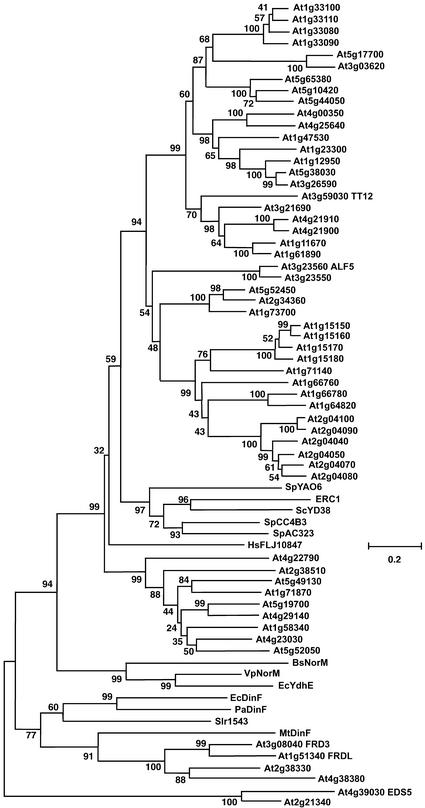
FRD3 is predicted to be a member of the MATE family, an extensive group of membrane proteins of largely unknown function. There are MATE family members in humans, in the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, in Escherichia coli and other bacteria, and in archaea. Arabidopsis has 56 MATE family members, which is 10 times more than any other sequenced organism (Arabidopsis Genome Initiative, 2000).
Figure 7 shows a dendrogram of all 56 Arabidopsis proteins, 1 human protein, 5 proteins from yeast, and selected bacterial members. The Arabidopsis genes fall into two main groups. The top group in Figure 7 contains 50 Arabidopsis members and is associated loosely with the yeast and human family members. The other, smaller group contains FRD3 and the bacterial NorM and DinF proteins.
Figure 7.
Dendrogram Showing Amino Acid Sequence Similarity Relationships among Selected MATE Family Members.
Multiple sequence alignments were determined using the BCM Search Launcher, and the dendrogram was produced using MEGA 2.1. Bootstrap values are shown next to each junction.
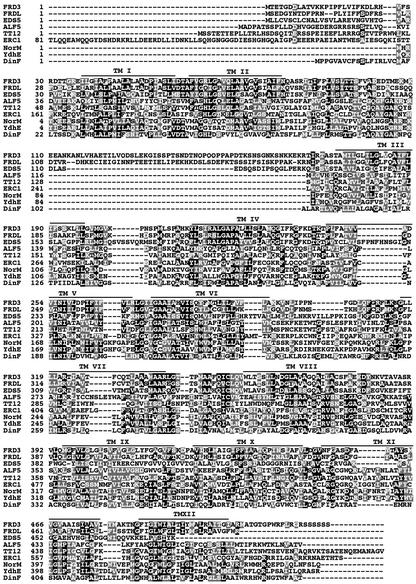
Figure 8 shows an alignment of nine MATE proteins: five from Arabidopsis, ERC1 from yeast, and three from bacterial family members. These nine proteins share sequence similarity along their entire lengths, except for the very N-terminal portion. As expected, the transmembrane domains are the most conserved. FRD3 is 57.8% identical to another Arabidopsis protein, FRD3-like or FRDL. FRD3 and FRDL are unique among the MATE family members shown in Figure 8 in possessing an enlarged cytoplasmic loop between transmembrane domains II and III.
Figure 8.
Alignment of the Amino Acid Sequences of Selected MATE Family Members.
Identical residues are shown on a black background, and conservative substitutions are shown on a gray background. Lines depict FRD3 transmembrane domains (TM I to TM XIII) as predicted by HMMTOP. Multiple sequence alignments were determined using the BCM Search Launcher, and residues were shaded using BoxShade 3.21.
DISCUSSION
The frd3 mutant phenotype includes chlorosis, expression of iron deficiency responses under conditions of iron sufficiency, and an overaccumulation of iron and other metals. There are two models that most easily explain this phenotype. First, frd3 mutant plants could have an iron-signaling defect. Specifically, frd3 mutant plants might be unable to sense iron levels, communicate information about iron status between various parts of the plant, or repress the expression of iron deficiency responses. Alternatively, the frd3 mutant phenotype could result from incorrect localization of iron in the shoot. If the iron in the shoots was unavailable to the cells or organelles that generate the iron deficiency signal, the roots would be appropriately responding to a need for additional iron in portions of the shoot.
Relatively little is known about the regulation of iron deficiency responses in Arabidopsis or other plant species. The pea mutants brz (bronze) and dgl (degenerative leaves) both exhibit constitutive expression of strategy I responses and overaccumulation of iron (Gottschalk, 1987; Kneen et al., 1990; Welch and LaRue, 1990; Grusak and Pezeshgi, 1996). Grafting studies, in which wild-type shoots were grafted onto mutant roots and vice versa, indicate that there is a shoot-derived signal that controls the iron uptake responses in the root (Grusak and Pezeshgi, 1996). When wild-type shoots were grafted onto brz roots, the roots regained the wild-type phenotype and expressed iron uptake responses only under conditions of iron deficiency. And when brz or dgl shoots were grafted onto wild-type roots, the roots expressed iron deficiency responses constitutively.
The fact that the behavior of the roots is determined by the genotype of the grafted shoots implies that a signal from the shoot controls the expression of iron deficiency responses in the root. frd3 mutant plants have phenotypes similar to those of brz and dgl mutant plants. However, because FRD3 expression has not been detected in the shoots, FRD3 is more likely to be involved in the perception of this shoot-derived signal.
However, it is possible that iron deficiency signaling is intact in the frd3 mutant and that iron localization is altered. Although frd3 mutant plants have high levels of iron in their shoot tissue (Figure 3), it is difficult to measure iron levels in specific cell types or subcellular organelles. The lack of ferritin protein in the shoots of frd3 mutant plants indicates that their chloroplasts may have lower levels of iron than the chloroplasts of wild-type plants. Conceivably, mislocalization of iron in the mutant could result in certain cells or organelles, such as the chloroplast, becoming iron deficient, even though the shoots as a whole have more iron than in the wild type.
If these iron-deficient cells or organelles were the source of the shoot-to-root iron deficiency signal mentioned above, the roots of frd3 mutants would simply be responding appropriately to a shoot iron deficiency signal and constitutively expressing the three strategy I responses. Because the FRD3 gene is not expressed in shoot tissue, its gene product could only have an indirect effect on iron localization in the shoot. For example, the wild-type FRD3 protein could efflux, into the vascular system, an iron chelator that is synthesized only in the roots and that is necessary for iron transport into certain cells or organelles in the shoot. In-depth characterization of the frd3 mutant phenotype and the role of the wild-type FRD3 protein is in progress and will distinguish between these hypotheses.
Genetically, the frd3 mutant phenotype is recessive to the wild type, indicating a loss of function. The severely truncated FRD3 proteins predicted by the DNA sequences of the frd3-2 and frd3-3 alleles certainly are consistent with a loss-of-function phenotype. It is unclear if the single amino acid substitution predicted by the frd3-1 DNA sequence would lead to a total loss of function. However, it is easy to imagine how a nonconservative amino acid substitution, such as frd3-1's Ala to Asp in the first transmembrane domain, could have significant effects on the protein's localization, stability, or function. Additionally, there were no significant differences in the expression of iron deficiency responses among mutants carrying any of the three frd3 alleles. Therefore, it may be assumed that all three frd3 alleles are equally nonfunctional.
The only differences among the three alleles observed to date are in the expression levels of the FRD3 gene itself. Plants carrying any of the three alleles expressed higher levels of FRD3 mRNA than wild-type plants. This increase varied from 10-fold higher in frd3-2 to almost 100-fold higher in frd3-3 (Figure 6). FRD3 also was expressed at higher levels under iron sufficiency than under iron deficiency in mutants carrying any of the three alleles. This finding is in contrast to what was seen in the wild type, in which FRD3 was induced twofold by iron deficiency. Wild-type FRD3 acted under conditions of iron sufficiency because that is the situation in which the mutant phenotype is most apparent. An autoregulatory mechanism may sense a lack of FRD3 function more acutely under iron sufficiency and induce FRD3 mRNA to higher levels in the mutants.
The biochemical function of FRD3 is not clear. The NorM gene from Vibrio parahaemolyticus is the best-characterized MATE family member to date. NorM has been shown to encode a Na+/drug antiport efflux system for structurally unrelated antibiotics such as norfloxacin, kanamycin, and streptomycin and small toxic molecules such as ethidium (Morita et al., 1998, 2000). Although biochemical functions for Arabidopsis MATEs have not been demonstrated, the ALF5 (Diener et al., 2001) and TT12 (Debeaujon et al., 2001) mutant phenotypes are consistent with the involvement of these genes in transporting small, organic molecules.
The ALF5 gene is expressed in Arabidopsis root epidermis and when mutated leads to increased root sensitivity to a variety of inhibitory compounds, including a contaminant of commercial agar and tetramethylammonium (Diener et al., 2001), suggesting that ALF5 transports these inhibitory compounds either out of the epidermal cells or into the vacuole. The TT12 mutant phenotype is an alteration in seed coat (or testa) pigmentation (Debeaujon et al., 2001). It is probable that the TT12 protein functions to transport flavonoids into the vacuoles of the seed coat endothelium.
A biochemical function for EDS5 is less clear from its mutant phenotype. eds5 mutant plants are more susceptible to certain bacterial pathogens and have lower levels of salicylic acid than wild-type plants after pathogen attack (Nawrath and Metraux, 1999). It is possible that EDS5 functions to transport salicylic acid or a precursor of salicylic acid (Nawrath et al., 2002). Another Arabidopsis MATE gene, AtDTX1 (At2g04070), was cloned recently by functional complementation of an E. coli acrAB mutant deficient in multidrug resistance (Li et al., 2002). AtDTX1 functions in E. coli as an efflux carrier of plant-derived alkaloids, antibiotics, and other toxic compounds (Li et al., 2002).
The demonstrated and proposed biochemical functions of various MATE family members make it likely that FRD3 also functions to transport a low molecular mass organic compound. Because iron overaccumulates in the shoots of frd3 mutant plants, FRD3 cannot be a major factor in iron translocation between Arabidopsis roots and shoots. Therefore, it is unlikely that iron is a substrate for FRD3.
FRD3 may transport the metal chelator NA, a polyamine synthesized from the condensation of three molecules of S-adenosyl Met. NA has been suggested to function in the vascular transport of transition metals. Much of what we know about the role of NA in plants comes from studies of the tomato mutant chloronerva (chln), which lacks NA. chln shows constitutive iron deficiency responses (King, 1991), is chlorotic, and accumulates more iron than the wild type in its tissues (Stephan and Scholz, 1993). The chln gene was cloned recently using a map-based approach and shown to encode a functional NA synthase (Ling et al., 1999).
It has been hypothesized that NA is necessary for proper iron storage and intracellular localization (Becker et al., 1995). This would explain the chln mutant phenotype. Without NA, iron does not reach the locations where it is needed in the leaves, causing chlorosis and constitutive expression of the iron deficiency responses. NA synthase genes also have been cloned from barley, rice, and Arabidopsis (Herbik et al., 1999; Higuchi et al., 1999, 2001; Suzuki et al., 1999). Although we have shown that frd3 mutant plants contain NA, nothing is known about the transport or localization of NA in frd3 mutant plants. It is possible that mislocalization of NA would cause phenotypes similar to the total lack of NA, as seen in chln.
The product of the maize ys1 gene was identified recently as a transporter of phytosiderophores that are structurally similar to NA (Curie et al., 2001) and is likely to transport NA as well. The primary sequences for ys1 and FRD3 are unrelated, implying that FRD3 has a different function than ys1. Indeed, there are eight Arabidopsis genes that encode proteins similar to the ys1 gene product (Curie et al., 2001). These proteins are better candidates for Arabidopsis NA transporters than is FRD3. Additionally, frd3 mutant plants contain higher levels of NA in both the shoots and the roots than do wild-type plants. This feature is more likely to be related to its iron overaccumulation defect than to a defect in NA transport.
In the pea mutants brz and dgl and in iron-overloaded wild-type pea, NA levels have been shown to parallel iron levels, implying that NA synthesis is induced by high iron content (Pich et al., 2001). It is possible that FRD3 is not a transporter and is instead a receptor. Several proteins have sequence similarity to transporters but appear to function as receptors. The Arabidopsis gene EIN2 is a central component of the ethylene signal transduction pathway (Alonso et al., 1999). Although the N-terminal portion of EIN2 shows similarity to the Nramp family of metal ion transporters, the C-terminal cytoplasmic domain does not. Expression of only the C-terminal domain is sufficient to constitutively activate ethylene responses, further implicating EIN2 in ethylene signal transduction (Alonso et al., 1999). No metal-transporting activity has been detected for EIN2 (Alonso et al., 1999).
This situation is similar to that of the yeast Glc sensors Snf3 and Rgt2. These proteins also possess N-terminal portions similar to Glc transporters and C-terminal hydrophilic domains that can activate Glc responses when overexpressed (Coons et al., 1997; Ozcan et al., 1998). EIN2, Snf3, and Rgt2 have in common a large cytoplasmic C-terminal domain that is not conserved in their transporter relatives. Although FRD3 does not have a C-terminal extension, it does have an ∼60–amino acid addition between transmembrane domains 2 and 3. However, this loop is much smaller than the signaling domains of EIN2, Snf3, and Rgt2.
Experiments are ongoing to further characterize FRD3 and to elucidate FRD3's role in iron deficiency signaling and homeostasis. It is of crucial importance to determine the biochemical function of FRD3 and to identify additional proteins that act in this pathway. Whether FRD3 is involved in iron deficiency signaling or in iron localization, the phenotype of the frd3 mutant plants indicates that it is an important component of the iron homeostatic mechanism in Arabidopsis. The cloning of FRD3 provides a start for the characterization of the iron deficiency response pathway and the identification of novel pathway components.
METHODS
Arabidopsis Lines and Growth Conditions
The Arabidopsis thaliana mutants frd3-1 and frd3-2 and the corresponding Columbia gl-1 wild type have been described previously (Yi, 1995). man1 was obtained from the ABRC (http://www.biosci.ohio-state.edu/~plantbio/Facilities/abrc/abrchome.htm). Unless specified otherwise, plants were grown under sterile conditions as described previously (Yi and Guerinot, 1996). Briefly, seeds were sown on Petri plates containing Gamborg's B5 medium (Sigma) and grown until the four– to six–true leaf stage. Plants then were transferred to plates with or without 50 μM Fe(III) EDTA for iron-sufficient or iron-deficient conditions, respectively, for 3 days before analysis. Both Fe(III) chelate reductase assays and the pH plates also were described previously (Yi and Guerinot, 1996).
RNA Gel Blot Hybridization
RNA isolation, RNA gel blot analysis, and all molecular biology procedures were performed using standard protocols (Ausubel et al., 2002). RNA gel blots (5 μg of total RNA per lane) were visualized by exposure to either x-ray film for 1 to 2 days or to a Typhoon PhosphorImager screen (Molecular Dynamics, Sunnyvale, CA) for 4 to 24 h. IRT1 and FRO2 probes were made from previously described cDNA clones (Eide et al., 1996; Robinson et al., 1999); the UBQ5 probe was a PCR product amplified as described previously (Rogers and Ausubel, 1997). Probe DNA containing ∼30 μC of 32P was used for each blot.
Immunoblot Analysis
Immunoblot analysis was performed as described previously (Connolly et al., 2002). Total protein was prepared from the roots and shoots of plants grown axenically on plates that were either iron deficient or iron sufficient. Extracts were prepared by grinding tissue (1 to 2 mL of buffer per 1 g of wet tissue) on ice in extraction buffer (50 mM Tris, pH 8.0, 5% glycerol, 4% SDS, 1% polyvinylpolypyrrolidone, and 1 mM phenylmethylsulfonyl fluoride), followed by centrifugation at 4°C for 15 min at 14,000g. The supernatant was recovered, and total protein was estimated using the bicinchoninic acid protein assay (Pierce, Rockford, IL). Samples for SDS-PAGE were diluted with an equal volume of 2 × sample prep buffer (Ausubel et al., 2002) and boiled for 2 min.
Total protein (10 to 30 μg) was separated by SDS-PAGE (Laemmli, 1970) and transferred to either polyvinylidene fluoride or nitrocellulose membranes by electroblotting (Towbin et al., 1979). Membranes were blocked in 1 × PBST (0.1% Tween 20 in 1 × PBS) with 5% nonfat dry milk for 3 h at 37°C and then washed two times in 1 × PBST for 5 min each. The membranes then were incubated overnight at 4°C with either anti-IRT1 or anti-ferritin antibody (1:1000 dilution in 1 × PBST and 1% nonfat dry milk for both). The IRT1 antibody was raised and affinity purified against a synthetic peptide (PANDVTLPIKEDDSSN) that corresponds to amino acids 162 to 177 of the IRT1 deduced protein sequence and that is unique to IRT1 (Quality Controlled Biochemicals, Hopkinton, MA).
The ferritin antibody was raised against purified pea seed ferritin (Van Wuytswinkel et al., 1995) and was a kind gift from Jean-François Briat (Centre National de la Recherche Scientifique, Montpellier, France). It has been shown to cross-react with Arabidopsis ferritin (Gaymard et al., 1996). Next, the membranes were washed in 1 × PBST four times for 15 min each. Membranes then were incubated for 1 h with goat anti-rabbit IgG conjugated to horseradish peroxidase (1:5000 dilution in 1 × PBST and 1% nonfat dry milk) (Pierce) followed by four washes for 15 min each in 1 × PBST. Chemiluminescence was performed using the Renaissance protein gel blot chemiluminescence reagent according to the instructions of the manufacturer (DuPont–New England Nuclear Life Science Products).
Elemental Analysis
Approximately 200 plants grown under iron-sufficient or iron-deficient conditions were pooled and subjected to elemental analysis. The metal content of the tissue was determined by the Dartmouth Superfund Trace Metal Core Facility using a magnetic sector inductively coupled plasma mass spectrometer (Finnigan MAT, Bremen, Germany) as described previously (Chen et al., 2000). All values obtained were within the linear sensitivity range for this instrument.
Detection of Nicotianamine
Nicotianamine was extracted from Arabidopsis tissue as described previously (Pich et al., 2001). Briefly, samples were ground in liquid nitrogen, extracted in water at 80°C, and centrifuged, and the supernatant was dried by lyophilization. Extracts were redissolved in water and spotted on thin layer chromatography plates, which were developed in butanol:acetic acid:water (4:1:1) (Shojima et al., 1989). Nicotianamine was visualized after reaction with ninhydrin. Samples were chromatographed alongside chemically synthesized nicotianamine (a kind gift of Axel Pich, Oldenburg, Germany) for identification purposes.
frd3 Mapping and Complementation
Cleaved amplified polymorphic sequence, simple sequence length polymorphism, and restriction fragment length polymorphism markers, which are available publicly on the Arabidopsis Information Resource World Wide Web page (http://www.arabidopsis.org/home.html), were used where possible to obtain an approximate map position of frd3. AtMLP3, F6, and F7 are simple sequence length polymorphism markers constructed around simple sequence repeats in the corresponding BAC sequence. F9 is a restriction fragment length polymorphism marker identified experimentally.
The polymorphism covered by F11 is from the Cereon Arabidopsis Polymorphism Collection (available on the Arabidopsis Information Resource World Wide Web page) and was scored by sequencing PCR products of that region. The complementing clone was constructed by digesting BAC T8G24 and ligating the total digest into the binary vector pCambia2300 (http://www.cambia.org.au/) according to standard molecular biology procedures (Ausubel et al., 2002).
The resulting clones were screened by PCR for the construct of interest. The complementing clone was introduced into the frd3-1 mutant by Agrobacterium tumefaciens–mediated transformation (Clough and Bent, 1998). 5′ rapid amplification of cDNA ends was performed according to the instruction manual for the 5′ System for Rapid Amplification of cDNA Ends, version 2.0 (Life Technologies, Bethesda, MD).
DNA and Protein Sequence Analysis
DNA sequencing was performed at the Dartmouth Molecular Biology Core Facility on an ABI Prism 3100 automated DNA sequencer (Applied Biosystems, Foster City, CA). Sequences were analyzed with the Genetics Computer Group (Madison, WI) software package and by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/). Multiple sequence alignments were determined using the BCM Search Launcher (http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html). These alignments were transformed into dendrograms using MEGA version 2.1 (http://www.megasoftware.net/) or colored using BoxShade (http://www.ch.embnet.org/software/BOX_form.html)
Accession Number
The GenBank accession number for the cDNA clone from the FRD3 gene is AF448231.
Acknowledgments
The authors thank Manny Delhaize for man1 seed, Tama Fox for preliminary experimentation, Bjorn Klaue for help with inductively coupled plasma mass spectrometry, Mark McPeek for help with MEGA 2.1, and David Eide, Rob McClung, and Laura Green for critical reading of the manuscript. This work was supported by a National Science Foundation grant to M.L.G. E.E.R. was a Department of Energy Energy BioSciences Fellow of the Life Sciences Research Foundation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001495.
References
- Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Askwith, C., and Kaplan, J. (1998). Iron and copper transport in yeast and its relevance to human disease. Trends Biochem. Sci. 23, 135–138. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (2002). Current Protocols in Molecular Biology. (New York: John Wiley & Sons).
- Becker, R., Fritz, E., and Manteuffel, R. (1995). Subcellular localization and characterization of excessive iron in the nicotianamine-less tomato mutant chloronerva. Plant Physiol. 108, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat, J.F., Lobreaux, S., Grignon, N., and Vansuyt, G. (1999). Regulation of plant ferritin synthesis: How and why. Cell. Mol. Life Sci. 56, 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.Y., Stemberger, R.S., Klaue, B., Blum, J.D., Pickhardt, P.C., and Folt, C.L. (2000). Accumulation of heavy metals in food web components across a gradient of lakes. Limnol. Oceanogr. 45, 1525–1536. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Connolly, E.C., Fett, J., and Guerinot, M.L. (2002). Transgenic plants engineered to overexpress the IRT1 metal transporter reveal post-transcriptional regulation by metals. Plant Cell 14, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coons, D.M., Vagnoli, P., and Bisson, L.P. (1997). The C-terminal domain of Snf3p is sufficient to complement the growth defect of snf3 null mutations in Saccharomyces cerevisiae: SNF3 functions in glucose recognition. Yeast 13, 9–20. [DOI] [PubMed] [Google Scholar]
- Curie, C., Panaviene, Z., Loulergue, C., Dellaporta, S.L., Briat, J.-F., and Walker, E.L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409, 346–349. [DOI] [PubMed] [Google Scholar]
- Debeaujon, I., Peeters, A.J.M., Leon-Kloosterziel, K.M., and Koornneef, M. (2001). The TRANSPARENT TESTA12 gene of Arabidopsis encodes a multidrug secondary transporter–like protein required for flavonoid sequestration in vacuoles of the seed coat endothelium. Plant Cell 13, 853–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize, E. (1996). A metal-accumulator mutant of Arabidopsis thaliana. Plant Physiol. 111, 849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener, A.C., Gaxiola, R.A., and Fink, G.R. (2001). Arabidopsis ALF5, a multidrug efflux transporter gene family member, confers resistance to toxins. Plant Cell 13, 1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide, D., Broderius, M., Fett, J., and Guerinot, M.L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 93, 5624–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Foley, K.P., Leonard, M.W., and Engel, J.D. (1993). Quantitation of RNA using the polymerase chain reaction. Trends Genet. 9, 380–385. [DOI] [PubMed] [Google Scholar]
- Gaymard, F., Boucherez, J., and Briat, J.F. (1996). Characterization of ferritin mRNA from Arabidopsis thaliana accumulated in response to iron through an oxidative pathway independent of abscisic acid. Biochem. J. 318, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk, W. (1987). Improvement in the selection value of gene dgl through recombination. Pisum Newsl. 19, 9–11. [Google Scholar]
- Grusak, M.A., and Pezeshgi, S. (1996). Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol. 110, 329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot, M.L. (1994). Microbial iron transport. Annu. Rev. Microbiol. 48, 743–772. [DOI] [PubMed] [Google Scholar]
- Halliwell, B., and Gutteridge, J.M.C. (1992). Biologically relevant metal ion-dependent hydroxyl radical generation. FEBS Lett. 307, 108–112. [DOI] [PubMed] [Google Scholar]
- Herbik, A., Koch, G., Mock, H.-P., Dushkov, D., Czihal, A., Thielmann, J., Stephan, U.W., and Baeumlein, H. (1999). Isolation, characterization and cDNA cloning of nicotianamine synthase from barley: A key enzyme for iron homeostasis in plants. Eur. J. Biochem. 265, 231–239. [DOI] [PubMed] [Google Scholar]
- Higuchi, K., Suzuki, K., Nakanishi, H., Yamaguchi, H., Nishizawa, N.K., and Mori, S. (1999). Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol. 119, 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, K., Wantanabe, S., Takahashi, M., Kawasaki, S., Nakanishi, H., Nishizawa, N.K., and Mori, S. (2001). Nicotianamine synthase gene expression differs in barley and rice under Fe-deficient conditions. Plant J. 25, 159–167. [DOI] [PubMed] [Google Scholar]
- Jeon, J.-S., Lee, S., Jung, K.-H., Jun, S.-H., Kim, C., and An, G. (2000). Tissue-preferential expression of a rice α-tubulin gene, OsTubA1, mediated by the first intron. Plant Physiol. 123, 1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, J. (1991). The Genetic Basis of Plant Physiological Processes. (London: Oxford University Press).
- Kneen, B.E., LaRue, T.A., Welch, R.M., and Weeden, N.F. (1990). Pleiotropic effects of brz. Plant Physiol. 93, 717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Korshunova, Y., Eide, D., Clark, G., Guerinot, M., and Pakrasi, H. (1999). The Irt1 protein from Arabidopsis thaliana is a metal transporter with broad specificity. Plant Mol. Biol. 40, 37–44. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Li, L., He, Z., Pandey, G.K., Tsuchiya, T., and Luan, S. (2002). Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 277, 5360–5368. [DOI] [PubMed] [Google Scholar]
- Ling, H.-Q., Koch, G., Baumlein, H., and Ganal, M.W. (1999). Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc. Natl. Acad. Sci. USA 96, 7098–7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott, J.N.A., and West, M.M. (2001). Elements present in mineral nutrient reserves in dry Arabidopsis thaliana seeds of wild type and pho1, pho2, and man1 mutants. Can. J. Bot. 79, 1292–1296. [Google Scholar]
- Morita, Y., Kataoka, A., Shiota, S., Mizushima, T., and Tsuchiya, T. (2000). NorM of Vibrio parahaemolyticus is a Na+-driven multidrug efflux pump. J. Bacteriol. 182, 6694–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita, Y., Kodama, K., Shiota, S., Mine, T., Kataoka, A., Mizushima, T., and Tsuchiya, T. (1998). NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob. Agents Chemother. 42, 1778–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, K., and Kanehisa, M. (1992). A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., Heck, S., Parinthawong, N., and Metraux, J.-P. (2002). EDS5, an essential component of SA-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE-transporter family. Plant Cell 14, 275–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath, C., and Metraux, J.-P. (1999). Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11, 1393–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan, S., Dover, J., and Johnston, M. (1998). Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 17, 2566–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren, M.G. (2001). Plant plasma membrane H+-ATPases: Powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–845. [DOI] [PubMed] [Google Scholar]
- Pich, A., Manteuffel, R., Hillmer, S., Scholz, G., and Schmidt, W. (2001). Fe homeostasis in plant cells: Does nicotianamine play multiple roles in the regulation of cytoplasmic Fe concentration? Planta 213, 967–976. [DOI] [PubMed] [Google Scholar]
- Robinson, N.J., Procter, C.M., Connolly, E.L., and Guerinot, M.L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697. [DOI] [PubMed] [Google Scholar]
- Rogers, E.E., and Ausubel, F.M. (1997). Arabidopsis enhanced disease susceptibility mutants exhibit enhanced susceptibility to several bacterial pathogens and alteration in PR-1 gene expression. Plant Cell 9, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld, V. (1987). Different strategies for iron acquisition in higher plants. Physiol. Plant. 70, 231–234. [Google Scholar]
- Shojima, S., Nishizawa, N.-K., and Mori, S. (1989). Establishment of a cell-free system for the biosynthesis of nicotianamine. Plant Cell Physiol. 30, 673–677. [Google Scholar]
- Stephan, U.W., and Scholz, G. (1993). Nicotianamine: Mediator of transport of iron and heavy metals in the phloem? Physiol. Plant. 88, 522–529. [Google Scholar]
- Suzuki, K., Higuchi, K., Nakanishi, H., Nishizawa, N.K., and Mori, S. (1999). Cloning of nicotianamine synthase genes from Arabidopsis thaliana. Soil Sci. Plant Nutr. 45, 993–1002. [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady, G.E., and Simon, I. (2001). Principles governing amino acid composition of integral membrane proteins: Applications to topology prediction. J. Mol. Biol. 283, 489–506. [DOI] [PubMed] [Google Scholar]
- Van Wuytswinkel, O., Savino, G., and Briat, J.-F. (1995). Purification and characterization of recombinant pea-seed ferritins expressed in Escherichia coli: Influence of N-terminus deletions on protein solubility and core formation in vitro. Biochem. J. 305, 253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert, G., Grotz, N., Dédaldéchamp, F., Gaymard, F., Guerinot, M., Briat, J.-F., and Curie, C. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and plant growth. Plant Cell 14, 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, R.M., and LaRue, T.A. (1990). Physiological characteristics of Fe accumulation in the ‘Bronze’ mutant of Pisum sativum L., cv ‘Sparkle’ E107 (brz brz). Plant Physiol. 93, 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, Y. (1995). Iron Uptake in Arabidopsis thaliana. PhD dissertation (Hanover, NH: Dartmouth College).
- Yi, Y., and Guerinot, M.L. (1996). Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 10, 835–844. [DOI] [PubMed] [Google Scholar]
- Yun, C.-W., Ferea, T., Rashford, J., Ardon, O., Brown, P.O., Botstein, D., Kaplan, J., and Philpott, C.C. (2000. a). Desferrioxamine-mediated iron uptake in Saccharomyces cerevisiae: Evidence for two pathways of iron uptake. J. Biol. Chem. 275, 10709–10715. [DOI] [PubMed] [Google Scholar]
- Yun, C.-W., Tiedman, J.S., Moore, R.E., and Philpott, C.C. (2000. b). Siderophore-iron uptake in Saccharomyces cerevisiae: Identification of ferrichrome and fusarine transporters. J. Biol. Chem. 275, 16354–16359. [DOI] [PubMed] [Google Scholar]