Abstract
The soil-borne fungus Fusarium oxysporum causes vascular wilts of a wide variety of plant species by directly penetrating roots and colonizing the vascular tissue. The pathogenicity mutant B60 of the melon wilt pathogen F. oxysporum f. sp. melonis was isolated previously by restriction enzyme–mediated DNA integration mutagenesis. Molecular analysis of B60 identified the affected gene, designated FOW1, which encodes a protein with strong similarity to mitochondrial carrier proteins of yeast. Although the FOW1 insertional mutant and gene-targeted mutants showed normal growth and conidiation in culture, they showed markedly reduced virulence as a result of a defect in the ability to colonize the plant tissue. Mitochondrial import of Fow1 was verified using strains expressing the Fow1–green fluorescent protein fusion proteins. The FOW1-targeted mutants of the tomato wilt pathogen F. oxysporum f. sp. lycopersici also showed reduced virulence. These data strongly suggest that FOW1 encodes a mitochondrial carrier protein that is required specifically for colonization in the plant tissue by F. oxysporum.
INTRODUCTION
Fungal plant pathogens use diverse strategies to infect their host plants (Schäfer, 1994; Oliver and Osbourn, 1995; Knogge, 1998). Recently, pathogenicity and virulence genes of fungi have been identified at an exponential rate. Idnurm and Howlett (2001) tabulated 79 genes described to date and divided them into several categories, depending on their involvement in the formation of infection structures, cell wall degradation, responses to the host environment, toxin biosynthesis, signal cascades, and novel functions. Knowledge of the genes that control pathogenicity or virulence will not only increase our overall understanding of the disease process but also will contribute to the search for a target for disease control.
The vascular wilt fungus Fusarium oxysporum is a soil-borne facultative parasite that causes economically important losses in a wide variety of crops. Individual pathogenic strains within the species have a limited host range, and strains with similar or identical host ranges are assigned to intraspecific groups, called formae speciales (f. sp.) (Armstrong and Armstrong, 1981). Some of the formae speciales are further divided into subgroups, named races, on the basis of virulence to a set of differential cultivars within the same plant species (Armstrong and Armstrong, 1981).
Although F. oxysporum pathogens cause severe wilts in ∼80 botanical species, the mechanisms of pathogenicity and symptom induction by this fungus are poorly understood (Beckman, 1987). The genes that encode cell wall–degrading enzymes and plant saponin-detoxifying enzymes, which have been implicated as possible key factors in pathogenicity or virulence, have been cloned from F. oxysporum (Arie et al., 1998; Di Pietro and Roncero, 1998; Huertas-González et al., 1999; Roldán-Arjona et al., 1999; Ruiz-Roldán et al., 1999; García-Maceira et al., 2000). Their exact roles, however, are undefined (Arie et al., 1998; Di Pietro and Roncero, 1998; Huertas-González et al., 1999; Roldán-Arjona et al., 1999; Ruiz-Roldán et al., 1999; García-Maceira et al., 2000).
Recently, Di Pietro et al. (2001) isolated the fmk1 gene of F. oxysporum, which encodes a mitogen-activated protein kinase (MAPK) belonging to the yeast and fungal extracellular signal-regulated kinase family (Xu, 2000). The fmk1 gene was isolated from the tomato wilt pathogen F. oxysporum f. sp. lycopersici by PCR amplification with primers derived from the Fusarium solani MAPK cDNA (Li et al., 1997; Di Pietro et al., 2001). MAPKs of this family have been shown to play a key role in infection-related morphogenesis and pathogenicity in several plant pathogenic fungi (Xu, 2000). Transformation-mediated targeting revealed the essential role of fmk1 in root penetration by F. oxysporum (Di Pietro et al., 2001).
To isolate genes required for pathogenicity and symptom induction by F. oxysporum, we adopted a mutagenic approach. We previously described a mutant screen of F. oxysporum f. sp. melonis using restriction enzyme–mediated integration (REMI) mutagenesis (Kuspa and Loomis, 1992; Lu et al., 1994; Inoue et al., 2001; Namiki et al., 2001). This forma specialis causes vascular wilt of melon and has been divided into four races (races 0, 1, 2, and 1,2) on the basis of virulence to differential cultivars (Leach and Currence, 1938; Risser et al., 1976).
We transformed strain Mel02010 (race 2) with the plasmid pSH75, conferring resistance to hygromycin B (Kimura and Tsuge, 1993), by the REMI method and tested 2929 transformants for pathogenicity to three melon cultivars, Amus, Ogon 9, and Ohi (Inoue et al., 2001; Namiki et al., 2001). The race 2 strain causes wilt in Amus and Ogon 9 but not in Ohi (Namiki et al., 1998). Pathogenicity testing selected 43 mutants that showed reduction or complete loss of virulence on susceptible melon cultivars (Inoue et al., 2001; Namiki et al., 2001). Among the mutants, we found an Arg auxotroph and identified the ARG1 gene, which encodes argininosuccinate lyase essential for the Arg biosynthesis and virulence of this pathogen (Namiki et al., 2001).
Here, we describe the characterization of the mutant B60, which was isolated previously by REMI mutagenesis (Inoue et al., 2001). B60 showed markedly reduced virulence to melon plants (Inoue et al., 2001). The affected gene, named FOW1, encodes a protein with high similarity to mitochondrial carrier proteins (MCPs) of yeast (Nelson et al., 1998; Belenkiy et al., 2000). MCPs are small transport proteins of the mitochondrial inner membrane that catalyze the transport of metabolites across the inner membrane with a high degree of substrate specificity (Palmieri, 1994; Nelson et al., 1998; Belenkiy et al., 2000).
The REMI mutant and the FOW1-targeted mutants of F. oxysporum f. sp. melonis were defective in vascular colonization in host plants, although they showed normal growth and conidiation in culture. The FOW1-targeted disruption in F. oxysporum f. sp. lycopersici also caused marked reduction of virulence to tomato plants. Thus, it appears that FOW1 is essential for the virulence of F. oxysporum. Our findings provide evidence that a specific MCP may play a role in fungal pathogenesis, probably as a response to the host environment.
RESULTS
The Tagged Locus in the REMI Mutant B60
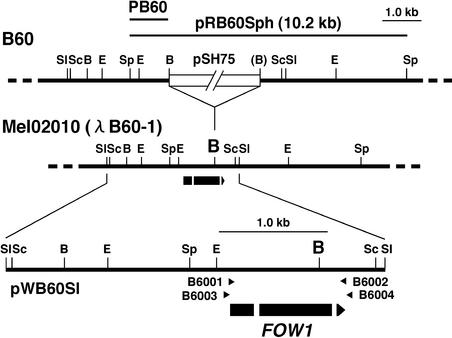
The REMI mutant B60 was generated by transformation of the wild-type strain Mel02010 with the plasmid pSH75 in the presence of BamHI (Inoue et al., 2001). DNA gel blot analysis using pSH75 as a probe revealed a single copy of pSH75 in B60 and identified a 10.2-kb SphI fragment, which included pSH75 and the flanking chromosomal DNA (Figure 1). The 10.2-kb SphI fragment was recovered by plasmid rescue. The recovered plasmid, named pRB60Sph, contained 1.1- and 3.6-kb chromosomal DNA upstream and downstream of pSH75, respectively (Figure 1). Restriction mapping revealed that the BamHI site in one of the junctions of pSH75 and chromosomal DNA had been lost (Figure 1).
Figure 1.
Tagged Locus in the REMI Mutant B60.
The 10.2-kb SphI fragment was recovered from total DNA of B60 by plasmid rescue as pRB60Sph. Restriction sites of the tagged locus in B60 were deduced by restriction mapping of pRB60Sph and DNA gel blot analysis of B60 with the pSH75 probe. The BamHI site, which had been lost in one of the junctions of pSH75 and chromosomal DNA, is shown in parentheses. Fragment PB60 from pRB60Sph was used as a probe to screen a genomic library of the wild-type strain Mel02010, and a clone, λB60-1, was isolated. The 3.5-kb SalI fragment from λB60-1 was cloned in pBluescript KS+ as pWB60Sl. The arrow indicates a putative ORF of FOW1, and white segments indicate the positions of introns. Arrowheads above the ORF denote the orientations and locations of oligonucleotide primers used in PCR and RT-PCR experiments. B, BamHI; E, EcoRI; Sc, SacI; Sl, SalI; Sp, SphI.
The chromosomal DNA fragment PB60 from pRB60Sph (Figure 1) was used as a probe to screen a genomic library of Mel02010. A positive clone, named λB60-1, was partially restriction mapped (Figure 1). Restriction sites upstream and downstream of the tagged BamHI site in B60 were identical to those in λB60-1 (Figure 1). A 3.5-kb SalI fragment from λB60-1 was cloned in pBluescript KS+ as pWB60Sl (Figure 1). Comparison of sequences of pRB60Sph and pWB60Sl revealed that 68 nucleotides at one end of plasmid pSH75, including the BamHI site, had been deleted during the integration process of plasmid in B60.
A Putative Open Reading Frame at the Tagged Locus
Sequencing of pWB60Sl found a putative open reading frame (ORF) from the tagged site in B60 (Figure 1). This ORF consists of three exons (229, 673, and 52 bp) divided by two introns (60 and 54 bp) and potentially encodes a 318–amino acid protein. The introns were deduced initially on the basis of consensus sequences for 5′ and 3′ splice signals typical of fungal genes (Bruchez et al., 1993). To confirm the introns, the cDNA was prepared from total RNA of Mel02010 by reverse transcription–PCR (RT-PCR) with primers B6001 and B6002 (Figure 1). RT-PCR amplification produced a 0.96-kb fragment of DNA. Comparison of the genomic sequence with that of the RT-PCR product confirmed that the two introns are spliced. Sequence analysis of pRB60Sph and pWB60Sl showed that the insertion of the transformation vector pSH75 in B60 had occurred at the BamHI site in the second exon (Figure 1).
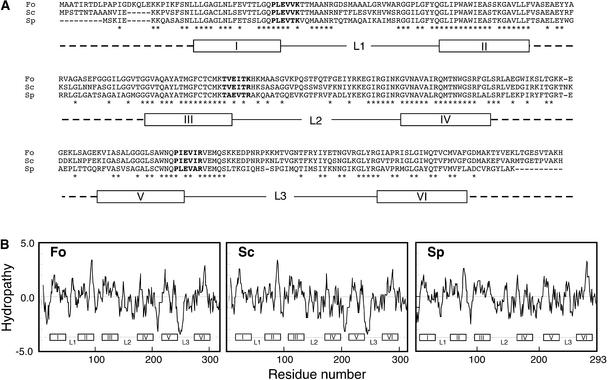
The deduced amino acid sequence encoded by the ORF reveals high similarity to MCPs of yeast: 69 and 51% identical to an MCP encoded by YMR241w of Saccharomyces cerevisiae and a putative MCP of Schizosaccharomyces pombe, respectively (Figure 2A). The YMR241w-encoded MCP was suggested to transport citrate or other tricarboxylates (Mayer et al., 1997). However, the function of the S. pombe MCP is unknown. MCPs generally consist of three homologous domains, each of ∼100 amino acids, that contain the conserved motif PX(D/E)XX(K/R) (Palmieri, 1994; Nelson et al., 1998; Belenkiy et al., 2000). Each domain carries two hydrophobic, transmembrane α-helices linked by a hydrophilic, extramembrane loop (Palmieri, 1994; Nelson et al., 1998; Belenkiy et al., 2000).
Figure 2.
Putative MCP Encoded by the Tagged ORF in B60.
(A) Amino acid alignment of putative MCPs from F. oxysporum (Fo), S. cerevisiae (Sc), and S. pombe (Sp). The amino acid sequence of the ORF tagged in B60 was aligned with the sequences of YMR241w-encoded MCP of S. cerevisiae and a putative MCP of S. pombe. Identical amino acids among three sequences are indicated with asterisks. Amino acid residues that are similar to the conserved motif PX(D/E)XX(K/R) of MCPs are shown as boldface letters. Segments I to VI and segments L1 to L3 represent putative hydrophobic, transmembrane stretches and hydrophilic, extramembrane loops, respectively, as suggested by Nelson et al. (1998).
(B) Hydrophobicity plots of amino acid sequences of putative MCPs of F. oxysporum (Fo), S. cerevisiae (Sc), and S. pombe (Sp). Hydropathy analysis was performed using the method of Kyte and Doolittle (1982). Hydrophobic amino acids have scores of >0, whereas hydrophilic amino acids have scores of <0.
The ORF product of F. oxysporum also consists of three homologous domains (Figure 2A). Each domain contains the two segments homologous with transmembrane segments of MCPs of S. cerevisiae and S. pombe suggested by Nelson et al. (1998) (segments I to VI in Figure 2A). Extramembrane loops also were deduced between putative transmembrane segments in each domain (segments L1 to L3 in Figure 2A). Hydrophobicity plots of these MCPs suggest that segments I to VI and segments L1 to L3 correspond to hydrophobic, transmembrane stretches and hydrophilic, extramembrane loops, respectively (Figure 2B). The first and third domains contain the conserved motif sequences PLEVVK and PIEVIR, respectively (Figure 2A). The second domain contains TVEITK, which is similar to the conserved motif (Figure 2A). Thus, the tagged ORF in B60 possibly encodes a protein that has structural characteristics typical of MCPs.
FOW1 Is Required for Virulence
To determine whether mutation of the MCP ORF was responsible for the reduced virulence of the REMI mutant B60, a genetic complementation experiment was performed. We introduced pWB60Sl, which includes the entire candidate ORF (Figure 1), into B60 by cotransformation with plasmid pII99, which confers resistance to geneticin (Namiki et al., 2001) and isolated 75 transformants. Two plants of cv Amus, which had a single true leaf, were inoculated with bud cells of each transformant by the root dip method (Namiki et al., 1998). Disease symptoms were assessed 3 weeks after inoculation.
Of 75 transformants, 37 induced severe wilt or death on all inoculated plants, as did the wild-type strain. We analyzed the integration mode of pWB60Sl in five virulent transformants and verified that they contained a single copy of the plasmid. This result showed that pWB60Sl complemented the mutation of B60. Thus, the gene encoding a putative MCP was designated FOW1 (Fusarium oxysporum gene required for wilt symptom induction).
To further characterize the essential role of FOW1 in the virulence of this pathogen, the rescued plasmid pRB60Sph (Figure 1) was used to disrupt FOW1 in the wild-type strain Mel02010. Mel02010 was transformed with SphI-linearized pRB60Sph, and 163 transformants were isolated. Each transformant was tested for virulence using four plants with a single true leaf. Of 163 transformants, 85 caused severe wilt or death on all plants. However, the remaining 78 transformants did not cause severe wilt or death on any plants, similar to B60.
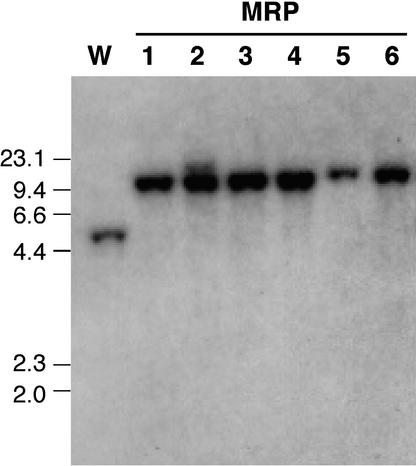
The integration mode of pRB60Sph in six transformants (MRP1 to MRP6) that showed reduced virulence was analyzed by DNA gel blot analysis (Figure 3). Total DNA of Mel02010 and the transformants was digested with SphI, and the blot was probed with the PB60 fragment from pRB60Sph (Figure 1). The probe hybridized to an expected band of 4.7 kb in Mel02010 (Figure 3). However, all of the MRP transformants lacked 4.7-kb bands and had 10.2-kb bands, resulting from homologous integration of pRB60Sph (Figure 3). This hybridization showed that FOW1 was inactivated by the same mode in B60 and MRP transformants (Figures 1 and 3). RT-PCR amplification of FOW1 cDNA with primers B6001 and B6002 (Figure 1) produced no DNA fragments from total RNA of B60 and MRP transformants (data not shown).
Figure 3.
DNA Gel Blot Analysis of pRB60Sph Transformants.
Total DNA of each strain was digested with SphI and electrophoresed on a 0.8% agarose gel. The blot was probed with PB60 fragment (Figure 1). Sizes (in kilobases) of marker DNA fragments (HindIII-digested λDNA) are indicated at left. Lane W, wild-type strain Mel02010; lanes MRP1 to MRP6, pRB60Sph transformants.
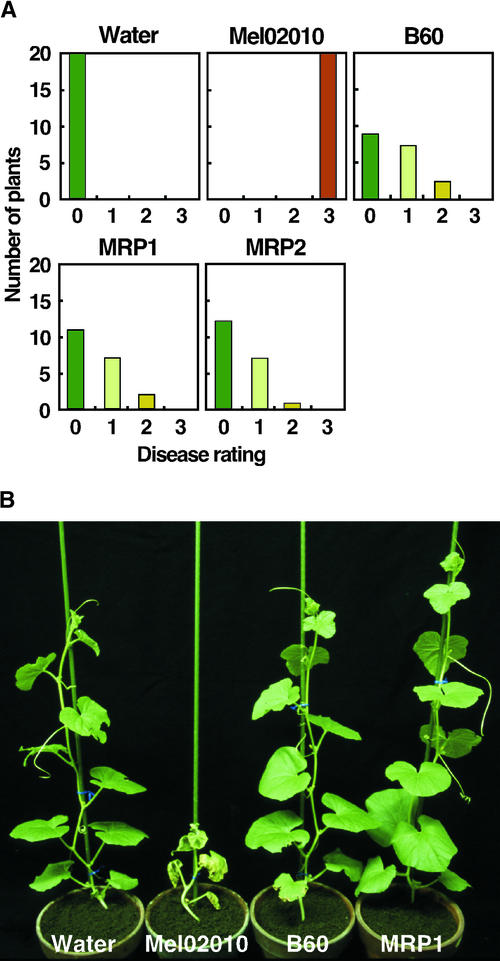
To further characterize the pathological role of FOW1 in this fungus, we performed a detailed comparative analysis of virulence in wild-type and fow1 mutant strains. Twenty young plants with a single true leaf were inoculated with bud cells of each strain by the root dip method, and symptoms were scored 3 weeks after inoculation according to the following disease ratings: 0 = no symptoms, 1 = yellowing, 2 = wilting, and 3 = death (Inoue et al., 2001). The wild-type strain Mel02010 caused death on all plants (Figure 4A). The REMI mutant B60 caused yellowing and wilt on eight and three plants, respectively, but it never caused death on any plants (Figure 4A). The FOW1-targeted transformants MRP1 and MRP2 also did not cause death, and their levels of virulence were similar to that of B60 (Figure 4A).
Figure 4.
Virulence of fow1 Mutants.
(A) Twenty melon plants that had a single true leaf were inoculated with bud cell suspension (∼1 × 107 cells/mL) of each strain by the root dip method. Control plants were immersed in water. Disease symptoms were evaluated 3 weeks after inoculation according to the following disease ratings: 0 = no symptoms, 1 = yellowing, 2 = wilting, and 3 = death. Mel02010, wild-type strain; B60, REMI mutant; MRP1 and MRP2, FOW1-targeted transformants.
(B) Plants with four true leaves were inoculated with bud cell suspension (∼1 × 107 cells/mL) of each strain by the root dip method. The photograph was taken 3 weeks after inoculation.
The fow1 mutants also were tested for virulence using older plants. Five plants with four true leaves were inoculated with bud cells of wild-type and mutant strains, and symptoms were observed 3 weeks after inoculation. The wild type caused typical wilt symptoms on all plants (Figure 4B). In contrast, the mutants B60 and MRP1 caused no visible symptoms on inoculated plants (Figure 4B). To assess the colonization of wild-type and mutant strains in the plant tissue, fungi were isolated from inoculated plants. After observation of the symptoms shown in Figure 4B, stem segments (∼1.5 cm) immediately above the crowns were cut from the plants, surface sterilized, and incubated on potato dextrose agar (PDA). Fusarium colonies appeared from stem segments of all plants inoculated with the wild type but not from those of plants inoculated with B60 and MRP1.
Colonization of the fow1 Mutant
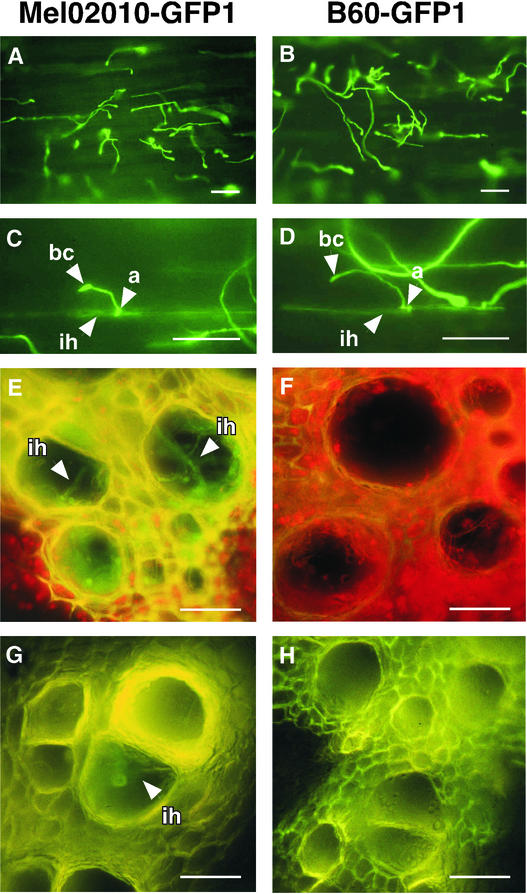
To observe colonization of the plant tissue by wild-type and fow1 mutant strains, we used transformants constitutively expressing green fluorescent protein (GFP). The GFP expression vector pTEFEGFP (Vanden Wymelenberg et al., 1997) was introduced into Mel02010 and B60 by cotransformation with plasmid pII99, which confers geneticin resistance. We observed the mycelia of transformants grown on PDA by fluorescence microscopy. Of 40 transformants from Mel02010, 10 expressed GFP; of 10 transformants from B60, 2 expressed GFP. We used transformants Mel02010-GFP1 and B60-GFP1 from Mel02010 and B60, respectively, in the following experiments.
By DNA gel blot analysis, Mel02010-GFP1 and B60-GFP1 were verified to contain one and two copies of pTEFEGFP, respectively (data not shown). Radicles of germinated seeds of cv Amus were dipped in bud cell suspensions of the GFP-expressing transformants. The radicle surface was observed 24 h after inoculation. Most bud cells from both Mel02010-GFP1 and B60-GFP1 germinated and elongated hyphae on the root surface (Figures 5A and 5B). Elongated mycelia of both strains frequently differentiated appressorium-like small structures (Figures 5A to 5D) and occasionally formed infection hyphae growing within the epidermal cells (Figures 5C and 5D). This observation suggests that the FOW1 mutation does not affect hyphal growth on the root surface and penetration of the epidermal cells under the conditions tested.
Figure 5.
Infection Behavior and Plant Colonization of Wild-Type and fow1 Mutant Strains
Mel02010-GFP1 and B60-GFP1, which constitutively expressed GFP, were prepared by transformation of the wild-type strain Mel02010 and the REMI mutant B60, respectively, with the GFP expression vector pTEFEGFP. a, appressorium-like structure; bc, bud cell; ih, infection hyphae. Bars = 50 μm.
(A) to (D) Germination, hyphal growth, and penetration of Mel02010-GFP1 ([A] and [C]) and B60-GFP1 ([B] and [D]) on the root surface. Radicles of germinated melon seeds were inoculated with bud cell suspension (∼1 × 106 cells/mL) of each transformant by the root dip method and incubated for 24 h. Radicle samples were observed with a fluorescence microscope.
(E) to (H) Infection hyphae of Mel02010-GFP1 ([E] and [G]) and B60-GFP1 ([F] and [H]) in the xylem vessel. Plants with four true leaves were inoculated with bud cell suspension (∼1 × 107 cells/mL) of each transformant by the root dip method and incubated for 10 days. Cross-sections from crowns ([E] and [F]) and roots ([G] and [H]) were observed with a fluorescence microscope.
Three plants with four true leaves were inoculated with bud cells of each transformant by the root dip method. At 10 days after inoculation, plants were removed, and cross-sections from crowns and roots were made to observe fungal structures in the plant tissue by fluorescence microscopy. More than 10 sections each from crown and main root of each plant were observed. Plants inoculated with Mel02010-GFP1 showed wilt symptoms, but those inoculated with B60-GFP1 showed no visible symptoms. Infection hyphae of Mel02010-GFP1 growing within the xylem vessels were observed in almost all sections from crowns and roots (Figures 5E and 5G). However, no fungal structures were detected in any samples inoculated with B60-GFP1 (Figures 5F and 5H). Along with the reisolation experiments of F. oxysporum colonies from inoculated plants, this cytological observation clearly shows that FOW1 is essential for the colonization of plant tissue by F. oxysporum f. sp. melonis.
Vegetative Growth of fow1 Mutants
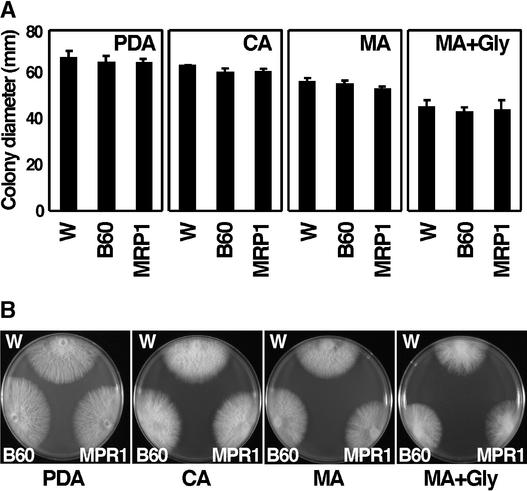
The vegetative growth of fow1 mutants was evaluated by measuring colony diameter grown on PDA, complete agar (Sanderson and Srb, 1965), and minimal agar (MA) (Sanderson and Srb, 1965) at 25°C for 5 days. All of these media contained Glc as a carbon source. Colony diameters of the REMI mutant B60 and the FOW1-targeted mutant MRP1 were not significantly different from those of the wild-type strain (at the P < 0.05 level according to Fisher's protected LSD test) (Figure 6A). The colony morphology of the wild type and these mutants also was indistinguishable (Figure 6B). Mutations of nuclear genes that encode mitochondrial proteins often cause defects in growth on nonfermentable carbon sources such as glycerol, so they are called gly− mutants (Tzagoloff and Dieckmann, 1990; Nelson et al., 1998). However, fow1 mutants showed normal growth on MA supplemented with glycerol instead of Glc, showing that they were not gly− (Figure 6).
Figure 6.
Vegetative Growth of fow1 Mutants.
Wild-type and fow1 mutant strains were grown on PDA, complete agar (CA), minimal agar (MA), and MA containing glycerol instead of Glc (MA+Gly) at 25°C for 5 days. W, wild-type strain Mel02010; B60, REMI mutant; MRP1, FOW1-targeted transformant.
(A) Colony diameter. Data represent means ± sd of three replications.
(B) Colony morphology.
When F. oxysporum is grown in liquid medium on an orbital shaker, it elongates mycelia and forms bud cells. When wild-type and fow1 mutant strains were grown in potato dextrose broth on an orbital shaker at 25°C for 3 days, the weight of mycelia and the number of bud cells were not significantly different between the wild type and the mutants (data not shown). To observe conidiation, they were grown on carnation leaf agar, a medium for the conidiation of F. oxysporum (Togawa, 1992). B60 and MRP1 normally formed macroconidia, microconidia, and chlamydospores (data not shown). Thus, FOW1 is dispensable for the growth and conidiation of this fungus in culture.
Expression of FOW1
In vitro expression of FOW1 was determined by RNA gel blot analysis. The wild-type strain Mel02010 was grown in potato dextrose broth on an orbital shaker at 25°C for 3 days, and total RNA was isolated from mycelia and bud cells. The RNA gel blot was probed with PB60 containing the FOW1 fragment (Figure 1). The probe hybridized to an ∼1.3-kb band in RNA from both mycelia and bud cells (data not shown), indicating that FOW1 is expressed by this fungus during vegetative growth in a nutrition-rich medium.
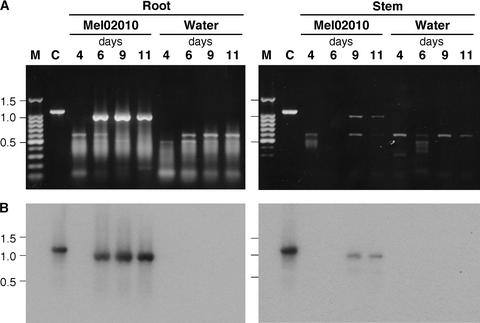
To determine whether FOW1 is expressed by this pathogen during infection of the host plants, RT-PCR with primers B6001 and B6002 (Figure 1) was used to detect the transcripts in melon plants infected with Mel02010. Melon plants with a single true leaf were inoculated with the bud cells. Control plants were immersed in water. Total RNA was extracted from roots and stems at 4, 6, 9, and 11 days after inoculation. Inoculated plants showed no symptoms, yellowing, wilting, and severe wilting at 4, 6, 9, and 11 days, respectively, after inoculation. Total DNA of Mel02010 was used as a template for PCR to compare the sizes of the amplified fragments with or without the introns (1071 and 957 bp, respectively).
Electrophoresis of the RT-PCR products from infected roots and stems showed a fragment of the expected size (0.96 kb) that was absent in the water-treated controls (Figure 7A). In roots, the 0.96-kb DNA was detected at 6, 9, and 11 days after inoculation; in stems, it was detected at 9 and 11 days after inoculation (Figure 7A). Because the smaller-sized bands also were detected in inoculated and control plants (Figure 7A), the gel blot was hybridized with PB60 probe (Figure 1). This probe hybridized to the 0.96-kb DNA but not to the smaller DNAs (Figure 7B), indicating that the smaller DNAs were attributable to nonspecific amplification of plant cDNA.
Figure 7.
In Planta Expression of FOW1 by F. oxysporum f. sp. melonis.
(A) Melon plants that had a single true leaf were inoculated with bud cell suspension (∼1 × 107 cells/mL) of strain Mel02010 by the root dip method. Control plants were immersed in water. Plants were harvested at 4, 6, 9, and 11 days after inoculation, and total RNA was extracted separately from roots and stems. FOW1 cDNA was amplified from total RNA (1 μg) by RT-PCR with primers B6001 and B6002 (Figure 1). Total DNA of Mel02010 was used as a control (lane C). Aliquots (5 μL) of RT-PCR mixtures (100 μL) were electrophoresed on 1.0% agarose gels.
(B) Blots were probed with the PB60 fragment (Figure 1).
Sizes (in kilobases) of marker DNA fragments (a 100-bp ladder; lane M) are indicated at left.
Intracellular Localization of GFP-Tagged Fow1
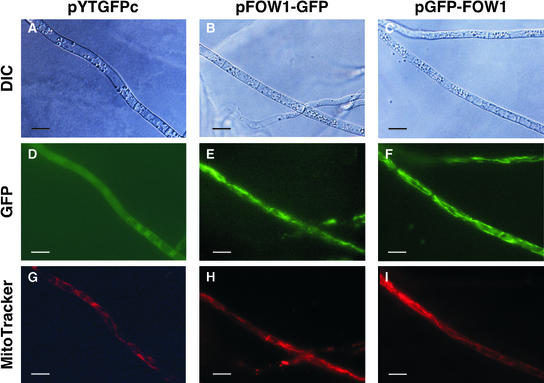
The predicted amino acid sequence of Fow1 suggests that Fow1 is an MCP (Figure 2). To determine whether Fow1 localizes to mitochondria, we made strains expressing Fow1-GFP and GFP-Fow1 fusion proteins and observed the intracellular distribution of GFP fluorescence in the strains by fluorescence microscopy. We constructed the FOW1-GFP and GFP-FOW1 gene fusions under the control of the Aspergillus nidulans trpC promoter (Mullaney et al., 1985) as pFOW1-GFP and pGFP-FOW1, respectively. Both constructs were introduced into Mel02010 by cotransformation with the plasmid pSH75, which confers hygromycin B resistance. As a control, Mel02010 was transformed with the plasmid pYTGFPc, which carries GFP under the control of the trpC promoter.
Transformants were grown on PDA, and their mycelia were observed by fluorescence microscopy. Of 45 pYTGFPc transformants, 12 expressed GFP. In these transformants, GFP fluorescence did not localize in any cell components throughout mycelia (Figure 8). Of 37 pFOW1-GFP transformants, 28 emitted GFP fluorescence; of 11 pGFP-FOW1 transformants, 3 emitted GFP fluorescence. In all transformants expressing Fow1-GFP and GFP-Fow1 fusions, GFP fluorescence was targeted in reticular components in the cells (Figure 8).
Figure 8.
Intracellular Localization of the GFP-Tagged Fow1.
Transformants with pYTGFPc ([A], [D], and [G]), pFOW1-GFP ([B], [E], and [H]), and pGFP-FOW1 ([C], [F], and [I]) were grown on PDA at 25°C for 5 days. Mycelia were stained with MitoTracker Red CMXRos and observed by fluorescence microscopy. Bars = 10 μm.
(A) to (C) Differential interference contrast (DIC) images.
(D) to (F) GFP fluorescence images.
(G) to (I) MitoTracker Red CMXRos straining images.
Figure 8 shows microscopic images of three transformants, each of which had a single copy of pYTGFPc, pFOW1-GFP, or pGFP-FOW1. The same mycelia were stained with the mitochondrial probe MitoTracker Red CMXRos. The red fluorescence of MitoTracker Red and the green fluorescence of Fow1-GFP and GFP-Fow1 localized in the same reticular compartments, indicating that these internal components were mitochondria (Figure 8).
FOW1 Conservation in F. oxysporum
The distribution of FOW1 in other formae speciales of F. oxysporum was analyzed by DNA gel blot analysis. We used strains from four other formae speciales, cucumerinum, niveum, lycopersici, and raphani, which cause vascular wilts of cucumber, watermelon, tomato, and radish, respectively. PB60 probe (Figure 1) hybridized to DNA bands of all strains with similar signal intensity (data not shown), suggesting that FOW1 is conserved in F. oxysporum.
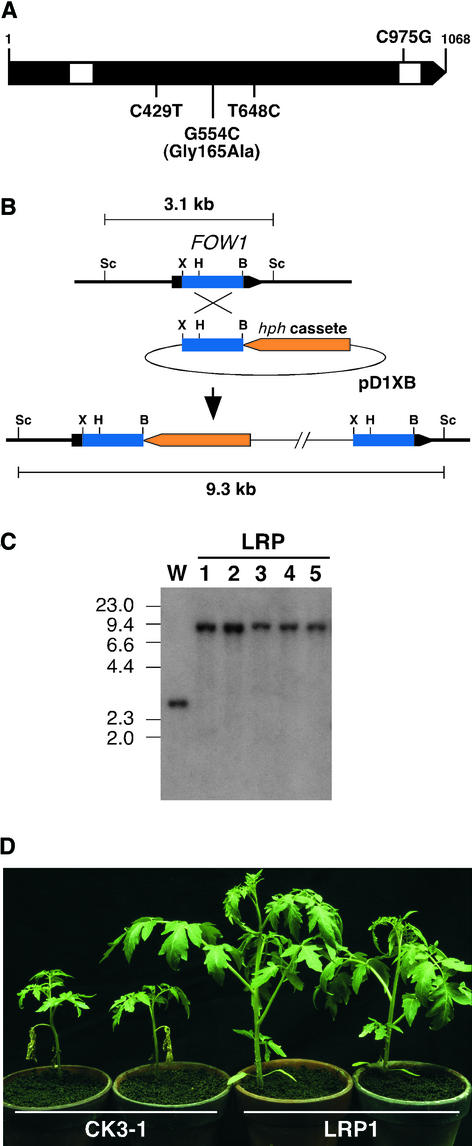
We isolated the FOW1 homologs from strains Cucu05-015, CK3-1, and MAFF305123 of the formae speciales cucumerinum, lycopersici, and raphani, respectively. The FOW1 homologs were amplified from total DNA of each strain by PCR with primer pair B6003-B6004 (Figure 1). Sequence analysis of the PCR products revealed that they have the same sizes of 1114 bp, identical to that of the corresponding region of FOW1. The sequence of forma specialis cucumerinum is identical to that of FOW1. The sequence of forma specialis raphani has a single nucleotide difference in the second intron (Figure 9A). The sequence of forma specialis lycopersici has three nucleotide differences in the second exon, one of which causes an amino acid substitution (Gly to Ala) at position 165 (Figure 9A).
Figure 9.
FOW1 Homologs of F. oxysporum Pathogens.
(A) FOW1 homologs of F. oxysporum f. sp. raphani and f. sp. lycopersici. The arrowed bar indicates a protein coding region of FOW1 with introns (white segments). Compared with the f. sp. melonis FOW1, nucleotides different in the raphani and lycopersici sequences are shown above and beneath the bar, respectively.
(B) FOW1-targeted disruption in F. oxysporum f. sp. lycopersici. Structure of the FOW1 locus before and after homologous integration of the targeting vector pD1XB. To make pD1XB, the 0.7-kb XhoI-BamHI fragment internal to the lycopersici FOW1 was cloned in pSH75. B, BamHI; H, HindIII; Sc, SacI; X, XhoI.
(C) DNA gel blot analysis of pD1XB transformants. Total DNA of the wild-type strain CK3-1 (W) and the pD1XB transformants (LRP1 to LRP5) was digested with SacI and electrophoresed on a 0.8% agarose gel. The blot was probed with the FOW1 fragment from pD1XB. Sizes (in kilobases) of marker DNA fragments (HindIII-digested λDNA) are indicated at left.
(D) Virulence of the pD1XB transformant. Tomato plants of cv Ponderosa that had two true leaves were inoculated with bud cell suspensions (∼1 × 107 cells/mL) of CK3-1 and LRP1 by the root dip method. The photograph was taken 3 weeks after inoculation.
To assess the pathological role of FOW1 in F. oxysporum f. sp. lycopersici, the wild-type strain CK3-1 was transformed with plasmid pD1XB, which contains the 0.7-kb XhoI-BamHI fragment of the lycopersici FOW1 in pSH75 (Figure 9B). The integration mode of pD1XB in transformants was analyzed by DNA gel blot analysis, and five transformants (LRP1 to LRP5) that result from the homologous integration of pD1XB were selected (Figure 9C). Total DNA of CK3-1 and the transformants was digested with SacI, and the blot was hybridized with the 0.7-kb XhoI-BamHI fragment from pD1XB. This probe hybridized to an expected 3.1-kb band in CK3-1 and to 9.3-kb bands in the transformants, resulting from the homologous integration of pD1XB (Figures 9B and 9C).
Tomato plants of cv Ponderosa, which had two true leaves, were inoculated with bud cells of wild-type and fow1 mutant strains by the root dip method, and symptoms were observed 3 weeks after inoculation. Figure 9D shows symptoms of tomato plants caused by CK3-1 and the fow1 mutant LRP1 (Figure 9C) as an example. CK3-1 caused growth inhibition and wilt of all plants (Figure 9D). However, fow1 mutants caused no symptoms on inoculated plants (Figure 9D). Ectopic integration transformants caused wilt on all plants, as did the wild type (data not shown). Thus, this gene also is required for the virulence of F. oxysporum f. sp. lycopersici.
To determine whether Fow1 is related to the YMR241w-encoded MCP of S. cerevisiae, we constructed plasmid pEYMR241w containing the entire YMR241w ORF under the control of the A. nidulans trpC promoter for expression in the REMI mutant B60. The YMR241w expression vector was introduced into B60 by cotransformation with plasmid pII99, and 77 transformants were selected. Two melon plants with a single true leaf were inoculated with bud cells of each transformant. No transformants caused severe wilt or death on any plants. DNA gel blot analysis of 10 transformants showed that at least 3 transformants contained the entire YMR241w ORF fused to the trpC promoter (data not shown). Thus, introduction of YMR241w in B60 did not restore virulence.
DISCUSSION
Molecular analysis of the REMI mutant B60 of the melon wilt pathogen F. oxysporum f. sp. melonis identified the MCP-encoding gene FOW1. fow1 mutants occasionally caused slight wilt symptoms on melon plants when they were inoculated on young plants with a single true leaf. These mutants, however, caused no symptoms when they were inoculated on older plants with four true leaves. The wild-type strain caused death on both young and older plants. The FOW1 gene is conserved in other formae speciales of F. oxysporum, causing wilt of different plants. We observed that FOW1 targeting in the tomato wilt pathogen F. oxysporum f. sp. lycopersici also resulted in the marked reduction of virulence to tomato plants. Thus, it appears that FOW1 acts as a virulence determinant in F. oxysporum.
Fow1 shows high similarity to the YMR241w-encoded MCP of S. cerevisiae and a putative MCP of S. pombe and has structural characteristics typical of MCPs. MCPs are small transport proteins of the mitochondrial inner membrane (Palmieri, 1994; Nelson et al., 1998; Belenkiy et al., 2000). They are essential for communication between the matrix and the cytosol, and they occupy a central role in eukaryotic metabolism (Palmieri, 1994; Nelson et al., 1998; Belenkiy et al., 2000). Mitochondrial import of Fow1 was verified using strains expressing GFP-tagged Fow1. We observed that the Fow1-GFP and GFP-Fow1 fusions both were targeted to mitochondria. Thus, the mitochondrial localization of Fow1 is not affected by N- or C-terminal fusions of GFP to this protein. Import of MCPs into mitochondria may depend on the internal regions of MCPs (Palmieri, 1994; Nelson et al., 1998).
The structural similarity between Fow1 and MCPs and the mitochondrial localization of Fow1 strongly suggest that it belongs to this family of proteins. Many pathogenicity and virulence genes of fungi were identified recently and divided into several categories, depending on their functions in plant pathogenesis (Idnurm and Howlett, 2001). However, FOW1 is the first mitochondrial protein gene that has been identified as a virulence determinant of plant pathogenic fungi.
Search of the S. cerevisiae genome identified 35 mitochondrial carrier homologs, and 12 of these sequences have known or inferred functions (Nelson et al., 1998; Belenkiy et al., 2000; Palmieri et al., 2000). The YMR241w-encoded protein and Fow1 are highly similar (69% identity in amino acid sequence), suggesting that they have the same function. Mayer et al. (1997) reported that YMR241w might transport citrate or other tricarboxylates. We attempted genetic complementation of the FOW1 mutation of the REMI mutant B60 with the YMR241w expression vector, in which the YMR241w ORF is fused to the A. nidulans trpC promoter. However, the transformants carrying the entire trpC promoter–YMR241w construct did not restore full virulence to melon plants. This observation indicates that Fow1 may differ in function from the YMR241w-encoded MCP.
The major role of mitochondria in eukaryotic cells is oxidative phosphorylation. In S. cerevisiae, mutation of nuclear genes encoding mitochondrial proteins often causes a petite phenotype, so named for the small colonies that grow on Glc medium, as a result of respiration deficiency (Tzagoloff and Dieckmann, 1990; Nelson et al., 1998). Such mutants cannot grow on nonfermentable carbon sources, such as glycerol, and are called gly− mutants (Tzagoloff and Dieckmann, 1990; Nelson et al., 1998). The majority of well-characterized MCPs are involved in oxidative phosphorylation (Palmieri, 1994; Nelson et al., 1998), and their mutations generally cause the gly− phenotype (Nelson et al., 1998).
The fow1 mutants, however, had normal mycelial growth on both Glc and glycerol media. They formed macroconidia, microconidia, and chlamydospores on carnation agar medium, similar to the wild type. These results indicate that FOW1 is dispensable for the saprophytic growth and conidiation of this fungus, although the fungus expresses this gene during growth in culture. Thus, fow1 mutants are indistinguishable from the wild type except for their marked reduction of virulence.
In addition to the major MCPs, such as those required for oxidative phosphorylation, many others must exist in the inner mitochondrial membrane for the import of various nucleotides, cofactors, and compounds that are not synthesized in mitochondria. Eighteen MCP genes of S. cerevisiae have been disrupted or mutated, but only six show the gly− phenotype (Nelson et al., 1998). This observation implies that the majority of these genes are not required for growth, at least under the specific conditions used.
The dif-1 gene from Caenorhabditis elegans, which possibly encodes an MCP, is needed specifically during embryonic tissue differentiation (Ahringer, 1995). In mammals, certain MCPs are tissue specific and have limited distribution, reflecting their importance in special functions (Palmieri, 1994). Thus, some MCPs are not required for the biogenesis, maintenance, and primary function of mitochondria and may have novel functions important under specific conditions.
The infection process of F. oxysporum involves the following steps: spores germinate in response to root exudates, produce penetration hyphae that attach to the root surface and penetrate it directly, and grow invasively in host plant tissue (Rodríguez-Gálvez and Mendgen, 1995). We observed the infection behavior of wild-type and fow1 mutant strains using transformants that constitutively express GFP. When bud cells of the GFP-expressing transformants were inoculated on radicles of germinated seeds, germination rate and hyphal elongation were comparable in both strains. Within 24 h after inoculation, elongated mycelia of both strains frequently differentiated appressorium-like small structures and occasionally formed infection hyphae growing in the epidermal cells. This observation indicates that Fow1 is not essential for hyphal growth on the root surface and penetration of the epidermal cells under the conditions tested.
To observe the invasive growth and colonization of wild-type and mutant strains, cross-sections from crowns and roots of plants inoculated with the GFP-expressing transformants were prepared 10 days after inoculation. Fluorescence microscopy observation of the sections readily detected hyphae of the wild type in the vascular tissue of crowns and roots. In contrast, no mycelial structures of the mutant were observed within the plant tissue. F. oxysporum colonies were isolated from plants inoculated with the wild type, but not from plants inoculated with fow1 mutants. RT-PCR experiments revealed that FOW1 is expressed by this pathogen in inoculated plants. These results strongly suggest that FOW1 has a function that is required for the colonization of plant tissue by F. oxysporum.
The molecular mechanisms of pathogenicity and symptom induction by F. oxysporum remain largely undefined, although this fungus is an economically important plant pathogen with worldwide distribution (Beckman, 1987). We previously isolated 43 pathogenicity mutants from 2929 REMI transformants (Inoue et al., 2001; Namiki et al., 2001). Successful isolation of ARG1 (Namiki et al., 2001) and FOW1 from two of the mutants suggests that the isolation and characterization of pathogenicity mutants through REMI mutagenesis will provide a powerful tool to investigate genes that encode pathogenicity or virulence factors of F. oxysporum. We expect the characterization of the tagged sites in our mutants to allow us to identify more genes of interest.
METHODS
Fungal Strains and Transformation
Strain Mel02010 (JCM9288) and its restriction enzyme–mediated integration mutant B60 of Fusarium oxysporum forma specialis (f. sp.) melonis (Namiki et al., 1994; Inoue et al., 2001) were used to isolate FOW1. Strains Cucu05-015, MAFF305543, CK3-1, and MAFF305123 of the formae speciales cucumerinum, niveum, lycopersici, and raphani, respectively (Namiki et al., 1994), were used for the analysis of FOW1 distribution. These strains were maintained routinely on potato dextrose agar (PDA; Difco, Detroit, MI).
Protoplast preparation and transformation of F. oxysporum were performed using methods described previously (Inoue et al., 2001; Namiki et al., 2001). Transformants carrying the hph gene (Gritz and Davies, 1983) and the nptII gene (Beck et al., 1982) were selected on regeneration media containing hygromycin B (Wako Pure Chemicals, Osaka, Japan) at 60 μg/mL and geneticin (Gibco BRL/Life Technologies, Gaithersburg, MD) at 180 μg/mL, respectively (Inoue et al., 2001; Namiki et al., 2001).
Plasmids and Genomic Library
The integrative transformation vectors pSH75 (Kimura and Tsuge, 1993) and pII99 (Namiki et al., 2001) were used to transform F. oxysporum. These vectors carry hph (Gritz and Davies, 1983) and nptII (Beck et al., 1982), respectively, fused to the Aspergillus nidulans trpC promoter and terminator (Mullaney et al., 1985). The green fluorescent protein (GFP) expression vector pTEFEGFP (Vanden Wymelenberg et al., 1997) was used to make strains of F. oxysporum that constitutively expressed GFP. This vector carries the EGFP gene (Cormack et al., 1996) fused to the Aureobasidium pullulans TEF promoter and the Aspergillus awamori gla terminator (Vanden Wymelenberg et al., 1997).
A genomic library of Mel02010, constructed in λDASHII (Stratagene, La Jolla, CA), has been described (Namiki et al., 2001). Screening of the library by plaque hybridization was conducted using standard methods (Sambrook et al., 1989).
Nucleic Acid Manipulations
Isolation of total DNA and RNA from F. oxysporum and DNA and RNA gel blot hybridization were performed as described previously (Okuda et al., 1998; Inoue et al., 2001; Namiki et al., 2001). Plasmid DNA and recombinant λ phage DNA were extracted using standard methods (Sambrook et al., 1989).
DNA sequences were determined with the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Warrington, UK) and an automated fluorescent DNA sequencer (model 373A; Applied Biosystems) according to the manufacturer's instructions. DNA sequences were analyzed with BLAST (Altschul et al., 1997). Alignment of nucleotide and amino acid sequences was performed with the CLUSTAL W program (Thompson et al., 1994). Hydropathy of amino acid sequences was analyzed by the method of Kyte and Doolittle (1982).
Isolation of FOW1
Restriction mapping of the pSH75 insertion site in B60 revealed the presence of an SphI site that was absent in the vector. Total DNA of B60 was digested with SphI, religated, and transformed to Escherichia coli DH5α. Ampicillin-resistant transformants were selected, and the rescued plasmid, named pRB60Sph, was isolated (Figure 1). PB60 fragment from pRB60Sph (Figure 1) was used as a probe to screen a genomic library of Mel02010, and a positive clone, λB60-1, containing FOW1 was isolated. The 3.5-kb SalI fragment in λB60-1 was cloned in pBluescript KS+ (Stratagene) as pWB60Sl (Figure 1).
The FOW1 cDNA was isolated by reverse transcription–PCR (RT-PCR) using the RNA PCR Kit version 2.1 (Takara, Ohtsu, Japan). The cDNA was amplified from total RNA (1 μg) of Mel02010 with primers B6001 (5′-ATGGCTGCTACTATCCGTAC-3′) and B6002 (5′-CTAATGCTTAGCGGTGACCG-3′) (Figure 1) according to the manufacturer's instructions. B6001 and B6002 contain the FOW1 initiation and termination codons (underlined), respectively. RT-PCR products were cloned in pGEM-T Easy vector (Promega, Madison, WI) to determine the sequences.
The FOW1 homologs of other formae speciales were isolated by PCR amplification with primers B6003 (5′-TCTACGACACTCCCAAAGTC-3′) and B6004 (5′-CAAACCAGATTCCTAAACGC-3′) (Figure 1). B6003 and B6004 locate upstream and downstream of the FOW1 open reading frame (ORF), respectively (Figure 1). Total DNA (100 ng) of each strain was used as a template, and PCR amplification was performed with Taq DNA polymerase (Takara) according to the manufacturer's instructions. PCR products were cloned in pGEM-T Easy. The XhoI-BamHI internal fragment of the FOW1 homolog of F. oxysporum f. sp. lycopersici was cloned in the XhoI-BamHI site of pSH75 to make pD1XB (Figure 9B).
Construction of the FOW1-GFP Fusion Vectors
The A. nidulans trpC promoter was amplified from pSH75 by PCR with primers P-NotI (5′-ATAGCGGCCGCATGCCAGTTGTTCCAGTGA-3′) and P-XbaI (5′-GCCCTCTAGAGCTTGGGTAGAATAGGTAAG-3′), which contain N-terminal NotI and XbaI sites (underlined), respectively. The trpC terminator was amplified from pSH75 by PCR with primers T-EcoRI (5′-CCCGAATTCTGATTTAATAGCTCCATGTC-3′) and T-XhoI (5′-GCCCCTCGAGAAAGAAGGATTACCTCTA-3′), which contain N-terminal EcoRI and XhoI sites (underlined), respectively. Amplified promoter and terminator fragments were digested with NotI-XbaI and EcoRI-XhoI, respectively, and cloned into the NotI-XbaI and EcoRI-XhoI sites of pBluescript KS+. The resulting plasmid, pEC2, has XbaI, BamHI, SmaI, PstI, and EcoRI sites between the promoter and terminator sequences.
The 0.73-kb XbaI-BamHI fragment containing the complete GFP (EGFP) ORF was cut from pCB16EGFP (Kimura et al., 2001). The last codon of the ORF in pCB16EGFP is fused to 15 nucleotides encoding a Gly linker followed by a BamHI site (Kimura et al., 2001). The XbaI-BamHI fragment was cloned into the XbaI-BamHI site of pEC2 to make pYTGFP-N. The FOW1 cDNA was amplified from total RNA of Mel02010 by RT-PCR with primers FOW1N-f (5′-ACCTGATCATGGCTGCTACTATCCGTAC-3′) and FOW1N-r (5′-GACGAATTCTAATGCTTAGCGGTGACCG-3′). FOW1N-f has a BclI site (underlined) with the initiation codon (italics); FOW1N-r has an EcoRI site (underlined) with the termination codon (italics). The amplified DNA was digested with BclI and EcoRI and cloned into the BamHI-EcoRI site of pYTEGFP-N to make pGFP-FOW1, resulting in a N-terminal fusion of GFP to Fow1.
The GFP ORF was amplified from pCB16EGFP with primers Ps-Gly-GFP (5′-AAACTGCAGggtggtggtggtggtATGGTGAGCAAGGGCGAG-3′) and GFP-St-Ec (5′-CCCGAATTCTTACTTGTACAGCTCGTC-3′). Ps-Gly-GFP contains a PstI site (underlined) and 15 nucleotides encoding a Gly linker (lowercase letters) fused to the initiation codon (italics); GFP-St-Ec contains an EcoRI site (underlined) fused to the termination codon (italics). The PCR product was digested with PstI and EcoRI and cloned into the PstI-EcoRI site of pEC2 to make pYTGFP-C.
The FOW1 cDNA was amplified from total RNA of Mel02010 by RT-PCR with primers FOW1C-f (5′-ACCTCTAGATGGCTGCTACTATCCGTAC-3′) and FOW1C-r (5′-AGACTGCAGATGCTTAGCGGTGACCG-3′). FOW1C-f contains an XbaI site (underlined) with the initiation codon (italics); FOW1C-r contains a PstI site (underlined) fused to the last codon of FOW1. The amplified DNA was digested with XbaI and PstI and cloned into the XbaI-PstI site of pYTGFP-C to make pFOW1-GFP, resulting in a C-terminal fusion of GFP to Fow1.
The GFP ORF was amplified from pCB16EGFP by PCR with primers T3 (5′-AATTAACCCTCACTAAAGGG-3′) and GFP-r (5′-TTGTTGATCAACCACCACCACCACCCTT-3′). The T3 sequence is upstream of the GFP ORF in pCB16EGFP; GFP-r has a BclI site (underlined) carrying the termination codon (italics). The PCR product was digested with XbaI and BclI and cloned into the XbaI-BamHI site of pEC2 to make the GFP expression vector pYTGFPc.
All PCR products cloned in the vectors were sequenced to confirm the fact that no nucleotide substitution had occurred during amplification.
Construction of the YMR241w Expression Vector
Total DNA of strain YPH102 of Saccharomyces cerevisiae was isolated with the Yeast DNA Isolation Kit (Takara) according to the manufacturer's instructions. The entire ORF of YMR241w, which has no intron (Mayer et al., 1997), was amplified from total DNA by PCR with primers Sc-Xb-f (5′-ACCTCTAGATGCCATCTACCACTAATAC-3′) and Sc-EI-r (5′-AGAGAATTCTAATGTTTGGCAACTGG-3′). Sc-Xb-f contains a terminal XbaI site (underlined) with the initiation codon (italics); Sc-EI-r contains a terminal EcoRI site (underlined) with the termination codon (italics). The PCR product was digested with XbaI and EcoRI and cloned into the XbaI-EcoRI site of pEC2 to make pEYMR241w carrying the trpC promoter-YMR241w-trpC terminator construct. The YMR241w sequence amplified was verified to be same as that reported previously.
Test for Pathogenicity and Vegetative Growth
The pathogenicity of F. oxysporum f. sp. melonis and f. sp. lycopersici was tested by the root dip method using melon (Cucumis melo cv Amus) and tomato (Lycopersicon esculentum cv Ponderosa), respectively, as described previously (Namiki et al., 1994, 1998; Inoue et al., 2001). Bud cells were suspended in sterilized water at a concentration of ∼1 × 107 cells/mL, and plant roots were dipped in the suspension for 15 s. Plants were grown in pots filled with sterilized soil. Melon plants with a single true leaf or with four true leaves were used for inoculation, and disease symptoms were assessed 3 weeks after inoculation. Tomato plants with two true leaves were inoculated, and disease symptoms were assessed 3 weeks after inoculation.
To determine colonization by F. oxysporum in inoculated plants, fungi were isolated from stems of inoculated plants. Stem segments (∼1.5 cm) immediately above crowns were cut from inoculated plants, surface-sterilized by soaking in 1% sodium hypochlorite for 5 min, and rinsed with sterilized water. The segments were incubated on PDA at 25°C for 7 days, and the appearance of F. oxysporum colonies was observed.
To test for vegetative growth of F. oxysporum strains, they were grown on PDA, minimal agar (MA; 10 g/L KNO3, 5 g/L KH2PO4, 2.5 g/L MgSO4·7H2O, 0.02 g/L FeCl3, 10 g/L Glc, and 20 g/L agar) (Sanderson and Srb, 1965) and complete agar (MA supplemented with 1 g/L yeast extract and 1 g/L peptone) (Sanderson and Srb, 1965) at 25°C for 5 days, and colony diameters were measured. To determine the gly− phenotype of fow1 mutants, they were grown on MA supplemented with 3% ethanol and 3% glycerol instead of Glc (Nelson et al., 1993). Strains also were grown in potato dextrose broth at 25°C for 3 days on an orbital shaker (110 rpm). The resulting culture was passed through four layers of cheesecloth to separate mycelia and bud cells. Mycelia on cheesecloth were dried and weighed. Bud cells in filtrates were counted with a light microscope (BX50; Olympus, Tokyo, Japan). The formation of macroconidia, microconidia, and chlamydospores was evaluated on carnation leaf agar by the method of Togawa (1992).
Microscopic Observation of Infection Behavior
Melon seeds were placed on moist filter papers on Petri plates in sterile conditions and allowed to germinate for 3 days. Germinated seeds with radicles (∼1.5 cm) were selected, and radicles were dipped in bud cell suspensions (1 × 106 cells/mL) of the pTEFEGFP transformants of Mel02010 and B60 (Mel02010-GFP1 and B60-GFP1, respectively). After incubation in a mist box at 25°C for 24 h, radicles were observed with a fluorescence microscope (BX50) using a U-MWIG filter (Olympus).
Three melon plants with four true leaves were inoculated with bud cells (1 × 107 cells/mL) of each transformant by the root dip method. At 10 days after inoculation, plants were removed and washed carefully with water to remove adhering soil particles. More than 10 sections each were made from the crown and the main root of each plant and were observed with a fluorescence microscope (BX50) using a U-MWIG filter.
RT-PCR of RNA from Infected Melon Plants
Melon plants with a single true leaf were inoculated with bud cell suspensions (∼1 × 107 cells/mL) of Mel02010 by the root dip method. Control plants were immersed in water. Plants were grown in plastic pots filled with sterilized soil. At 4, 6, 9, and 11 days after inoculation, 10 plants from each treatment were removed and washed carefully with water to remove adhering soil particles. The roots and stems were separated with a sterile scalpel.
Total RNA was isolated from each tissue by the method of Yoshioka et al. (1996) and treated with RQ1 RNase-Free DNase (1000 units/mL; Promega) at 37°C for 15 min. FOW1 cDNA was amplified from total RNA (1 μg) from plant tissues with primers B6001 and B6002 (Figure 1). PCR was performed for 36 cycles with an initial 2 min at 94°C for denaturation and a final 10 min at 72°C for extension. Each cycle consisted of 30 s at 94°C, 30 s at 60°C, and 1.5 min at 72°C. Aliquots (5 μL) of the RT-PCR mixtures (100 μL) were electrophoresed on 1.0% agarose gels, and the blots were probed with PB60 fragments (Figure 1).
Observation of Intracellular Localization of the GFP-Tagged Fow1
Transformants of the wild-type strain with pYTGFPc, pFOW1-GFP, and pGFP-FOW1 were grown on PDA at 25°C for 5 days. The resulting mycelia were stained with 2.5 μM MitoTracker Red CMXRos (Molecular Probes, Eugene, OR) and observed with a fluorescence microscope (BX50) using a U-MWIG filter for GFP fluorescence and a U-MNIBA2 filter (Olympus) for MitoTracker Red CMXRos fluorescence.
Upon request, all novel material described in this article will be made available in a timely manner for noncommercial research purposes. No restriction or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The GenBank accession number for FOW1 is AB078975. The accession numbers for the sequences of the YMR241w mitochondrial carrier protein of S. cerevisiae and a putative mitochondrial carrier protein of S. pombe mentioned in Figure 2 are Z48756 and D89111, respectively.
Acknowledgments
We are grateful to Yoshitaka Takano and Hirofumi Aiba for providing pCB16EGFP and yeast strains, respectively, and for valuable suggestions. We thank Daniel Cullen and John Andrews for providing pTEFEGFP. We also thank Hirofumi Yoshioka, Kazuhito Kawakita, Noriyuki Doke, and Kazufumi Nishi for valuable suggestions; the Radioisotope Research Center at Nagoya University for technical assistance; and the Microorganisms Section of the Ministry of Agriculture, Forestry, and Fisheries Gene Bank for providing fungal strains. This work was supported by grants-in-aid from the Japanese Society for the Promotion of Sciences (T.T.) and by a research fellowship of the Japan Society for the Promotion of Science for Young Scientists (I.I.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002576.
References
- Ahringer, J. (1995). Embryonic tissue differentiation in Caenorhabditis elegans requires dif-1, a gene homologous to mitochondrial solute carriers. EMBO J. 14, 2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arie, T., Goutha, S., Shimazaki, S., Kamakura, T., Kimura, N., Inoue, M., Takio, K., Ozaki, A., Yoneyama, K., and Yamaguchi, I. (1998). Immunological detection of endopolygalacturonase secretion by Fusarium oxysporum in plant tissue and sequencing of its encoding gene. Ann. Phytopathol. Soc. Jpn. 64, 7–15. [Google Scholar]
- Armstrong, G.M., and Armstrong, J.K. (1981). Formae speciales and races of Fusarium oxysporum causing wilt disease. In Fusarium: Disease, Biology, and Taxonomy, P.E. Nelson, T.A. Toussoun, and R.J. Cook, eds (University Park, PA: Pennsylvania State University Press), pp. 391–399.
- Beck, E., Ludwig, G., Auerswald, E.A., Reiss, B., and Schaller, H. (1982). Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene 19, 327–336. [DOI] [PubMed] [Google Scholar]
- Beckman, C.H. (1987). The Nature of Wilt Diseases of Plants. (St. Paul, MN: American Phytopathological Society Press).
- Belenkiy, R., Haefele, A., Eisen, M.B., and Wohlrab, H. (2000). The yeast mitochondrial transport proteins: New sequences and consensus residues, lack of direct relation between consensus residues and transmembrane helices, expression patterns of the transport protein genes, and protein-protein interactions with other proteins. Biochem. Biophys. Acta 1467, 207–218. [DOI] [PubMed] [Google Scholar]
- Bruchez, J.J.P., Eberle, J., and Russo, V.E. (1993). Regulatory sequences in the transcription of Neurospora crassa gene: CAAT box, TATA box, intron, poly(A) tail formation sequences. Fungal Genet. Newsl. 40, 89–96. [Google Scholar]
- Cormack, B.P., Valdivia, R.H., and Falkow, S. (1996). FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173, 33–38. [DOI] [PubMed] [Google Scholar]
- Di Pietro, A., and Roncero, M.I.G. (1998). Cloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum. Mol. Plant-Microbe Interact. 11, 91–98. [DOI] [PubMed] [Google Scholar]
- Di Pietro, A.D., García-Maceira, F.I., Méglecz, E., and Roncero, I.G. (2001). A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39, 1140–1152. [PubMed] [Google Scholar]
- García-Maceira, F.I., Di Pietro, A., and Roncero, M.I. (2000). Cloning and disruption of pgx4 encoding an in planta expressed exopolygalacturonase from Fusarium oxysporum. Mol. Plant-Microbe Interact. 13, 359–365. [DOI] [PubMed] [Google Scholar]
- Gritz, L., and Davies, J. (1983). Plasmid-encoded hygromycin B resistance: The sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25, 179–188. [DOI] [PubMed] [Google Scholar]
- Huertas-González, M.D., Ruiz-Roldán, M.C., Maceira, F.I.G., Roncero, M.I.G., and Di Pietro, A. (1999). Cloning and characterization of pl1 encoding an in planta-secreted pectate lyase of Fusarium oxysporum. Curr. Genet. 35, 36–40. [DOI] [PubMed] [Google Scholar]
- Idnurm, A., and Howlett, B.J. (2001). Pathogenicity genes of phytopathogenic fungi. Mol. Plant Pathol. 2, 241–255. [DOI] [PubMed] [Google Scholar]
- Inoue, I., Ohara, T., Namiki, F., and Tsuge, T. (2001). Isolation of pathogenicity mutants of Fusarium oxysporum f. sp. melonis by insertional mutagenesis. J. Gen. Plant Pathol. 67, 191–199. [Google Scholar]
- Kimura, A., Takano, Y., Furusawa, I., and Okuno, T. (2001). Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 13, 1945–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, N., and Tsuge, T. (1993). Gene cluster involved in melanin biosynthesis of the filamentous fungus Alternaria alternata. J. Bacteriol. 175, 4427–4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knogge, W. (1998). Fungal pathogenicity. Curr. Opin. Plant Biol. 1, 324–328. [DOI] [PubMed] [Google Scholar]
- Kuspa, A., and Loomis, W.F. (1992). Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc. Natl. Acad. Sci. USA 89, 8803–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Leach, J.G., and Currence, T.M. (1938). Fusarium wilt of muskmelon in Minnesota. Minn. Agric. Exp. Stn. Tech. Bull. 129, 32. [Google Scholar]
- Li, D., Rogers, L., and Kolattukudy, P.E. (1997). Cloning and expression of cDNA encoding a mitogen-activated protein kinase from a phytopathogenic filamentous fungus. Gene 195, 161–166. [DOI] [PubMed] [Google Scholar]
- Lu, S., Lyngholm, L., Yang, G., Bronson, C., Yoder, O.C., and Turgeon, B.G. (1994). Tagged mutations at the Tox1 locus of Cochliobolus heterostrophus by restriction enzyme-mediated integration. Proc. Natl. Acad. Sci. USA 91, 12649–12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, J.A., Kakhniashvili, D., Gremse, D.A., Campbell, C., Krämer, R., Schroers, A., and Kaplan, R.S. (1997). Bacterial overexpression of putative yeast mitochondrial transport proteins. J. Bioenerg. Biomembr. 29, 541–547. [DOI] [PubMed] [Google Scholar]
- Mullaney, E.J., Hamer, J.E., Roberti, K.A., Yelton, M.M., and Timberlake, W.E. (1985). Primary structure of the trpC gene from Aspergillus nidulans. Mol. Gen. Genet. 199, 37–45. [DOI] [PubMed] [Google Scholar]
- Namiki, F., Matsunaga, M., Okuda, M., Inoue, I., Nishi, K., Fujita, Y., and Tsuge, T. (2001). Mutation of an arginine biosynthesis gene causes reduced pathogenicity in Fusarium oxysporum f. sp. melonis. Mol. Plant-Microbe Interact. 14, 580–584. [DOI] [PubMed] [Google Scholar]
- Namiki, F., Shiomi, T., Kayamura, T., and Tsuge, T. (1994). Characterization of the formae speciales of Fusarium oxysporum causing wilt of cucurbits by DNA fingerprinting with nuclear repetitive DNA sequences. Appl. Environ. Microbiol. 60, 2684–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namiki, F., Shiomi, T., Nishi, K., Kayamura, T., and Tsuge, T. (1998). Pathogenic and genetic variation in the Japanese strains of Fusarium oxysporum f. sp. melonis. Phytopathology 88, 804–810. [DOI] [PubMed] [Google Scholar]
- Nelson, D.R., Felix, C.M., and Swanson, J.M. (1998). Highly conserved charge-pair networks in the mitochondrial carrier family. J. Mol. Biol. 277, 285–308. [DOI] [PubMed] [Google Scholar]
- Nelson, D.R., Lawson, J.E., Klimgenberg, M., and Douglas, M.G. (1993). Site-directed mutagenesis of the yeast mitochondrial ADP/ATP translocator: Six arginines and one lysine are essential. J. Mol. Biol. 230, 1159–1170. [DOI] [PubMed] [Google Scholar]
- Okuda, M., Ikeda, K., Namiki, F., Nishi, K., and Tsuge, T. (1998). Tfo1: An Ac-like transposon from the plant pathogenic fungus Fusarium oxysporum. Mol. Gen. Genet. 258, 599–607. [DOI] [PubMed] [Google Scholar]
- Oliver, R., and Osbourn, A. (1995). Molecular dissection of fungal phytopathogenicity. Microbiology 141, 1–9. [DOI] [PubMed] [Google Scholar]
- Palmieri, F. (1994). Mitochondrial carrier proteins. FEBS Lett. 346, 48–54. [DOI] [PubMed] [Google Scholar]
- Palmieri, L., Runswick, M.J., Fiermonte, G., Walker, J.E., and Palmieri, F. (2000). Yeast mitochondrial carriers: Bacterial expression, biochemical identification and metabolic significance. J. Bioenerg. Biomembr. 32, 67–77. [DOI] [PubMed] [Google Scholar]
- Risser, G., Banihashemi, Z., and Davis, D.W. (1976). A proposed nomenclature of Fusarium oxysporum f. sp. melonis races and resistance genes in Cucumis melo. Phytopathology 66, 1105–1106. [Google Scholar]
- Rodríguez-Gálvez, E., and Mendgen, K. (1995). The infection process of Fusarium oxysporum in cotton root tips. Protoplasma 189, 61–72. [DOI] [PubMed] [Google Scholar]
- Roldán-Arjona, T., Pérez-Espinosa, A., and Ruiz-Rubio, M. (1999). Tomatinase from Fusarium oxysporum f. sp. lycopersici defines a new class of saponinases. Mol. Plant-Microbe Interact. 12, 852–861. [DOI] [PubMed] [Google Scholar]
- Ruiz-Roldán, M.C., Di Pietro, A., Huertas-González, M.D., and Roncero, M.I.G. (1999). Two xylanase genes of the vascular wilt pathogen Fusarium oxysporum are differentially expressed during infection of tomato plants. Mol. Gen. Genet. 261, 530–536. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sanderson, K.E., and Srb, A.M. (1965). Heterokaryosis and parasexuality in the fungus Ascochyta imperfecta. Am. J. Bot. 42, 72–81. [PubMed] [Google Scholar]
- Schäfer, W. (1994). Molecular mechanisms of fungal pathogenicity to plants. Annu. Rev. Phytopathol. 32, 461–477. [Google Scholar]
- Thompson, J.D., Higgins, D.C., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa, M. (1992). Effect of sterilization methods, plant varieties and leaf stages on conidia and perithecia formation in the genus Fusarium in CLA-culture. Trans. Mycol. Soc. Jpn. 33, 385–393. [Google Scholar]
- Tzagoloff, A., and Dieckmann, C.L. (1990). PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 54, 211–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanden Wymelenberg, A.J., Cullen, D., Spear, R.N., Schoenike, B., and Andrews, J.H. (1997). Expression of green fluorescent protein in Aureobasidium pullulans and quantification of the fungus on leaf surfaces. Biotechniques 23, 686–690. [DOI] [PubMed] [Google Scholar]
- Xu, J.-R. (2000). MAP kinases in fungal pathogens. Fungal Genet. Biol. 31, 137–152. [DOI] [PubMed] [Google Scholar]
- Yoshioka, H., Miyabe, M., Hayakawa, Y., and Doke, N. (1996). Expression of genes for phenylalanine ammonia-lyase and 3-hydroxy- 3-methylglutaryl CoA reductase in aged potato tubers infected with Phytophthora infestans. Plant Cell Physiol. 37, 81–90. [Google Scholar]