Abstract
The Arabidopsis ABSCISIC ACID–INSENSITIVE3 (ABI3) protein plays a crucial role during late seed development and has an additional function at the vegetative meristem, particularly during periods of growth-arresting conditions and quiescence. Here, we show that the ABI3 homolog of poplar (PtABI3) is expressed in buds during natural bud set. Expression occurs clearly after perception of the critical daylength that initiates bud set and dormancy in poplar. In short-day conditions mimicking natural bud set, the expression of a chimeric PtABI3::β-glucuronidase (GUS) gene occurred in those organs and cells of the apex that grow actively but will undergo arrest: the young embryonic leaves, the subapical meristem, and the procambial strands. If PtABI3 is overexpressed or downregulated, bud development in short-day conditions is altered. Constitutive overexpression of PtABI3 resulted in apical buds with large embryonic leaves and small stipules, whereas in antisense lines, bud scales were large and leaves were small. Thus, PtABI3 influences the size and ratio of embryonic leaves and bud scales/stipules that differentiate from the primordia under short-day conditions. These observations, together with the expression of PtABI3::GUS in embryonic leaves but not in bud scales/stipules, support the idea that wild-type PtABI3 is required for the relative growth rate and differentiation of embryonic leaves inside the bud. These experiments reveal that ABI3 plays a role in the cellular differentiation of vegetative tissues, in addition to its function in seeds.
INTRODUCTION
Plants continuously adjust the growth rate of their organs in response to developmental and environmental signals. One possible means of integrating the growth of different organs consists of imposing dormancy on some of the meristems along the plant. This strategy is elaborated most evidently by trees that arrest many of their meristems in axillary buds to achieve their typical architecture. Bud dormancy is exploited similarly when plants have to persist through periods that are unfavorable for growth or that are potentially hazardous, such as the winter season. In this case, growth inactivity is imposed on all meristems.
Despite many physiological studies, only fragmentary knowledge exists regarding the endogenous processes that direct the creation of a dormant state (Lang, 1996). Although seed dormancy has become better understood, our knowledge of bud dormancy remains scarce. However, because seed and bud dormancy share a number of characteristics, they were assumed to be founded on similar physiological processes (Wareing, 1956). Arguments for a common basis of both processes include the following: (1) a similar chilling requirement to overcome dormancy in seeds and buds for a given genotype in some species; (2) similar influences of the plant hormones abscisic acid (ABA) and gibberellins on dormancy induction and dormancy release; (3) similar changes in water availability in the cells together with the acquisition of desiccation tolerance; and (4) a similar accumulation of reserve proteins and lipids (Saure, 1985; Powell, 1987; Dennis, 1996). Moreover, these analogies suggest that both seed and bud dormancy are regulated, at least in part, by similar molecular mechanisms.
One ideal candidate to test this hypothesis is the ABSCISIC ACID–INSENSITIVE3 (ABI3) gene, which plays a crucial role during late seed development (Bonetta and McCourt, 1998; Leung and Giraudat, 1998). Although the abi3 mutant was isolated originally for its insensitivity to ABA during seed germination (Koornneef et al., 1984), the reduced seed dormancy seems to arise to only some extent from an ABA-related defect. Phenotypes such as the premature activation of the germination program and the failure to break down chlorophyll and to accumulate reserves are not observed in other aba or abi mutants (Nambara et al., 1995). Therefore, ABI3 may have broader functions than ABA signaling; rather, it may act as a global regulator of cell fate that allows cellular maturation (Bonetta and McCourt, 1998).
Despite the facts that the most prominent phenotype of the abi3 mutant is its reduced seed dormancy and the prevalent function of ABI3 is in seeds (Parcy et al., 1994), ABI3 is not seed specific (Rohde et al., 2000c). Processes during the vegetative and reproductive growth of Arabidopsis, such as flowering time, inflorescence morphology, and plastid differentiation in seedlings, are affected by ABI3 as well (Robinson and Hill, 1999; Kurup et al., 2000; Rohde et al., 2000a; Suzuki et al., 2001). The fact that ABI3 functions in vegetative Arabidopsis meristems, particularly during periods of growth-arresting conditions and quiescence (Rohde et al., 1999), suggests that ABI3 could be involved in the regulation of bud dormancy in trees.
Here, we have investigated the function of the ABI3 homolog from poplar (Populus trichocarpa), PtABI3, throughout bud development and dormancy. Although poplars do not develop a very deep dormancy in buds and seeds, they have been used successfully to study the genetics and physiology of bud dormancy (Bradshaw and Stettler, 1995; Olsen et al., 1997; Frewen et al., 2000; Rohde et al., 2000b). We demonstrate that PtABI3 is expressed in autumn buds at the time of vegetative growth arrest. Accordingly, the expression of a chimeric PtABI3::β-glucuronidase (GUS) construct occurred primarily in young embryonic leaves of the bud during bud set–inducing short days.
In short days, Populus tremula × Populus alba trees transformed with sense and antisense constructs of PtABI3 showed altered bud morphology. Constitutive overexpression of PtABI3 resulted in apical buds with large embryonic leaves and small stipules, whereas in antisense lines, bud scales were large and leaves were small. Thus, PtABI3 influences the size and ratio of embryonic leaves and bud scales/stipules that differentiate from the primordia under short-day conditions.
These observations, together with the expression of PtABI3::GUS in embryonic leaves but not in bud scales/stipules, suggest that wild-type PtABI3 is required for the relative growth rate and differentiation of embryonic leaves during bud set. ABA levels peaked in buds concomitantly with PtABI3::GUS expression, suggesting that ABA and PtABI3 act simultaneously in bud set. We conclude that PtABI3 is an essential component of successful bud set that in turn is a precondition for the establishment of dormancy.
RESULTS
PtABI3 Is Expressed during Autumnal Bud Set under Natural Conditions
To delineate the seasonal expression pattern of PtABI3, its expression was analyzed in various tissues and in apical buds. A bud, surrounded by protective bud scales, contains a densely packed series of young leaves and leaf primordia that do not elongate but undergo growth arrest (approximately from August until March). By contrast, an active apex produces leaves that elongate simultaneously (approximately from April until August).
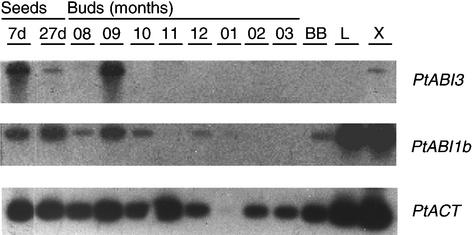
PtABI3 expression, as shown in Figure 1, was observed in developing poplar seeds, consistent with the prominent expression of ABI3 in Arabidopsis seeds. In apical buds, PtABI3 was found to be expressed exclusively in September, which corresponds to the time of vegetative bud set under natural conditions (Figure 1). Because bud set covers a 2-month period in field conditions (Belgium, 50°N) from the perception of the critical daylength (15.5 h) by mid August until the formation of the terminal bud by mid October, these data suggest that PtABI3 is not involved directly in the perception of the critical daylength. PtABI3 expression has not been analyzed from April until July because no apical buds are made during these months. However, PtABI3::GUS analyses never revealed expression in actively growing apices, which are equivalent to summer month apices (see below).
Figure 1.
Seasonal Expression of PtABI3, PtABI1b, and PtACT in Poplar.
Expression was analyzed in seeds (7 and 27 days after pollination), in buds (from August until March), at bud break (BB), in young leaves (L; harvested in April), and in xylem (X). Fragments of the genes were amplified by reverse transcriptase–mediated (RT)–PCR using gene-specific primers, hybridized with 32P-labeled probes of the respective genes, and visualized through autoradiography. Actin fragments could be amplified from all samples, indicating that cDNA was generated successfully, although the amount of actin transcripts varied and was particularly low in January. This experiment was repeated with similar results with independent bud material.
Interestingly, the expression of PtABI3 in autumn buds coincided with that of PtABI1b, as it did in seeds (Figure 1). ABI1 is a negative regulator of ABA signal transduction in Arabidopsis seeds. The simultaneous expression of PtABI3 and PtABI1b in buds at the time of growth arrest shows that seed and bud dormancy involve, at least in part, similar molecular components. However, unlike PtABI3, PtABI1b was expressed in buds at other times and in leaves and xylem of actively growing plants. Weak PtABI3 expression also was found in the xylem of greenhouse-grown P. tremula × P. alba (Figure 1). Because the expression in buds of PtABI3, but not of PtABI1b, appeared exclusively during autumnal bud set, we characterized its expression with a chimeric promoter-GUS fusion (PtABI3::GUS) and studied its function during bud set in transgenic P. tremula × P. alba upregulated and downregulated for PtABI3 expression.
Bud Set under Controlled Short-Day Conditions
Poplar belongs to the group of trees that show continuous growth under long days and that react to short days with growth arrest and dormancy induction (Nitsch, 1957). Experimentally, natural dormancy induction can be mimicked by short-day treatments in controlled greenhouses. As illustrated in Figures 2A to 2C, bud set is achieved within 5 weeks in P. tremula × P. alba under a photoperiod of 8 h/day. We analyzed the morphogenetic fates of the primordia in P. tremula × P. alba during short day–induced bud set (Figures 2 and 3). Figure 3 presents the corresponding morphogenetic fates of the individual primordia that together accomplish the change from an actively growing apex to a bud. The organs are termed according to Goffinet and Larson (1981)(1982).
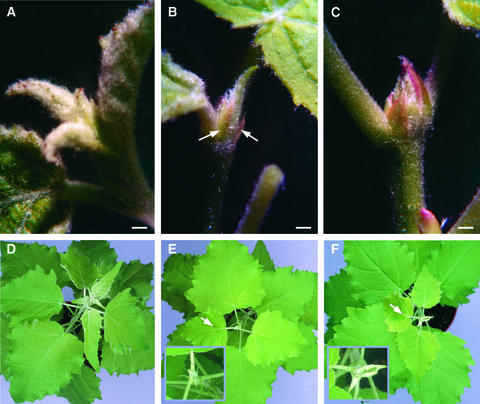
Figure 2.
P. tremula × P. alba Plants during Short Day–Induced Bud Set (8 h/Day Photoperiod).
(A) Close-up of the apex of an actively growing wild-type plant under long-day conditions. Two stipules are formed together with each leaf but are too small to be seen at this stage.
(B) Close-up of the apex of a bud-setting wild-type plant with formation of the first bud scales after 4 weeks under short-day conditions. Arrows point to the initiated bud scales.
(C) Close-up of the apical bud of a wild-type plant grown for 6 weeks under short-day conditions. Arrows point to the bud scales that have enlarged and started to encase the embryonic leaves.
(D) Apical part of a wild-type plant grown under long-day conditions.
(E) Apical part of a wild-type plant grown for 6 weeks under short-day conditions. The arrow marks the last mature leaf below the bud. The inset shows an enlargement of the apex.
(F) Apical part of a PtABI3-overexpressing plant grown for 6 weeks under short-day conditions. The arrow marks the last mature leaf below the bud. The inset shows an enlargement of the apex.
Bars in (A) to (C) = 1 mm.
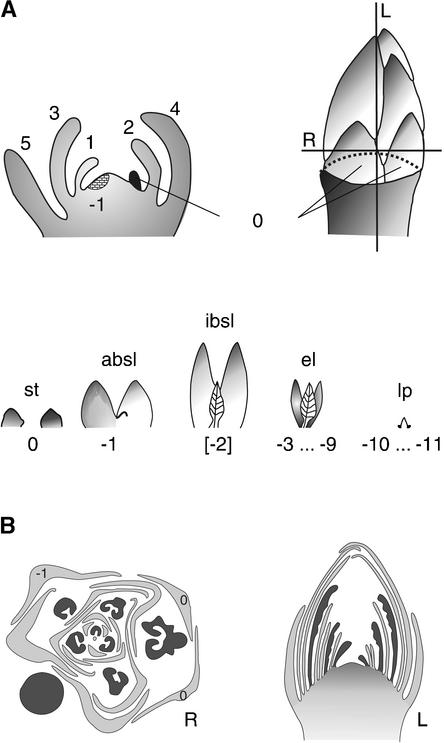
Figure 3.
Schemes of the Fate of Successive Primordia during Short Day–Induced Bud Set in Poplar and of the Morphology of the Bud at Accomplished Bud Set.
(A) Dormancy induction is illustrated with an apex at the perception of the critical daylength (top left) and a bud after bud set has been completed (top right and bottom). The planes of sectioning for longitudinal (L) and radial (R) sections are indicated by solid lines, and the apical meristem is indicated by the dashed line through the bud. At the perception of the critical daylength, the apex of the plant already contains committed primordia that will develop into leaves (primordia 5 to 0). Only those primordia initiated after the perception of the critical daylength will change morphogenetic fate (primordium −1 and younger ones). After bud set has been completed, the following units of organs are found inside a P. tremula × P. alba bud (in successive order from outside to inside): one abortive bud scale leaf, consisting of two sometimes fused bud scales (primordium −1; absl); one incipient bud scale leaf, consisting of two bud scales and a small embryonic leaf lamina (primordium −2; ibsl [not always found]); approximately seven embryonic leaves sensu stricto, consisting of two stipules and an embryonic leaf lamina (primordia −3 to −9; el); and approximately two not yet developed leaf primordia (primordia −10 and −11; lp).
(B) Morphology of the bud after bud set has been completed. As a result of the helical phyllotaxis of the bud, radial sections (R) reveal approximately one-third more organs than longitudinal sections (L). The plane of radial sections is situated just above the apical meristem (see bud in [A]); therefore, the enlarged stipules contributing to bud scales (primordia 1 and 0) usually are not revealed. In the schemes of radial and longitudinal sections, embryonic leaf laminae are dark gray and stipules and bud scales are light gray. In the radial section, organs 0 and −1 out of primordia 0 and −1 are numbered (to conform with the primordium numbers in [A]) to illustrate the fact that at the end of bud formation, a different number of organs may persist from different primordia.
During the first 3 weeks under short days, all primordia that had been committed before the onset of short days will develop into leaves, although internode elongation between them ceases gradually and becomes fully arrested after 3 to 4 weeks (Figure 3A, primordia 5 to 0). The last primordium that had been committed before the onset of short days is the last leaf to mature, often not to its full size, and to subtend the bud (Figure 3A, organ 0). The stipules of this leaf may persist after leaf abscission and enlarge to serve as bud scales for the developing bud.
Only the primordia initiated after the perception of the critical daylength will change morphogenetically from the beginning of their development (Figure 3A, primordium −1 and younger ones). After the 4th week, the bud scales are apparent (Figure 2B). They originate from the first primordium initiated after the onset of short days: the lamina of this primordium senesces and abscises prematurely, so that the stipular domains of the primordium enlarge to form bud scales (Figure 3A, abortive bud scale leaf). Embryonic leaves immediately distal to the abortive bud scale leaf do not abort their lamina. Nevertheless, the development of their midrib is suppressed so that the lamina appears broad and short (Figure 3A, incipient bud scale leaf). Inside the forming bud, organogenesis proceeds to form embryonic leaves sensu stricto, each with two stipules, and leaf primordia without yet distinct stipules (Figure 3A, embryonic leaves sensu stricto and leaf primordia).
Although all organs inside a bud often are commonly called leaf primordia, Goffinet and Larson (1981)(1982) introduced for all but the youngest organs other terms to account for their already apparent differences in differentiation and future fate. In their terminology, each primordium potentially gives rise to a unit of a leaf lamina and two stipules. Stipules can differentiate further into bud scales if the lamina is abscised or stays inferior. At the completion of bud set, all three organs from each primordium are not always present. As illustrated for primordium 0 in Figure 3A, only the stipules of the last leaf that matured below the bud persist and serve as bud scales. Similarly, the abortive bud scale leaf consists at the end of its development of two sometimes fused bud scales (primordium −1 in Figure 3A and organ −1 in Figure 3B).
For convenience, we refer to bud scales, stipules, and embryonic leaves as separate organs, although they may have arisen from a single primordium. Moreover, the term “embryonic leaves” is used in a broader sense than that suggested by Goffinet and Larson (1981)(1982): all leaf laminae (of the incipient bud scale leaf, the embryonic leaves sensu stricto, and the leaf primordia) are referred to as embryonic leaves.
After these organs have been made, the activity of the apical meristem ceases and the bud is formed (Figure 2C). Besides the described morphogenetic processes, some aspects of natural bud set are not accomplished under short-day treatments. P. tremula × P. alba plants, for example, do not shed leaves and do not acclimate to cold under the conditions used (see Methods). Bud set is a precondition for the establishment of dormancy in the meristem; still, considerable time may elapse before full winter dormancy is achieved.
PtABI3::GUS Expression in Buds under Short-Day and Long-Day Conditions
To establish the temporal and spatial expression pattern driven by the PtABI3 promoter, transgenic P. tremula × P. alba carrying a PtABI3::GUS construct were generated and analyzed (see Methods). In actively growing apices, no PtABI3::GUS expression was detected during growth in vitro or under standard (long-day) greenhouse conditions. In contrast, under short-day conditions, which are known to inhibit elongation growth and to promote bud set in poplars, strong PtABI3::GUS expression was observed. In short days, internode elongation was arrested gradually within the first 3 to 4 weeks, after which the first bud scales were initiated (Figure 2B).
Soon after the visible appearance of bud scales, PtABI3::GUS expression was detected in apical buds. As shown in the micrographs in Figure 4, the highest expression of PtABI3::GUS was seen in the youngest embryonic leaves inside the setting apical bud, starting at ∼22 days after the onset of short days (Figures 4A and 4D). PtABI3:: GUS expression then was observed repeatedly in every newly initiated embryonic leaf until meristematic activity ceased at ∼42 days. This GUS activity seemed to persist in the embryonic leaves but faded with increasing age and differentiation (Figure 4C). In lines with overall weaker GUS activity, PtABI3::GUS expression was concentrated at the tips of the embryonic leaves. By contrast, PtABI3::GUS expression was not detected in the bud scales or the stipules (Figures 4A and 4D). PtABI3-driven GUS activity also was found, albeit less intensely, in other very young or meristematic cells in the subapical region (Figure 4B). Overall PtABI3::GUS expression pattern was similar, albeit often with more pronounced subapical expression, in the uppermost five axillary buds that reside in the leaf axils along the shoot (Figure 4E).
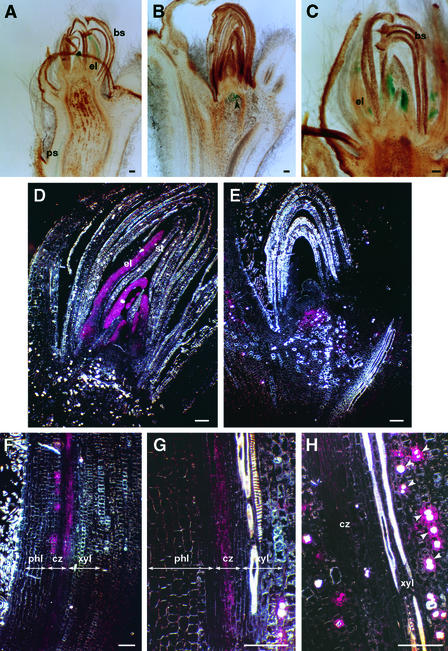
Figure 4.
Histochemical Localization of PtABI3::GUS Expression in P. tremula × P. alba after 22 to 42 days under Short-Day Conditions (See Text).
(A) to (C) PtABI3::GUS expression in apical buds under bright-field optics. The arrowhead in (B) points to a group of subapical cells expressing PtABI3::GUS.
(D) PtABI3::GUS expression in an apical bud under dark-field optics.
(E) PtABI3::GUS expression in an axillary bud under dark-field optics.
(F) to (H) PtABI3::GUS expression in the first internode beneath the apical bud under dark-field optics. Arrowheads in (H) point to single PtABI3::GUS–expressing cells containing oxalate crystals.
The scheme in Figure 3B indicates the plane of sectioning through the bud as well as the type and order of organs seen in longitudinal sections. Similar results were obtained with seven independent PtABI3::GUS lines (see Methods). bs, bud scales; cz, cambial zone; el, embryonic leaves; phl, phloem; ps, procambial strand; st, stipules; xyl, xylem. Bars = 100 μm.
Furthermore, PtABI3::GUS activity was observed in the procambial strands and in the cambium of the first two internodes below the apical bud (Figures 4A, 4F, and 4G). The expression in these tissues coincided with young, actively growing cells whose growth has to be retarded and arrested to ultimately meet short-day conditions. Additionally, expression was detected in single cells or small groups of cells close to the vascular cambium (Figure 4H). These cells often contained a large oxalate crystal, indicating their marked differentiation compared with neighboring cells. This aspect of PtABI3::GUS expression is interesting because it might highlight a link with cell-autonomous differences in the state of cellular differentiation.
PtABI3::GUS expression lasted from the 4th week after the onset of short days until the end of the short-day experiment (6 weeks). When plants were returned to long days after various periods of short-day treatment, PtABI3::GUS expression persisted in the inactive buds, presumably because of GUS protein stability, but disappeared as soon as growth had resumed (data not shown).
Axillary buds on long-day plants also change during their development from an active phase of organ initiation to an inactive, dormant state. In accordance with the fact that these buds also go through a developmental phase that prepares their arrest and dormancy in long-day plants, PtABI3::GUS activity also was detected in the axillary buds (data not shown). Similarly, cambial PtABI3::GUS expression also occurred in actively growing long-day plants (data not shown). In both cases, the expression pattern was completely comparable to that of axillary buds and cambium in the short-day plants shown in Figure 4.
However, in axillary buds and cambium of long-day plants, the expression was found at a greater distance from the apex than in plants grown under short days. In both cases, moreover, this distance varied greatly among individual plants, probably depending on vigor and environmental conditions. None of these stages of PtABI3::GUS expression under long days could be correlated with obvious phenotypic alterations in PtABI3-downregulated or PtABI3-overexpressing P. tremula × P. alba, suggesting that PtABI3 acts along with additional factors that are present only in growth-arresting short days.
In conclusion, the cessation of internode elongation and bud scale initiation happened well before the time when PtABI3::GUS expression was detected in the embryonic leaves. Therefore, PtABI3 functions during embryonic leaf growth and differentiation but not during leaf initiation. Moreover, the expression occurred in organs and cells in which growth still occurs but ultimately needs to be stopped: the young embryonic leaves, the subapical meristem, and the procambial strands.
PtABI3 Overexpression Prevents Correct Bud Formation in Response to Short Days
To understand the function of PtABI3 during bud set in poplar, transgenic P. tremula × P. alba plants were generated that overexpressed PtABI3. No phenotypic alterations were noted in these transgenic plants when grown under long-day conditions. However, aberrant phenotypes were observed under growth-arresting short days.
The growth response of overexpressing and control plants was comparable during the first 4 weeks after transfer to short-day conditions. On average, four additional internodes, which accounted for an ∼12-cm increase in height, were generated until the 4th week under short days. As shown in Figure 2, wild-type and PtABI3-overexpressing plants started to differ 4 weeks after transfer to short days. The apical shoot tip remained “open” in PtABI3-overexpressing plants (Figure 2F), whereas the embryonic leaves and leaf primordia in wild-type plants were positioned close to each other and were encased by bud scales (Figure 2E).
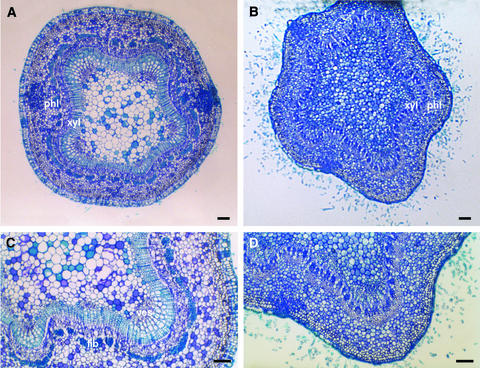
Figure 5 gives a detailed analysis of the correspondingly aberrant bud morphology in terms of embryonic leaves versus stipules/bud scales. After 6 weeks in short days, wild-type buds formed typical bud scales that encased entirely the embryonic leaves and their stipules (Figures 5A and 5E). By contrast, overexpression of PtABI3 led to plants with larger embryonic leaves with clear vascular bundles and small stipules, leaving the arrested apex exposed in an open bud (Figures 5B and 5F). There were no bud scales encasing the bud (Figures 5B and 5F). This apex morphology of PtABI3-overexpressing plants resembled that of actively growing plants (cf. Figures 2D and 2F with 5D and 5F).
Figure 5.
Morphology of Apical Buds of P. tremula × P. alba after 6 Weeks of Short-Day Treatment in Wild-Type, PtABI3-Overexpressing, and PtABI3 Antisense Lines.
(A) Wild-type line, longitudinal section.
(B) PtABI3-overexpressing line, longitudinal section.
(C) PtABI3 antisense line, longitudinal section.
(D) Actively growing wild-type plant (long-day plant) for comparison, radial section.
(E) Wild-type line, radial section.
(F) PtABI3-overexpressing line, radial section.
(G) PtABI3 antisense line, radial section. Resinous inclusions give rise to greenish blue staining in the bud scales of antisense lines and are highlighted by arrowheads.
Bud scales and stipules can be distinguished from embryonic leaves by their lighter blue staining with toluidine blue and by their morphology. For an easier interpretation of the micrographs, corresponding schemes with identical lowercase letters ([a] to [g]) indicate the types of organs seen in the sections. Bud scales and stipules are shown in light gray, and embryonic leaves and leaf primordia are shown in dark gray. Similar results were obtained with five PtABI3-overexpressing and four PtABI3 antisense lines. AS, PtABI3 antisense; LD, long day; OE, PtABI3 overexpression; SD, short day; wt, wild type. Bars = 100 μm.
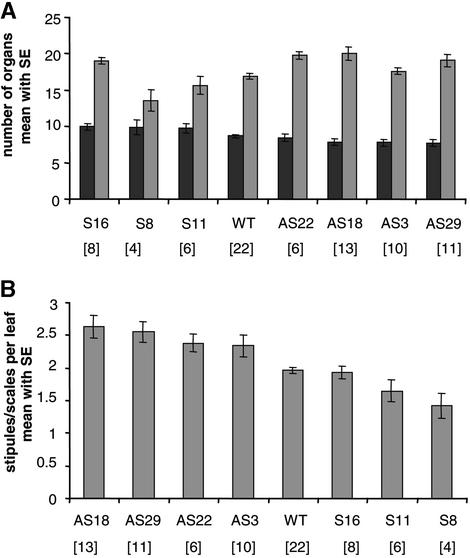
As shown in Figure 6, not only embryonic leaf growth and differentiation had been favored in PtABI3-overexpressing plants, but the number of embryonic leaves also had increased slightly (Figure 6A). Compared with the wild type, the increased number of leaf organs underscores the significance of the morphological observation that embryonic leaf differentiation is favored in the PtABI3-overexpressing lines. In its target tissue, the young embryonic leaves in which wild-type PtABI3 normally is expressed, PtABI3 overexpression allowed prolonged growth and differentiation compared with the wild type. On the other hand, in nontarget tissues in which wild-type PtABI3 normally is not expressed, the growth of bud scales and stipules was suppressed to varying degrees in different overexpression lines (Figures 5F and 6A).
Figure 6.
Number of Leaf-Like and Stipule/Scale-Like Organs in Apical Buds of P. tremula × P. alba after 6 Weeks of Short-Day Treatment in Wild-Type, PtABI3-Overexpressing (S8, S11, and S16), and Antisense (AS3, AS18, AS22, and AS29) Lines.
The number of organs irrespective of size was counted in serial microscopic sections of a bud and is expressed as average number ± se. The number of buds analyzed per line is given in brackets. wt, wild type.
(A) Number of leaf-like (embryonic leaves; dark gray) and stipule/scale-like (bud scales and stipules; light gray) organs per bud, given with decreasing number of leaf-like organs.
(B) Index of stipules per leaf as calculated by dividing the number of stipules by the number of embryonic leaves, given with decreasing index.
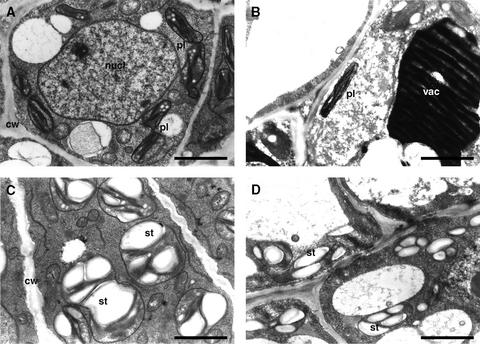
Another distinct characteristic of the PtABI3-overexpressing plants is that an enormous amount of starch accumulated in the buds, as seen in the electron micrographs shown in Figure 7. Numerous amyloplasts were observed in the PtABI3-overexpressing plants, whereas wild-type plants had only small chloroplasts with a few membranes and occasionally a tiny starch granule (Figures 7A to 7D). This observation shows once more that PtABI3 overexpression alters the metabolic state of both the embryonic leaves and the stipules.
Figure 7.
Transmission Electron Microscopy of Embryonic Leaves and Bud Scales in Apical Buds of P. tremula × P. alba after 6 Weeks of Short-Day Treatment in Wild-Type and PtABI3-Overexpressing Lines.
(A) Plastids with thylakoid membranes in embryonic leaves of the wild-type line.
(B) Small plastids with thylakoid membranes in stipule/scale-like organs of the wild-type line. Phenolic compounds give rise to the dark staining in the vacuole.
(C) Amyloplasts with thylakoid membranes in embryonic leaves of a PtABI3-overexpressing line.
(D) Amyloplasts with thylakoid membranes in stipule/scale-like organs of a PtABI3-overexpressing line.
Similar results were obtained with two PtABI3-overexpressing lines. cw, cell wall; nucl, nucleus; pl, plastid; st, starch; vac, vacuole. Bars = 1 μm.
Concomitant with the changes in bud morphology, changes in the vascular bundle immediately beneath the bud were noted. The stem tissue of the first internode below the bud of PtABI3-overexpressing plants clearly was different from that of wild-type plants. Figure 8 illustrates the morphological differences found in comparable stem segments. As stated above, wild-type and PtABI3-overexpressing plants differed neither in height nor in time of elongation arrest. Although the wild type achieved vascular maturity up to the apical bud, the vascular bundle of overexpressing plants remained in a less developed state (Figures 8A and 8B).
Figure 8.
Morphology of the First Internode Subtending the Bud after 6 Weeks of Short-Day Treatment of Wild-Type and PtABI3-Overexpressing P. tremula × P. alba Plants.
(A) and (C) Wild-type plants.
(B) and (D) PtABI3-overexpressing P. tremula × P. alba plants.
Five individual plants of the wild type, two PtABI3-overexpressing plants, and two PtABI3 antisense plants (sections not shown) were investigated. fib, phloem fibers; phl, phloem; ves, xylem vessels; xyl, xylem. Bars = 100 μm.
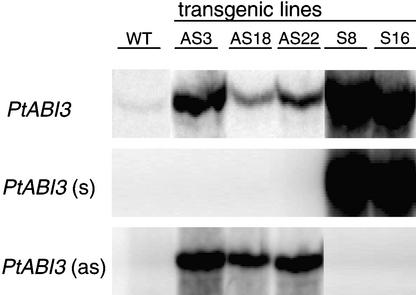
In the wild type, the differentiation of xylem vessel cells and phloem fibers was found ∼2800 and 3200 μm below the apex, respectively (Figure 8C). Neither morphological characteristic was found at a distance of <4000 μm below the apex in the two overexpressing lines (n = 5 for both) that were investigated in detail (Figure 8D). Because wild-type PtABI3 was expressed in procambial strands (Figures 1, 4A, and 4F), the less mature vascular bundle of PtABI3-overexpressing plants has to be attributed to an altered, probably slower, differentiation of this tissue under overexpression conditions. However, the possibility cannot be excluded that this vascular phenotype is a consequence of altered leaf development in the open bud of the PtABI3-overexpressing lines or vice versa. The phenotypic changes in the overexpression lines correlated with a significant overexpression of the PtABI3 transcript in the apical buds, as illustrated for lines S8 and S16 in Figure 9.
Figure 9.
Expression of Endogenous PtABI3 and Transgenes in Apical Buds of P. tremula × P. alba Grown for 4 Weeks under Short Days of 8 h/Day Photoperiod.
Fragments were amplified by RT-PCR with specific primers and 33P-labeled dATP. Products were separated in denaturing polyacrylamide gels and visualized by autoradiography. Primers for PtABI3 amplify both the endogenous and transgene transcripts. Primers for PtABI3 (s) and PtABI3 (as) amplify the sense and antisense transgene transcripts, respectively (see Methods). WT, wild type.
PtABI3 Antisense Lines Display Pronounced Bud Scale Development
Because a pronounced growth and differentiation of embryonic leaves out of a primordium had been observed in PtABI3-overexpressing plants, PtABI3 antisense plants were expected to display opposite phenotypes. Although RT-PCR easily revealed the expression of the PtABI3 antisense transcript, it remained difficult to demonstrate the downregulation of the endogenous PtABI3 transcript, because it was impossible to design primers that specifically detected the endogenous transcript (Figure 9). Nevertheless, 4 of 16 tested independent antisense lines showed similar and reproducible phenotypic alterations that were not observed in the wild type.
In these PtABI3 antisense lines, the growth of bud scales was very pronounced, occasionally even leading to a greater number of bud scales (Figures 5C, 5G, and 6A). The embryonic leaves tended to be smaller and fewer than those of the wild type (Figures 5C and 6A). However, the bud scales and embryonic leaves did not differ from those of the wild type in starch accumulation or in ultrastructure (data not shown). That the growth and differentiation of embryonic leaves cannot be finalized because of the absence of PtABI3 and are arrested prematurely may be the reason why the bud scales grow and differentiate more extensively than in the wild type. The advanced bud scale differentiation can be inferred from the accumulation of resinous compounds, as is apparent from the heavily staining inclusions in the outer bud scales of the antisense plants (Figure 5G).
Furthermore, unlike what was seen in PtABI3-overexpressing poplars, the formation of xylem vessels and phloem fibers in the vascular bundle beneath the bud took place earlier than in the wild type. Xylem vessels and phloem fibers, although morphologically similar to those of the wild type (Figures 8A and 8C), were found closer to the apex in the two antisense lines (n = 5 for both) studied in detail (at 2000 and 2500 μm instead of 2800 and 3200 μm in the wild type). Thus, in the absence of PtABI3, vascular differentiation proceeds faster than in the wild type.
If the alterations in organ number in PtABI3-overexpressing and antisense plants are expressed as an index of stipules per leaf, it becomes evident that PtABI3 influences the ratio of embryonic leaves and bud scales/stipules that differentiate out of the primordia under short-day conditions (Figure 6B). Comparing the bud morphology of PtABI3-overexpressing and antisense plants (Figures 5F and 5G) suggests the most obvious differences in bud scales. However, the expression of PtABI3::GUS occurs in embryonic leaves and not in bud scales/stipules, leading to the conclusion that wild-type PtABI3 is required primarily for the relative growth rate and differentiation of the embryonic leaves and, as a compensating effect, for the growth and differentiation of the stipules and bud scales.
ABA Concentration Shows a Distinct Peak during Short Day–Induced Bud Set
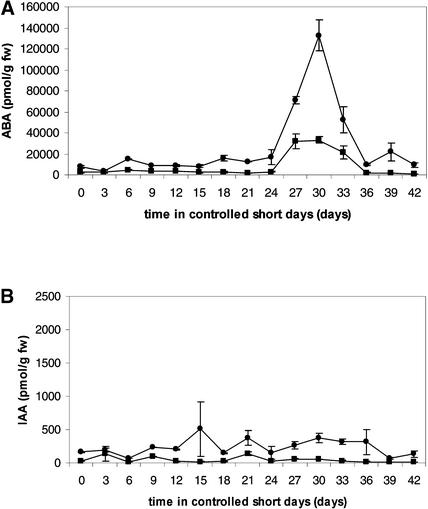
To determine whether ABA could be at least one of the expected growth-retarding factors that potentially act along with PtABI3, the ABA concentrations in apical buds and the first leaf subtending the bud were determined in short-day experiments. As shown in Figure 10A, the ABA concentration increased sevenfold in the apical buds after ∼24 to 27 days in short-day conditions. The ABA concentration in the subtending leaf showed a similar, albeit weaker, increase at this stage. The indole-3-acetic acid concentration remained fairly constant throughout the period of bud set (Figure 10B).
Figure 10.
Evolution of ABA (A) and IAA (B) Concentrations in the Apical Bud (Circles, Top Line) and the First Leaf Subtending the Bud (Rectangles, Bottom Line) during Short Day–Induced Bud Set.
Values given are means ± se of three individual plants. This experiment was repeated twice with similar results. Although the magnitude of ABA concentration was different, a similar increase was observed after 24 days.
The timing of the increase in ABA concentration could not be correlated with changes in environmental factors, because photoperiod, temperature, humidity, and watering of the plants had been kept constant. For that reason, this ABA peak was inherent to the developmental program of bud set. Moreover, it coincided with the inception of bud scales and the onset of PtABI3::GUS expression. Therefore, PtABI3 may act with ABA in preparing bud set.
DISCUSSION
Bud and Seed Dormancy Share ABI1- and ABI3-Mediated Signal Transduction Pathways
Seed and bud dormancy have common physiological characteristics that have led to the assumption that both are based on similar processes (Wareing, 1956). Extending the physiological analogies, both seed and bud dormancy also could involve similar cellular and molecular mechanisms. In support of this hypothesis, three of five identified PtABI1 genes and the PtABI3 gene, their Arabidopsis homologs being crucial regulators during late seed development (Leung and Giraudat, 1998), were found to be expressed both in seeds and during bud set in poplar (Figure 1; A. Rohde, unpublished data). The map position of one of the three PtABI1 genes expressed at bud set, PtABI1b (Figure 1), was found to coincide with a quantitative trait locus affecting bud set and bud flush in a poplar mapping pedigree designed to maximize the segregation of dormancy-related traits (Frewen et al., 2000). From these results, the idea emerges that at least ABI1 and ABI3 play a role during both bud set and seed dormancy.
Moreover, the PtABI3 expression data clearly suggest a function of ABI3 in meristem differentiation in both seed and vegetative tissues that are prepared for dormancy. Although seeds are regarded as the most prevalent dormant organ, the bud probably was the first dormant organ to evolve during evolution (Crabbé, 1994). In this rationale, the function of ABI3 in the preparation of meristem arrest may have been reused during the evolution of the seed to allow embryo maturation as a precondition of seed dormancy.
Seed and bud dormancy both involve ABI3 expression, but the analogy with ABI3 function applies only at the molecular level, not at the level of the whole dormancy process. Obviously, ABI3 is integrated with different molecular networks in the two processes, implying that ABI3 may have different functions. It remains to be determined whether ABI3 expression in buds and seeds depends on the same or different developmental and environmental factors.
PtABI3 Promotes the Growth and Differentiation of Embryonic Leaves in the Bud before Growth Arrest
In general, the winter bud contains the apical meristem and a portion or all of the next year's shoot organs. The formation of such a dormant bud is not a rapid process (Figure 3). PtABI3 is expressed halfway through the bud formation process: approximately 1 month after perception of the critical daylength under natural conditions and 3 to 4 weeks after the onset of controlled short days (Figures 1 and 4). This “late” expression implies that PtABI3 is not involved in the perception of the short-day signal, as is, for example, the phytochrome PHYA (Olsen et al., 1997). Overexpression of an oat PHYA in Populus tremula × Populus tremuloides results in changes in the critical daylength and prevents the cessation of internode elongation and the initiation of bud scales.
Both of these morphogenetic events happen well before the time that PtABI3::GUS expression is first detected. PtABI3 is expressed exclusively in organs and cells that grow actively but will undergo growth arrest: the young embryonic leaves, the subapical meristem, and the procambial strands (Figure 4). Growth in these tissues has to be finalized and arrested and acclimation has to be initiated to prepare the plant for the expected environmental changes signaled by short days. As revealed in the embryonic leaves, PtABI3 impinges on the embryonic leaf development that has been initiated before. Most likely, in the other tissues in which PtABI3 is expressed, PtABI3 also functions during differentiation toward a particular fate, perhaps through slowing of growth cessation.
That PtABI3 has a role in embryonic leaf growth and differentiation is supported by the phenotypes of PtABI3-overexpressing and antisense plants. Overexpression of PtABI3 resulted in plants with larger embryonic leaves and smaller bud scales than those in the wild type, leaving the apex exposed in an open but nevertheless arrested bud (Figures 2 and 5). The relative growth and differentiation rate of an embryonic leaf out of a primordium is favored over that of a bud scale or a stipule (Figure 6).
By contrast, in antisense lines, bud scale growth and differentiation were more pronounced and the embryonic leaves inside the bud tended to be smaller than in the wild type (Figure 5). Thus, the downregulation of PtABI3 leads to the suppression of part of the leaf differentiation program and is compensated for by bud scale development. These observations suggest that wild-type PtABI3 is required for the relative growth rate and differentiation of the embryonic leaves and, correlated with it, of the bud scales and stipules, well before their eventual growth arrest.
Therefore, the expression of PtABI3 controls the final differentiation state that is achieved in the embryonic leaves before their further development is arrested and interrupted by dormancy. At first glance, the fact that the downregulation of PtABI3 leads to an apparently dormant bud, whereas bud formation is not accomplished by PtABI3 overexpression (Figure 5), suggests that PtABI3 promotes leaf growth rather than dormancy. However, meristematic activity finally is arrested in both PtABI3 overexpression and antisense lines, but the degree of embryonic leaf growth and differentiation is different. Proper leaf differentiation is as much a part of successful bud set as is the development of encasing bud scales.
Is the Function of ABI3 Conserved in Poplar and Arabidopsis?
The phenotypes associated with mutant, antisense, and overexpression plants of ABI3 and PtABI3, as well as part of the spatial expression pattern of both genes, are conflicting and raise the question of whether the function of ABI3 is conserved between the annual Arabidopsis and the perennial poplar.
For example, although the expression of ABI3 and PtABI3 seems to be well conserved in seeds (Figure 1) (Parcy et al., 1994), vegetative ABI3::GUS expression in Arabidopsis has been found in stipules (Rohde et al., 1999). By contrast, PtABI3::GUS expression was not observed in stipules or bud scales but was observed in embryonic leaves in poplar (Figure 4). During poplar bud formation, leaf development undergoes a specific program to create the perennial organ, a developmental program that is not found in Arabidopsis. Embryonic leaf development is interrupted at an early stage and resumes after dormancy only if the tissue is sufficiently prepared for dormancy. PtABI3 contributes to this preparation of the embryonic leaves for dormancy through the integration of a differentiation step that does not occur in leaf development of Arabidopsis and does not occur in bud scales of poplar; the development of stipules and bud scales is almost accomplished until bud set and does not continue much after the dormant period. This example demonstrates that the expression of the ABI3 and PtABI3 genes is associated with particular stages of development that can take place in different organs and at different times rather than being associated with a particular organ and time.
Moreover, because of the duration of bud set, the expression of PtABI3 could be correlated with a particular phase during bud set, namely the preparation of embryonic leaf tissues for dormancy before bud set and dormancy. These functional aspects cannot be revealed during the establishment of vegetative quiescence in Arabidopsis. Because quiescence is short and difficult to control in Arabidopsis, ABI3 expression was correlated only with meristematic growth arrest in general (Rohde et al., 1999).
At the functional level, PtABI3-overexpressing poplars seem to proceed with “growth” and thus to resemble the prematurely germinating abi3 mutant much more than PtABI3 antisense poplars do. However, a closer look reveals that the growth of the abi3 mutant is a consequence of skipped seed maturation as much as the “nongrowth” of PtABI3 antisense poplars results from the omission of part of embryonic leaf differentiation. Overexpression phenotypes remain conflicting. PtABI3 overexpression led to prolonged embryonic leaf differentiation, whereas ABI3 overexpression had no phenotypic consequences during seed germination (Parcy et al., 1994).
ABI3 overexpression renders Arabidopsis plants hypersensitive to ABA but does not lead to constitutive ABA-related responses (Parcy et al., 1994). It is possible that PtABI3 overexpression coincides sufficiently with the endogenous increase in ABA during short day–induced bud set to result in phenotypic consequences (Figure 10). This speculation is supported by the fact that in poplar no overexpression phenotypes were observed in actively growing plants.
In conclusion, a functional analogy of PtABI3 and ABI3 exists at the molecular and cellular level, but it cannot be taken to the gross phenotypic level. Yet, like ABI3, PtABI3 also acts in the maturation and differentiation of cells and tissues or allows the time for it to occur. PtABI3 acts on appropriate growth and differentiation of embryonic leaves before bud set and dormancy, as ABI3 acts on the appropriate accumulation of storage proteins and the acquisition of desiccation tolerance during seed dormancy.
PtABI3 and ABA Act Simultaneously on Bud Set
PtABI3 overexpression led to a clear promotion of embryonic leaf growth and differentiation and to a suppression of bud scales; in the wild type, however, the action of PtABI3 has to be counterbalanced by factors that promote the formation of bud scales. Assuming that PtABI3 acts to slow growth cessation to allow appropriate differentiation of tissues, growth-arresting factors are prime candidates for causing counterbalancing effects.
One such candidate could be ABA. Indeed, ABA often has been found in dormant tissues and has growth-arresting effects when applied exogenously. ABA concentrations in apical buds increased significantly after ∼3 to 4 weeks of short days (Figure 10). The need for ABA biosynthesis during bud set is further supported by the expression of the poplar homolog of VIVIPAROUS14, which encodes the dioxygenase that is thought to perform the first committed and rate-limiting step in ABA biosynthesis (Schwartz et al., 1997) in September and October, the period of bud set under field conditions (A. Rohde, unpublished data).
The fact that ABA alone cannot provoke ABI3/PtABI3 expression in Arabidopsis (Parcy et al., 1994) or in poplar (A. Rohde, unpublished data) suggests that ABI3/PtABI3 may interact with ABA rather than being simply a component in the signal transduction chain of ABA. Still, the temporal correlation of PtABI3 expression and the increase in ABA concentration suggest that both PtABI3 and ABA affect an ongoing differentiation program in embryonic leaves (Figures 4 and 10). In this context, PtABI3 and ABA could act, at least in part, antagonistically. ABA, which generally is considered to be a potent growth inhibitor, would slow or stop growth. In PtABI3 antisense lines, embryonic leaf growth and differentiation would be arrested prematurely (Figure 5). The presence of wild-type PtABI3 might counteract ABA to accommodate processes that prepare the tissues sufficiently for later arrest. Likewise, overexpression of PtABI3 would prolong embryonic leaf growth and differentiation (Figure 5).
However, the rather close interaction between ABA and PtABI3 presumed in this scenario will remain hypothetical until the precise relationship of ABI3 and ABA is elucidated. Interesting in this respect is the fact that Arabidopsis ABI3 interacts with ABI5, an ABA-inducible TRAB homolog (transcription factor responsible for ABA regulation), and identifies a cross-point of ABI3-mediated and ABA-mediated signal transduction (Finkelstein and Lynch, 2000; Nakamura et al., 2001).
Another important observation is the prominent starch accumulation found exclusively in PtABI3-overexpressing plants (Figure 7). Similarly, Arabidopsis overexpressing ABI3 plants were shown recently to be hypersensitive to Glc (Finkelstein and Gibson, 2001). Additionally, a number of other experiments in Arabidopsis suggest a cross-connection of ABA and sugar signaling (Gazzarini and McCourt, 2001). ABI4, first isolated for its ABA insensitivity during germination (Finkelstein, 1994), now has been identified in several independent screens for sugar-related mutants (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Rook et al., 2001). It is possible that ABA signaling alters the tissue state so that cells become more sensitive to a separate sugar signal that induces, among others, starch biosynthesis genes (Rook et al., 2001). Although ABI4 does not interact directly with ABI3, full ABI4 expression requires ABI3 expression (Söderman et al., 2000; Nakamura et al., 2001), making it likely that ABI3 also affects the sugar-signaling processes governed by ABI4. Although all of these observations suggest a connection of ABI3 and ABA signal transduction with sugar homeostasis, the nature of these interactions is not yet fully understood.
PtABI3 Is an Essential Component of Bud Set
The assertion that PtABI3 is essential for correct embryonic leaf differentiation during bud set lends credence to the earlier speculation that ABI3 plays a permissive role in plant development to allow cellular maturation (Bonetta and McCourt, 1998). Achieving bud set, which involves a certain degree of differentiation in the embryonic leaves, likely bears a twofold ecological advantage for the buds: an embryonic leaf with a certain degree of differentiation probably will perform better in terms of frost tolerance and survival and has greater growth potential for the growth flush of next spring. These advantages are obtained at the cost of a relatively slow and elaborate bud-setting procedure. It will be interesting to evaluate transgenic plants with altered PtABI3 expression and inappropriate bud morphology for their properties in cold acclimation and survival.
METHODS
Plant Material and Growth Conditions
Apical buds were harvested monthly (1995 and 1998) and additionally weekly from August 21 until October 14 (1998) from 3-year-old trees of poplar (Populus trichocarpa cv Trichobel) grown in the field (Institute of Forestry and Game Management, Geraardsbergen, Belgium). At each sampling time, the apical buds of all branches (∼20 buds) of three trees were harvested and pooled per tree. Poplar seeds were generated from a controlled cross in the greenhouse between clone 70,039/115 and clone 70,039/109 of Populus trichocarpa and harvested at the indicated days after pollination (Figure 1). The pollen of the male clone was collected and stored at 4°C until female flower buds were perceptive for pollination.
For all experiments with transgenic plants, Populus tremula × Populus alba clone INRA No. 717.1B4 was used. Selected transgenic lines were multiplied clonally in vitro on Murashige and Skoog (1962) medium. The multiplied shoots were rooted on hormone-free Murashige and Skoog (1962) medium. Plantlets were acclimated and transferred to a greenhouse (∼18°C without additional light). All experiments were performed with at least three independent transgenic lines for each construct. For the bud set experiments, P. tremula × P. alba of ∼40 to 50 cm in height were exposed to two contrasting photoperiods, 16 or 8 h of light, in controlled Weiss chambers. The other growth conditions during experiments in Weiss rooms were as follows: 21°C day and 18°C night temperatures, 100 μmol·m−2·s−1 photosynthetic active radiation at plant level, and 55 to 70% RH.
Gene Isolation and Reverse Transcriptase–Mediated PCR
The poplar ABI3 homolog (PtABI3) is a single gene and has been described elsewhere (Rohde et al., 1998). In reverse transcriptase–mediated (RT)–PCR (Figure 1), a 910-bp PtABI3 fragment was amplified with 5′-ATGGGTCCAAACGAATAC-3′ as the sense primer and 5′-GGGCCAAAACCTATAACGCAT-3′ as the antisense primer, with the antisense primer spanning an intron to avoid genomic contamination.
For the amplification of ABI1 homologs from poplar, degenerated oligonucleotide primers for nested PCR amplification were designed based on the ABI1 sequence of Arabidopsis thaliana (Leung et al., 1994). In the first PCR, the following primers were used: 5′-AGR-CCNGARATGGARGAYGC-3′ and 5′-TCRCANGCYTCYTCRTC-3′, which are complementary in the sense and antisense directions to the sequences that encode amino acids RPEMEDA and DEEACE, respectively. This PCR product was used as a template for a second amplification with two internal degenerated primers: 5′-GTNTAYGAY-GGNCAYGGNGG-3′ and 5′-CATNACRTCCCANACNCCRTC-3′, which are complementary in the sense and antisense directions to the sequences that encode amino acids VYDGHGG and DGVWDVM, respectively.
Homologous primers were synthesized on the basis of the poplar sequence and led to the identification of the PtABI1b homolog. Its predicted protein is 64% similar to the ABI1 and ABI2 proteins. In total, five poplar ABI1 homologs have been identified to date, with PtABI1b being the most homologous with the ABI1 and ABI2 proteins. For each of these homologs, specific primers for PCR amplification have been developed. The following sense and antisense primers were used for PtABI1b: 5′-CGTATCCATTCGGCTTTGT-3′ and 5′-GATCCACTGATAATGCCATT-3′, resulting in a fragment of 303 bp on cDNA.
For the amplification of the actin homolog, degenerated oligonucleotide primers were designed based on an alignment of several known actin genes from plants. The degenerated primers 5′-GGG-AYGAYATGGARAARACNTGG-3′ and 5′-GGNGCNCANACYTTN-GTYTTCAT-3′ were complementary in the sense and antisense directions to the sequences that encode the conserved amino acids WDDMEKTW and MKTKVVAP, respectively. A 582-bp fragment was amplified from genomic DNA of poplar and sequenced. On the basis of this sequence, homologous primers (5′-TGGCATCACACCTTCTAC-3′ and 5′-CGACCACTGGCATAAAGG-3′ for the sense and antisense directions, respectively) were used for the amplification of a 190-bp fragment.
Total RNA from various tissues of poplar was extracted according to Goormachtig et al. (1995), except for xylem RNA that was extracted from greenhouse-grown P. tremula × P. alba using hot phenol (Verwoerd et al., 1989). Poly(A)+ RNA was purified with oligo(dT) beads (Dynal, Oslo, Norway) and subsequently reverse transcribed into double-stranded cDNA (cDNA Synthesis System Plus; Amersham Pharmacia Biotech, Little Chalfont, UK). PCR (reaction buffer as supplied with the Taq polymerase, deoxynucleotide triphosphate at 200 pmol, primers at 100 ng) was performed under the following conditions: 30 cycles at 94°C for 30 s, at 50°C for 30 s, and at 72°C for 2 min, and a final extension step at 72°C for 10 min. PCR products were run on agarose gels and hybridized to the respective 32P-labeled probes (Figure 1).
In the other RT-PCR experiments (Figure 9), reactions in 25 μL (50 ng of each primer) contained a modified nucleotide mix: dCTP, dTTP, and dGTP were at 200 pmol, whereas dATP was reduced to 20 pmol. To each reaction, 0.1 μL of 33P-labeled dATP (10 mCi/mL, 2500 Ci/mmol) was added, resulting in a hot-to-cold dATP ratio of 1:2500. Products were separated on 4.5% polyacrylamide gels and visualized on dried gels by means of autoradiography.
ABI3 Constructs and the Production of Transgenic Poplar
For the PtABI3::β-glucuronidase (GUS) construct, a 1267-bp promoter fragment from poplar was amplified from the appropriate genomic clone using the 1224 universal vector primer of pUC19 at the 5′ end and an antisense primer at the ATG. This primer was complementary to the first 18 bp of the coding sequence of PtABI3, starting at the ATG and including the second in-frame ATG. To the 5′ end of this primer, a SalI restriction site and four extra nucleotides for better digestibility were added. After amplification, the PCR fragment was digested with SalI and HindIII and cloned into the SalI-HindIII–digested binary vector pBI101.2 (Jefferson et al., 1987).
For the generation of PtABI3 sense and antisense constructs, the PtABI3 cDNA (Rohde et al., 1998) was excised with SalI and BamHI from pUC19 and cloned in the antisense direction into a SalI-BamHI–digested pLBR19 vector between the 35S promoter with a double enhancer and the 3′ sequences of Cauliflower mosaic virus (CaMV). For the sense construct, the cDNA was excised with EcoRI and BamHI and cloned in the sense direction into the EcoRI-BamHI–digested pLBR19. Because the last 21 bp of the PtABI3 cDNA were missing, it was necessary to sequence the 3′ end of the sense chimeric PtABI3 construct to verify in-frame stop codons in the 3′ CaMV sequences (stop codons at 108 and 123 bp after the last nucleotide of the PtABI3 cDNA). Both constructs then were excised with XbaI and KpnI from pLBR19 and inserted into the XbaI-KpnI–digested binary vector pBIN19, giving rise to the constructs 35S::ABI3s and 35S:: ABI3as.
Constructs 35S::ABI3s, 35S::ABI3as, and PtABI3::GUS were transformed into Agrobacterium tumefaciens strain C58RifR(pMP90) by heat shock (Zahm et al., 1984). Internodes and petioles of P. tremula × P. alba INRA No. 717.1B4 plants grown in vitro were transformed with Agrobacterium as described by Leplé et al. (1992). Independent transformants (28, 29, and 32, respectively) have been isolated and propagated clonally for the sense, antisense, and promoter-GUS constructs, respectively. Plants of 17 sense lines, 16 antisense lines, and 22 PtABI3::GUS lines were characterized under controlled growth conditions known to influence the growth and dormancy of buds. Plants were tested for growth cessation under short days (8-h photoperiod) and resumption of growth when transferred from short to long days.
In these experiments, five overexpression lines (S8, S11, S16, S19, and S24) and four antisense lines (AS3, AS18, AS22, and AS29) were selected for further detailed study. Of the 22 PtABI3::GUS lines tested under short days, 16 expressed PtABI3::GUS. Seven of these lines with high and temporally and spatially consistent expression (4B, 11A, 14A, 15B, 16A, 17A, and 17B) were selected for the experiments.
To correlate either the overexpression or the reduced expression of PtABI3 with the observed phenotypic changes, transcripts of the endogenous PtABI3 and the transgenes were followed by RT-PCR (Figure 9). 33P-dATP PCR conditions were as described above. The following primers were used as sense and antisense primers for the amplification of the endogenous, sense, and antisense transcripts, respectively: 5′-TCTTACCACAGGAATCTGAATCAT-3′ and 5′-GGGCCAAAACCTATAACGCAT-3′ (460 bp); 5′-TGCGTTATAGGT-TTTGGCCCA-3′ and 5′-CGAGCTCTCCCATATGGTCGACCTG-3′ (310 bp); and 5′-GCCCAAGCAGCCGCTGAAGAAG-3′ and 5′-GTG-ATTTCAGCGTACCGAATTCC-3′ (370 bp). For the sense and antisense transcripts, one of the primers was constructed so that the sequence was complementary to the junction of the PtABI3 cDNA, with the subsequent vector sequence in front of the CaMV 3′ untranslated region.
Histochemical GUS Assays
Vibroslices of agarose-embedded fresh material were stained immediately for GUS activity using the histochemical procedure described by Jefferson (1987). After the histochemical reaction, the material was fixed with 3% glutaraldehyde in phosphate buffer for 1 h, washed twice with phosphate buffer, and passed over 30, 50, 70, and 95% ethanol to remove the chlorophyll. Slices were cleared using chlorallactophenol and examined with a light microscope.
Light Microscopy
For morphological observations, fresh bud and stem material was fixed overnight in 4% formaldehyde, 5% iceacetic acid, and 50% ethanol and subsequently passed over a graded ethanol series. Ethanol was replaced by Histo-Clear (National Diagnostics, Hull, UK), and Histo-Clear was replaced by paraffin. Paraffin-embedded sections of 10 μm were cut on a rotary microtome. After paraffin was removed, sections were stained with 0.05% toluidine blue and mounted. Sections were examined with a light microscope.
For GUS-stained material, selected vibroslices were treated as described above and embedded in Technovit 7100 (Heraeus Kulzer, Wehrheim, Germany) after the 95% ethanol step according to the manufacturer's instructions. Sections (4 μm) were cut on a rotary microtome and mounted. Sections were examined with dark-field optics.
Transmission Electron Microscopy
Apical buds of wild-type, two overexpression, and two antisense lines were harvested after 6 weeks under short days and trimmed smaller before fixation. These pieces were fixed with a mixture of 4% paraformaldehyde and 3% glutaraldehyde and postfixed with 1% OsO4 and 1.5% K3Fe(CN)6 in 0.1 M Na-cacodylate buffer, pH 7.2. Samples were dehydrated through a graded ethanol series, including bulk staining with 2% uranyl acetate at the 50% ethanol step. Ethanol was replaced subsequently by LR White hard-grade embedding medium (London Resin, Basingstoke, UK), and samples were embedded in LR White.
Ultrathin sections of gold interference color were cut using an ultramicrotome and collected on collodion-coated copper grids of 200 mesh. The sections were poststained in an ultrostainer (Leica, Herburgg, Switzerland) for 15 min in 2% uranyl acetate (Ultrostain 1; Leica) at 40°C and for 4 min in lead citrate (Ultrostain 2; Leica) at 20°C. The sections were examined by transmission electron microscopy (Elmiskop 101; Siemens, Karlsruhe, Germany).
Analysis of Endogenous Abscisic Acid and Indole-3-Acetic Acid
Apical bud and leaf material from three plants was harvested separately within the first hour of the daily photoperiod during a 6-week short-day experiment. The material was immediately weighed, frozen in liquid nitrogen, and ground in liquid nitrogen using a rotary-ball mill (Kurt Retsch GmbH, Haan, Germany).
Abscisic acid (ABA) and indole-3-acetic acid (IAA) in the apical bud and the first leaf of individual plants were analyzed using a combined solid-phase extraction procedure (Prinsen et al., 1991, 2000). 13C6-IAA (200 pmol; Cambridge Isotope Laboratories, Andover, MA) and 18O1-ABA (200 pmol; prepared according to Gray et al., 1974) were used for isotope dilution purposes. After pentafluorobenzyl (PFB) esterification with α-bromopentafluorotoluene (Netting and Milborrow, 1988), PFB-IAA and PFB-ABA were analyzed by negative ion chemical ionization gas chromatography–coupled mass spectrometry (HP 5890 series II coupled to a VG FISONS TRIO 2000 quadrupole mass spectrometer; column 15m BD-XLB, 0.25 mm i.d. [J&W Scientific, Folsom, CA]; gas phase, He at 1.5 mL/min; gas chromatography oven gradient, 175 to 300°C at 15°/min; injection temperature, 150 to 325°C in 2 min). The source temperature was 140°C by constant filament emission; source vacuum pressure was 7 to 8 × 10−5 mbar; ionization potential was 35 eV; ionization current was 100 μA. The following diagnostic ions were used: mass-to-charge ratio (m/z) 263 (PFB-ABA), m/z 265 (PFB-18O1-ABA), m/z 174 (PFB-IAA), and m/z 180 (13C-PFB-IAA).
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are AJ003165 (PtABI3 gene) and AJ003166 (PtABI3 cDNA).
Upon request, all novel material described in this article will be made available in a timely manner for noncommercial purposes. No restrictions or conditions will be placed on the use of any material described in this article that would limit their use for noncommercial research purposes.
Acknowledgments
The Institute of Forestry and Game Management (Geraardsbergen, Belgium) is gratefully acknowledged for all field material of poplar. We thank Bart Burggraeve and Sevgi Öden for technical assistance, Martine De Cock for help in preparing the manuscript, and Rebecca Verbanck and Karel Spruyt for artwork. A.R. is indebted to the Vlaams Instituut voor de Bevordering van het Wetenschappelijk-Technologisch Onderzoek in de Industrie (Brussels, Belgium) for a postdoctoral fellowship.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003186.
References
- Arenas-Huertero, F., Arroyo, A., Zhou, L., Sheen, J., and León, P. (2000). Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 14, 2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Bonetta, D., and McCourt, P. (1998). Genetic analysis of ABA signal transduction pathways. Trends Plant Sci. 3, 231–235. [Google Scholar]
- Bradshaw, H.D., Jr., and Stettler, R.F. (1995). Molecular genetics of growth and development in Populus. IV. Mapping QTLs with large effects on growth, form, and phenology traits in a forest tree. Genetics 139, 963–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbé, J.J. (1994). Dormancy. In Encyclopedia of Agricultural Science, Vol. 1, C.J. Arntzen and E.M. Ritter, eds (San Diego, CA: Academic Press), pp. 597–611.
- Dennis, F.G., Jr. (1996). A physiological comparison of seed and bud dormancy. In Plant Dormancy: Physiology, Biochemistry and Molecular Biology, G.A. Lang, ed (Wallingford, UK: CAB International), pp. 47–56.
- Finkelstein, R.R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 5, 765–771. [Google Scholar]
- Finkelstein, R.R., and Gibson, S.I. (2001). ABA and sugar interaction regulating development: Cross-talk or voices in a crowd? Curr. Opin. Plant Biol. 5, 26–32. [DOI] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frewen, B.E., Chen, T.H.H., Howe, G.T., Davis, J., Rohde, A., Boerjan, W., and Bradshaw, H.D., Jr. (2000). Quantitative trait loci and candidate gene mapping of bud set and bud flush in Populus. Genetics 154, 837–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarini, S., and McCourt, P. (2001). Genetic interactions between ABA, ethylene and sugar signaling pathways. Curr. Opin. Plant Biol. 4, 387–391. [DOI] [PubMed] [Google Scholar]
- Goffinet, M.C., and Larson, P.R. (1981). Structural changes in Populus deltoides terminal buds and in the vascular transition zone of the stems during dormancy induction. Am. J. Bot. 68, 118–129. [Google Scholar]
- Goffinet, M.C., and Larson, P.R. (1982). Lamina abortion in terminal bud-scale leaves of Populus deltoides during dormancy induction. Bot. Gaz. 143, 331–340. [Google Scholar]
- Goormachtig, S., Valerio-Lepiniec, M., Szczyglowski, K., Van Montagu, M., Holsters, M., and de Bruijn, F.J. (1995). Use of differential display to identify novel Sesbania rostrata genes enhanced by Azorhizobium caulinodans infection. Mol. Plant-Microbe Interact. 8, 816–824. [DOI] [PubMed] [Google Scholar]
- Gray, R.T., Mallaby, R., Ryback, G., and Williams, V.P. (1974). Mass spectra of methyl abscisate and isotopically labelled analogues. J. Chem. Soc. Perkin Trans. 2, 919–924. [Google Scholar]
- Huijser, C., Kortstee, A., Pego, J., Weisbeek, P., Wisman, E., and Smeekens, S. (2000). The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: Involvement of abscisic acid in sugar responses. Plant J. 23, 577–585. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Reuling, G., and Karssen, C.M. (1984). The isolation and characterization of abscisic acid-insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 61, 377–383. [Google Scholar]
- Kurup, S., Jones, H.D., and Holdsworth, M.J. (2000). Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J. 21, 143–155. [DOI] [PubMed] [Google Scholar]
- Laby, R.J., Kincaid, M.S., Kim, D., and Gibson, S.I. (2000). The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 23, 587–596. [DOI] [PubMed] [Google Scholar]
- Lang, G.A., ed (1996). Plant Dormancy: Physiology, Biochemistry and Molecular Biology. (Wallingford, UK: CAB International).
- Leplé, J.C., Brasileiro, A.C.M., Michel, M.F., Delmotte, F., and Jouanin, L. (1992). Transgenic poplars: Expression of chimeric genes using four different constructs. Plant Cell Rep. 11, 137–141. [DOI] [PubMed] [Google Scholar]
- Leung, J., Bouvier-Durand, M., Morris, P.-C., Guerrier, D., Chefdor, F., and Giraudat, J. (1994). Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science 264, 1448–1452. [DOI] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nakamura, S., Lynch, T.J., and Finkelstein, R.R. (2001). Physical interactions between ABA response loci of Arabidopsis. Plant J. 26, 627–635. [DOI] [PubMed] [Google Scholar]
- Nambara, E., Keith, K., McCourt, P., and Naito, S. (1995). A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana. Development 121, 629–636. [Google Scholar]
- Netting, A.G., and Milborrow, B.V. (1988). Methane chemical ionization mass spectrometry of the pentafluorobenzyl derivatives of abscisic acid, its metabolites and other plant growth regulators. Biomed. Environ. Mass Spectrom. 17, 281–286. [Google Scholar]
- Nitsch, J.P. (1957). Photoperiodism in woody plants. Proc. Am. Soc. Hortic. Sci. 70, 526–544. [Google Scholar]
- Olsen, J.E., Junttila, O., Nilsen, J., Eriksson, M.E., Martinussen, I., Olsson, O., Sandberg, G., and Moritz, T. (1997). Ectopic expression of oat phytochrome A in hybrid aspen changes critical day length for growth and prevents cold acclimatization. Plant J. 12, 1339–1350. [Google Scholar]
- Parcy, F., Valon, C., Raynal, M., Gaubier-Comella, P., Delseny, M., and Giraudat, J. (1994). Regulation of gene expression programs during Arabidopsis seed development: Roles of the ABI3 locus and of endogenous abscisic acid. Plant Cell 6, 1567–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, L.E. (1987). Hormonal aspects of bud and seed dormancy in temperate-zone woody plants. HortScience 22, 845–850. [Google Scholar]
- Prinsen, E., Rüdelsheim, P., and Van Onckelen, H. (1991). Extraction, purification and analysis of endogenous indole-3-acetic acid and abscisic acid. In BioMethods, Vol. 4: A Laboratory Guide for Cellular and Molecular Plant Biology, I. Negrutiu and G.B. Gharti-Chhetri, eds (Basel, Switzerland: Birkhäuser Verlag), pp. 198–209.
- Prinsen, E., Van Laer, S., Öden, S., and Van Onckelen, H. (2000). Auxin analysis. Methods Mol. Biol. 141, 49–65. [DOI] [PubMed] [Google Scholar]
- Robinson, C.K., and Hill, S.A. (1999). Altered resource allocation during seed development in Arabidopsis caused by the abi3 mutation. Plant Cell Environ. 22, 117–123. [Google Scholar]
- Rohde, A., Ardiles-Diaz, W., Van Montagu, M., and Boerjan, W. (1998). Isolation and expression analysis of an abscisic acid-insensitive (ABI3) homologue from Populus trichocarpa. J. Exp. Bot. 49, 1059–1060. [Google Scholar]
- Rohde, A., De Rycke, R., Beeckman, T., Engler, G., Van Montagu, M., and Boerjan, W. (2000. a). ABI3 affects plastid differentiation in dark-grown Arabidopsis seedlings. Plant Cell 12, 35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, A., Howe, G.T., Olsen, J.E., Moritz, T., Van Montagu, M., Junttila, O., and Boerjan, W. (2000b). Molecular aspects of bud dormancy in trees. In Molecular Biology of Woody Plants, Vol. 1, S.M. Jain and S.C. Minocha, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 89–134.
- Rohde, A., Kurup, S., and Holdsworth, M. (2000. c). ABI3 emerges from the seed. Trends Plant Sci. 5, 418–419. [DOI] [PubMed] [Google Scholar]
- Rohde, A., Van Montagu, M., and Boerjan, W. (1999). The ABSCISIC ACID-INSENSITIVE 3 (ABI3) gene is expressed during vegetative quiescence processes in Arabidopsis. Plant Cell Environ. 22, 261–270. [Google Scholar]
- Rook, F., Corke, F., Card, R., Munz, G., Smith, C., and Bevan, M.W. (2001). Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 26, 421–433. [DOI] [PubMed] [Google Scholar]
- Saure, M.C. (1985). Dormancy release in deciduous fruit trees. Hortic. Rev. 7, 239–300. [Google Scholar]
- Schwartz, S.H., Tan, B.C., Gage, D.A., Zeevaart, J.A.D., and McCarty, D.R. (1997). Specific oxidative cleavage of carotenoids by VP14 of maize. Science 276, 1872–1874. [DOI] [PubMed] [Google Scholar]
- Söderman, E.M., Brocard, I.M., Lynch, T.J., and Finkelstein, R.R. (2000). Regulation and function of the Arabidopsis ABA-insensitive4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 124, 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, M., Kao, C.Y., Cocciolone, S., and McCarty, D.R. (2001). Maize VP1 complements Arabidopsis abi3 and confers a novel ABA/auxin interaction in roots. Plant J. 28, 409–418. [DOI] [PubMed] [Google Scholar]
- Verwoerd, T.C., Dekker, B.M.M., and Hoekema, A. (1989). A small-scale procedure for rapid isolation of plant RNAs. Nucleic Acids Res. 17, 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wareing, P.F. (1956). Photoperiodism in woody plants. Annu. Rev. Plant Physiol. 7, 191–214. [Google Scholar]
- Zahm, P., Hohmeyer, C., and Geider, K. (1984). Site-specific mutagenesis of the Ti plasmid by transformation of Agrobacterium tumefaciens with mutagenized T-DNA fragments cloned in E. coli plasmids. Mol. Gen. Genet. 194, 188–194. [Google Scholar]