Abstract
Genes that confer defense against pathogens often are clustered in the genome and evolve via diverse mechanisms. To evaluate the organization and content of a major defense gene complex in cereals, we determined the complete sequence of a 261-kb BAC contig from barley cv Morex that spans the Mla (powdery mildew) resistance locus. Among the 32 predicted genes on this contig, 15 are associated with plant defense responses; 6 of these are associated with defense responses to powdery mildew disease but function in different signaling pathways. The Mla region is organized as three gene-rich islands separated by two nested complexes of transposable elements and a 45-kb gene-poor region. A heterochromatic-like region is positioned directly proximal to Mla and is composed of a gene-poor core with 17 families of diverse tandem repeats that overlap a hypermethylated, but transcriptionally active, gene-dense island. Paleontology analysis of long terminal repeat retrotransposons indicates that the present Mla region evolved over a period of >7 million years through a variety of duplication, inversion, and transposon-insertion events. Sequence-based recombination estimates indicate that R genes positioned adjacent to nested long terminal repeat retrotransposons, such as Mla, do not favor recombination as a means of diversification. We present a model for the evolution of the Mla region that encompasses several emerging features of large cereal genomes.
INTRODUCTION
Plant resistance genes often belong to large, clustered families. Proteins encoded by these host resistance determinants (R) are thought to interact directly or indirectly with cognate pathogen avirulence factors (Avr) in a gene-for-gene manner (Dixon et al., 2000). Large arrays of similar sequences allow for equal or unequal recombination events, resulting in the formation of several gene family members with new recognition specificities (Michelmore and Meyers, 1998; Ellis et al., 2000; Hulbert et al., 2001). Defense signaling proteins encoded by these genes are known to share conserved amino acid domains with functionally similar proteins across diverse taxa.
For example, the NBS (Nucleotide Binding Site) domain of most plant disease resistance genes is highly similar to the NBS of apoptosis-related CED4/APAF1 from Caenorhabditis elegans and human (van der Biezen and Jones, 1998). Likewise, the TIR (Toll/Interleukin Receptor) domain of some plant R genes is analogous to the Drosophila Toll or human interleukin receptor, which is essential for the development of the immune response (Baker et al., 1997). These shared protein domains indicate the presence of common functions in the way that cells respond to attack in eukaryotic organisms. In addition, the overall organization of genomes sometimes favors the clustering of genes that are structurally unique but functionally related (Beck and Trowsdale, 2000; Hulbert et al., 2001).
Approximately 30 alleles of the barley Mla locus specify resistance to the obligate fungal biotroph Blumeria (Erysiphe) graminis f. sp. hordei (Bgh), which is the causal agent of powdery mildew disease. Genetic variants of the Mla locus are found in cultivars worldwide and thus provide a unique experimental system in which to explore the evolution of the allelic diversity of R genes in monocot species (Jørgensen, 1994; Wise, 2000). Cloned alleles of the Mla locus belong to the coiled-coil, nucleotide binding site, Leu-rich repeat (CC-NBS-LRR) class of genes implicated in specific recognition between host and pathogen (Halterman et al., 2001; Zhou et al., 2001; for review, see Jones, 2001). To function, the MLA6 protein requires the zinc binding protein RAR1 (Shirasu et al., 1999) and a subunit of the SCF ubiquitin ligase complex SGT1 (Kitagawa et al., 1999; Azevedo et al., 2002). MLA1 does not appear to use the RAR1–SGT1 pathway (Zhou et al., 2001; Azevedo et al., 2002), although the phenotypes of the Mla-Bgh host–pathogen interactions are indistinguishable (Wise and Ellingboe, 1983; Boyd et al., 1995) and the two MLA proteins are 92.2% similar (Halterman et al., 2001).
Limited contiguous DNA sequences from grain cereal plants, especially those with genomes larger than that of human, have precluded a full understanding of how their genomes are organized. At the macrogenome level, recent reports have postulated the existence of gene-dense islands interrupted by gene-poor space (Barakat et al., 1997; Moore, 2000). The major impetus for sequencing large contigs was to provide a platform for the map-based cloning of genes identified by their phenotype, and less focus has been given to the overall architecture of the gene-poor space (Meyers et al., 2001; Wicker et al., 2001).
To evaluate the structural evolution of a major disease defense complex in the 5000-Mb barley genome, we determined the complete sequence of a 261-kb BAC contig from barley cv Morex that spans the Mla (powdery mildew) resistance locus on chromosome 5 (1H). Computational analysis revealed that 15 of the 32 predicted genes are associated with plant defense responses. These 15 protein-coding sequences are grouped into five families, including three CC-NBS-LRR resistance gene homolog (RGH) families, one CI2 family, and one chemically induced family. Predicted protein-coding sequences are organized as three gene-rich islands separated by two nested complexes of transposable elements and a gene-poor region. Detailed analysis of the gene-poor region and the adjacent 40-kb tandem duplication revealed an extraordinary array of repeats in which DNA is hypermethylated yet the transcription of genes is common.
RESULTS
The Mla Region Contains Multiple Classes of Genes Associated with Plant Defense Responses
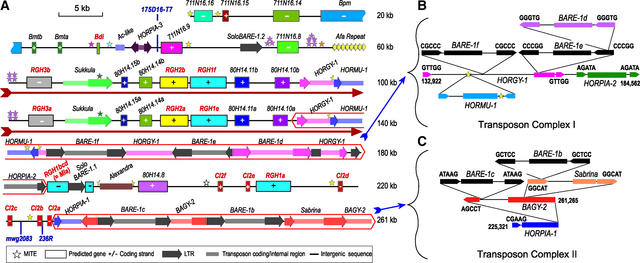
As illustrated in Figure 1A, 32 protein-coding and 2 duplicated tRNAser genes were predicted by a combination of BLASTx and BLASTp, GENSCAN, GeneMark, and Grail (see supplemental data online). Eight RGHs containing CC, NBS, and LRR domains constitute the major defense-related group. The CC-NBS-LRR class is the most prevalent among cloned plant resistance genes and is known to function in the recognition of bacterial, fungal, viral, and nematode pathogens (Baker et al., 1997; van der Biezen and Jones, 1998).
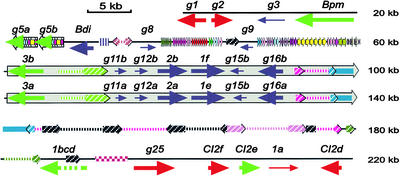
Figure 1.
Sequence Annotation of the Barley Mla Locus from cv Morex.
BAC 711N16 (top; 112,178 nucleotides) was sequenced at ×12 redundancy, and BAC 80H14 (bottom; 158,773 nucleotides) was sequenced at ×15 redundancy; sequence error is <1 bp/10 kb. The two BACs are joined by 9686 nucleotides of overlapping sequence encompassing RGH3a and the 5′ end of the Sukkula retrotransposon. The GC content of 261,265 nucleotides of finished sequence is 44.9%, the CpG ratio is 4.2%, the observed/expected CpG ratio is 0.83, and the CpG/GpC ratio is 1.21. These values are consistent with barley genomic contigs encompassing barley Mlo (Panstruga et al., 1998) and Rar1 (Shirasu et al., 2000). + indicates forward strand transcription, and − designates complementary strand transcription.
(A) Annotation of the 261,265-nucleotide region. The dark-red text indicates genes associated with defense responses, and the dark-red arrow is below the 40-kb tandem duplication. Different colors show different families of genes or transposable elements.
(B) Nested transposon complex I on BAC 80H14 between 132,922 and 184,562 nucleotides.
(C) Nested transposon complex II on BAC 80H14 between 225,321 and 261,265 nucleotides.
Retrotransposon placement was determined by the 5-bp LTR inverted repeat insertion signature (6-bp direct repeat shown in letters) at the two ends of each element. This direct repeat marks the boundary for each retroelement.
The eight RGHs are classified into three dissimilar families, with <43% amino acid sequence similarity between families and 78 to 100% similarity within families (Table 1). The only marked similarity among the three RGH families is the P-loop (GKTTL), kinase 2a (RYLVIIDDI), and the GGVPLA conserved domains found in all NBS-LRR–encoding genes. These results suggest functional similarity but evolutionary independence.
Table 1.
DNA and Amino Acid Sequence Similarities of Mla6 from C.I. 16151 and Eight Resistance Gene Homologs (RGHs) in the Morex Mla Complex
|
RGH Designation
|
|||||||
|---|---|---|---|---|---|---|---|
| RGH | Mla6 | RGH1a | RGH1bcd | RGH1e | RGH1f | RGH2a/b | RGH3a/b |
| Mla6 | – | 82 | 87 | 84 | 84 | 45 | 43 |
| RGH1a | 78 | – | 84 | 83 | 83 | 46 | 44 |
| RGH1bcd | 84 | 81 | – | 86 | 86 | 47 | 44 |
| RGH1e | 79 | 79 | 81 | – | 100a | 47 | 44 |
| RGH1f | 79 | 79 | 81 | 100 | – | 47 | 44 |
| RGH2a/b | 43 | 42 | 42 | 42 | 42 | – | 41 |
| RGH3a/b | 41 | 41 | 38 | 39 | 39 | 36 | – |
Comparisons were done with the Genetics Computer Group GAP program. Numbers above the dashes designate nucleic acid similarities of the coding region, whereas numbers below the dashes indicate predicted amino acid similarities.
There is a single nucleic acid sequence difference between RGH1e and 1f that does not result in an amino acid change.
RGH1a and 1e were used recently as probes to isolate the Mla1 (Zhou et al., 2001), Mla6 (Halterman et al., 2001), and Mla13 (D. Halterman, F. Wei, and R. Wise, unpublished data) alleles that confer resistance specificity to Bgh. Comparative sequence analysis now indicates that RGH1bcd is a susceptible allele of Mla from cv Morex. This conclusion is based on the fact that RGH1bcd shares the highest nucleotide and amino acid similarity with the Mla1, Mla6, and Mla13 resistance alleles derived from Cereal Introduction (C.I.) 16137, C.I. 16151, and C.I. 16155, respectively (Halterman et al., 2001) (Table 1). In addition, the Alexandra LINE element on the 5′ side of RGH1bcd is also in the same orientation adjacent to the Mla6 sequence, but not Mla1. The HORPIA-2 element on the 3′ side of RGH1bcd is on the 3′ side of Mla1, but appears to be missing from the 3′ side of Mla6. This is not surprising, considering that this Copia-like element inserted at this location only 0.48 million years ago (see below).
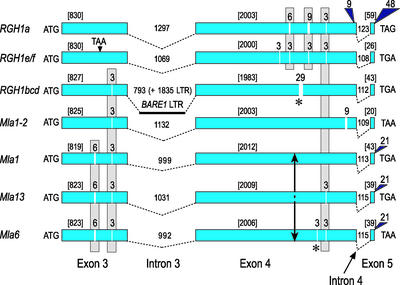
As shown in Figure 2, comparison of Morex Mla-RGH1 paralogs with cloned Mla specificities demonstrates that almost all insertions or deletions (InDels) have occurred in multiples of three nucleotides. These InDel triplets have maintained a full-length open reading frame during evolution and occur in both the NBS and LRR domains. InDels in the LRR of RGH1a, e, and f occur in identical positions even though the overall sequence similarity between RGH1a and RGH1e/f is only 83% (Table 1). Similarly, InDels also occur in like positions 5′ to the NBS of RGH1bcd, Mla1, Mla1-2, Mla6, and Mla13; the overall similarity of these genes is no more than 87%. Hence, the presence of shared InDels among various Mla family members indicates that they evolved from a common ancestor and that InDel events occurred early in, or before, sequence divergence.
Figure 2.
Structural Comparison of Morex RGH1 Paralogs with Functional Mla Specificities from Accessions C.I. 16137 (Mla1), C.I. 16151 (Mla6), and C.I. 16155 (Mla13).
Deletions in the coding regions are designated by vertical white bars, and insertions are designated by navy blue triangles. Above each vertical white bar or blue triangle is the number of nucleotides for each InDel. InDels that delete an amino acid in the solvent-exposed region are designated with asterisks below the InDel. The number of nucleotides in each exon are shown in brackets. The number of nucleotides in each intron are specified above the intron. The divergence point in the predicted LRR domain of MLA1/MLA6 is marked by an arrow. The BARE-1 LTR in the major intron of RGH1bcd is designated by a solid horizontal line. RGH1e and 1f are both within the 40-kb duplication and differ by one nucleotide but no amino acid change.
Closer inspection revealed a rather complicated scenario during the evolution of the Mla family. There is a clear point of divergence in the predicted LRR domain of MLA1 and MLA6, beginning at the solvent-exposed Ala/Ser at position 683 of the Mla6 open reading frame (Figure 2). Diagnostic amino acid similarities between RGH1bcd and MLA1, as opposed to RGH1bcd and MLA6, are evident 3′ to this point of divergence, indicating a possible recombination between paralogs, as opposed to a direct linear descent between alleles (Halterman et al., 2001). A single Ser (compared with MLA1) or Thr (compared with MLA13 [D. Halterman, F. Wei, and R. Wise, unpublished data]) residue is deleted at position 801 in the predicted solvent-exposed region of the MLA6 LRR domain. The deletion of these Ser and Thr residues, both of which can be phosphorylated, may change the topography of the LRR.
In lettuce Dm3 homologs, trinucleotide InDels also occur in the LRR backbone, indicating that such genetic events could be significant to the evolution of specificity encoded by these genes (R.W. Michelmore, personal communication). In the flax L and M loci, 5- to 20-bp InDels have resulted from transposon insertion/excision events. Two spontaneous rust-susceptible alleles involved the insertion of flax transposon Lute-1; rust-resistant revertants contain an in-frame, 9-bp InDel as a result of imprecise excision of Lute-1 (Ellis et al., 1997).
In addition to small InDels during the evolution of the Mla-RGH families, intron 3 of RGH1bcd has been host to a BARE-1 retrotransposon that was excised through unequal recombination (Figures 1 and 2). Thus, the present-day RGH1bcd contains only a solo long terminal repeat (LTR) of BARE-1 in the major intron in addition to a 29-bp deletion in the LRR domain, resulting in premature termination of the open reading frame. RGH1e and 1f reside in each of two copies of a 40-kb tandem duplication; thus, both have the same mutation in amino acid 151, resulting in early stop codons in their NBS domains. The RGH3a and 3b pseudogenes contain an early stop codon in amino acid 582 of their LRR regions, whereas RGH2a and 2b are predicted to encode full-length CC-NBS-LRR proteins.
A family of six intact chymotrypsin inhibitor 2 (CI2) genes range in size from 55 to 85 amino acids and flank RGH1a (Figure 1, Table 2). CI2 genes belong to the potato inhibitor I family of Ser protease inhibitors and function in various biochemical pathways, notably, to provide resistance to insect pathogens (Heath et al., 1997; Koiwa et al., 1997), wounding (Lee et al., 1986), and the ethylene-regulated expression of fruit ripening (Margossian et al., 1988). In addition, CI2 genes can be induced by salicylic acid (SA) mimics and also by jasmonic acid (JA), resulting in increased disease resistance in graminaceous plants such as barley and maize (Cordero et al., 1994; Besser et al., 2000).
Table 2.
DNA and Amino Acid Similarities of Six Chymotrypsin Inhibitor (CI2) Genes Flanking RGH1a
| CI2 | CI2a | CI2b | CI2c | CI2d | CI2e | CI2f |
|---|---|---|---|---|---|---|
| CI2a | – | 87 | 74 | 75 | 65 | 57 |
| CI2b | 85 | – | 80 | 88 | 69 | 57 |
| CI2c | 69 | 77 | – | 84 | 77 | 57 |
| CI2d | 71 | 85 | 78 | – | 72 | 56 |
| CI2e | 62 | 68 | 71 | 61 | – | 59 |
| CI2f | 62 | 61 | 70 | 61 | 58 | – |
Comparisons were done with the Genetics Computer Group GAP program. Numbers above the dashes designate nucleic acid similarities of the coding region, whereas numbers below the dashes indicate predicted amino acid similarities.
CI2c has been cloned independently as BCI7, which upon induction with 2,6-dichloroisonicotinic acid (DCINA; a SA mimic) is associated with enhanced resistance to Bgh. BCI9, a homolog of Bdi (barley DCINA-induced), also can be induced by DCINA or JA, and concomitantly, barley plants show increased resistance to Bgh (Besser et al., 2000). Nonetheless, the current data indicate that BCI7 or BCI9 cannot be induced only by inoculation with Bgh.
The last complete gene in the Mla contig is Bpm, a barley Pum/Mpt5/FBF-like gene. The products of Pum/Mpt5/ FBF genes (Puf family) have been shown to repress mRNA translation by binding to the 3′ untranslated region of the RNAs in Drosophila, C. elegans, and yeast (Tadauchi et al., 2001). In plants, putative Puf homologs have been reported only in Arabidopsis (Lin et al., 1999) and rice (see supplemental data online), but their function in plants remains untested.
The Mla Region Contains All Classes of Transposable Elements
All major classes of transposable elements are present in the Mla region (Figures 1A to 1C). Among them, the largest class in the region is the LTR class of retrotransposons, including four Copia-like families (BARE-1 and HORPIA-1, 2, and 3), three Gypsy-like families (BAGY-2, HORGY-1, and Sukkula), and one Athila-like element (Sabrina). Sukkula and Sabrina were reported previously only as solo LTRs (Shirasu et al., 2000); however, we report the complete description of the elements with both polyproteins and LTRs, enabling their classification into the Gypsy and Athila families, respectively. The Athila family is represented by a full-length Sabrina element, whereas there is a deletion of the entire 3′ LTR in the Sukkula element.
BARE-1 (Manninen and Schulman, 1993) is the major retrotransposon, accounting for 17.5% of the sequence. Five of the seven BARE-1 elements are full length (Figures 1B and 1C). In these five, point mutations resulting in premature stop codons interrupt the polyprotein open reading frames. The remaining two (solo LTR) BARE-1 elements are a consequence of intraelement recombination, as confirmed by their 5-bp (direct repeat) insertion signatures.
Eight MITE (Miniature Inverted Transposable Element) families that range in size from 29 to 528 bp (see supplemental data online) are positioned near the 5′ or 3′ ends of the predicted genes. As the most abundant MITE family in the region, eight Stowaway elements (Bureau and Wessler, 1994) are present, ranging from 128 to 243 bp in size. The 16-member, 39-bp HORMITE-3 family possesses a unique clustered arrangement of three to five members interspersed with short sequences. This tandem arrangement predicts that the transposable unit could be flexible in size.
An apparent full-length, 6459-bp MuDR-like transposon, HORMU-1, is inserted into the Stowaway.4 MITE element. Its 179-bp terminal inverted repeats are 94% sequence identical, and its pseudogene encodes a 654–amino acid transposase with an early stop codon at amino acid residue 489. DNA gel blot analysis using an internal portion of the transposase-encoding sequence demonstrated that there are at least 50 copies of HORMU-1 in the barley genome (data not shown). Mutator transposons are widespread in grasses (Lisch et al., 2001); thus, it is likely that there is an intact HORMU-1 element in the barley genome that still possesses transposase activity.
A new 3578-bp, non-LTR LINE element, designated Alexandra, is inserted into Stowaway.7 and was identified via BLASTx search for polyprotein, its 3′ poly(A) signal, and the 7-bp ATACGGA insertion signature. Finally, a truncated Ac-like transposon is located adjacent to the BCI-9–like gene. Thus, transposon-like sequences constitute 44.9% of the sequence, with retroelements accounting for 40.2%, transposons accounting for 3.6%, and MITE elements accounting for 1.1%.
Chronology of Retrotransposon Insertion
Because of the mechanism of transposition, the two LTRs of a retrotransposon are identical at the time of their insertion. Thus, calculation of the base substitution rate between the two LTRs can be used to determine the chronology of retrotransposon activity (SanMiguel et al., 1998). We used the average base substitution rate derived from the grass (Gramineae) adh1-adh2 region of 6.5 × 10−9 per site per year (Gaut et al., 1996), given the assumption of a uniform mutation rate. Using this figure combined with the calculated base substitution rates listed in Table 3, we estimate that the five BARE-1 elements inserted into their present location between 0.6 and 2 million years ago. Except for the HORGY-1 element insertion at ∼7.3 million years ago, all other retroelements inserted in the past 2.5 million years, which is comparable to the <3 million year time frame of most retrotransposon insertions in maize (SanMiguel et al., 1998).
Table 3.
Chronology of Retrotransposon Insertion in the Mla Region
| Elementa | Length of LTRb | Total Substitutionb | Base Substitution Ratec | Time (million years)d |
|---|---|---|---|---|
| BARE-1b | 1813 | 15 | 0.0083 | 0.64 |
| BARE-1c | 1717 | 20 | 0.0116 | 0.89 |
| Sabrina | 1526 | 48 | 0.0315 | 2.42 |
| BARE-1f | 1817 | 22 | 0.0121 | 0.93 |
| BARE-1d | 1801 | 42 | 0.0233 | 1.79 |
| BARE-1e | 1761 | 47 | 0.0267 | 2.05 |
| HORGY-1 | 286 | 27 | 0.0944 | 7.26 |
| HORPIA-2 | 321 | 2 | 0.0062 | 0.48 |
| HORPIA-3 | 521 | 14 | 0.0269 | 2.07 |
Full length retrotransposon with two LTRs.
Deletion or insertion was counted as a single event, including a retrotransposon insertion. The substitution was calculated as the total nucleotide difference between the two LTRs.
For <10% base substitution rate, the rate was estimated as the number of substitutions per nucleotide (k) for calculation of divergence time.
Divergence time as calculated as k/2 × knus. knus designates the base substitution rate per nucleotide per year.
Arrays of Diverse Tandem and Simple Sequence Repeats in the 45-kb Gene-Poor Region Proximal to the Mla Locus
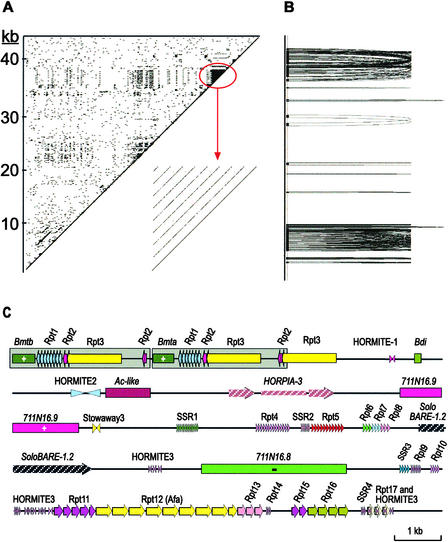
Using total barley DNA to probe filters containing EcoRI-digested BACs spanning the Mla region (Wei et al., 1999), we were able to determine that the gene-poor region proximal to Mla consisted of middle to high repetitive fractions of the genome (data not shown). Therefore, we used a self-comparison DotPlot program to conduct a detailed analysis of the 45-kb gene-poor region for duplication events and repetitive sequences (Figure 3A). A 2.5-kb tandem duplication, several satellite DNA sequences, and a block of nine tandem repeats were identified by this analysis. The positions of these repeats can be visualized easily with Miropeat analysis (Parsons, 1995), which cross-connects all of the related repeats (Figure 3B). In total, there are 4 simple sequence repeats (SSRs) and 17 tandem repeat families, ranging from 2 to 29 copies (Figure 3C; see supplemental data online).
Figure 3.
Tandem Repeats and SSRs in the 45-kb Gene-Poor Region between Nucleotides 20,000 and 65,000 of the Morex Mla Locus.
(A) Self-comparison of the sequence by DotPlot analysis. A 2.5-kb tandem duplication (straight lines at bottom left), several satellite DNA sequences (dense dots at center), and a block of nine tandem repeats (dense lines at top right and enlarged region) were identified by this analysis. The angled lines indicate duplication, and the clustering of lines indicates multiple tandem duplications. The interval between two lines shows the size of a single repeat element. The clustering of dots suggests a SSR region.
(B) Miropeat analysis illustrates the sequence relationship among repeats.
(C) Representation of the position of each repeat element.
The most significant tandem repeat is the Afa family, consisting of eight 338-bp members. The Afa repeat, designated for its conserved AfaI restriction site, was identified originally in Aegilops squarrosa and then was found to be widespread in the Triticeae (Nagaki et al., 1995). Upon closer inspection, we identified a 24-bp inverted repeat within the Afa sequence. These data support the hypothesis that the Afa repeat could be a MITE element, which have terminal inverted repeats and usually are associated with the 3′ or 5′ ends of transcriptionally active genes (Bureau and Wessler, 1994).
BLASTn searches with the Afa repeat sequence detected barley ESTs (5′ reads) from testa/pericarp, post-anthesis spike, and shoot libraries. Fluorescence in situ hybridization analysis has demonstrated that the Afa repeat is distributed in subtelomeric and interstitial regions of chromosomes (Nagaki et al., 1995; Tsujimoto et al., 1997). This is consistent with the location of the Mla locus, which is positioned at the telomeric end of barley chromosome 1HS.
Fifty-four SSRs were identified in the 261-kb Mla contig (see supplemental data online). Nineteen of the SSRs are associated with transposable elements, including 14 in LTR retroelements and 5 in the Mutator transposon, and 35 SSRs are located in the nontransposable element region. These nontransposable element SSRs include 10 in gene-coding regions and 25 in the intergenic regions. Twenty-two SSRs are present in the 40-kb duplication (six of these are associated with genes). Thus, in contrast to the genome-wide estimates by Morgante et al. (2002), SSRs in the 261 kb spanning the Mla locus were associated preferentially with high-copy intergenic regions and retrotransposons.
DNA Methylation Associated with the Mla Locus
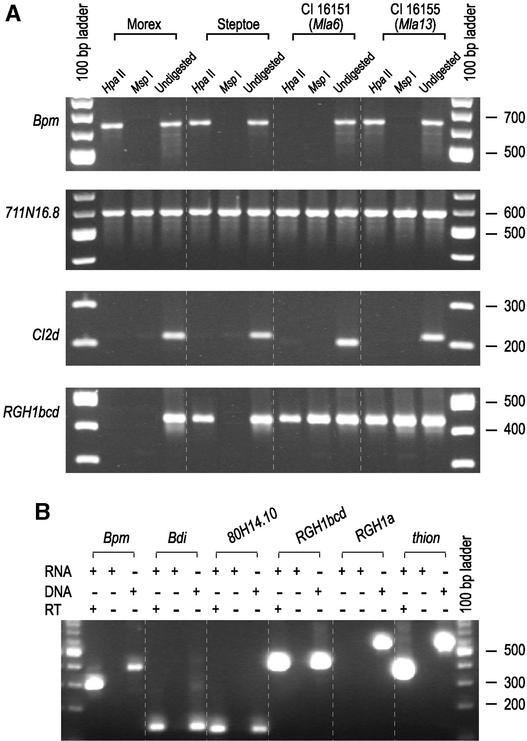
The cytosine of CpG islands is a common target for methylation in the genome. Because DNA often is methylated in repetitive fractions, a PCR-based method was used to determine the methylation state of predicted genes within and adjacent to the gene-poor, repetitive fraction of the Mla locus. The isoschizomers HpaII and MspI were chosen for this analysis because of their differential sensitivity to cytosine methylation. MspI does not cleave DNA when the 5′ C residue in the 5′-C/CGG-3′ restriction site is methylated, but it does cleave DNA when the 3′ C residue is methylated. HpaII does not cleave DNA when either C residue is methylated.
Thus, if the 5′ C residue is methylated, the DNA is not cleaved by either HpaII or MspI, and primers flanking the restriction site can be used to amplify a fragment from digested genomic DNA. However, if only the 3′ C is methylated and the 5′ C is not methylated, a PCR product can be amplified from the HpaII-digested DNA template but not from the template digested with MspI. The results of these experiments are presented in Table 4. A subset of these data presented in Figure 4A illustrates the amplification of DNA digested with HpaII or MspI from four barley accessions. Morex (origin of the primer sequence) and Steptoe are parents of a common barley mapping population (Kleinhofs et al., 1993), whereas C.I. 16151 and C.I. 16155 are parents of a high-resolution recombinant population spanning the Mla region (Wei et al., 1999).
Table 4.
Results of Methylation and Expression Tests of Predicted Genes in the Barley Mla Region
| Methylation Testb
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene Abbreviation (in Figure 5) |
Gene | Primer Pair | Primer Positiona |
Annealing Temperature |
Size (bp) |
Morex | Steptoe | C.I. 16151 |
C.I. 16155 |
Transcription |
| g1 | 711N16.16 | CGTAGATAGACAACGGAATCGAGG-GGGCGGGGTTCAACATATTG | CDS | 58 | 271 | − | + | − | − | + |
| g2 | 711N16.15 | CGGACGAACAGTTGGTGTGCACCA-GGAGGAAGGAACACGC | CDS | 62 | 215 | − | − | − | − | + |
| g3 | 711N16.14 | CGATCACAGTCACCGAGCAATCTG-AAGATAGCCAACGCCCTG | CDS | 58 | 138 | ++ | ++ | ++ | ++ | − |
| Bpm | Bpmb | ATAAGCCGCCTTGCATAATACGAA-CTACCACTTCACACCTTGCAGG | Intron | 55 | 625 | + | + | − | + | + |
| g5a&b | Bmtb | TTCTAGCAAGCATCTAAAGGTTTCG-ACATCTGTTAGTCGGGCAAGAGAC | 3′ UTR | 54 | 576 | + | + | + | + | − |
| Bdi | Bdi | AGGGCATCTTCAATGGTCCTATGC-GGAGCTTGTGCGTGAACC | Predicted CDS | 51 | 136 | ++ | ++ | ++ | ++ | + |
| g8 | 711N16.9 | CGACATTGTCCTGCTCTGTTGCCG-GTTTGAATTGGACGAGCTTG | CDS | 58 | 134 | ++ | ++ | NP | ++ | + |
| g9 | 711N16.8 | TGTTGACTCCTTGATTCCATCCACC-TGCTAGATAAAGGCATCGTACCTG | CDS | 57 | 582 | ++ | ++ | ++ | ++ | + |
| 3a&3b | RGH3a/b | GGTGTGTGATTTCGATGCCCAGGA-GCCTGCACCGTCT | CDS | 54 | 146 | + | + | + | NP | + |
| g11a&b | 80H14.15 | ACCCTGCTGCCGATCTACTGATAG-ATCGTTCCTCTTCCTCGTCGTC | CDS | 59 | 157 | ++ | ++ | ++ | ++ | − |
| g12a&b | 80H14.14c | GAGCACACTGGATTGTTGAAATGG-CACCCATGCCTTCCATAGTAGC | CDS | 58 | 374 | ++ | ++ | ++ | ++ | − |
| 2a&b | RGH2a/b | ACCCTCGCCAGACAAGTTTACCGA-AAAGCAGCTTATGCACGTCG | CDS | 57 | 165 | ++ | NP | ++ | NP | + |
| 1e&f | RGH1e/f | CATGGTTAGCCTGATCTCCAAGCT-GGAATTGCTCCGTGATTTCC | CDS | 57 | 372 | ++ | NP | ++ | NP | + |
| g15a&b | 80H14.11 | TCACACCATACGCCGCAACAGGTG-CTCACCGCAACGTC | CDS+Intron | 64 | 378 | ++ | ++ | ++ | ++ | − |
| g16a&b | 80H14.10 | CCTTTGTCGTTAGGATCGCATTCGT-TGGTAGGGTTATTGTTCATGCAC | CDS | 56 | 131 | ++ | ++ | ++ | ++ | + |
| 1bcd | RGH1bcd | GAGTAATTGTCGCCGTTTGTCCGC-AACACACCAACTAGAGGAAACAC | CDS | 54 | 434 | − | + | ++ | ++ | + |
| g25 | 80H14.8b | TCGGTGACGGTGTGACAATTCCTT-GATCTTCCCCTTGCTTGC | Intron+CDS +3′ UTR |
58 | 493 | − | NP | − | − | + |
| CI2f | CI2f | GGACCTGGGCAATGTTGTCGCAAA-GGAAGCAAGCGTAACAAGG | 5′ UTR+CDS | 58 | 264 | − | − | − | + | + |
| CI2e | CI2e | GCAACCCAACTAGCCAACGTGGC-CAAGGAGATCATTCTCAAGGAC | CDS+3′ UTR | 59 | 150 | + | + | + | ++ | + |
| 1a | RGH1a | TTGGGAAAAATAGCAGCCTGCATG-CAAGGGGTCTCTCATTATCC | CDS | 56 | 572 | − | ++ | − | − | − |
| CI2d | CI2d | GCTGACCCAAAAGCACCCGCTAG-CCGATGTGAGGAGTGGTC | CDS | 59 | 216 | − | − | − | − | + |
The position of the primer pair is designated as follows: CDS, coding sequence; UTR, untranslated region.
Methylation test: +, only HpaII sensitive, internal C methylated; ++, both HpaII and MspI sensitive, external or both Cs methylated; −, not methylated; NP, no PCR product amplified from unrestricted genomic DNA.
These primers were used only for methylation tests. For RT-PCR, primers 5′-GAGCACACTGGATTGTTGAAATGG-3′ and 5′-GGAGCTTGATAGGTGTTGGCAAG-3′ were used at an annealing temperature of 55°C to amplify a 169-bp 80H14.14 product, primers 5′-AGAAAAGTTCCAGAAGATG-3′ and 5′-AAAAGACTATTTCGATTCC-3′ were used at an annealing temperature of 44°C to amplify a 117-bp Bmt product, primers 5′-ACTTCTTCAGTGTGTTCAGGTGAG- C-3′ and 5′-GCAAGCCACATGAGAGAACTGC-3′ were used at an annealing temperature of 55°C to amplify a 387-bp Bpm product, and primers 5′-GCTACTGGAGCCATCACATGAAG-3′ and 5′-CTTGATCTTCCCCTTGCTTGC-3′ were used at an annealing temperature of 58°C to amplify a 209-bp 80H14.8 product.
Figure 4.
Methylation and Transcription Analysis of Genes Associated with the Barley Mla Region.
Primers, amplification conditions, and a summary of results are provided in Table 4.
(A) Different patterns of DNA methylation of genes adjacent to and within the Mla locus. Positive amplification occurs if a particular 5′-C/CGG-3′ site is methylated and cannot be cut by the restriction enzyme. Likewise, amplification cannot occur if the particular 5′-C/CGG-3′ site is not methylated and is digested to completion by the restriction enzyme.
(B) RT-PCR analysis of predicted genes adjacent to and within the Mla locus. The templates in each one-step RT-PCR analysis are shown at top. RNA or DNA indicates the use of RNA or genomic DNA as a template for the reaction. In the analysis of each gene, the first lane shows the result of the RT-PCR. The second and the third lanes represent a DNA-free RNA negative control without RT and a positive DNA control, respectively.
In summary, the tested 5′-C/CGG-3′ restriction sites within genes between 711N16.15 and RGH1bcd are methylated, whereas the 5′-C/CGG-3′ restriction sites within genes both proximal and distal to this region are not methylated, except for CI2e (Figure 4A, Table 4). The 5′-C/CGG-3′ sequence between Bmtb and RGH1bcd is hypermethylated except within RGH3a/b. This hypermethylated region includes promoters, 5′ or 3′ termini, coding sequences, or intergenic regions. The genes Bpm, Bmta/b, RGH3a/b, and CI2e are methylated only at the internal C and thus are not hypermethylated. Interestingly, several genes displayed line-specific methylation patterns.
The most dramatic differences among lines were in sequences flanking, but not within the region between, genes Bmta and RGH1bcd. However, because C.I. 16151 and C.I. 16155 are congenic lines (94% similar) (Moseman, 1972), methylation status may not be correlated with a particular genetic background; more likely, it is inherited with specific alleles (Cocciolone et al., 2001).
Genes Are Transcribed Actively within and Adjacent to Methylated Regions
EST BLAST searches, cDNA plaque screening, and reverse transcription (RT)–PCR analyses were performed to assay the transcription of sequences within and adjacent to the Mla locus. The combined results of these three analyses are summarized in Table 4. As of March 28, 2002, BLASTn searches of 155,400 barley ESTs indicated that Bpm, Bdi, 80H14.8, CI2f, CI2d, and CI2c are transcribed. Only a few transcripts of Mla-RGH1 family members were detected in EST BLASTn searches (see supplemental data online).
In addition to EST BLAST searches, 110 Mla-RGH1 family clones were identified from screening 1.2 million plaques of a cDNA library from barley accession C.I. 16155 that contains the Mla13 allele of RGH1bcd (D. Halterman, F. Wei, and R. Wise, unpublished data). Similarly, 29 RGH1 family clones were identified from screening 600,000 plaques of a cDNA library of barley accession C.I. 16151 that contains the Mla6 allele (Halterman et al., 2001). No clones were recovered by hybridization with RGH2 or RGH3 family probes in either library screen.
Because of the limited sample sizes of the barley EST database and cDNA plaque hybridizations, transcripts of some genes were not detected. Thus, to increase the sensitivity for rare transcripts, RT-PCR analysis was used. RNA was extracted from 7-day-old seedlings, the same tissue used for DNA isolation for the methylation tests described above. As shown in Table 4 and Figure 4B, transcripts were detected for 711N16.15, Bpm, Bdi, 711N16.9, 711N16.8, RGH3a/b, RGH2a/b, RGH1e/f, 80H14.10a/b, RGH1bcd, CI2e, and CI2d. From the three analyses, we conclude that most genes in the region are transcribed at least at a basal level.
Thus, in contrast to the long-observed interdependence between methylation and gene silencing (Cocciolone et al., 2001; Paszkowski and Whitham, 2001), there was no correlation between methylation state and transcription in the 261-kb region (Figure 5). Genes Bdi, 711N16.9, 711N16.8, RGH2a/b, RGH1e/f, RGH1bcd, and 80H14.10a/b are hypermethylated but transcribed. Genes Bpm, RGH3a/b, and CI2e are methylated, but not hypermethylated, and transcribed. No transcripts were detected for 711N16.16 and RGH1a, even though they are not methylated.
Figure 5.
Overview of Methylation and Transcription Patterns of the Barley Mla Region.
Predicted genes are specified by horizontal arrows: unmethylated genes are coded red; methylated genes are coded navy; and partially methylated genes are coded green. Transcribed genes are designated with thick arrows, whereas nontranscribed genes are designated with thin arrows. The gray-filled rectangle (lines 3 and 4) illustrates the 40-kb duplication.
DISCUSSION
Molecular Evolution of the Mla Region Provides Evidence for Genome Expansion in Barley
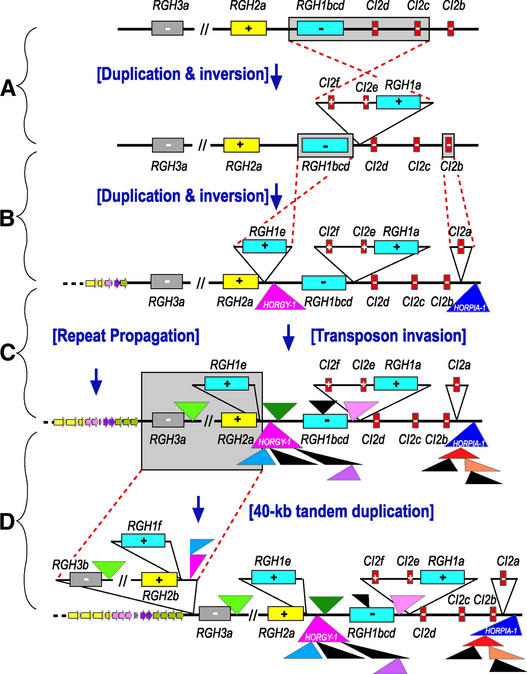
To construct a model for the molecular evolution of the Mla region, we evaluated large-scale rearrangements among gene families in addition to the chronology of retrotransposon insertion and repeat propagation. Based on these analyses, the structure of this segment is compatible with the following scenario. As shown in Figure 6A, ∼30 kb of progenitor sequence, including RGH1bcd, CI2d, and CI2c, sustained duplication and inversion events to generate CI2f, CI2e, and RGH1a. After these events, a second duplication and inversion of RGH1bcd gave rise to RGH1e, and CI2a arose from a duplication of CI2b (Figure 6B).
Figure 6.
Evolution of the Barley Mla Complex.
Of the 34 predicted genes in the Mla region, 24 have at least one homolog. The RGH1 and CI2 families represent the major gene duplications in the region and account for 16% of the sequence. The remaining gene duplications have arisen from tandem fragment duplications, of which 14 are located in the 40-kb gene-rich tandem repeat and 2 are located in the 2.6-kb tandem repeat.
(A) DNA sequence similarities of RGH1bcd, CI2d, and CI2c compared with RGH1a, CI2f, and CI2e in the opposite orientation indicate an ancient inversion event followed by divergent evolution.
(B) A more recent duplication and inversion to create RGH1e is indicated by the higher sequence similarity between RGH1bcd and RGH1e than that between RGH1bcd and RGH1a (Table 1). Similarly, CI2a possesses the highest sequence similarity with CI2b within the CI2 family (Table 2).
(C) Within the last 3 million years, extensive transposon insertions in addition to repeat propagation increased the size of the region approximately threefold.
(D) A 40-kb tandem duplication of the region containing RGH1e, RGH2a, and RGH3a is the most recent addition to the present-day Mla region.
Genes are shown in rectangles with different colors representing different families, and transposable elements are designated by triangles. Designated colors match those in Figures 1 and 3 for each element. + indicates forward strand transcription, and − designates complementary strand transcription.
Subsequent to the gene duplications and inversions described above, our data indicate that the “founder” element HORGY-1 of complex I (Figure 1B) inserted between RGH1bcd and RGH1e >7 million years ago and acted as a receiver for 51,641 bp of nested transposons (Figure 6C). Seven additional insertion events took place, including MITE Stowaway.5 into retro-HORGY-1, transposon HORMU-1 into Stowaway.5, and Stowaway.4 into HORMU-1.
Similarly, in complex II (Figure 1C), HORPIA-1 functioned as a receiver for five retrotransposon insertion events totaling 35,945 bp. Insertion of HORPIA-2 into complex I and BARE-1b into complex II occurred only 0.48 and 0.64 million years ago, respectively, indicating that these are actively evolving complexes. All of the retrotransposons in complex I or II have both LTRs and polyproteins. This is in contrast to the nested insertion of only solo LTRs remaining in the barley Rar1 region (Shirasu et al., 2000) but is consistent with the retrotransposon structure at the maize Adh1 locus (SanMiguel et al., 1996, 1998).
However, in the adh1 locus, the nested elements are only retrotransposons, whereas transposon and MITE elements, in addition to retrotransposons, are found at the Mla locus. Subsequent to transposon invasion, a final tandem duplication of 40 kb of gene-rich sequence expanded the original 30 kb (Figure 6D), including the ancient Mla allele, RGH1bcd, to the >200 kb we see today (for time-lapse animation of Figure 6, see supplemental data online).
The Mla Region Possesses Similar Gene Density to Arabidopsis
The average gene density in the genome of the model plant Arabidopsis is one per 4.5 kb. The genomes of grain cereal plants are orders of magnitude greater in size than that of Arabidopsis, yet as eukaryotic flowering plants, they would be expected to encode a similar number of low-copy genes. There is increasing evidence for gene-dense islands interrupted by gene-poor, repetitive DNA in large cereal genomes (Barakat et al., 1997; Panstruga et al., 1998; Feuillet and Keller, 1999; Moore, 2000; Shirasu et al., 2000; Wicker et al., 2001). Our results reveal that the overall gene density in the 261-kb Mla contig is one per 8.1 kb. However, the density in the gene-rich islands is one per 4.6 kb, statistically identical to the gene density of Arabidopsis.
Clustering of Genes Associated with Defense Responses
Of the 32 protein-coding genes detected in the Mla region, 15 associated with plant defense responses can be classified into five structurally unique, non-cross-hybridizing families. These include three CC-NBS-LRR RGH families, one CI2 family, and one chemically induced family. Three of the five families likely function in at least two independent pathways associated with powdery mildew defense responses, including the Mla gene-for-gene pathway and an induced systemic resistance pathway.
The Mla signal cascade has been reported to be SA independent in resistant Mla–Bgh interactions (Huckelhoven et al., 1999). By contrast, BCI-7 of the CI2 family and the BCI-9–like gene Bdi are predicted to function via SA induction in the induced systemic resistance pathway. More interestingly, BCI-7 and BCI-9 cannot be induced by direct interaction of barley with Bgh, indicating that chemically induced systemic resistance to Bgh operates via a different pathway than gene-for-gene specificity (Besser et al., 2000).
There are several examples of clustered genes with related functions in plant systems (Hulbert et al., 2001). For example, Tip1 and Tip2 are two additional genes adjacent to barley Rar1, which is required for some, but not all, Mla-specified resistance (Shirasu et al., 2000). Tip homologs in Arabidopsis are inducible by both SA and JA (Schenk et al., 2000). Tip and Rar1 genes are predicted to function in two different pathways, similar to CI2 and Mla genes.
Adjacent to the barley Mlo (broad-spectrum powdery mildew resistance) gene is a ring finger protein (Panstruga et al., 1998); the tobacco homolog of this gene can be elicited rapidly in Avr9–Cf9 interactions. There also are five Ser/Thr kinase Pto paralogs adjacent to the Prf gene, which is required for Pto-specified resistance to bacterial speck disease in tomato (Jia et al., 1997).
In Arabidopsis, 115 of the 166 NBS-LRR–like R genes are clustered in 40 loci, and six of these clusters are heterogeneous (Richly et al., 2002). On Arabidopsis chromosome 4, a 180-kb region encodes six NBS-LRR RGH families, a nine-member PR-2 (chitinase) gene family, a NPR1 homolog, and several potential SA- or JA-inducible gene families. The NPR1 gene is a major signal transduction component of the NBS-LRR resistance pathway and is required for SA-dependent R gene function (Cao et al., 1997).
Likewise, there is a 40-kb region on Arabidopsis chromosome 1 that contains a RPM1 disease resistance gene homolog, a mlo resistance gene homolog, and two distinct Ser/Thr kinase genes. In the Arabidopsis RPP5 locus, a Ser/Thr kinase pseudogene is adjacent to the RPP5 TIR-NBS-LRR gene (Noel et al., 1999).
On a more global scale, it has long been known that a majority of the mildew resistance genes in barley are positioned near the telomeric end of chromosome 5S (1HS). This also is the case with the syntenic region in wheat, in which the receptor-like kinase gene Lrk10 and the Pm3b powdery mildew resistance locus are just 0.7 centimorgan distal to the Mla homolog TaMla (Zhou et al., 2001). Thus, at least on Triticeae group 1 chromosomes, evolutionary forces appear to have positioned functionally similar sequences in physically linked groups (Feuillet and Keller, 1999).
Dynamics of the Mla Region
The interesting feature of the gene-poor region proximal to Mla is that it contains many diverse repeats, including 4 SSRs and 17 tandem repeats. A plausible hypothesis for the origin of these small duplications is that they have resulted from DNA replication slippage (Trinh and Sinden, 1991; Weston and Berg, 1991; Morgante et al., 2002). Palindromic or quasipalindromic sequences present in the founder sequence are essential for this slippage mechanism to form complex secondary structures in single-stranded DNA (Awadalla and Ritland, 1997). For example, all 15 MITE elements in the region immediately proximal to Mla can form hairpin structures. Subsequent to the formation of a hairpin structure, DNA replication would continue to produce another copy of the palindromic sequence, resulting in a tandem duplication with inverted terminal repeats. Once tandem arrays of repeats are established, stretches of homologous sequence can facilitate pairing and further repeat propagation by unequal recombination.
Recently, Morgante et al. (2002) demonstrated that microsatellites were associated primarily with low-copy fractions of plant genomes and that their distribution was a function of genome dynamics coupled with selective constraints. As we have observed for the gene-poor, high-copy region proximal to the Mla locus, repetitive regions in plant genomes can be under mutational (slippage) pressure that increases microsatellite frequency. Furthermore, Morgante et al. (2002) speculated that although sevenfold lower (677 versus 99 imperfect microsatellites per megabase), the equilibrium frequency in the repetitive fraction could be dependent on the timing of the amplification of repeats.
Given the assumption of a uniform mutation rate within repeats, significantly greater DNA similarities would indicate that the distal side of the Mla region has evolved more recently than the proximal side. We postulate that the proximal end has degenerated and that the region has shifted to a more distal position via repeat amplification. Indeed, within the 2.5-kb Bmt duplication, there is a family of eight repeats that share 70% identity (tandem repeat 1 [Rpt1]; Figure 3C). Further distal, the Rpt11 family is up to 89% sequence identical, whereas Rpt12 (Afa) shares up to 98% identity, Rpt13 shares up to 96% identity, Rpt15 shares up to 90% identity, Rpt16 shares up to 95% identity, and Rpt17 shares up to 100% identity. This sequence similarity analysis indicates that the propagation of these repeats is coincident with most of the retrotransposon insertion in the past 3 million years (Figures 1 and 6).
Subsequent to the addition of 87 kb of transposons interspersed within the Mla complex, a 40-kb gene-rich duplication became the most recent addition to the region. This would appear advantageous, because duplication of gene-rich regions propagates genetic diversity through multiple cycles of selection and the creation of new gene family members. Unequal crossover among new gene family members has been shown to be a driving force in the generation of new resistance specificities (Hulbert et al., 2001). However, R genes such as Mla, which are positioned adjacent to or within retrotransposon complexes, would seem to favor modes of diversification other than recombination, such as point mutations, duplications, and insertion/deletion events (Ellis et al., 2000).
Previously, we reported recombination suppression at the Mla locus by analysis of a high-resolution mapping population with RAPD–, AFLP–, BAC end–, and YAC end–derived markers (Wei et al., 1999). With the current detailed sequence of the Mla region, we are able to add significant information to our previous study with regard to other recent findings in eukaryotes. Recombination suppression (>5.6 Mb/centimorgan) occurs between 236R and RGH3b, two markers 160 kb apart that cosegregate genetically with Mla resistance specificity (see supplemental data online).
As illustrated in Figure 1, the 236R-to-RGH3b interval includes the Mla gene family interrupted by a 51-kb complex of nested LTR retrotransposons. Indeed, recombination suppression also has been associated with LTR retrotransposon complexes in maize (Fu et al., 2002), Drosophila (Rizzon et al., 2002), and C. elegans (Duret et al., 2000). By contrast, a 40-fold higher rate of recombination (140 kb/centimorgan) occurs directly proximal to Mla in the gene-poor, repeat-dense region between RGH3b and 175D16.T7. These data further substantiate the notion that high rates of recombination can occur in repeat-dense, gene-poor regions in higher eukaryotes, similar to that documented for C. elegans (Barnes et al., 1995; Duret et al., 2000).
In more general terms, the propensity for wide distribution of the retroelements among gene-rich contigs may complicate strategies for isolating low-copy BAC clones for large-genome sequencing efforts (Meyers et al., 2001). Furthermore, the lack of correlation between methylation state and transcription in the Mla region also may have implications for strategies to sequence the transcribed regions of complex cereal genomes. It now seems probable that strategies to isolate gene-enriched fractions of the genome using methylation-sensitive host strains (Rabinowicz et al., 1999; Mayer and Mewes, 2002) could exclude several agriculturally valuable genes.
METHODS
DNA Sequencing
Shotgun sequencing libraries were constructed as described previously with randomly sheared BACs ligated into the pUC18 vector (Wei et al., 1999). DNA sequence templates were prepared by the alkaline lysis method (Sambrook et al., 1989) with minor modifications. Escherichia coli cells were incubated in 96-well culture blocks (Qiagen, Valencia, CA) with 1.2 mL/well TB medium (1.2% bacto-tryptone, 2.4% bacto-yeast extract, 0.4% [v/v] glycerol, 0.017 M KH2PO4, and 0.072 M K2HPO4) at 37°C for at least 16 h at 325 rpm. Subsequent to centrifugation, cell pellets were resuspended in 100 μL of solution I (50 mM Glc, 10 mM EDTA, pH 8.0, 25 mM Tris-HCl, pH 8.0, and 0.2 mg of RNase A), lysed with 100 μL of solution II (0.2 N NaOH and 1% SDS), and neutralized in 100 μL of solution III (3 M KOAc, pH 4.8).
The lysate was transferred to 96-well MultiScreen-NA filter plates (Millipore, Bedford, MA) positioned on 96-well receiver plates (Whatman Polyfiltronics, Rockland, MA). The paired plates were centrifuged at 4000 rpm (4°C) for 15 min to filter cell debris, and DNAs were precipitated by adding 200 μL of isopropanol at room temperature for 10 min and centrifuged at 4000 rpm (4°C) for 15 min. All of the solutions described above were mixed by gentle vortexing.
Sequence-ready DNA pellets were washed with 70% ethanol, air dried for 10 min, and dissolved in 40 μL of sterile double-distilled water. Sequence data were obtained with 4 μL of the DNA described above using ABI BigDye terminators (Perkin-Elmer Biosystems, Foster City, CA) at the Clemson University Genomics Institute DNA Sequencing Center and the DNA Sequencing and Synthesis Facility at Iowa State University.
Sequence Assembly, Quality Assessment, and Contig Gap Filling
Sequence reads for the 261,265-nucleotide contig were assembled with the Phred and Phrap software packages (Ewing et al., 1998; Ewing and Green, 1998) and edited with the Consed program (Gordon et al., 1998). At seven to eight times redundancy, internal low-quality regions of each contig were identified and treated as gaps or different contigs. To fill gaps in the sequence, end clones of contigs were sequenced with the M13 reverse primer. These data were used for reassembly, and the same protocol was repeated two more times. For further gap filling, a primer-walking procedure was used to sequence clones whose sequences are located at the ends of different contigs.
Sequencing primers were designed with the MacVector software package (Oxford Molecular Group, Campbell, CA) and synthesized at the DNA Sequencing and Synthesis Facility at Iowa State University. GTG Primer Mix (Perkin-Elmer Biosystems) was added to resolve the potential secondary structure in remaining gaps consisting of AT- or GC-rich regions. Sequence-based restriction maps were compared with HindIII, EcoRI, and MluI digests for confirmation of the accuracy of the final assembled sequences.
Sequence Annotation and Computational Analysis
The World Wide Web–based programs GENSCAN (Burge and Karlin, 1997) and GeneMark (Lukashin and Borodovsky, 1998) were used for general gene mining. BLASTx, BLASTn, and BLASTp from the National Center for Biotechnology Information (Altschul et al., 1997) were used to identify known genes or repetitive elements at intervals of 500 bp for BLASTx and 1000 bp for BLASTn. The full lengths of these BLAST-identified genes were determined by ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), and BLAST aligned the results.
The long terminal repeat (LTR) class of retrotransposons was identified by their LTRs, the inverted repeats of the LTRs, and their insertion signatures. The DotPlot program was used for self-comparison to determine MITE elements at a 600-bp interval with a 400-bp rolling window (http://alces.med.umn.edu/rawdot.html; University of Minnesota). Simple sequence repeats were analyzed with the RepeatMasker program (A.F.A. Smit and P. Green, unpublished data; http://repeatmasker.genome.washington.edu/cgi-bin/RM2_req.pl).
The Wisconsin Genetics Computer Group package (Madison, WI) GAP program was used for similarity analysis, and MacVector version 6.0 was used for translation analysis. Repeat sequences were identified by the World Wide Web–based Large DotPlots program (http://alces.med.umn.edu/rawdot.html) in combination with Miropeat software (Parsons, 1995). The Genetics Computer Group Pileup and Prettybox programs were used for multiple sequence comparison and visualization. PAUP 4.0 software (http://www.lms.si.edu/PAUP/) was used for phylogenetic analysis of tandem repeats. EST searches were performed with the National Center for Biotechnology Information BLASTn program.
Methylation Tests
Barley (Hordeum vulgare) genomic DNA isolated from 7-day-old seedlings was digested with either MspI or HpaII. The restricted DNA then was used as a template for PCR amplification. PCR primers were designed from the coding regions of genes such that a product could not be amplified from the completely restricted DNA, whereas a product should be amplified from the methylated template. If there was no restriction site in the coding region, primers were designed in regions within 2 kb upstream or downstream of the respective gene. DNA was digested with HpaII or MspI (10 units of enzyme per μg of DNA; New England Biolabs, Beverly, MA) at 37°C for 10 h, and the enzymes were inactivated at 65°C for 20 min. PCR (Taq DNA polymerase from Life Technologies [Gibco BRL]) was performed to amplify the sequence. PCR primers and annealing temperatures are shown in Table 4.
Reverse Transcription–PCR Analysis
Total RNA was isolated from 7-day-old barley seedlings as instructed in the RNeasy Plant Mini Kit (Qiagen). Isolated total RNA was digested further with DNase I (Ambion, Austin, TX) and purified using the clean-up application of the Qiagen kit. Reverse transcription (RT)–PCR primer pairs are the same as those used in the methylation test except those from genes that do not have a MspI or HpaII restriction site in the coding region. In the latter situation, new RT-PCR primer pairs were designed according to the predicted coding region. The sequences of PCR primers and their annealing temperatures are shown in Table 4.
Because most primer pairs are within one exon of their respective genes, strict controls were performed to ensure that the total RNA was DNA free. DNase-cleaned total RNA was first tested with primer pairs from genes whose transcription was confirmed by barley EST database searches. Genomic DNA was used as a positive PCR control. In the RT-PCR analysis of each gene, a negative control with only Taq polymerase but no reverse transcriptase, and a positive genomic DNA control, were used. RT-PCR was conducted according to the Superscript One-Step RT-PCR Kit (Gibco BRL) with 0.5 μg of total RNA per reaction. The amplified DNA fragments were resolved on 2% Low EEO agarose gels (FMC Bioproducts, Rockland, ME) with ethidium bromide.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are AF427791 (Hordeum vulgare Mla locus from cv Morex), AJ302293 (H. vulgare Mla6 gene), AY009938 (H. vulgare Mla1 gene), AAG43550 (Nicotiana tabacum Avr9/Cf-9 rapidly elicited protein 132), AL161551 (Arabidopsis chromosome 4, contig fragment No. 51), U95973 (Arabidopsis chromosome 1, BAC T19D16), BG417789 (barley EST from HVSMEk testa/pericarp library), BM374865 (barley EST from EBma05 post-anthesis spike library), and BG310375 (barley EST from HVSMEc shoot library).
NOTE ADDED IN PROOF
Tanaka et al. (2002) reported recently that short inverted repeats initiate gene amplification through the formation of a large DNA palindrome in mammalian cells. This finding suggests that short inverted repeats can have a critical role in the initiation of gene amplification.
Tanaka, H., Tapscott, S.J., Trask, B.J., and Yao, M.-C. (2002). Short inverted repeats initiate gene amplification through the formation of a large DNA palindrome in mammalian cells. Proc. Natl. Acad. Sci. USA 99, 8772–8777.
Supplementary Material
Acknowledgments
The authors thank D. Voytas and S. Whitham for critical review of the manuscript, X. Gu for helpful advice regarding the paleontology analysis, D. Halterman for the animation of Figure 6, P. Schulze-Lefert for providing the Mla1-2 sequence, and L. Mao, S.-S. Woo, M. Sasinowski, Y. Yu, R. Kingsbury, T. Rambo, and the staff at the Clemson University Genomics Institute Bioinformatics and Sequencing Centers for technical assistance. This research was supported in part by U.S. Department of Agriculture National Research Initiative Competitive Grants Program Grants 98-35300-6169 and 00-35300-9213 to R.P.W. and National Science Foundation Grant MRI #9724557 to R.A.W. This is a joint contribution of the Corn Insects and Crop Genetics Research Unit, the U.S. Department of Agriculture Agricultural Research Service, and the Iowa Agriculture and Home Economics Experiment Station. This is Journal Paper No. J-19346 of the Iowa Agricultural and Home Economics Experiment Station, Ames, IA, Project No. 3368, supported by Hatch Act and State of Iowa funds.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002238.
Footnotes
Online version contains Web-only data.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awadalla, P., and Ritland, K. (1997). Microsatellite variation and evolution in the Mimulus guttatus species complex with contrasting mating systems. Mol. Biol. Evol. 14, 1023–1034. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Baker, B., Zambriski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant-microbe interactions. Science 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Barakat, A., Carels, N., and Bernardi, G. (1997). The distribution of genes in the genomes of Gramineae. Proc. Natl. Acad. Sci. USA 94, 6857–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes, T.M., Kohara, Y., Coulson, A., and Hekimi, S. (1995). Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141, 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, S., and Trowsdale, J. (2000). The human major histocompatibility complex: Lessons from the DNA sequence. Annu. Rev. Genom. Hum. Genet. 1, 117–137. [DOI] [PubMed] [Google Scholar]
- Besser, K., Jarosch, B., Langen, G., and Kogel, K.H. (2000). Expression analysis of genes induced in barley by chemical activation reveals distinct disease resistance pathways. Mol. Plant Pathol. 1, 277–286. [DOI] [PubMed] [Google Scholar]
- Boyd, L.A., Smith, P.H., Foster, E.M., and Brown, J.K.M. (1995). The effects of allelic variation at the Mla resistance locus in barley on the early development of Erysiphe graminis f. sp. hordei and host responses. Plant J. 7, 959–968. [Google Scholar]
- Bureau, T.E., and Wessler, S.R. (1994). Stowaway: A new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell 6, 907–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- Cao, H., Glazebrook, J., Clarke, J.D., Volko, S., and Dong, X. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. [DOI] [PubMed] [Google Scholar]
- Cocciolone, S.M., Chopra, S., Flint-Garcia, S.A., McMullen, M.D., and Peterson, T. (2001). Tissue-specific patterns of a maize Myb transcription factor are epigenetically regulated. Plant J. 27, 467–478. [DOI] [PubMed] [Google Scholar]
- Cordero, M.J., Raventos, D., and San Segundo, B. (1994). Expression of a maize proteinase inhibitor gene is induced in response to wounding and fungal infection: Systemic wound-response of a monocot gene. Plant J. 6, 141–150. [DOI] [PubMed] [Google Scholar]
- Dixon, M.S., Golstein, C., Thomas, C.M., van der Biezen, E.A., and Jones, J.D.G. (2000). Genetic complexity of pathogen perception by plants: The example of Rcr3, a tomato gene required specifically by Cf-2. Proc. Natl. Acad. Sci. USA 97, 8807–8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret, L., Marais, G., and Biémont, C. (2000). Transposons but not retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics 156, 1661–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J., Dodds, P., and Pryor, T. (2000). The generation of plant disease resistance gene specificities. Trends Plant Sci. 5, 373–379. [DOI] [PubMed] [Google Scholar]
- Ellis, J., Lawrence, G., Ayliffe, M., Anderson, P., Collins, N., Finnegan, J., Frost, D., Luck, J., and Pryor, T. (1997). Advances in the molecular genetic analysis of the flax-flax rust interaction. Annu. Rev. Phytopathol. 35, 271–291. [DOI] [PubMed] [Google Scholar]
- Ewing, B., and Green, P. (1998). Base calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8, 186–194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M., and Green, P. (1998). Base calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8, 175–185. [DOI] [PubMed] [Google Scholar]
- Feuillet, C., and Keller, B. (1999). High gene density is conserved at syntenic loci of small and large grass genomes. Proc. Natl. Acad. Sci. USA 96, 8265–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, H., Zheng, Z., and Dooner, H.K. (2002). Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. USA 99, 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B.S., Morton, B.R., McCaig, B.C., and Clegg, M.T. (1996). Substitution rate comparisons between grasses and palms: Synonymous rate differences at the nuclear gene Adh parallel rate differences at the plastid gene rbcL. Proc. Natl. Acad. Sci. USA 93, 10274–10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. (1998). Consed, a graphical tool for sequence finishing. Genome Res. 8, 195–202. [DOI] [PubMed] [Google Scholar]
- Halterman, D., Zhou, F., Wei, F., Wise, R.P., and Schulze-Lefert, P. (2001). The MLA6 Coiled-Coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. 25, 335–348. [DOI] [PubMed] [Google Scholar]
- Heath, R.L., McDonald, G., Christeller, J.T., Lee, M., Bateman, K., West, J., Van Heeswijck, R., and Anderson, M.A. (1997). Proteinase inhibitors from Nicotiana alata enhance plant resistance to insect pests. J. Insect Physiol. 43, 833–842. [DOI] [PubMed] [Google Scholar]
- Huckelhoven, R., Fodor, J., Preis, C., and Kogel, K.H. (1999). Hypersensitive cell death and papilla formation in barley attacked by the powdery mildew fungus are associated with hydrogen peroxide but not with salicylic acid accumulation. Plant Physiol. 119, 1251–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert, S., Webb, C.A., Smith, S.M., and Sun, Q. (2001). Resistance gene complexes: Evolution and utilization. Annu. Rev. Phytopathol. 39, 285–312. [DOI] [PubMed] [Google Scholar]
- Jia, Y., Loh, Y.T., Zhou, J., and Martin, G.B. (1997). Alleles of Pto and Fen occur in bacterial speck-susceptible and fenthion-insensitive tomato cultivars and encode active protein kinases. Plant Cell 9, 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G. (2001). Putting knowledge of plant disease resistance genes to work. Curr. Opin. Plant Biol. 4, 281–287. [DOI] [PubMed] [Google Scholar]
- Jørgensen, J.H. (1994). Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 13, 97–119. [Google Scholar]
- Kitagawa, K., Skowyra, D., Elledge, S.J., Harper, J.W., and Hieter, P. (1999). SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4, 21–33. [DOI] [PubMed] [Google Scholar]
- Kleinhofs, A., et al. (1993). A molecular, isozyme and morphological map of the barley (Hordeum vulgare) genome. Theor. Appl. Genet. 86, 705–712. [DOI] [PubMed] [Google Scholar]
- Koiwa, H., Bressan, R.A., and Hasegawa, P.M. (1997). Regulation of proteinase inhibitors and plant defense. Trends Plant Sci. 2, 379–384. [Google Scholar]
- Lee, J.S., Brown, W.E., Graham, J.S., Pearce, G., Fox, E.A., Dreher, T.W., Ahern, K.G., Pearson, G.D., and Ryan, C.A. (1986). Molecular characterization and phylogenetic studies of a wound-inducible proteinase inhibitor I gene in Lycopersicon species. Proc. Natl. Acad. Sci. USA 83, 7277–7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X., et al. (1999). Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402, 761–768. [DOI] [PubMed] [Google Scholar]
- Lisch, D.R., Freeling, M., Langham, R.J., and Choy, M.Y. (2001). Mutator transposase is widespread in the grasses. Plant Physiol. 125, 1293–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin, A.V., and Borodovsky, M. (1998). GeneMark.hmm: New solutions for gene finding. Nucleic Acids Res. 26, 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manninen, I., and Schulman, A.H. (1993). BARE-1, a copia-like retroelement in barley (Hordeum vulgare L.). Plant Mol. Biol. 22, 829–846. [DOI] [PubMed] [Google Scholar]
- Margossian, L.J., Federman, A.D., Giovannoni, J.J., and Fischer, R.L. (1988). Ethylene-regulated expression of a tomato fruit ripening gene encoding a proteinase inhibitor I with a glutamic residue at the reactive site. Proc. Natl. Acad. Sci. USA 85, 8012–8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K., and Mewes, H.-W. (2002). How can we deliver the large plant genomes? Strategies and perspectives. Curr. Opin. Plant Biol. 5, 173–177. [DOI] [PubMed] [Google Scholar]
- Meyers, B.C., Tingey, S.V., and Morgante, M. (2001). Abundance, distribution, and transcriptional activity of repetitive elements in the maize genome. Genome Res. 11, 1660–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R.W., and Meyers, B.C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130. [DOI] [PubMed] [Google Scholar]
- Moore, G. (2000). Cereal chromosome structure, evolution, and pairing. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 195–223. [DOI] [PubMed] [Google Scholar]
- Morgante, M., Hanafey, M., and Powell, W. (2002). Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 30, 194–200. [DOI] [PubMed] [Google Scholar]
- Moseman, J.G. (1972). Isogenic barley lines for reaction to Erysiphe graminis f. sp. hordei. Crop Sci. 12, 681–682. [Google Scholar]
- Nagaki, K., Tsujimoto, H., Isono, K., and Sasakuma, T. (1995). Molecular characterization of a tandem repeat, Afa family, and its distribution among Triticeae. Genome 38, 479–486. [DOI] [PubMed] [Google Scholar]
- Noel, L., Moores, T.L., van der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11, 2099–2112. [PMC free article] [PubMed] [Google Scholar]
- Panstruga, R., Büschges, R., Piffanelli, P., and Schulze-Lefert, P. (1998). A contiguous 60 kb genomic stretch from barley reveals molecular evidence for gene islands in a monocot genome. Nucleic Acids Res. 26, 1056–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, J.D. (1995). Miropeats: Graphical DNA sequence comparisons. Comput. Appl. Biosci. 11, 615–619. [DOI] [PubMed] [Google Scholar]
- Paszkowski, J., and Whitham, S.A. (2001). Gene silencing and DNA methylation processes. Curr. Opin. Plant Biol. 4, 123–129. [DOI] [PubMed] [Google Scholar]
- Rabinowicz, P.D., Schutz, K., Dedhia, N., Yordan, C., Parnell, L.D., Stein, L., McCombie, W.R., and Martienssen, R.A. (1999). Differential methylation of genes and retrotransposons facilitates shotgun sequencing of the maize genome. Nat. Genet. 23, 305–308. [DOI] [PubMed] [Google Scholar]
- Richly, E., Kurth, J., and Leister, D. (2002). Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol. Biol. Evol. 19, 76–84. [DOI] [PubMed] [Google Scholar]
- Rizzon, C., Marais, G., Gouy, M., and Biemont, C. (2002). Recombination rate and the distribution of transposable elements in the Drosophila melanogaster genome. Genome Res. 12, 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 20, 43–45. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., Tikhonov, A., Jin, Y.K., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P.S., Edwards, K.J., Lee, M., Avramova, Z., and Bennetzen, J.L. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science 274, 765–768. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97, 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu, K., Lahaye, T., Tan, M.W., Zhou, F., Azevedo, C., and Schulze-Lefert, P. (1999). A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99, 355–366. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., Schulman, A.H., Lahaye, T., and Schulze-Lefert, P. (2000). A contiguous 66-kb barley DNA sequence provides evidence for reversible genome expansion. Genome Res. 10, 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadauchi, T., Matsumoto, K., Herskowitz, I., and Irie, K. (2001). Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 20, 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh, T.Q., and Sinden, R.R. (1991). Preferential DNA secondary structure mutagenesis in the lagging strand of replication in Escherichia coli. Nature 352, 544–547. [DOI] [PubMed] [Google Scholar]
- Tsujimoto, H., Mukai, Y., Akagawa, K., Nagaki, K., Fujigaki, J., Yamamoto, M., and Sasakuma, T. (1997). Identification of individual barley chromosomes based on repetitive sequences: Conservative distribution of Afa-family repetitive sequences on the chromosomes of barley and wheat. Genes Genet. Syst. 72, 303–309. [DOI] [PubMed] [Google Scholar]
- van der Biezen, E.A., and Jones, J.D.G. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456. [DOI] [PubMed] [Google Scholar]
- Wei, F., Gobelman-Werner, K., Morroll, S.M., Kurth, J., Mao, L., Wing, R., Leister, D., Schulze-Lefert, P., and Wise, R.P. (1999). The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics 153, 1929–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston, H.K., and Berg, D.E. (1991). Limits to the role of palindromy in deletion formation. J. Bacteriol. 173, 315–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker, T., Stein, N., Albar, L., Feuillet, C., Schlagenhauf, E., and Keller, B. (2001). Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J. 26, 307–316. [DOI] [PubMed] [Google Scholar]
- Wise, R.P. (2000). Disease resistance: What's brewing in barley genomics. Plant Dis. 84, 1160–1170. [DOI] [PubMed] [Google Scholar]
- Wise, R.P., and Ellingboe, A.H. (1983). Infection kinetics of Erysiphe graminis f. sp. hordei on barley with different alleles at the Ml-a locus. Phytopathology 73, 1220–1222. [Google Scholar]
- Zhou, F., Kurth, J., Wei, F., Elliott, C., Vale, G., Yahiaoui, N., Keller, B., Somerville, S., Wise, R., and Schulze-Lefert, P. (2001). Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell 13, 337–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.