Abstract
Xie and colleagues previously isolated the Arabidopsis COI1 gene that is required for response to jasmonates (JAs), which regulate root growth, pollen fertility, wound healing, and defense against insects and pathogens. In this study, we demonstrate that COI1 associates physically with AtCUL1, AtRbx1, and either of the Arabidopsis Skp1-like proteins ASK1 or ASK2 to assemble ubiquitin-ligase complexes, which we have designated SCFCOI1. COI1E22A, a single amino acid substitution in the F-box motif of COI1, abolishes the formation of the SCFCOI1 complexes and results in loss of the JA response. AtRbx1 double-stranded RNA-mediated genetic interference reduces AtRbx1 expression and affects JA-inducible gene expression. Furthermore, we show that the AtCUL1 component of SCFCOI1 complexes is modified in planta, where mutations in AXR1 decrease the abundance of the modified AtCUL1 of SCFCOI1 and lead to a reduction in JA response. Finally, we demonstrate that the axr1 and coi1 mutations display a synergistic genetic interaction in the double mutant. These results suggest that the COI1-mediated JA response is dependent on the SCFCOI1 complexes in Arabidopsis and that the AXR1-dependent modification of the AtCUL1 subunit of SCFCOI1 complexes is important for JA signaling.
INTRODUCTION
Plant hormones influence many diverse developmental processes, ranging from seed germination to root, leaf, shoot, and flower formation. Jasmonic acid and its cyclopentanous derivatives (collectively referred to here as jasmonates [JAs]) are a new class of plant hormones that modulate the expression of numerous genes (Reymond et al., 2000) and regulate plant developmental processes, including root growth, pollen formation, tuberization, and fruit ripening. They also mediate responses to stress, wounding, insect attack, and pathogen infection (for reviews, see Staswick, 1992; Sembdner and Parthier, 1993; Blechert et al., 1995; Creelman and Mullet, 1997; Wasternack and Parthier, 1997; Reymond and Farmer, 1998; Zhao and Ma, 2000; Berger, 2001; Farmer, 2001).
The effects of JA on a variety of plant species were discovered initially through the application of exogenous JA to plants (for review, see Creelman and Mullet, 1997). Furthermore, plant responses to JA were defined mainly through genetic analysis of mutants, including fad3/fad7/fad8 (McConn and Browse, 1996), opr3 (Sanders et al., 2000; Stintzi and Browse, 2000; Stintzi et al., 2001), jar1 (Staswick et al., 1992, 1998), coi1 (Feys et al., 1994), jin1 and jin4 (Berger et al., 1996), cev1 (Ellis and Turner, 2001), cet (Hilpert et al., 2001), cex1 (Xu et al., 2001), and others (Howe et al., 1996; Petersen et al., 2000). Although the effect of JA on plants is well characterized, little is known about the mechanisms whereby JAs exert their biological functions.
The Arabidopsis mutants jar1, coi1, jin1, and jin4 have been used to study the signal transduction pathway that modulates JA action. The jar1, jin1, and jin4 mutants exhibit decreased sensitivity to JA, as assayed for the inhibition of root growth and for the inducible expression of the Arabidopsis vegetative storage protein (AtVSP) in response to JA treatment. The coi1-1 mutant is male sterile, insensitive to JA, and lacks the expression of JA-induced proteins, including AtVSP and the plant defense–related proteins Thi2.1 and PDF1.2 (Feys et al., 1994; Benedetti et al., 1995; Penninckx et al., 1998; Xie et al., 1998). The COI1 gene was found to be required for all JA-regulated plant fertility and defense processes (Feys et al., 1994; Xie et al., 1998) and is suggested to act upstream of JIN1 and JAR1/JIN4 in the JA signal transduction chain (Berger et al., 1996; Rojo et al., 1998; Berger, 2001).
Molecular analysis of the COI1 sequence indicated that COI1 might encode a protein containing Leu-rich repeats and a degenerate F-box motif (Xie et al., 1998). These structural features are characteristic of F-box proteins that function in ubiquitin-ligase complexes for the ubiquitylation of substrate proteins targeted for degradation. The F-box motif is present in many yeast and mammalian proteins that interact with Skp1 and Cdc53 (cullin) to assemble SCF ubiquitin-ligase complexes (Skp1-Cdc53-F-box protein) (for reviews, see Hershko and Ciechanover, 1998; Deshaies, 1999). In the ubiquitin-dependent proteolytic pathway, ubiquitin is linked to substrates through a well-organized process involving the sequential action of a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3). The ubiquitin-related proteins RUB/NEDD8 also are conjugated to target proteins by a mechanism similar to the ubiquitin conjugation process involving the action of a RUB-activating enzyme and a RUB-conjugating enzyme.
One target for RUB conjugation is the Cdc53/cullin component of SCF complexes (Lammer et al., 1998; del Pozo and Estelle, 1999; Gray et al., 1999), and this RUB modification of Cdc53/cullin is important in regulating the activity of SCF ubiquitin ligases (del Pozo and Estelle, 1999; Osaka et al., 2000; Podust et al., 2000; Lyapina et al., 2001). In Arabidopsis, the Cdc53-like protein AtCUL1 (del Pozo and Estelle, 1999) and the Skp1-like proteins ASK1 and ASK2 interact with F-box proteins such as TIR1 to assemble SCF complexes (Gray et al., 1999, 2001; del Pozo and Estelle, 2000; Schwechheimer et al., 2001). AtCUL1 is modified by the ubiquitin-related protein RUB1 in vivo (del Pozo and Estelle, 1999). RUB1 is activated for conjugation to AtCUL1 by a RUB-activating enzyme that is composed of AXR1 and ECR1 and the RUB-conjugating enzyme RCE1 (del Pozo et al., 1998, 2002; del Pozo and Estelle, 1999).
In this study, we demonstrate that the Arabidopsis F-box protein COI1 associates with AtCUL1, AtRbx1, and the Skp1-like proteins ASK1 and ASK2 to assemble SCFCOI1 ubiquitin-ligase complexes in planta. COI1E22A, a single amino acid substitution in the F-box motif of COI1, abrogates the formation of SCFCOI1 complexes and causes defects in JA response. Double-stranded AtRbx1 RNA-mediated genetic interference reduces AtRbx1 expression and affects the inducible accumulation of JA-inducible gene expression.
Furthermore, we find that the AtCUL1 component of SCFCOI1 complexes is modified in planta, where mutations in AXR1 decrease the abundance of the modified AtCUL1 of SCFCOI1 and lead to a reduction in JA response. Finally, we demonstrate that the axr1 and coi1 mutations display a synergistic interaction in double mutants. Together, these results indicate that the SCFCOI1 complexes are required for JA response in Arabidopsis and that the AXR1-dependent modification of AtCUL1 is important for JA signaling.
RESULTS
Two-Hybrid Screen for COI1-Interacting Proteins
To identify Arabidopsis proteins with which COI1 can interact in the yeast two-hybrid system, we constructed two COI1 “baits” with in-frame fusions to either the GAL4 or LexA DNA binding domains in pGBKT7 or pLexA, resulting in pGBKT7-COI1 and pLexA-COI1, respectively. pGBKT7-COI1 and pLexA-COI1 were used to exhaustively screen the corresponding GAL4- or LexA-based Arabidopsis cDNA libraries, leading to the identification of 137 positive colonies. Cross-hybridization analysis and restriction enzyme digestion of the PCR-amplified cDNA inserts of the 137 positive colonies indicated that they belonged to four distinct classes.
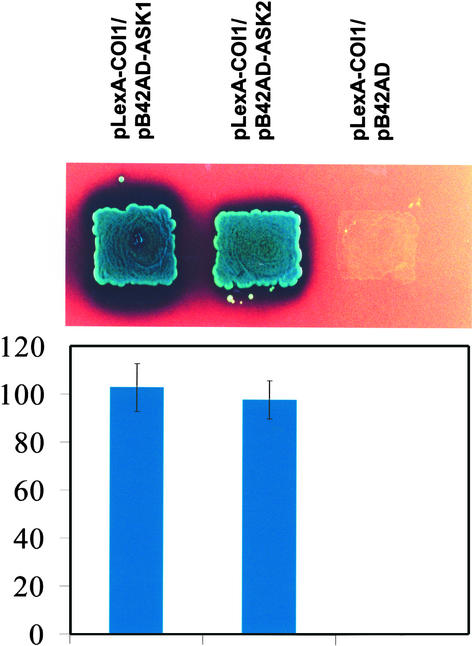
Representative cDNAs from each class were sequenced. Two of the four genes were identical to the Arabidopsis Skp1-like genes ASK1 and ASK2, represented by 37 and 24 colonies, respectively (Porat et al., 1998; Gray et al., 1999; Samach et al., 1999; Yang et al., 1999). ASK1 and ASK2 share strong similarity with each other and with the SKP1 proteins from human and yeast (Bai et al., 1996; Connelly and Hieter, 1996; Gray et al., 1999; Samach et al., 1999). Figure 1 shows that COI1 specifically interacted with ASK1 and ASK2 in yeast. The interactions of COI1 with ASK1 and ASK2 were further characterized in planta using coimmunoprecipitation assays.
Figure 1.
COI1 Interacts with ASK1 and ASK2 in the Yeast Two-Hybrid System.
COI1 was fused to the LexA DNA binding domain in pLexA and examined for interaction with ASK1 and ASK2 fused in frame to the activating domain in pB42AD. The two-hybrid reporter strain EGY48 coexpressing the indicated fusion proteins was grown on 2% Gal/1% raffinose/SD/-Ura/-His/-Trp/-Leu/X-β-Gal (top). Interactions of the indicated constructs were quantified by routine β-galactosidase assay and are expressed in Miller units (bottom).
COI1 Forms SCF Complexes with ASK1, ASK2, AtRbx1, and AtCUL1 in Arabidopsis
Many yeast and mammalian F-box proteins interact with Skp1 and Cdc53 (cullin) to assemble SCF ubiquitin-ligase complexes (for reviews, see Hershko and Ciechanover, 1998; Deshaies, 1999). Recent studies have identified a small RING finger protein (Roc1, Rbx1, or Hrt1) that is a new subunit of SCF complexes (Kamura et al., 1999; Ohta et al., 1999; Seol et al., 1999; Tan et al., 1999). In Arabidopsis, a DNA database search identified two Arabidopsis Rbx1-like proteins (AAL13435 and CAB87200, referred to here as AtRbx1a and AtRbx1b) that have strong identities to each other and to mammalian Rbx1/ROC1 (Figure 2). The Arabidopsis Skp1-like proteins ASK1 and ASK2 have been shown to associate with AtCUL1 and the F-box protein TIR1 in planta (Gray et al., 1999, 2001). Having demonstrated the interaction of COI1 with ASK1 and ASK2 in yeast, we examined whether COI1 interacts with ASK1 and ASK2 in planta and further explored the possible association of COI1 with AtCUL1 and AtRbx1.
Figure 2.
Alignment of Rbx1 Amino Acid Sequences of Arabidopsis, C. elegans, Drosophila, Mouse, and Human.
Identical amino acid residues are shaded in black. Dashes denote gaps introduced by the alignment program. Protein names are indicated at left, and amino acid residue positions are indicated at right.
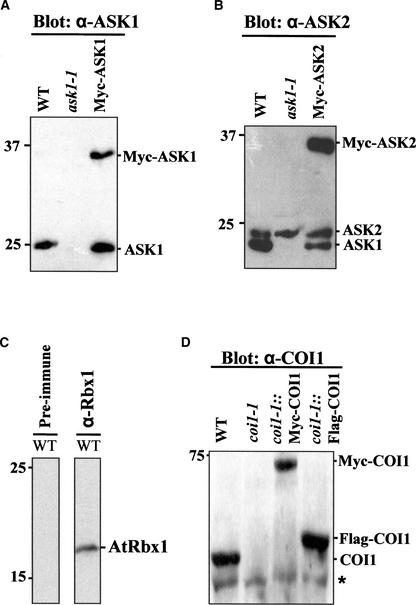
As a prelude to determining whether COI1 interacts with ASK1, ASK2, AtRbx1, and AtCUL1 using coimmunoprecipitation assays, we examined the specificity of polyclonal antibodies against ASK1, ASK2, AtCUL1, and COI1 and the possible recognition of AtRbx1 by the polyclonal antibody against human Rbx1. The results are shown in Figure 3. The anti- (α-)ASK1 antiserum detected an ∼21-kD band in the wild type but not in the mutant ask1-1 (Yang et al., 1999), suggesting that the α-ASK1 antiserum specifically recognized ASK1. This result was confirmed by protein gel blot analysis of transgenic plants expressing Myc-tagged ASK1 (Figure 3A). The α-ASK2 polyclonal antibody was shown previously to cross-react with ASK1 and to detect another larger band (∼23 kD) that presumably was the ASK2 protein (Gray et al., 1999).
Figure 3.
Protein Gel Blot Analysis of Extracts Prepared from the Indicated Arabidopsis Plants.
Blots were probed with polyclonal antisera against ASK1 (A), ASK2 (B), Rbx1 (C), or COI1 (D). The bands corresponding to each protein are indicated. The asterisk indicates the position of a nonspecific immunoreacting band. Molecular mass standards are given in kD (left). WT, wild type.
By protein gel blot analysis of Myc-tagged ASK2 transgenic plants and the ask1-1 mutant, we further demonstrated that α-ASK2 antiserum was able to detect both ASK2 and ASK1 (Figure 3B). The molecular masses of ASK1 and ASK2 detected in immunoblots were larger than their calculated masses, indicating that ASK1 and ASK2 might be modified in Arabidopsis, as SKP1 was modified by glycosylation in Dictyostelium (Teng-umnuay et al., 1998, 1999). The anti-Rbx1 antiserum, raised against the C-terminal region of the human Rbx1, detected an ∼18-kD band that presumably was AtRbx1a and/or AtRbx1b in Arabidopsis (Figure 3C).
The anti-AtCUL1 antibody used in this study was raised against the N-terminal 20–amino acid peptide of AtCUL1 and was found to specifically recognize two predominant bands (Shen et al., 2002), corresponding to AtCUL1 and the RUB-conjugated AtCUL1, as reported previously (Gray et al., 1998; del Pozo and Estelle, 1999). We also confirmed the specificity of this antibody by competition assays with the peptide (E. Lechner and P. Genschik, unpublished data) and through protein gel blot analysis of transgenic plants expressing Myc-tagged AtCUL1 (data not shown).
The α-COI1 antiserum detected a 65-kD band plus a slightly smaller band in protein extracts from the wild type, and the 65-kD band was absent from the null mutant allele (coi1-1) of COI1 (Figure 3D), indicating that the 65-kD band corresponds to the COI1 protein and the smaller band is a nonspecific protein cross-reacted with the α-COI1 antiserum. To avoid immunoprecipitation of nonspecific proteins by the α-COI1 antiserum, we generated transgenic plants expressing the epitope-tagged versions of COI1 for coimmunoprecipitation assays. The epitope-tagged versions of COI1 were expressed in coi1-1 mutant plants and detected specifically by the α-COI1 antiserum (Figure 3D).
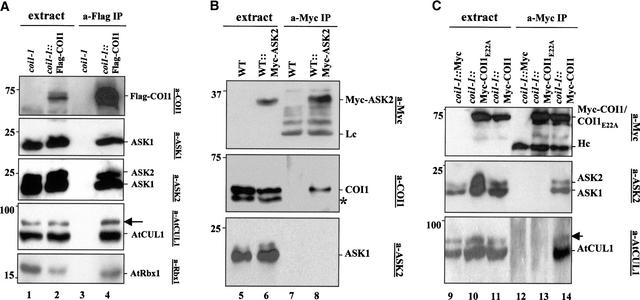
We used the α-Flag monoclonal antibody for immunoprecipitation from protein extracts of transgenic plants expressing Flag-tagged COI1, and the resulting immunoprecipitates were immunoblotted with α-COI1, α-ASK1, α-ASK2, α-Rbx1, and α-AtCUL1 antisera. The results are shown in Figure 4A. The Flag-tagged COI1 was coimmunoprecipitated with ASK1, ASK2, AtRbx1, and AtCUL1, suggesting that COI1 associates with ASK1, ASK2, AtRbx1, and AtCUL1 to assemble SCFCOI1 complexes in Arabidopsis. In the α-Flag immune complexes, the α-AtCUL1 antiserum detected both AtCUL1 and a slightly larger band that presumably was the RUB-modified AtCUL1 (Gray et al., 1998; del Pozo and Estelle, 1999). These results suggest that the AtCUL1 component of SCFCOI1 complexes was modified in planta, probably by RUB conjugation.
Figure 4.
Coimmunoprecipitation Assays.
(A) The Flag-tagged COI1 forms an SCF complex in Arabidopsis. Protein extracts from coi1-1 (lane 1) and transgenic coi1-1 expressing Flag-tagged COI1 (coi1-1::Flag-COI1; lane 2) were immunoprecipitated with α-Flag antibody (lanes 3 and 4).
(B) ASK2 associates physically with COI1 but not with ASK1 in planta. Protein extracts from a control wild-type plant (lane 5) and a transgenic plant expressing the Myc-tagged ASK2 (WT::Myc-ASK2; lane 6) were immunoprecipitated with α-Myc antibody (lanes 7 and 8).
(C) COI1E22A abrogates SCFCOI1. Protein extracts from coi1-1 plants transgenic for vector (coi1-1::Myc; lane 9), Myc-tagged COI1E22A (coi1-1:: Myc-COI1E22A; lane 10), or Myc-tagged COI1 (coi1-1::Myc-COI1; lane 11) were immunoprecipitated with α-Myc antibody (lanes 12 to 14).
The resulting immunoprecipitates were resolved by SDS-PAGE and detected with the indicated antibodies. Molecular mass standards are given in kD. Arrows in (A) and (C) indicate the modified version of AtCUL1. The asterisk in (B) indicates the position of a nonspecific immunoreacting band. Hc, immunoglobulin heavy chain; IP, immunoprecipitate; Lc, immunoglobulin light chain; WT, wild type.
To exclude the possibility that the Flag tag may affect the overall folding of COI1, which in turn may affect the association of COI1 with AtCUL1 and ASK1, we also used the α-COI1 antiserum to immunoprecipitate endogenous COI1-containing complexes from wild-type Arabidopsis and found that AtCUL1, ASK1, and ASK2 were present in the α-COI1 immune complexes (data not shown). These results are consistent with the data generated from epitope-tagged COI1 transgenic plants.
COI1 Forms Two Complexes Separately with ASK1 or ASK2
Because the Arabidopsis SKP1-like proteins ASK1 and ASK2 were present in the immunoprecipitated SCFCOI1 complex, we investigated whether COI1 assembled two complexes separately with ASK1 or ASK2. We made transgenic plants expressing Myc-tagged versions of ASK1 or ASK2, the α-Myc antiserum was used for immunoprecipitation from the transgenic plants, and the immunoprecipitated complexes were analyzed with α-COI1, α-Myc, and α-ASK2 or α-ASK1 antisera.
The results shown in Figure 4B indicate that Myc-tagged ASK2 was coimmunoprecipitated with COI1 but not with ASK1. Conversely, Myc-tagged ASK1 was coimmunoprecipitated with COI1 but not with ASK2 (data not shown). The observed association of COI1 separately with Myc-tagged ASK1 and ASK2 demonstrates that COI1 assembles two separate SCFCOI1 complexes with ASK1 and ASK2 in Arabidopsis. It remains unclear whether the two SCFCOI1 complexes have redundant functions.
A Single Amino Acid Replacement of Glu-22 with Ala in COI1 Reveals Essential Roles for SCFCOI1
To investigate whether the formation of the SCFCOI1 complexes correlated with function in planta and to elucidate the consequences of the disruption of the SCFCOI1 complexes, we introduced a single amino acid substitution from Glu to Ala at amino acid 22 (COI1E22A) in the COI1 F-box motif that is required for its binding to Skp1-like proteins. COI1E22A was Myc tagged and expressed subsequently in the null mutant coi1-1 (referred as to coi1::Myc-COI1E22A). The coi1-1 plants transgenic for the Myc-tagged COI1 (coi1::Myc-COI1) or Myc vector (coi1::Myc) were generated as controls.
Protein gel blot analysis revealed that the Myc-tagged COI1E22A was expressed in coi1-1 at a similar level to the control, the Myc-tagged COI1. As expected, the Myc-tagged COI1 was coimmunoprecipitated with ASK1, ASK2, and AtCUL1 (Figure 4C), consistent with the results obtained from Flag-tagged COI1 transgenic plants (Figure 4A). The Myc-COI1E22A protein failed to associate with AtCUL1, ASK1, and ASK2 (Figure 4C), indicating that the E22A mutation in COI1 abrogates the SCFCOI1 complexes.
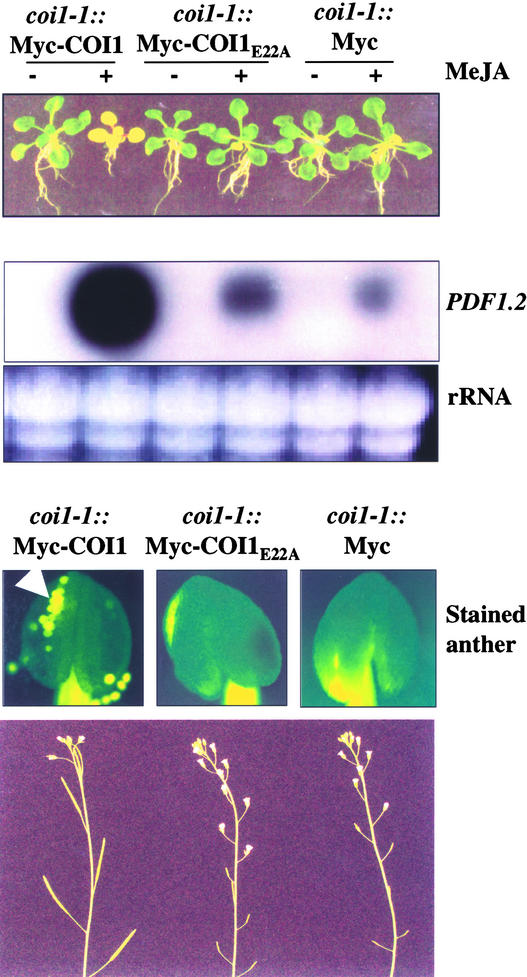
Functional analysis demonstrated that the COI1E22A mutation abolished JA response. The results shown in Figure 5 indicate that coi1::Myc-COI1E22A plants were resistant to JA, deficient in the production of viable pollen grains and seeds, and failed to express JA-inducible genes such as PDF1.2. In contrast, coi1::Myc-COI1 plants were fertile and exhibited normal induction of JA-inducible gene expression. The COI1E22A mutation disrupted the SCFCOI1 complexes and simultaneously abolished JA response, suggesting an essential role for the SCFCOI1 complexes in JA signaling.
Figure 5.
COI1E22A Abolishes JA Response.
coi1-1 plants transgenic for Myc-tagged COI1E22A, Myc-tagged COI1, or vector (see Figure 4C) were examined for JA-inhibitory root growth (top), JA-inducible expression of the defense gene PDF1.2 (middle), and JA-regulated pollen development and plant fertility (bottom). Seedlings were grown on Murashige and Skoog (1962) (MS) medium with (+) or without (−) 10 μM methyl jasmonate (MeJA; top). Total RNA was stained with ethidium bromide as an equal loading control (middle). The white arrow indicates the viable pollen grains in anther, which were stained with 1% fluorescein diacetate (catalog No. F-7378; Sigma) and visualized by fluorescence microscopy. The inflorescence was photographed in 6-week-old soil-grown plants (bottom).
AtRbx1 Is Required for the Normal Induction of JA-Inducible Gene Expression
To investigate JA response mediated by the AtCUL1, ASK1, and ASK2 components of SCFCOI1 complexes, T-DNA knockout mutants of AtCUL1 and ASK2 (W. Peng and D. Xie, unpublished data) and the Ds transposon mutant ask1-1 were examined. The null mutation in AtCUL1 caused early arrest during embryogenesis, and the homozygous mutant was lethal and unavailable for analysis of JA response (Shen et al., 2002; W. Peng and D. Xie, unpublished data). The ask1-1 or ask2 mutation did not obviously alter JA-inhibitory root growth and JA-induced gene expression (W. Peng and D. Xie, unpublished data), probably resulting from functional overlapping between ASK1 and ASK2.
We investigated the possible function of AtRbx1 in JA response through analysis of AtRbx1 double-stranded RNA interference (RNAi) mutant plants, generated by Genschik and colleagues (E. Lechner and P. Genschick, unpublished data), which contained a glucocorticoid-inducible AtRbx1 double-stranded RNA construct (Aoyama and Chua, 1997; Chuang and Meyerowitz, 2000; E. Lechner and P. Genschick, unpublished data).
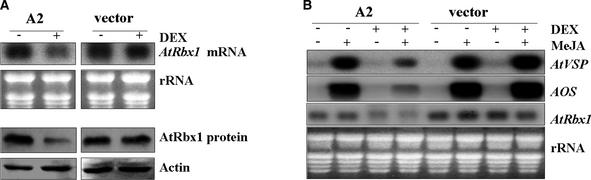
Figure 6A demonstrates that AtRbx1 double-stranded RNA–mediated genetic interference reduced AtRbx1 expression in AtRbx1(RNAi) mutant line A2. Dexamethasone (DEX; a strong synthetic glucocorticoid) treatment caused a decrease in AtRbx1 mRNA accumulation in glucocorticoid-inducible AtRbx1(RNAi) mutant plants. Protein gel blot analysis demonstrated that the decrease of AtRbx1 mRNA was correlated with a reduction of the AtRbx1 protein in DEX-treated AtRbx1(RNAi) mutant plants. As expected, AtRbx1 expression in control plants transgenic for a glucocorticoid-inducible vector was not affected by DEX treatment.
Figure 6.
Analysis of Glucocorticoid-Inducible AtRbx1(RNAi) Mutant Line A2.
(A) DEX treatment resulted in a decrease of both AtRbx1 mRNA and AtRbx1 protein in the glucocorticoid-inducible AtRbx1(RNAi) mutant line A2. Arabidopsis plants transgenic for the glucocorticoid-inducible AtRbx1(RNAi) construct (line A2) or vector were grown on MS medium for 3 weeks and treated with 0.5 μM DEX (+) (catalog No. D-1756; Sigma) or with water as a control (−) for 24 h. The plants were harvested for RNA gel blot analysis (top) and protein gel blot analysis (bottom). Total RNAs were stained with ethidium bromide as an equal loading control. The immunoblot was detected with α-actin antibody as a protein loading control.
(B) DEX treatment led to a reduction of JA-inducible accumulation of AtVSP and AOS transcripts. Three-week-old Arabidopsis plants were mock treated (−) or treated with 1 μM DEX for 24 h and then followed by mock treatment (−) or treatment with MeJA (+) for 9 h. The RNA gel blot was hybridized with digoxigenin-labeled AtVSP and AOS probes. Total RNAs were stained with ethidium bromide as an equal loading control. Consistent with the results shown in (A), the blot was probed with digoxigenin-labeled AtRbx1 to demonstrate that the DEX treatment led to a decrease of AtRbx1 expression in these glucocorticoid-inducible AtRbx1(RNAi) mutant plants.
The appearance of noninduced AtRbx1(RNAi) mutant plants (without DEX treatment) is normal. However, when germinated and grown continuously on medium containing DEX, AtRbx1(RNAi) mutant plants exhibited severe defects of growth and development (data not shown), which makes it difficult to examine the phenotype of JA-inhibitory root growth and JA-regulated fertility in these DEX-induced AtRbx1(RNAi) mutant plants. Therefore, we focused on analysis of the possible effect of AtRbx1 double-stranded RNA interference on JA-inducible gene expression.
AtRbx1(RNAi) mutant and empty vector control plants were grown for 3 weeks without DEX treatment. These 3-week-old plants then were treated with DEX (24 h) or JA (9 h) or both. Total RNAs were isolated from these plants, and JA-inducible genes, except for defense-related genes such as PDF1.2, induced by DEX in some transgenic plants containing a glucocorticoid-inducible vector (Kang et al., 1999), were used as probes in RNA gel blot analysis.
Results from AtRbx1(RNAi) mutant line A2 are shown in Figure 6B. Induction of AtVSP and allene oxide synthase (AOS) (Laudert and Weiler, 1998) by JA was normal in vector control and AtRbx1(RNAi) mutant plants not treated with DEX. However, in DEX-treated AtRbx1(RNAi) mutant plants, although these transcripts were still induced by JA, their accumulation declined in response to JA. Similar expression profiles were obtained for other JA-inducible genes, including lipoxygenase 2 (Bell and Mullet, 1993) (data not shown).
RNA gel blot analysis with AtRbx1 probe confirmed that AtRbx1 mRNA accumulation was decreased in DEX-treated AtRbx1(RNAi) mutant plants, which correlated with the observation that the accumulation of JA-inducible genes was reduced in these mutant plants. These results demonstrated that the reduction of AtRbx1 expression in DEX-induced AtRbx1(RNAi) mutants led to a severe decrease of JA-inducible gene expression, indicating a role for the AtRbx1 subunit of SCFCOI1 in JA response.
Mutations in AXR1 Affect Modification of the AtCUL1 Subunit of SCFCOI1 and Result in Reduced JA Response
The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1, and the formation of RUB-AtCUL1 is dependent on AXR1 and ECR1 in vivo (del Pozo et al., 1998, 2002; del Pozo and Estelle, 1999). Because we found that the AtCUL1 component of SCFCOI1 complexes was modified in vivo (Figure 4), we further demonstrated that the AtCUL1 subunit of SCFCOI1 was modified via AXR1 and that this modification was important for JA signaling.
Two mutant alleles, axr1-3 and axr1-12, of AXR1 were identified previously through a genetic screen for auxin-resistant mutants (Lincoln et al., 1990). To determine whether mutations in AXR1 affected modification of the AtCUL1 subunit of SCFCOI1, we generated transgenic axr1 plants expressing the Flag-tagged COI1 (axr1::Flag-COI1) and used the α-Flag antibody for immunoprecipitation. The α-Flag immunoprecipitates then were probed with α-AtCUL1, α-ASK1, and α-COI1 antisera.
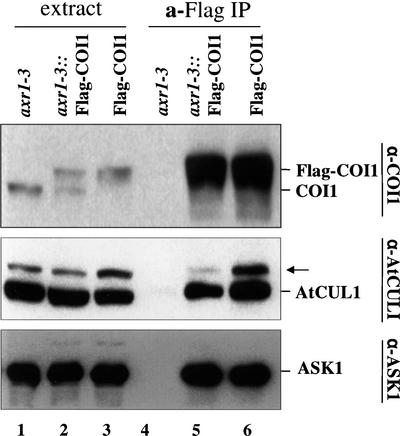
The results in Figure 7 show that a relatively low level of the modified AtCUL1 was detected in total proteins extracted from axr1-3 and axr1-3::Flag-COI1 plants (Figure 7, lanes 1 and 2) compared with that from AXR1::Flag-COI1 plants (Figure 7, lane 3). Quantitative analysis using a scanning densitometer showed that the abundance of the modified AtCUL1, normalized to the level of unmodified AtCUL1, in axr1-3 and axr1-3::Flag-COI1 was approximately one-third of that in AXR1::Flag-COI1 (data not shown). Figure 7 also demonstrates that Flag-COI1 was coimmunoprecipitated with ASK1 and AtCUL1 to assemble SCFCOI1 complexes in both axr1-3::Flag-COI1 and AXR1::Flag-COI1 (Figure 7, lanes 5 and 6). However, the modified AtCUL1 in the α-Flag immunoprecipitates from axr1-3::Flag-COI1 plants was reduced significantly compared with that from AXR1:: Flag-COI1 plants (Figure 7, lanes 5 and 6).
Figure 7.
The Abundance of the Modified AtCUL1 of SCFCOI1 Was Reduced in the axr1-3 Mutant.
Protein extracts from axr1-3 (lane 1), axr1-3 expressing Flag-tagged COI1 (axr1-3::Flag-COI1) (lane 2), and wild-type AXR1 plants expressing Flag-tagged COI1 (AXR1::Flag-COI1) (lane 3) were immunoprecipitated with α-Flag antibody (lanes 4 to 6). The α-Flag immune complexes were probed with α-COI1, α-AtCUL1, or α-ASK1 antisera. The arrow indicates the modified version of AtCUL1.
Previous studies have confirmed that RUB1 is activated by AXR1 and ECR1 for conjugation to AtCUL1 in vivo (del Pozo et al., 1998, 2002; del Pozo and Estelle, 1999). The observation that the abundance of the modified AtCUL1 of SCFCOI1 was decreased severely in axr1::Flag-COI1 demonstrates that the AtCUL1 subunit of SCFCOI1 was modified via AXR1, probably by RUB modification. A similar conclusion was reached from the results generated with transgenic axr1-12 plants expressing Flag-tagged COI1 (data not shown).
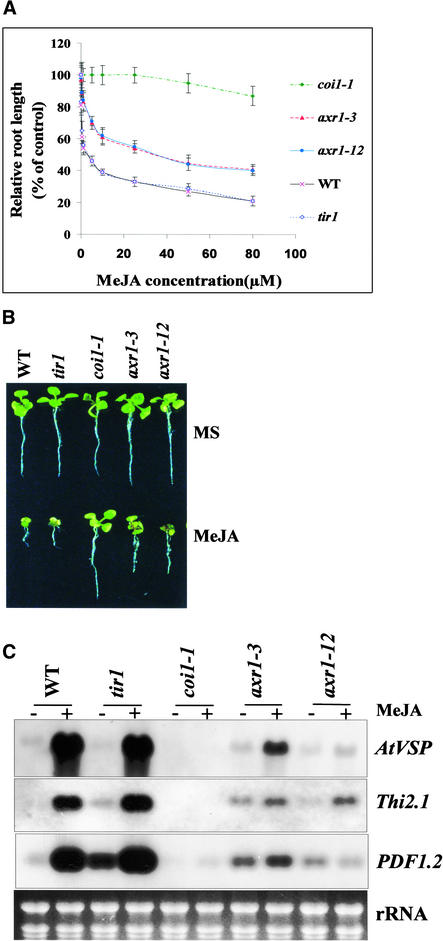
To further explore the possible role of AXR1-dependent modification of the AtCUL1 component of SCFCOI1 in JA signaling, we examined JA response in axr1-3 and axr1-12 compared with wild-type and JA-resistant mutant coi1-1 plants. The auxin transport inhibitor response mutant tir1-1, a mutant allele of TIR1 encoding an F-box protein required for auxin response (Gray et al., 1999), also was included as a control. The results shown in Figure 8 demonstrate that both axr1-3 and axr1-12 seedlings were moderately resistant to JA, and the JA inhibition rate for axr1-3 or axr1-12 was between that of the wild-type and that of coi1-1.
Figure 8.
axr1-3 and axr1-12 Exhibited Reduced JA Response.
(A) MeJA dose–response curve of root growth. axr1-3, axr1-12, tir1-1, coi1-1, and wild-type (WT) plants were grown for 1 week on MS medium containing various concentrations of MeJA. Root length of the seedlings grown on MS containing MeJA (0.1, 0.5, 1, 5, 10, 25, 50, or 80 μM) was expressed as a percentage of root length on MS medium. Each data point is the mean of >15 samples. The experiment was repeated four times, and the results were consistent. Error bars represent sd (n > 15).
(B) Phenotype of 10-day-old seedlings grown on MS medium or MS with 25 μM MeJA.
(C) RNA gel blot analysis of the JA-inducible expression of PDF1.2, Thi2.1, and AtVSP. Ethidium bromide staining of rRNAs shown at bottom indicates the equal loading amount of total RNA on the gel.
When seedlings were grown on MS medium containing 25 μM MeJA, ∼50% inhibition of root elongation was observed for axr1-3 and axr1-12, whereas ∼70% inhibition was observed for both the wild type and tir1-1 and no inhibition was observed for coi1-1. Furthermore, the induction of AtVSP, Thi2.1, and PDF1.2 by JA in the axr1-3 and axr1-12 mutants was reduced compared with that in the wild type and tir1. Collectively, the evidence that mutations in AXR1 reduced the abundance of the modified AtCUL1 subunit of SCFCOI1 and led to a reduction of JA response indicates that the AXR1-dependent modification of the AtCUL1 component of SCFCOI1 is important for JA signaling.
axr1 and coi1 Display a Synergistic Genetic Interaction
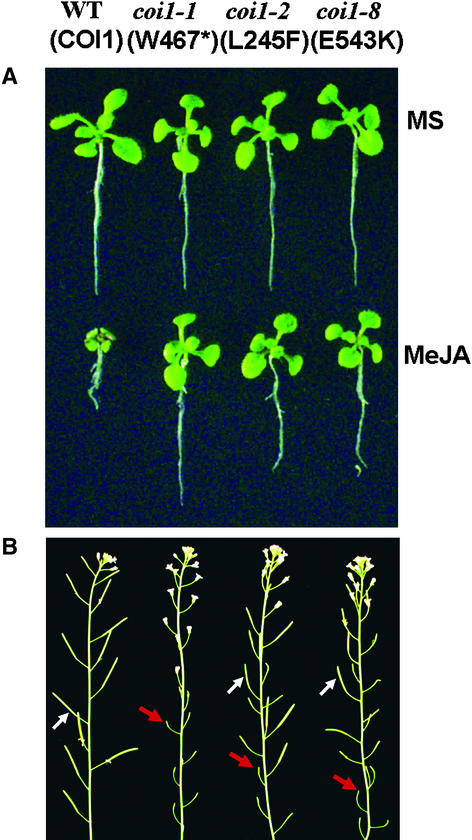
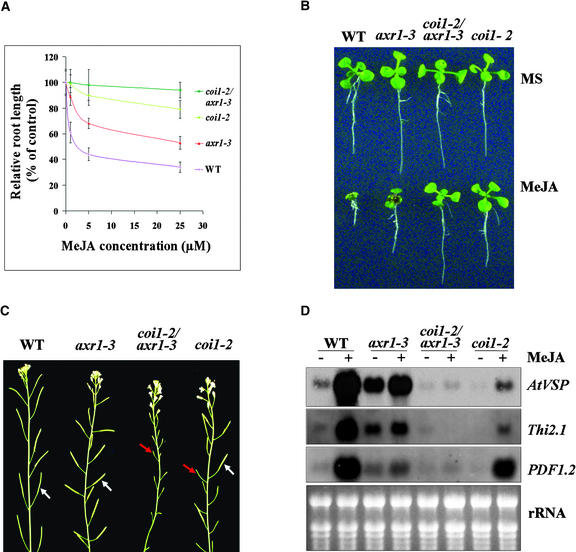
To conveniently explore the possible genetic interaction between AXR1 and COI1, we identified two leaky mutant alleles of COI1 (Figure 9): coi1-2, with missense mutation L245F, and coi1-8, with G543L, which exhibited reduced JA insensitivity and partial fertility. The null mutant coi1-1 was completely male sterile and resistant to JA. We generated coi1 axr1 double mutants homozygous for both coi1-2 and axr1-3 or axr1-12. The results shown in Figure 10 indicate that the coi1-2 axr1-3 double mutants displayed a significant reduction of JA response compared with their parent mutants coi1-2 and axr1-3.
Figure 9.
Identification of Leaky Mutant Alleles coi1-2 and coi1-8 of COI1.
(A) coi1-2 and coi1-8 exhibit insensitivity to JA-inhibitory root elongation compared with the wild type (WT), but it is less severe than that in coi1-1.
(B) coi1-2 and coi1-8 display a partial fertility phenotype, whereas the wild type is fertile and coi1-1 is male sterile. Shown are inflorescences of 8-week-old plants grown in soil under 16-h-light (21–23°C)/8-h-dark (16–19°C) growth conditions. The white arrows indicate fertile siliques, and the red arrows indicate sterile flowers.
Figure 10.
The coi1-2 axr1-3 Double Mutant Exhibited a Severe Defect in JA Response.
(A) MeJA dose–response curve of root growth. Root length of 1-week-old seedlings grown on MS medium containing 1, 5, or 25 μM MeJA is expressed as a percentage of root length on MS. Each data point represents the mean of >15 samples. The experiment was repeated three times. Error bars represent sd (n > 15).
(B) Phenotype of 8-day-old seedlings grown on MS or MS with 25 μM MeJA.
(C) The coi1-2 axr1-3 double mutation resulted in male sterility. Shown are inflorescences of 8-week-old plants. The white arrows indicate fertile siliques, and the red arrows indicate sterile flowers.
(D) The coi1-2 axr1-3 double mutation led to a significant reduction of JA-inducible accumulation of AtVSP, PDF1.2, and Thi2.1 transcripts. Ethidium bromide staining of rRNAs shown at bottom indicates the equal loading amount of total RNA on the gel.
WT, wild type.
The coi1-2 axr1-3 seedlings were fully resistant to JA, and no inhibition of root elongation was observed at 25 μM MeJA, whereas the single mutants coi1-2 and axr1-3 were partially resistant to JA: ∼20 and 50% inhibition on root elongation by 25 μM MeJA was observed for coi1-2 and axr1-3, respectively (Figures 10A and 10B). These results suggest that axr1-3 and coi1-2 display a synergistic genetic interaction in double mutants.
A similar conclusion was reached when the coi1-2 axr1-3 double mutants were examined for other JA responses, including JA-regulated fertility and inducible gene expression in response to JA. RNA gel blot analysis demonstrated that the inducible accumulation of AtVSP, PDF1.2, and Thi2.1 by JA was reduced significantly in the coi1 axr1 double mutants compared with that in the single mutants coi1-2 and axr1-3 (Figure 10D). Furthermore, a dramatic reduction in plant fertility was observed in the double mutants. The coi1-2 axr1-3 double mutation resulted in male sterility, whereas the single mutant axr1-3 produced normal seeds and coi1-2 was partially fertile (Figure 10C). The examination of double mutants homozygous for coi1-2 and axr1-12 reached the same conclusion, that the coi1 axr1 double mutation resulted in a more severe JA response defect than either of the single mutations (data not shown).
DISCUSSION
Xie et al. (1998) previously isolated the COI1 gene that is required for normal JA response, including JA-regulated plant fertility and defense. It was speculated that COI1 might be an F-box protein that recruits regulators of defense response and pollen development for modification by ubiquitylation (Xie et al., 1998). In this study, we showed biochemical and genetic evidence to demonstrate that COI1 associates physically with Arabidopsis AtCUL1, AtRbx1, and either of the SKP1 orthologs, ASK1 or ASK2, to assemble SCFCOI1 complexes in Arabidopsis.
The E22A mutation in the F-box motif of COI1 disrupted SCFCOI1 complexes and abolished the JA response. Downregulation of AtRbx1 led to a reduction of JA response in transgenic plants, as assayed for JA-inducible gene expression. In addition, we demonstrated that mutations in AXR1 decreased the abundance of the modified AtCUL1 component of SCFCOI1 and reduced JA response in planta. Furthermore, we found that the coi1 axr1 double mutations resulted in a severe defect of JA response. These results strongly suggest that the SCFCOI1 complexes are required for JA response and that the AXR1-dependent modification of the AtCUL1 subunit of SCFCOI1 is important for JA signaling in Arabidopsis.
Formation of the SCF Complex
The SCF ubiquitin-ligase complex is conserved structurally in yeast, Caenorhabditis elegans, fruit fly, and human, and the conserved Skp1, cullin (Cdc53), and Rbx1 (ROC1/Hrt1) proteins serve in multiple degradation pathways via the assembly of various SCF complexes involving different F-box proteins that recognize distinct target proteins and determine the specificity of protein ubiquitylation (Bai et al., 1996; Connelly and Hieter, 1996; Li and Johnston, 1997; Skowyra et al., 1997; Patton et al., 1998; Deshaies, 1999). In Arabidopsis, the cDNA and genome sequencing project revealed many putative F-box proteins (Xiao and Jang, 2000). Several F-box proteins, including FKF1/ZEITLUPE (Nelson et al., 2000; Somers et al., 2000), ORE9 (Woo et al., 2001), TIR1, COI1, and UFO have been shown to play major roles in various crucial biological processes.
AtCUL1 and the Skp1-like proteins ASK1 and ASK2 have been shown to interact with the F-box protein TIR1 to assemble the SCFTIR1 ubiquitin-ligase complex required for auxin response (Gray et al., 1999). ASK1 and ASK2 also have been shown, in the yeast two-hybrid system, to associate with UFO, which is required for normal patterning and growth in the floral meristem (Samach et al., 1999; Zhao et al., 1999). In this study, we demonstrate that AtCUL1, AtRbx1, ASK1, and ASK2 interact with COI1 in planta and that COI1 assembles two SCF complexes containing either ASK1 or ASK2. The two SCFCOI1 complexes may have different but overlapping functions in the JA signaling pathway.
Some F-box proteins, such as the yeast F-box proteins Pop1 and Pop2, form homodimers and heterodimers (Kominami et al., 1998). In this study, we found that COI1 functions as a monomer in the SCF complex. The results shown in Figure 7 (lane 5) demonstrated that COI1 was not present in Flag-COI1–containing SCF complexes that were coimmunoprecipitated by α-Flag antibody from the plants expressing both COI1 and the Flag-tagged COI1.
In F-box proteins, the F-box motif has been identified as the SKP1-interacting domain, and their variable protein–protein interaction domains (such as the Leu-rich repeat or WD40) function as substrate recognition domains (Bai et al., 1996; Connelly and Hieter, 1996; Li and Johnston, 1997; Schulman et al., 2000). Consistent with these conclusions, our results suggest that the F-box domain of COI1 is required for its association with AtCUL1 and the Skp1-like proteins. A single amino acid substitution of Glu-22 to Ala in the F-box domain of COI1 resulted in its dissociation with ASK1, ASK2, and AtCUL1 and abolished all JA response.
Function of SCF Components
It is envisaged that the Arabidopsis AtCUL1, AtRbx1, and ASK proteins are conserved subunits of various SCF complexes containing different F-box proteins, including COI1, TIR1, UFO, and others, that recruit different substrates and confer specificity on SCF complex function. It is speculated that mutations in one F-box protein would not affect the function mediated by other F-box proteins. However, mutations in these conserved components, including AtCUL1, AtRbx1, and ASK, would affect diverse phenotypes mediated by various SCF complexes. Because these SCF complexes may play crucial roles in plant growth and development, complete disruption of these SCF conserved components could result in a lethal phenotype or severe defects of plant growth and development.
Indeed, mutation in the F-box protein TIR1 led to a defect in the auxin response pathway (Ruegger et al., 1998) but not in the JA response pathway (Figure 8). Likewise, mutations in COI1 result in a defective JA response but have no effect on the TIR1-mediated auxin response (L. Xu and D. Xie, unpublished data).
In support of the speculation that mutations in the conserved components of SCF complexes would cause diverse and severe phenotypes, we found that mutation in AtCUL1 led to early arrest of embryogenesis (Shen et al., 2002; W. Peng and D. Xie, unpublished data). The mutation in ASK1, which has been shown to interact with UFO in the yeast two-hybrid system (Samach et al., 1999) and with TIR1 (Gray et al., 1999) and COI1 (Figure 4) in planta, caused diverse phenotypes. The ask1-1 mutation caused abnormality in floral morphology analogous to that caused by ufo (Zhao et al., 1999) and was resistant to auxin, like tir1 (Gray et al., 1999).
ask1-1 was male sterile and defective in male meiosis and failed to produce viable pollen grains (Yang et al., 1999), and the coi1-1 mutant also was male sterile (Feys et al., 1994; Xie et al., 1998) and failed to produce viable pollen grains. Surprisingly, the ask1 or ask2 mutation did not obviously alter JA response, as assayed for JA-inhibitory root growth and inducible gene expression (W. Peng and D. Xie, unpublished data). This finding might result from the functional redundancy of ASK1 and ASK2, correlated with our result that COI1 forms two SCFCOI1 complexes containing either ASK1 or ASK2. It would be interesting to examine the phenotype of ask1 ask2 double mutants.
Arabidopsis harbors two related AtRbx1 genes, AtRbx1a and AtRbx1b, that are highly identical to mammalian Rbx1/ROC1, particularly within internal and C-terminal regions (Figure 2). The α-Rbx1 antibody against the C-terminal region of human Rbx1 recognized Arabidopsis AtRbx1 but could not distinguish AtRbx1a and AtRbx1b. We demonstrated the association of COI1 with AtRbx1, but it remains unclear whether COI1 forms SCFCOI1 complexes with both or either AtRbx1a or AtRbx1b and whether both have distinctive or similar functions. Because of the high similarity between their sequences, AtRbx1 double-stranded RNA–mediated genetic interference probably would reduce the expression of both AtRbx1a and AtRbx1b.
Functional analysis of the glucocorticoid-inducible AtRbx1- (RNAi) mutant showed that AtRbx1 double-stranded RNA interference decreased AtRbx1 expression, and declining AtRbx1 expression is correlated with a reduction of JA-inducible gene expression, suggesting a role for AtRbx1 in JA signaling. The alteration of AtRbx1 expression also is correlated with other phenotypes, such as a decrease of some auxin-related phenotypes (E. Lechner and P. Genschik, unpublished results), indicating the possible association of AtRbx1 with other signaling complexes, including SCFTIR1.
AXR1-Dependent Modification of the AtCUL1 Subunit of SCFCOI1 Complexes
The ubiquitin-related proteins, including mammalian NEDD8 or the NEDD8 homologs of yeast and Arabidopsis (called RUB) (Kamitani et al., 1997; del Pozo et al., 1998; Lammer et al., 1998; Liakopoulos et al., 1998), and mammalian SUMO (Okura et al., 1996) and yeast Smt3 (Meluh and Koshland, 1995), were shown to modify other cellular proteins. In Arabidopsis, RUB1 is activated for conjugation to AtCUL1 via AXR1 and ECR1; this modification of AtCUL1 is required for the normal activity of SCFTIR1, and mutations in AXR1 lead to a defect of auxin response (del Pozo et al., 1998, 2002; del Pozo and Estelle, 1999; Gray and Estelle, 2000). The essentially antagonistic steps of RUB1 conjugation mediated by AXR1 and deconjugation promoted by the COP9 signalosome both are required for proper auxin response (Schwechheimer et al., 2001).
In this study, we demonstrate that the AtCUL1 subunit of SCFCOI1 complexes is modified and that this modification is required for a normal JA response. Mutations in AXR1 decreased the abundance of the modified AtCUL1 subunit of SCFCOI1 and led to a reduced JA response. The axr1 mutant is not completely deficient in RUB-AtCUL1 (del Pozo et al., 2002), because we also found that the relatively low level of the AtCUL1 subunit of SCFCOI1 still is modified in axr1 mutant plants (Figure 7, lane 5). The existing modification of the AtCUL1 subunit of SCFCOI1 in axr1 mutant plants may be caused by RUB conjugation independent of AXR1 and/or by conjugation of other Arabidopsis ubiquitin-related proteins. The low level of the modified AtCUL1 subunit of SCFCOI1 detected in the axr1 mutant may maintain moderate JA response.
Alternatively, the role of the RUB modification is to enhance the activity of SCFCOI1, as the NEDD8 pathway enhances E3 activity (Morimoto et al., 2000; Wu et al., 2000). Consistent with this suggestion, mutations in AXR1 were found to moderately but not severely reduce the SCFCOI1-mediated JA response, whereas the coi1-2 arx1-3 and coi1-2 axr1-12 double mutations significantly decreased the SCFCOI1-mediated JA response (Figure 10). It would be interesting to investigate whether other RUB modification–required components, such as ECR1 and RCE1 (del Pozo and Estelle, 1999), are involved in JA signaling.
Substrates of SCF Complexes
F-box proteins are the substrate recognition components of SCF complexes: they bind substrates through variable protein–protein interaction domains such as WD40 or Leu-rich repeats. In Arabidopsis, substrates for the SCFTIR1 complex were identified recently (Gray et al., 2001; Schwechheimer et al., 2001). SCFTIR1 recruits substrate auxin/indoleacetic acid proteins for degradation through the ubiquitin-proteasome pathway (Gray et al., 2001). SCFTIR1 also interacts with the COP9 signalosome, a multiple-protein complex that acts as a negative regulator of photomorphogenesis in Arabidopsis and that is required for efficient degradation of the target protein PSIAA6 (Schwechheimer et al., 2001).
The substrates for SCFCOI1 are unknown. Recently, several mutants (cev, cet, and cex) that confer constant expression of JA-inducible genes or that continuously activate the JA pathway were identified (Ellis and Turner, 2001; Hilpert et al., 2001; Xu et al., 2001). Some of these genes might encode negative regulators that are potential candidate substrates for SCFCOI1. The diverse phenotypes associated with coi1-1 suggest that SCFCOI1 might have multiple substrates. We propose that these multiple substrates could act as putative repressors to negatively regulate the expression of their downstream target genes that are involved in the JA response.
In response to JA, environmental, or developmental cues (such as wounding and flowering), the JA signal might be activated to modify some or all of the substrates, conceivably through phosphorylation or dephosphorylation. COI1 could recruit the modified substrates to SCFCOI1 for ubiquitylation. Subsequent degradation of the ubiquitylated substrates by the 26S proteasome would result in removal of the putative repressors, leading to the expression of the downstream genes that mediate the JA response. Identification and functional analysis of these substrates and the other potential factors involved in this signaling pathway will be essential to understanding the molecular mechanism by which JA regulates plant growth, development, and defense.
METHODS
Yeast Two-Hybrid Screen
Yeast (Saccharomyces cerevisiae) host strain AH109 (catalog No. K1612-1; Clontech, Palo Alto, CA) was cotransformed with the bait construct pGBKT7-COI1 (a Trp1 marker), and the GAL4-based Arabidopsis thaliana cDNA library was made from mRNAs isolated from Arabidopsis suspension cells, young seedlings, flowers, stems, roots, and leaves (in pGADT7, a Leu2 marker; a gift from N.H. Chua, Rockefeller University, New York, NY). Approximately 5 × 106 yeast transformants were screened on the selective medium Synthetic Dropout (SD)/-Ade/-His/-Leu/-Trp/X-α-Gal according to the manufacturer's instructions.
For the two-hybrid screen using pLexA-COI1 as bait, a LexA-based Arabidopsis cDNA library constructed from mRNAs isolated from methyl jasmonate-treated plants (in pB42AD; a gift of D.F. Huang) was transformed into yeast strain EGY48 according to the manufacturer's instructions (catalog No. K1609-1; Clontech). A total of 8 × 107 yeast transformants were selected on 2% Gal/1% raffinose/SD/-Ura/-His/-Trp/-Leu /X-β-Gal.
β-Galactosidase Assay
β-Galactosidase activities were measured using a modified method based on Samach et al. (1999). Yeast strains in the LexA-based two-hybrid system were grown in 2% Gal/1% raffinose/SD/-Ura/-His/-Trp. Yeast cells cultured in 2% Glc/SD/-Ura/-His/-Trp were used as a control.
Plasmid Constructs, Arabidopsis Transgenic Lines, and coi1 Mutant Alleles
The COI1E22A mutation was generated using the overlap PCR extension strategy (Vallejo et al., 1995) by introducing the desired amino acid substitution (from Glu to Ala at amino acid 22) into the specially designed mutagenizing complementary primers E22A-forward (5′-TTGATGATGTCATCGCGCAAGTCATGACCT-3′) and E22A-reverse (5′-AGGTCATGACTTGCGCGATGACATCATCAA-3′).
Six copies of the Myc epitope (MEQKLISEEDLNE) (Rupp et al., 1994; Turner and Weintraub, 1994) were cloned at the BamHI and SmaI sites into plasmid pROK2, which contains the 35S promoter of Cauliflower mosaic virus in the binary vector pBIN19, resulting in pMYC2. COI1E22A amplified by Pfu DNA polymerase (catalog No. 600,135; Stratagene) was fused in frame to the Myc epitope at the SmaI site in pMYC2, resulting in pMyc-COI1E22A. The coding region of ASK1, ASK2, or COI1 was amplified by PCR and fused in frame to the Myc epitope at the SmaI site in pMYC2, resulting in pMyc-ASK1, pMyc-ASK2, or pMyc-COI1.
To make Flag-tagged COI1, the Flag epitope was introduced into the specially designed primer with the Flag sequence (underlined) fused in frame to the COI1 5′ sequence starting from the ATG codon (5′-ATGGATTACAAGGATGATGATGATAAGATGGAGGATCCTGAT-ATCAAGAGGT-3′). The full length of COI1 was Flag tagged by PCR amplification and cloned into pROK2 at the SmaI site, resulting in pFlag-COI1.
The constructs described above were sequence verified and transferred into Arabidopsis plants by in planta Agrobacterium tumefaciens–mediated vacuum infiltration (Bechtold et al., 1993; Bent, 2000). Transgenic plants expressing epitope-tagged proteins at levels similar to those of their corresponding endogenous proteins were identified and used for further experiments.
AtRbx1 double-stranded RNA interference (RNAi) mutant plants were generated by E. Lechner and P. Genschick. A PCR-amplified BamHI-SpeI green fluorescent protein (GFP) fragment was first introduced into the pBluescript KS+ vector (Stratagene). The AtRbx1a coding region then was introduced at XhoI and EcoRI restriction sites in the antisense orientation upstream of GFP after PCR amplification with primers 5′-ATCTAGCTCGAGTGCGGTGGTAACCAAATGAAC-3′ and 5′-ACTAGTGAATTCGCGACACTAGACTCCGACGT-3′ and at SpeI and SacI restriction sites in the sense orientation downstream of GFP after PCR amplification with primers 5′-TGATCATCTAGAGCG-ACACTAGACTCCGACGT-3′ and 5′-TACGTTGAGCTCACTAGTTGC-GGTGGTAACCAAATGAAC-3′. Finally, the XhoI-SpeI DNA fragment containing the AtRbx1 double-stranded RNAi was subcloned into the glucocorticoid-inducible vector pTA7002 (Aoyama and Chua, 1997).
The leaky mutant alleles coi1-2 and coi1-8 of COI1 were identified from ethyl methanesulfonate–mutagenized Arabidopsis ecotype Columbia M2 seeds (stock No. M2E-02-03; Lehle Seeds, Round Rock, TX) using the method described by Staswick et al. (1992). Sequencing results revealed the missense mutations L245F in coi1-2 and G543L in coi1-8.
Antibodies
Polyclonal antibodies against the full length of COI1 and ASK1 were generated in rabbits. The antibodies against ASK2 and AtCUL1 have been described previously (Gray et al., 1999; Shen et al., 2002). The crude antisera were used at a dilution of 1:1000 except for anti-(α-)ASK2 antiserum, which was used at a dilution of 1:4000. The rabbit α-Rbx1 polyclonal antibody, raised against the synthetic peptide Cys-PLDNREWEFQKYGH, corresponding to the C terminus of human Rbx1, was purchased from BioSource (Camarillo, CA; catalog No. AHO0402).
The α-actin goat polyclonal antibody was purchased from Santa Cruz Biotechnology (catalog No. SC-1615; Santa Cruz, CA). The α-Myc monoclonal mouse antibody (9E10) and α-Myc affinity matrix were purchased from Berkeley Antibody Company (catalog No. MMS-150P; AFC-150P; Richmond, CA). The α-Flag monoclonal mouse antibody (M2) and the α-Flag affinity matrix were purchased from Sigma (catalog Nos. F3165 and A1205). The secondary antibodies (goat anti-rabbit IgG–horseradish peroxidase [HRP], rabbit anti-goat IgG-HRP, and goat anti-mouse IgG-HRP) were purchased from Pierce. The commercial antibodies were used as recommended by the suppliers.
Protein Gel Blot Analysis
Protein extracts were prepared by homogenizing Arabidopsis tissues in ice-cold extraction buffer (50 mM sodium phosphate, pH 7.0, 200 mM NaCl, 10 mM MgCl2, 0.2% β-mercaptoethanol, and 10% glycerol) supplemented with the protease inhibitor cocktail (catalog No. 1836170; Roche, Mannheim, Germany). Proteins were separated on SDS-PAGE and transferred onto Hybond enhanced chemiluminescence membranes (catalog No. RPN303D; Amersham). The blots were blocked in PBST (PBS plus 0.05% Tween 20) containing 5% nonfat milk for 2 h and incubated for an additional 2 h with primary antibody (the α-Rbx1 antibody was prepared in PBST with 2.5% nonfat milk). The blots were washed five times with PBST and then incubated with secondary antibody for 1 h. Immunoblotting bands were detected using the enhanced chemiluminescence system (catalog No. CK52563; Pierce).
Coimmunoprecipitation Assay
Extracts (1 mL) containing 5 to 10 mg of total proteins prepared from young seedlings (2 weeks old) and mature plants (flowers and leaves) were incubated with 50 to 200 μL of α-Flag or α-Myc affinity matrix for 4 h at 4°C with gentle rocking. The matrix was washed five times with the extraction buffer. The α-Flag matrix then was eluted with 0.1 M Gly, pH 3.0, and the α-Myc matrix was suspended in SDS-PAGE sample buffer and heated for 5 min at 100°C. The immunoprecipitates then were subjected to protein gel blot analysis with various antibodies.
RNA Gel Blot Analysis
The specific primers were designed based on their DNA sequences to PCR amplify RNA gel blot probes PDF1.2, Thi2.1, AtVSP, AOS, AtRbx1, and 18S rDNA. The fragments were labeled with the PCR digoxigenin probe synthesis kit according to the manufacturer's instructions (catalog No. 1-636-090; Roche). Plant RNA was isolated using Trizol regent (catalog No.15596; Gibco BRL). Ten to 20 μg of RNAs was separated on 1.5% formaldehyde agarose gel and transferred onto Hybond N+ membranes (catalog No. RNP203B; Amersham). Hybridization and detection were performed according to procedures that are standard for the digoxigenin system, as advised by the manufacturer (Roche).
Plant Growth Conditions and Treatment
Seeds were surface-sterilized, chilled at 4°C for 3 days, and then germinated and grown in a growth room under a 16-h-light (21–23°C)/8-h-dark (16–19°C) photoperiod. For jasmonate treatment experiments in RNA gel blotting, seedlings were grown on Murashige and Skoog (1962) medium with 2% Suc for 2 to 3 weeks and then treated with 100 μM methyl jasmonate (catalog No. 39270-7; Aldrich) or water for 8 h in daytime. For root length measurement experiments, seedlings were grown on Murashige and Skoog (1962) medium supplemented with various concentrations of methyl jasmonate for 1 week before measurement. To test plant fertility, seedlings were transferred into soil and grown under a 16-h-light (21–23°C)/8-h-dark (16–19°C) photoperiod.
Upon request, all novel material described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
The GenBank accession numbers for the sequences shown in Figure 2 are AAL13435 (AtRbx1a), CAB87200 (AtRbx1b), BAB83695 (C. elegans Rbx1), T13388 (Drosophila ROC1a), AAF32313 (Drosophila ROC1b), NP_062686 (mouse Rbx1), and NP_055063 (human Rbx1). Accession numbers of the sequences used as probes in RNA gel blot analysis are T04323 (PDF1.2), L41344 (Thi2.1), AY072506 (AtVSP), X92510 (AOS), AY052401 (AtRbx1), and X16077 (18S rDNA).
Acknowledgments
We thank Mark Estelle and William M. Gray (University of Texas at Austin) for tir1, axr1-3, and axr1-12 mutants and useful help and Nam-Hai Chua (Rockefeller University) for the glucocorticoid-inducible vector and the yeast two-hybrid libraries. This work was supported by the Singapore National Science and Technology Board. E.L. is supported by Action Concertee Incitive “Jeune Chercheur.” L.X. and D.H. are supported by the National Science Foundation of China and by the 863-High Tech program.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.003368.
References
- Aoyama, T., and Chua, N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Bai, C., Sen, P., Hofmann, K., Ma, L., Goebl, M., Harper, J.W., and Elledge, S.J. (1996). SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86, 263–274. [DOI] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Bell, E., and Mullet, J.E. (1993). Characterization of an Arabidopsis lipoxygenase gene responsive to methyl jasmonate and wounding. Plant Physiol. 103, 1133–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti, C.E., Xie, D., and Turner, J.G. (1995). COI1-dependent expression of an Arabidopsis vegetative storage protein in flowers and siliques and in response to coronatine or methyl jasmonate. Plant Physiol. 109, 567–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A.F. (2000). Arabidopsis in planta transformation: Uses, mechanisms, and prospects for transformation of other species. Plant Physiol. 124, 1540–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, S. (2001). Jasmonate-related mutants of Arabidopsis as tools for studying stress signaling. Planta 214, 497–504. [DOI] [PubMed] [Google Scholar]
- Berger, S., Bell, E., and Mullet, J.E. (1996). Two methyl jasmonate-insensitive mutants show altered expression of AtVSP in response to methyl jasmonate and wounding. Plant Physiol. 111, 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechert, S., Brodschelm, W., Hölder, S., Kammerer, L., Kutchan, T.M., Mueller, M.J., Xia, Z.Q., and Zenk, M.H. (1995). The octadecanoic pathway: Signal molecules for the regulation of secondary pathways. Proc. Natl. Acad. Sci. USA 92, 4099–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, C.-F., and Meyerowitz, E.M. (2000). Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97, 4985–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly, C., and Hieter, P. (1996). Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86, 275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 355–381. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., and Estelle, M. (1999). The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 96, 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., and Estelle, M. (2000). F-box proteins and protein degradation: A emerging theme in cellular regulation. Plant Mol. Biol. 44, 123–128. [DOI] [PubMed] [Google Scholar]
- del Pozo, J.C., Dharmasiri, S., Hellmann, H., Walker, L., Gray, W.M., and Estelle, M. (2002). AXR1-ECR1–dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell 14, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., Timpte, C., Tan, S., Callis, J., and Estelle, M. (1998). The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280, 1760–1763. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and cullin/ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Ellis, C., and Turner, J. (2001). The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13, 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, E.E. (2001). Surface-to-air signals. Nature 411, 854–856. [DOI] [PubMed] [Google Scholar]
- Feys, B.F., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., and Estelle, M. (2000). Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem. Sci. 25, 133–138. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of the Aux/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., Ostin, A., Sandberg, G., Romano, C.P., and Estelle, M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 7197–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko, A., and Ciechanover, A. (1998). The ubiquitin system. Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- Hilpert, B., Bohlmann, H., op den Camp, R., Przybyla, D., Miersch, O., Cuchala, A., and Apel, K. (2001). Isolation and characterization of signal transduction mutants of Arabidopsis thaliana that constitutively activate the octadecanoid pathway and form necrotic microlesions. Plant J. 26, 1–14. [DOI] [PubMed] [Google Scholar]
- Howe, G.A., Lightner, J., Browse, J., and Ryan, C.A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8, 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani, T., Kito, K., Nguyen, H.P., and Yeh, E.T.H. (1997). Characterization of NEDD8, a developmentally downregulated ubiquitin-like molecule. J. Biol. Chem. 272, 28557–28562. [DOI] [PubMed] [Google Scholar]
- Kamura, T., Koepp, D.M., Conrad, M.N., Skowyra, D., Moreland, R.J., Iliopoulos, O., Lane, W.S., Kaelin, W.G., Elledge, S.J., Conaway, R.C., Haper, J.W., and Conaway, J.W. (1999). Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 284, 657–661. [DOI] [PubMed] [Google Scholar]
- Kang, H.G., Fang, Y., and Singh, K.B. (1999). A glucocorticoid-inducible transcription system causes severe growth defects in Arabidopsis and induces defense-related genes. Plant J. 20, 127–133. [DOI] [PubMed] [Google Scholar]
- Kominami, K., Ochotorena, I., and Toda, T. (1998). Two F-box/WD-repeat protein Pop1 and Pop2 form hetero- and homo-complexes together with cullin-1 in the fission yeast SCF (Skp1-Cullin-1-Fox) ubiquitin ligase. Genes Cells 3, 721–735. [DOI] [PubMed] [Google Scholar]
- Lammer, D., Mathias, N., Laplaza, J.M., Jiang, W., Liu, Y., Callis, J., Goebl, M., and Estelle, M. (1998). Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12, 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudert, D., and Weiler, E.W. (1998). Allene oxide synthase: A major control point in Arabidopsis thaliana octadecanoid signalling. Plant J. 15, 675–684. [DOI] [PubMed] [Google Scholar]
- Li, F.N., and Johnston, M. (1997). Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: Coupling glucose sensing to gene expression and the cell cycle. EMBO J. 16, 5629–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos, D., Doenges, G., Matuschewski, K., and Jentsch, S. (1998). A novel protein modification pathway related to the ubiquitin system. EMBO J. 17, 2208–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the arx1 mutants of Arabidopsis. Plant Cell 2, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsuge, T., Zhou, C., Wolf, D.A., Wei, N., Shevchenko, A., and Deshiaes, R.J. (2001). Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., and Koshland, D. (1995). Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell 6, 793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, M., Nishida, T., Honda, R., and Yasuda, H. (2000). Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF (skp2) toward p27 (kip1). Biochem. Biophys. Res. Commun. 270, 1093–1096. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nelson, D.C., Lasswell, J., Rogg, L.E., Cohen, M.A., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331–334. [DOI] [PubMed] [Google Scholar]
- Ohta, T., Mickel, J.J., Schottelius, A.J., and Xiong, Y. (1999). ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell 3, 535–541. [DOI] [PubMed] [Google Scholar]
- Okura, T., Gong, L., Kamitani, T., Wada, T., Okura, I., Wei, C.F., Chang, H.M., and Yeh, E.T. (1996). Protection against Fas/APO-1- and tumor necrosis factor-mediated cell death by a novel protein, sentrin. J. Immunol. 15, 4277–4281. [PubMed] [Google Scholar]
- Osaka, F., Saeki, M., Katayama, S., Aida, N., Toh-e, A., Kominami, K., Toda, T., Suzuki, T., Chiba, T., Tanaka, K., and Kato, S. (2000). Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 19, 3475–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., Sa, D., Kuras, L., Thomas, D., Craig, K.L., and Tyers, M. (1998). Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box protein complexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 12, 692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A.M.A., Thomma, B.P.H.J., Buchala, A., Metraux, J.-P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, M., et al. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. [DOI] [PubMed] [Google Scholar]
- Podust, V.N., Brownell, J.E., Gladysheva, T.B., Luo, R.S., Wang, C., Coggins, M.B., Pierce, J.W., Lightcap, E.S., and Chau, V. (2000). A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc. Natl. Acad. Sci. USA 97, 4579–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porat, R., Lu, P., and O'Neil, S.D. (1998). Arabidopsis SKP1, a homologue of a cell cycle regulator gene, is predominantly expressed in meristematic cells. Planta 204, 345–351. [DOI] [PubMed] [Google Scholar]
- Reymond, P., and Farmer, E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. [DOI] [PubMed] [Google Scholar]
- Reymond, P., Weber, H., Damond, M., and Farmer, E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo, E., Titarenko, E., Leon, J., Berger, S., Vancanneyt, G., and Sanchez-Serrano, J.J. (1998). Reversible protein phosphorylation regulates jasmonic acid-dependent and -independent wound signal transduction pathways in Arabidopsis thaliana. Plant J. 13, 153–165. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp, R.A.W., Snider, L., and Weintraub, H. (1994). Xenopus embryos regulate the nuclear localization of XmyoD. Genes Dev. 8, 1311–1323. [DOI] [PubMed] [Google Scholar]
- Samach, A., Klenz, J.E., Kohalmi, S.E., Risseeuw, E., Haughn, G.W., and Crosby, W.L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20, 433–445. [DOI] [PubMed] [Google Scholar]
- Sanders, P.M., Lee, P.Y., Biesgen, C., Boone, J.D., Beals, T.P., Weiler, E.W., and Goldberg, R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12, 1041–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman, B.A., Carrano, A.C., Jeffrey, P.D., Bowen, Z., Kinnucan, E.R.E., Finnin, M.S., Elledge, S.J., Wade Harper, J., Pagano, M., and Pavletich, N.P. (2000). Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.W. (2001). Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Sembdner, G., and Parthier, B. (1993). The biochemistry and the physiological and molecular actions of jasmonates. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44, 569–589. [Google Scholar]
- Seol, J.H., et al. (1999). Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 13, 1614–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W.H., Parmentier, Y., Hellmann, H., Lechner, E., Dong, A., Masson, J., Granier, F., Lepiniec, L., Estelle, M., and Genschik, P. (2002). Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol. Biol. Cell 13, 1916–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skowyra, D., Craig, K.L., Tyers, M., Elledge, S.J., and Harper, J.W. (1997). F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91, 209–219. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329. [DOI] [PubMed] [Google Scholar]
- Staswick, P.E. (1992). Jasmonate, genes, and fragrant signals. Plant Physiol. 99, 804–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89, 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Yuen, G.Y., and Lehman, C.C. (1998). Jasmonate signaling mutants of Arabidopsis are susceptible to the soil fungus Pythium irregulare. Plant J. 15, 747–754. [DOI] [PubMed] [Google Scholar]
- Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97, 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi, A., Weber, H., Reymond, P., Browse, J., and Farmer, E.E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98, 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, P., Fuchs, S.Y., Chen, A., Wu, K., Gomez, C., Ronai, Z., and Panz, Q. (1999). Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Mol. Cell 3, 527–533. [DOI] [PubMed] [Google Scholar]
- Teng-umnuay, P., Morris, H.R., Dell, A., Panico, M., Paxton, T., and West, C.M. (1998). The cytoplasmic F-box binding protein SKP1 contains a novel pentasaccharide linked to hydroxyproline in Dictyostelium. J. Biol. Chem. 273, 18242–18249. [DOI] [PubMed] [Google Scholar]
- Teng-umnuay, P., van der Wel, H., and West, C.M. (1999). Identification of a UDP-GlcNAc:Skp1-hydroxyproline GlcNAc-transferase in the cytoplasm of Dictyostelium. J. Biol. Chem. 274, 36392–36402. [DOI] [PubMed] [Google Scholar]
- Turner, D.L., and Weintraub, H. (1994). Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev. 8, 1434–1447. [DOI] [PubMed] [Google Scholar]
- Vallejo, A.N., Pogulis, R.J., and Pease, L.R. (1995). PCR Primers: A Laboratory Manual, C.W. Dieffenbach and G.S. Dveksler, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 603–612.
- Wasternack, C., and Parthier, B. (1997). Jasmonate-signalled plant gene expression. Trends Plant Sci. 2, 302–307. [Google Scholar]
- Woo, H.R., Chung, K.M., Park, J.H., Oh, S.A., Ahn, T., Hong, S.H., Jang, S.K., and Nam, H.G. (2001). ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, K., Chen, A., and Pan, Z.Q. (2000). Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1-CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. 275, 32317–32324. [DOI] [PubMed] [Google Scholar]
- Xiao, W., and Jang, J. (2000). F-box proteins in Arabidopsis. Trends Plant Sci. 5, 454–457. [DOI] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu, L.H., Liu, F.Q., Wang, Z.L., Peng, W., Huang, R.F., Huang, D.F., and Xie, D.X. (2001). An Arabidopsis mutant cex1 exhibits constant accumulation of jasmonate-regulated AtVSP, Thi2.1 and PDF1.2. FEBS Lett. 494, 161–164. [DOI] [PubMed] [Google Scholar]
- Yang, M., Hu, Y., Lodhi, M., McCombie, W.R., and Ma, H. (1999). The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc. Natl. Acad. Sci. USA 96, 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, D., and Ma, H. (2000). Male fertility: A case of enzyme identity. Curr. Biol. 10, R904–R907. [DOI] [PubMed] [Google Scholar]
- Zhao, D., Yang, M., Solava, J., and Ma, H. (1999). The ASK1 gene regulates development and interacts with the UFO gene to control floral organ identity in Arabidopsis. Dev. Genet. 25, 209–223. [DOI] [PubMed] [Google Scholar]