Abstract
The expression of α-amylase genes in cereals is induced by both gibberellin (GA) and sugar starvation. All α-amylase genes isolated from cereals contain a TATCCA element or its variants at positions ∼90 to 150 bp upstream of the transcription start sites. The TATCCA element was shown previously to be an important component of the GA response complex and the sugar response complex of α-amylase gene promoters. In the present study, three cDNA clones encoding novel MYB proteins with single DNA binding domains were isolated from a rice suspension cell cDNA library and designated OsMYBS1, OsMYBS2, and OsMYBS3. Gel mobility shift experiments with OsMYBSs showed that they bind specifically to the TATCCA element in vitro. Yeast one-hybrid experiments demonstrated that OsMYBS1 and OsMYBS2 bind to the TATCCA element and transactivate a promoter containing the TATCCA element in vivo. Transient expression assays with barley half-seeds showed that OsMYBS1 and OsMYBS2 transactivate a promoter containing the TATCCA element when sugar is provided, whereas OsMYBS3 represses transcription of the same promoter under sugar starvation. Transient expression assays also showed that these three OsMYBSs cooperate with a GA-regulated transcription factor, HvMYBGa, in the transactivation of a low-pI barley α-amylase gene promoter in the absence of GA. Two-hybrid experiments with barley half-seeds showed that OsMYBS1 is able to form a homodimer. The present study demonstrates that differential DNA binding affinity, promoter transactivation ability, dimerization, and interactions with other protein factors determine the biological function of OsMYBSs. This study also suggests that common transcription factors are involved in the sugar and hormonal regulation of α-amylase gene expression in cereals.
INTRODUCTION
Sugar regulation of gene expression has profound effects at all stages of the plant life cycle, from germination and vegetative growth to reproductive development and seed formation. A variety of genes, whose products are involved in diverse metabolic pathways and cellular functions, are either induced or repressed by sugars (Koch, 1996). Although sugar signal transduction and regulation of gene expression are central control mechanisms that mediate energy homeostasis, carbohydrate distribution, and the growth and development of plants, our understanding of the mechanisms involved in sugar signal transduction and the regulation of gene expression in plants remains very limited.
Sugar regulation of α-amylase gene expression has been used as a model system for studying the molecular mechanisms that mediate sugar repression in plants (Yu, 1999a, 1999b). Expression of α-amylase genes in the embryos of germinating rice (Yu et al., 1996) and barley (Perata et al., 1997) seeds and in cultured rice suspension cells (Yu et al., 1991, 1992) is activated by sugar starvation and repressed by sugar provision. Sugar repression of α-amylase gene expression involves the control of both transcription rate and mRNA stability (Chan et al., 1994; Sheu et al., 1994, 1996; Chan and Yu, 1998a, 1998b).
A sugar response sequence (SRS) in the promoter of a highly sugar starvation–inducible rice α-amylase gene, αAmy3, has been shown to confer sugar responsiveness to a minimal promoter (Lu et al., 1998). This SRS contains three essential motifs: the GC box, the G box, and the TATCCA element, for a high level of sugar starvation–induced gene expression in rice protoplasts (Lu et al., 1998). All of the α-amylase genes isolated from rice, barley, and wheat contain a TATCCA element or its variants at positions ∼90 to 150 bp upstream of the transcription start sites (Yu, 1999a). Mutations of the TATCCA element in the promoters of the barley high-pI and low-pI α-amylase genes Amy-pHV19 (Gubler and Jacobsen, 1992) and Amy32b (Lanahan et al., 1992), respectively, decreased expression to ∼20% of the maximal level but maintained gibberellin (GA) responsiveness in barley aleurone cells.
The TATCCA element is duplicated in the rice αAmy3 promoter, and mutation of either of the duplicated TATCCA elements also reduced αAmy3 promoter activity to 12 and 8%, respectively, of that of the wild-type sequence, but sugar starvation inducibility in rice protoplasts was maintained (Lu et al., 1998). Because the TATCCA element is an important component of the GA response complex (GARC) and the sugar response complex of α-amylase genes, the regulation of α-amylase gene expression by hormone and sugar signals may share a common regulatory protein(s) that binds to the TATCCA element.
MYBs are a group of transcription factors with conserved DNA binding domains. The cellular myb protooncogene (c-myb) is involved in the proliferation and/or differentiation of hematopoietic cells (Graf, 1992). Common to all MYB gene products is a strongly conserved DNA binding domain located at the N-terminal end. In animals, this DNA binding domain consists of three imperfect repeats of ∼51 to 52 amino acids each (designated R1, R2, and R3) with three highly conserved Trp residues regularly spaced by 18 or 19 amino acids (Weston, 1999). In plants and yeast, the predominant MYBs have two repeats (R2 and R3) (Martin and Paz-Ares, 1997; Jin and Martin, 1999). However, MYBs that contain only one repeat (Baranowskij et al., 1994; Kirik and Bäumlein, 1996; Feldbrügge et al., 1997; Wang et al., 1997) or three repeats (Braun and Grotewold, 1999) also have been identified in plants.
The structural characteristics of all plant R2R3 MYBs are very similar to those of c-Myb (Solano et al., 1997). Studies with c-Myb indicate that R2 and R3 are sufficient for the recognition of specific DNA sequences (Sakura et al., 1989). Each repeat of the DNA binding domain contains three helices and folds into a helix-turn-helix motif, and the third helix in both R2 and R3 is a recognition helix (Frampton et al., 1991; Ogata et al., 1994). R2 and R3 intercalate in the major groove of target DNA, so the two recognition helices contact each other and bind to the specific base sequence cooperatively (Ogata et al., 1994).
Arabidopsis contains 125 R2R3 MYB genes (Stracke et al., 2001). The R2R3 MYBs have been studied extensively, and their biological functions have been assigned for the regulation of secondary metabolism, cellular morphogenesis, cell cycle, development, signal transduction, and disease resistance (Martin and Paz-Ares, 1997; Jin and Martin, 1999). Functions of the less-studied one-repeat (1R) MYBs were assigned for circadian and light regulation, cell differentiation, and telomeric DNA binding (Jin and Martin, 1999). A barley R2R3 MYB, HvMYBGa, whose expression is induced by GA, is postulated to be part of the GA-responsive pathway leading to GA-inducible gene expression during seed germination (Gubler et al., 1995).
The binding of nuclear proteins from rice suspension cells to the TATCCA element in a sequence-specific and sugar-dependent manner was demonstrated previously by gel mobility shift assay (Lu et al., 1998). In this report, we describe the isolation and characterization of three OsMYBS genes that encode MYBs with single DNA binding repeats. The three OsMYBSs interact with the TATCCA element in vitro and in vivo and possess differential DNA binding affinity and promoter activity in both yeast and plant cells. These OsMYBs were found to play essential roles in both the sugar and GA regulation of α-amylase gene expression.
RESULTS
Three OsMYBS Genes Encode MYBs with One DNA Binding Repeat
To obtain cDNAs encoding proteins that bind specifically to the TATCCA element, the protein gel blot method was used to screen a cDNA library constructed from poly(A)+ mRNA prepared from 4-h Suc-starved rice suspension cells. A 416-bp probe consisting of eight tandem repeats of the −133 to −82 DNA fragment of the αAmy3 promoter (Lu et al., 1998), which contains the TATCCA element, was used. cDNA clones that interacted specifically with the probe were isolated, and their sequences were analyzed.
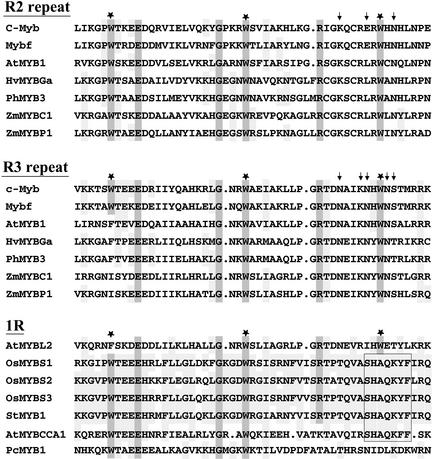
Three different genes were identified and designated OsMYBS1, OsMYBS2, and OsMYBS3. The open reading frames of OsMYBS1, OsMYBS2, and OsMYBS3 encode polypeptides of 306, 276, and 318 amino acid residues, respectively. The three OsMYBSs contain a highly conserved single DNA binding domain that is very similar to the DNA binding domains of other mammalian, Drosophila, and plant MYBs (Figure 1).
Figure 1.
Highly Conserved Amino Acid Residues Are Present among the R2, R3, and 1R Domains of the Plant MYBs.
The amino acid sequences of the R2 and R3 domains were derived from the DNA binding domains of the following sources: c-Myb from human (Majello et al., 1986); Mybf from Drosophila melanogaster (Katzen et al., 1985); AtMYB1 from Arabidopsis (Shinozaki et al., 1992); HvMYBGa from barley (Gubler et al., 1995); PhMYB3 from petunia (Solano et al., 1995); and ZmC1 and ZmP1 from maize (Paz-Ares et al., 1987; Grotewold et al., 1994). The amino acid sequences of the 1R domains were derived from the DNA binding domains of the following sources: AtMYBL2 and AtMYBCCA1 from Arabidopsis (Kirik and Bäumlein, 1996; Wang et al., 1997); StMYB1 from potato (Baranowskij et al., 1994); and PcMYB1 from parsley (Feldbrügge et al., 1997). The three regularly spaced Trp residues present in each repeat of the MYBs are labeled with asterisks. The amino acid sequences are aligned, and gaps (dots) have been introduced to maximize the alignment. The highly conserved amino acid residues in each repeat are highlighted in dark gray. The amino acid residues conserved within 1R and between 1R and the R2 and R3 repeats are highlighted in light gray. The positions corresponding to base-contacting residues of the human MYBs are marked with arrows. The highly conserved SHAQK(Y/F)F motifs in 1R are boxed.
Compared with other MYB proteins that contain a 1R in their DNA binding domains, the 1R residues of the three OsMYBSs are most closely related to MybSt1 (StMYB1) and are less closely related to AtMYBL2, PcMYB1, and CCA1 (AtMYBCCA1) (Figure 1). StMYB1 transactivates the 35S RNA gene of the Cauliflower mosaic virus (CaMV35S) promoter and is expressed in various organs of potato (Baranowskij et al., 1994). AtMYBL2 is expressed in Arabidopsis leaf, but its function has not been characterized (Kirik and Bäumlein, 1996). PcMYB1 interacts in vivo with a light-regulatory promoter unit and is expressed in parsley cell cultures and seedlings (Feldbrügge et al., 1997). AtMYBCCA1 binds to the promoter of an Arabidopsis light-harvesting chlorophyll a/b binding protein gene, Lhcb, and mediates the phytochrome regulation of Lhcb (Wang et al., 1997).
The 1R sequence of OsMYBS3 is most homologous with that of StMYB1 (92% identity) and is somewhat homologous with that of OsMYBS1 (87% identity) and OsMYBS2 (85% identity), and the 1R sequences of OsMYBS1 and OsMYBS2 are least homologous (77% identity) with each other. There is very low homology among the N- and C-terminal regions outside of the 1R regions of all of the MYBs with 1R DNA binding domains, except that OsMYBS3 and StMYB1 have 71% identity at the N-terminal 90–amino acid region and 62% identity at the C-terminal 70–amino acid region.
Comparison of amino acid residues between the 1R regions of the three OsMYBSs and the R2 and R3 repeats of the animal and plant MYBs shows that the 1R regions contain conserved Trp (W), Glu (E), Gly (G), and Arg (R) at the same positions (Figure 1). The Trp residue, which plays a critical role in stabilizing the DNA binding domain of animal MYBs (Ogata et al., 1992), is conserved at the first and second positions, but not at the third position, among most of the 1R regions. The putative base-contacting residues in the third helix of the animal MYBs are conserved in the plant R2R3 MYBs but not in the plant 1R MYBs (Figure 1).
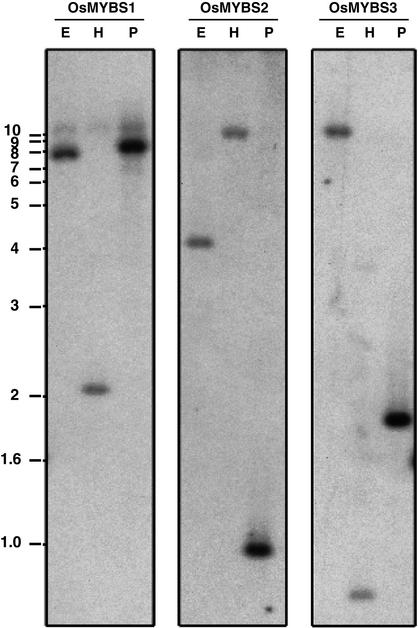
The 3′ untranslated regions of the three OsMYBS genes share very low homology (33 to 35% identity) and were used as gene-specific DNA probes for rice genomic DNA gel blot analysis. Only one band hybridized with each of the three gene-specific DNA probes (Figure 2). This result indicates that there is only one copy of each of the three OsMYBS genes in the rice MYB gene family.
Figure 2.
Each of the Three OsMYBS Genes Is a Single-Copy Gene in the Rice Genome.
Rice genomic DNA was digested with EcoRI (E), HindIII (H), and PstI (P) and subjected to gel blot analysis. OsMYBS1, OsMYBS2, or OsMYBS3 gene-specific DNA was used as a probe for hybridization of each of the three blots, which were prepared in parallel. Molecular mass markers (in kb) are shown at left.
OsMYBS Genes Are Expressed in Various Rice Tissues and Barley Aleurone Cells
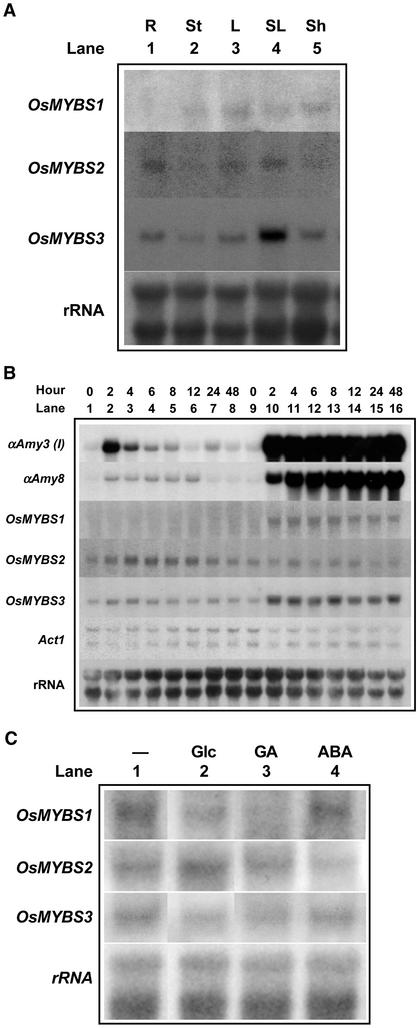
The expression patterns of OsMYBS in rice were analyzed. Total RNA was purified from various rice tissues and subjected to gel blot analysis using OsMYBS-specific DNA as a probe. Figure 3A shows that OsMYBS1 was expressed in aboveground tissues, with the highest level in leaves. OsMYBS2 was expressed mainly in roots, leaves, and senescent leaves at similar levels, and OsMYBS3 was expressed in all tissues, with the highest level in senescent leaves.
Figure 3.
OsMYBS Genes Are Expressed in Various Rice Tissues and Barley Aleurone Cells.
Roots (R), stems (St), leaves (L), senescent leaves (SL), and leaf sheaths (Sh) were collected from 2-month-old rice plants. Rice suspension cells were cultured in medium containing Suc or lacking Suc for various lengths of time. Barley embryoless half-seeds were allowed to imbibe in water for 4 days, treated with buffer (20 mM each CaCl2 and sodium succinate, pH 5.0) without Glc (−) or with 0.3 M Glc, 1 μM GA, or 20 μM abscisic acid (ABA) for 20 h. Total RNA was purified from rice tissues, cultured cells, or barley half-seeds and subjected to gel blot analysis using αAmy3, αAmy8, OsMYBS1, OsMYBS2, and OsMYBS3 gene-specific DNA, Act1 cDNA, and rDNA as probes.
(A) Expression of OsMYBS in rice tissues.
(B) Expression of OsMYBS in cultured rice cells.
(C) Expression of OsMYBS homologs in barley aleurone cells.
To determine whether OsMYBS gene expression was regulated by sugars, total RNA was isolated from rice suspension cells cultured in the presence or absence of Suc for 48 h and subjected to gel blot analysis using OsMYBS-specific DNAs as probes. Figure 3B shows that the levels of OsMYBS1 and OsMYBS3 mRNAs were low in the presence of Suc (lanes 1 to 8) but increased in the absence of Suc (lanes 9 to 16), which were parallel to the mRNA levels of αAmy3 and another rice α-amylase gene, αAmy8. By contrast, the levels of OsMYBS2 were higher in the presence than in the absence of Suc.
Transient expression assays with barley half-seeds were used as a system to study the function of OsMYBSs in sugar and hormone regulation in later experiments. To determine whether OsMYBSs are expressed in barley aleurone cells, total RNA was isolated from barley half-seeds and subjected to gel blot analysis using OsMYBS-specific DNAs as probes. Figure 3C shows that all of these OsMYBSs were expressed in barley aleurone cells in the absence of Glc. The expression levels of OsMYBS1 and OsMYBS3 were suppressed in the presence of Glc or GA but remained unchanged in the presence of abscisic acid. The expression of OsMYBS2 was enhanced in the presence of Glc and slightly suppressed by abscisic acid treatment.
OsMYBSs Interact Specifically with the TATCCA Element in Vitro
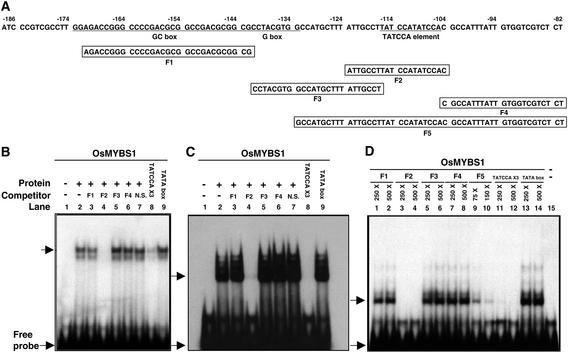
To determine whether the three OsMYBSs bind specifically to the TATCCA element, they were expressed in Escherichia coli. Interaction between affinity-purified recombinant OsMYBS and five DNA fragments encompassing the −171 to −82 region of the αAmy3 SRS was assayed by gel mobility shift assay (Figure 4). The five DNA fragments, designated F1 through F5, were used previously in gel mobility shift assays for their interaction with nuclear protein extract from rice suspension cells (Lu et al., 1998). F2, which contains the duplicated TATCCA element with some flanking sequence in SRS (Figure 4A), was used as the probe for gel mobility shift assays.
Figure 4.
The Three OsMYBSs Bind Specifically to the TATCCA Element in Vitro.
(A) Sequences and positions of oligonucleotides F1 through F5 within the SRS that were used for gel mobility shift assay.
(B) Interaction between recombinant OsMYBS1 and γ-32P-labeled F2.
(C) Interaction between recombinant OsMYBS2 and γ-32P-labeled F2.
(D) Interaction between recombinant OsMYBS3 and γ-32P-labeled F2.
The unlabeled oligonucleotides used for competitive binding in (B) and (C) were in 500-fold excess (w/w), and those in (D) were in 250- to 500-fold excess (w/w), of the F2 probe. The sequence of DNA fragment containing the TATA box is 5′-GTTCTATATATGCCCCC-3′ (TATA box underlined). The sequence of 3xTATCCA and 6xTATCCA is 5′-TATCCATATCCATATCCATATCCATATCCATATCCA-3′. Arrows indicate the positions of the DNA–protein complexes and free probes.
Figures 4B through 4D show that the three recombinant OsMYBSs bound to probe F2, resulting in the formation of low-mobility complexes. The interaction between OsMYBS1 or OsMYBS2 and F2 was competed only by F2 and the DNA fragment containing six tandem repeats of the 6-bp TATCCA element but not by 500-fold excess of other DNA fragments (Figures 4B and 4C, respectively). The interaction between OsMYBS3 and F2 also was competed only by F2, by F5, which contains the TATCCA element, and by the DNA fragment containing six tandem repeats of the 6-bp TATCCA element (Figure 4D). A 17-bp DNA fragment containing the TATA box sequence (TATATA), which differs from the TATCCA element by only two nucleotides, did not compete for binding (Figures 4B and 4C).
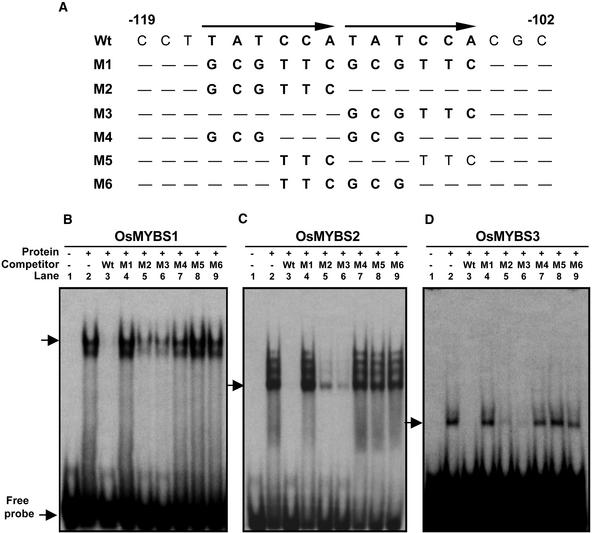
To further verify the specificity of binding between OsMYBSs and the TATCCA element, a series of site-directed mutations spanning region −119 to −102 containing the TATCCA tandem repeat were constructed (Figure 5A). The wild-type sequence was used as the probe for gel mobility shift assays. Figures 5B through 5D show that the three recombinant OsMYBSs bound to the wild-type probe, resulting in the formation of low-mobility complexes (lane 2). Figures 5B through 5D also show that the binding of each of these OsMYBSs was completely competed by unlabeled wild-type sequence.
Figure 5.
The Three OsMYBSs Bind Specifically to Both TATCCA Repeats with Different Sequence Binding Preferences.
(A) Sequences of oligonucleotides wild type (Wt) and M1 through M6. Mutated sequences are presented in boldface. Sequences identical to the wild-type sequence are indicated with dashes. Arrows indicate direct repeats of TATCCA.
(B) Interaction between recombinant OsMYBS1 and γ-32P-labeled wild type.
(C) Interaction between recombinant OsMYBS2 and γ-32P-labeled wild type.
(D) Interaction between recombinant OsMYBS3 and γ-32P-labeled wild type.
The unlabeled oligonucleotides used for competitive binding were in 500-fold excess (w/w) of the wild-type probe. Arrows indicate the positions of the DNA–protein complexes and free probes.
The DNA sequence with mutations throughout both repeats (M1) failed to compete, and sequences with either of the repeats replaced (M2 and M3) competed less effectively. Mutation of three of the six bases in both of these repeats (M4, M5, and M6) led to only slight competition. These results suggest that all of these OsMYBSs bind specifically to two copies of the TATCCA element. However, the binding affinity decreases if only one copy of TATCCA exists.
OsMYBSs Interact with the TATCCA Element in Vivo
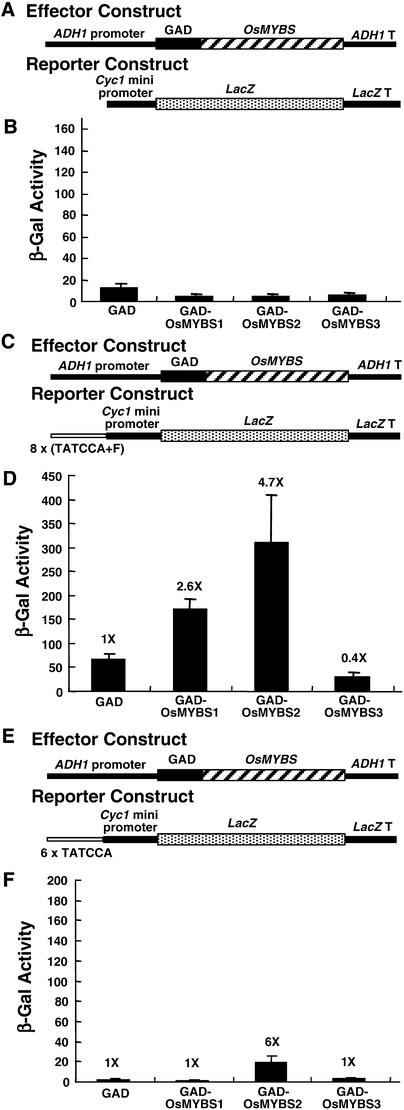
The in vivo binding activity of OsMYBSs to the TATCCA element was assayed using a yeast one-hybrid system. The full-length OsMYBS sequences were fused individually at the C terminus with the activation domain of the yeast GAL4 transcriptional activator (GAD). The GAD-OsMYBS chimeric gene then was fused downstream of the yeast ADH1 promoter and used as an effector construct (Figure 6A). A yeast strain containing a Cyc1 minimal promoter–LacZ fusion reporter construct then was transformed with the effector construct (Figure 6A). The presence of GAD alone or together with the GAD-OsMYBS fusion protein did not lead to the expression of LacZ (Figure 6B).
Figure 6.
All Three OsMYBSs Bind to the TATCCA Element in Vivo.
OsMYBS binding activity assay using the yeast one-hybrid system.
(A) The ADH1-GAD-OsMYBS chimeric gene was used as the effector construct. The Cyc1-LacZ chimeric gene was used as the reporter construct.
(B) β-Galactosidase activity in a yeast strain transformed with the ADH1-GAD-OsMYBS and Cyc1-LacZ constructs.
(C) The ADH1-GAD-OsMYBS chimeric gene was used as the effector construct. The 8x(TATCCA+F)-Cyc1-LacZ chimeric gene was used as the reporter construct.
(D) β-Galactosidase activity in a yeast strain transformed with the ADH1-GAD-OsMYBS and 8x(TATCCA+F)-Cyc1-LacZ constructs.
(E) The ADH1-GAD-OsMYBS chimeric gene was used as the effector construct. The 6xTATCCA-Cyc1-LacZ chimeric gene was used as the reporter construct.
(F) β-Galactosidase activity in a yeast strain transformed with the ADH1-GAD-OsMYBS and 6xTATCCA-Cyc1-LacZ constructs.
In (D) and (F), the value of β-galactosidase activity in yeast cells in the absence of OsMYBS was assigned a value of 1×, and the other values were calculated relative to this value. The experiments shown in (B) and (D) were performed simultaneously.
Eight tandem repeats of the −133 to −82 DNA sequence of αAmy3, designated 8x(TATCCA+F) (Table 1), were fused upstream of the Cyc1 minimal promoter–LacZ coding region and served as a reporter construct (Figure 6C). The reporter construct was delivered into yeast first, and the yeast was transformed with the effector constructs containing the ADH1-GAD-OsMYBS fusion gene (Figure 6C). The presence of GAD alone increased LacZ expression fivefold (cf. Figures 6B and 6D), indicating that the TATCCA element with flanking sequences slightly enhanced the Cyc1 minimal promoter activity even in the absence of OsMYBS. The presence of GAD-OsMYBS1 and GAD-OsMYBS2 increased LacZ expression threefold and fivefold, respectively, compared with LacZ expression in the absence of OsMYBS (Figure 6D). By contrast, the presence of OsMYBS3 reduced LacZ expression twofold (Figure 6D).
Table 1.
Sequence of the αAmy3 Promoter Used for Promoter Construction
| −186 | |
|---|---|
| SRS | ATCCCGTCGCCTTGGAGACCGGGCCCCGACGCGGCCGACGCGGCGCCTACGTGGCCATGCTTTATTGCCT |
| GC box G-box | |
| −116 −105 −82 | |
| TATCCATATCCACGCCATTTATTGTGGTCGTCTCT | |
| TATCCA element | |
| −134 −116 −105 −82 | |
| 8x(TATCCA+F) | Eight tandem repeats of GGCCATGCTTTATTGCCT TATCCATATCCACGCCATTTATTGTGGTCGTCTCT |
| TATCCA element | |
| −134 −82 | |
| 6x(TATCCA+F) | Six tandem repeats of GGCCATGCTTTATTGCCT TATCCATATCCACGCCATTTATTGTGGTCGTCTCT |
| TATCCA element | |
| −116 −105 | |
| 6xTATCCA | Three tandem repeats ofTATCCATATCCA |
| TATCCA element |
To determine whether the flanking sequence of the TATCCA element affects the binding affinity of OsMYBS1 and OsMYBS2, six tandem repeats of the 6-bp TATCCA element, designated 6xTATCCA (Table 1), were fused upstream of the Cyc1 minimal promoter–LacZ coding region and served as a reporter construct (Figure 6E). The reporter construct was delivered into yeast, and the yeast was transformed with the effector construct that contained the ADH1-GAD-OsMYBS fusion gene (Figure 6E). Only the presence of GAD-OsMYBS2 increased LacZ expression (Figure 6F).
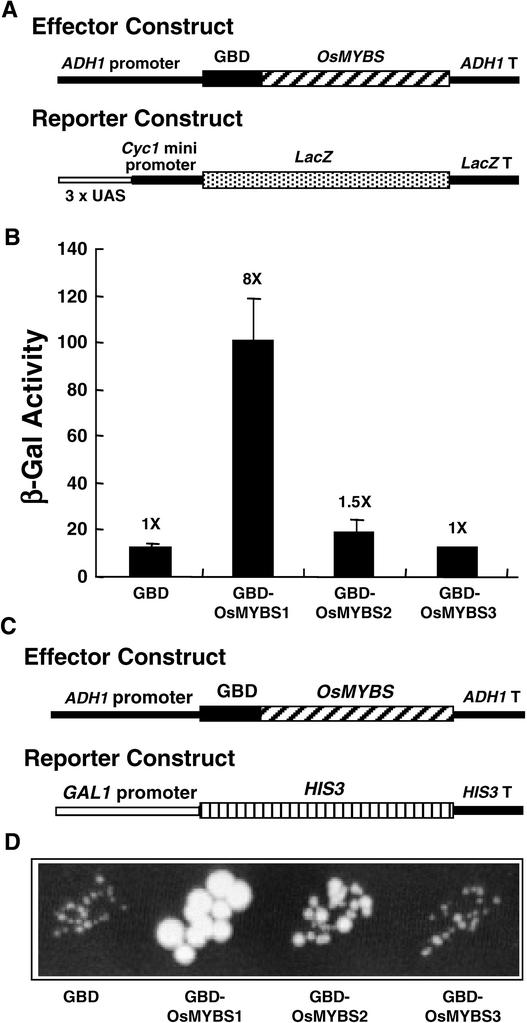
OsMYBS1 and OsMYBS2 Are Transcriptional Activators
To determine whether OsMYBS is a transcriptional activator, the full-length OsMYBS sequence was fused at the C terminus of the DNA binding domain of GAL4 (GBD). The chimeric gene then was fused downstream of the yeast ADH1 promoter and used as an effector construct (Figure 7A). A yeast strain hosting the 3xUAS–Cyc1 minimal promoter–lacZ fusion reporter construct (Figure 7A) was transformed with the effector construct. The presence of the GBD-OsMYBS1 fusion protein increased LacZ expression eightfold, GBD-OsMYBS2 increased LacZ expression only slightly, and GBD-OsMYBS3 did not increase LacZ expression (Figure 7B).
Figure 7.
OsMYBS1 and OsMYBS2 Are Transcriptional Activators in Yeast.
OsMYBS transcriptional activity assay using the yeast one-hybrid system.
(A) The ADH1-GBD-OsMYBS chimeric gene was used as the effector construct. The 3xUAS-Cyc1-LacZ chimeric gene was used as the reporter construct.
(B) β-Galactosidase activity in a yeast strain transformed with the ADH1-GBD-OsMYBS and 3xUAS-Cyc1-LacZ constructs.
(C) The ADH1-GBD-OsMYBS chimeric gene was used as the effector construct. The GAL1-HIS3 chimeric gene was used as the reporter construct.
(D) Growth of yeast cells in a medium lacking His in the presence of 3-aminotriazole.
The value of β-galactosidase activity in yeast cells in the absence of OsMYBS was assigned a value of 1×, and the other values were calculated relative to this value.
A yeast strain harboring the GAL1 promoter–HIS3 fusion reporter construct also was transformed with the effector construct that contained the ADH1-GBD-OsMYBS fusion gene (Figure 7C). The presence of the GBD-OsMYBS1 fusion protein allowed yeast cells to grow well on medium lacking His in the presence of 3-aminotriazole (Figure 7D). The GBD-OsMYBS2 fusion protein allowed yeast cells to grow slowly, but the GBD-OsMYBS3 fusion protein did not allow yeast cells to grow on the selective medium (Figure 7D). These results suggest that OsMYBS1 is a strong transcriptional activator, OsMYBS2 is a weak transcriptional activator, and OsMYBS3 is not a transcriptional activator in yeast.
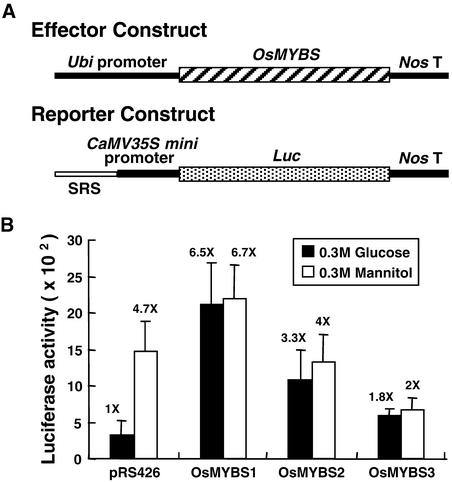
OsMYBS1 and OsMYBS2 Derepress the Sugar-Repressed Transcription of a Promoter Containing SRS
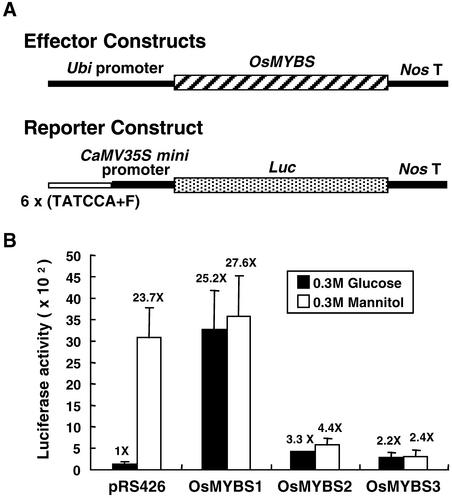
To determine the function of OsMYBS in the sugar regulation of α-amylase gene expression in plant cells, the transactivation ability of OsMYBS on SRS (Table 1) fused to the CaMV35S minimal promoter (−46 bp upstream of the transcription start site), forming a SRS-CaMV35S chimeric promoter, was analyzed in barley aleurone cells, which yield more consistent results than rice protoplasts. The three OsMYBS cDNAs were fused individually downstream of the maize ubiquitin gene (Ubi) promoter, forming Ubi-OsMYBS chimeric genes, which served as effectors (Figure 8A). The coding region of the luciferase gene (Luc) was fused downstream of the SRS-CaMV35S chimeric promoter and served as a reporter (Figure 8A).
Figure 8.
OsMYBS1 and OsMYBS2 Derepress the Sugar-Repressed Transcription of a Promoter Containing SRS.
Barley aleurone cells were transfected with plasmids and incubated with 300 mM each Glc or mannitol, and luciferase and GUS activity were assayed as described in Methods.
(A) The Ubi-OsMYBS fusion gene was used as the effector construct. The SRS-CaMV35S-Luc fusion gene was used as the reporter construct.
(B) Luciferase activity in barley aleurone cells after cotransfection with the effector, reporter, and control plasmids. pUGI served as the internal control plasmid. The amount of effector, reporter, and control plasmid DNA was at a ratio of 1:1:0.25.
The value of β-galactosidase activity in barley aleurone cells bombarded with pRS426 in the presence of Glc was assigned a value of 1×, and the other values were calculated relative to this value.
The barley half-seeds were particle bombarded simultaneously with the effector and reporter plasmids. The bombarded half-seeds were divided into two halves, each half was incubated with 0.3 M Glc or 0.3 M mannitol for 24 h, and luciferase activity was determined. Figure 8B shows that luciferase activity was higher in the absence of Glc when the half-seeds were cobombarded with the reporter plasmid and plasmid pRS426, an unrelated yeast plasmid with a molecular mass similar to that of the effector plasmid, which was used as a negative control. This result is consistent with our previous finding that the SRS-CaMV35S chimeric promoter was activated in rice protoplasts in the absence of Glc and was repressed in the presence of Glc (Lu et al., 1998).
When the half-seeds were cobombarded with the reporter and effector plasmids, OsMYBS1 significantly enhanced luciferase activity in the presence of Glc to a level similar to that in the absence of Glc. OsMYBS1 also enhanced luciferase activity in the absence of Glc, but to a lesser extent. OsMYBS2 enhanced luciferase activity in the presence of Glc but did not affect the activity in the absence of Glc. OsMYBS3 did not significantly enhance luciferase activity in the presence of Glc and repressed luciferase activity in the absence of Glc. These results indicate that OsMYBS1 and OsMYBS2 function as a strong and a weak activator, respectively, and that OsMYBS3 may serve as a repressor, for transcription of the promoter containing SRS.
OsMYBS1 Derepresses the Sugar-Repressed Transcription of a Promoter Containing the TATCCA Element with Adjacent Flanking Sequences
To determine whether OsMYBS transactivates a promoter containing only the TATCCA element and adjacent flanking sequences, a DNA fragment containing six tandem repeats of the −133 to −82 sequence of SRS, designated 6x(TATCCA+F) (Table 1), was fused to the CaMV35S minimal promoter. A plasmid containing the Ubi-OsMYBS construct served as an effector (Figure 9A), and Luc cDNA fused downstream of the 6x(TATCCA+F)-CaMV35S promoter served as a reporter (Figure 9A).
Figure 9.
OsMYBS1 Derepresses the Sugar-Repressed Transcription of a Promoter Containing the TATCCA Element with Adjacent Flanking Sequences.
Barley aleurone cells were transfected with plasmids and incubated with 300 mM each Glc or mannitol, and luciferase and GUS activities were assayed.
(A) The Ubi-OsMYBS fusion gene was used as the effector construct. The 6x(TATCCA+F)-CaMV35S-Luc fusion gene was used as the reporter construct.
(B) Luciferase activity in barley aleurone cells after cotransfection with the effector, reporter, and control plasmids. pUGI served as the internal control plasmid. The amount of effector, reporter, and control plasmid DNA was at a ratio of 1:1:0.25.
The value of β-galactosidase activity in barley aleurone cells bombarded with pRS426 in the presence of Glc was assigned a value of 1×, and the other values were calculated relative to this value.
The barley half-seeds were cobombarded with the effector and reporter plasmids. The bombarded half-seeds were divided into two halves, each half was incubated with 0.3 M Glc or 0.3 M mannitol for 24 h, and luciferase activity was determined. Figure 9B shows that OsMYBS1 significantly enhanced luciferase activity in the presence of Glc to the same level as in the absence of Glc. Although OsMYBS2 and OsMYBS3 seemed to enhance luciferase expression slightly in the presence of Glc, they significantly suppressed luciferase activity in the absence of Glc. These results indicate that OsMYBS1 activates, whereas OsMYBS2 and OsMYBS3 repress, the transcription of a promoter containing only the TATCCA element and adjacent flanking sequences.
OsMYBSs Cooperate with HvMYBGa in the Transactivation of a Barley Amy32b Promoter
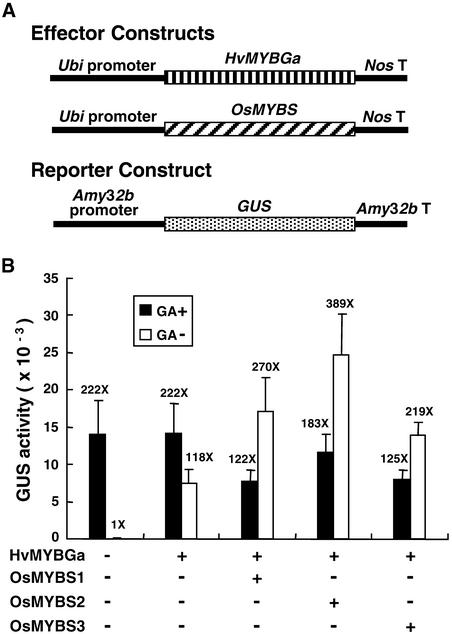
The GARC present in α-amylase gene promoters includes a GARE sequence and the TATCCA element (Gubler and Jacobsen, 1992; Lanahan et al., 1992). HvMYBGa has been shown to bind specifically to GARE in vitro and to activate the transcription of barley high-pI α-amylase and Cys proteinase gene promoters in the absence of GA in vivo (Gubler et al., 1995; Cercós et al., 1999). Because the TATCCA element is essential for the high-level GA-activated transcription of α-amylase gene promoters (Gubler and Jacobsen, 1992; Lanahan et al., 1992), we determined whether the OsMYBSs cooperate with HvMYBGa in enhancing the transcription of the α-amylase gene promoter.
The barley half-seeds were cobombarded with an effector plasmid containing the Ubi-HvMYBGa construct, another effector plasmid containing the Ubi-OsMYBS construct, and the reporter plasmid containing the Amy32b-GUS construct (Figure 10A). As shown in Figure 10B, β-glucuronidase (GUS) was expressed only in the presence of GA when no HvMYBGa was present, and overexpression of HvMYBGa significantly enhanced GUS activity in the absence of GA. This result was consistent with other results reported in the literature (Gubler et al., 1995; Cercós et al., 1999). Interestingly, coexpression of HvMYBGa with OsMYBS1, OsMYBS2, or OsMYBS3 further enhanced GUS activity by approximately twofold to threefold the activity with HvMYBGa alone in the absence of GA. These results suggest that the three OsMYBSs cooperate with HvMYBGa in the transactivation of the barley α-amylase gene promoter.
Figure 10.
OsMYBSs Cooperate with HvMYBGa in the Transactivation of the Barley Amy32b Promoter.
Barley aleurone cells were transfected with plasmids and incubated with or without 10−6 M GA3, and luciferase and GUS activities were assayed.
(A) The Ubi-HvMYBGa and Ubi-OsMYBS fusion genes were used as the effector constructs. The Amy32b-GUS fusion gene was used as the reporter construct.
(B) GUS activity in barley aleurone cells after cotransfection with the effector, reporter, and control plasmids. pAHC18 served as the internal control plasmid. The amount of effector, reporter, and control plasmid DNA was at a ratio of 1:1:0.25.
The value of GUS activity in barley aleurone cells without bombardment with any construct and treatment with GA was assigned a value of 1×, and the other values were calculated relative to this value.
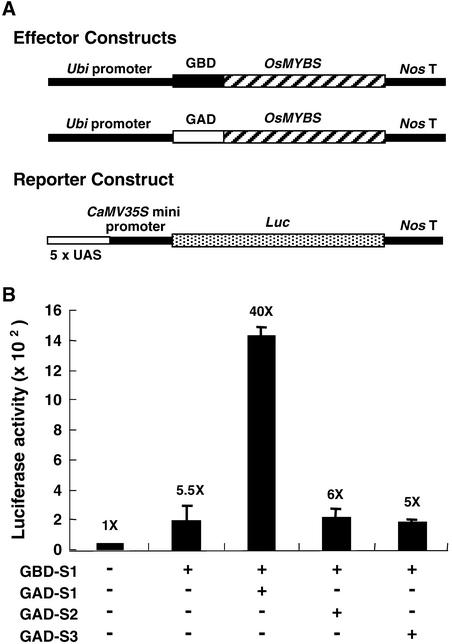
OsMYBS1 Forms a Homodimer in Barley Aleurone Cells
In c-Myb, R2 and R3 bind to specific DNA sequences cooperatively. To test the possibility that OsMYBS binds DNA as a dimer, we assayed the homologous and heterologous interactions among OsMYBSs using a two-hybrid system in barley aleurone cells. The full-length OsMYBS sequences were fused individually at the C terminus of GAD or GBD. The GAD-OsMYBS and GBD-OsMYBS chimeric genes were fused downstream of the Ubi promoter and used as effector constructs (Figure 11A). A DNA fragment containing five tandem repeats of UAS (5xUAS) was fused upstream of the CaMV35S minimal promoter–Luc chimeric gene and used as a reporter construct (Figure 11A).
Figure 11.
OsMYBS1 Forms a Homodimer in Barley Aleurone Cells.
Barely aleurone cells were transfected with plasmids and incubated in buffer without Glc for 24 h, and luciferase and GUS activities were assayed.
(A) The Ubi-GBD-OsMYBS and Ubi-GAD-OsMYBS fusion genes were used as the effector constructs. The 5xUAS-CaMV35S-Luc fusion gene was used as the reporter construct.
(B) Luciferase activity in barley aleurone cells after cotransfection with the effector, reporter, and control plasmids. pUGI served as the internal control plasmid. The amount of two effectors, reporter, and control plasmid DNA was at a ratio of 1:1:1:0.25.
The value of luciferase activity in barley aleurone cells bombarded with the reporter construct alone was assigned a value of 1×, and the other values were calculated relative to this value.
The barley half-seeds were particle bombarded simultaneously with two effector plasmids, each containing GAD-OsMYBS and GBD-OsMYBS fusion genes, and the reporter plasmid. The bombarded half-seeds were incubated in buffer without Glc for 24 h, and luciferase activity was determined. Figure 11B shows that coexpression of GBD-OsMYBS1 and GAD-OsMYBS1 significantly enhanced luciferase expression compared with expression of GBD-OsMYBS1 alone. However, coexpression of GBD-OsMYBS1 and GAD-OsMYBS2 or GAD-OsMYBS3 did not further enhance luciferase activity compared with expression of GBD-OsMYBS1 alone. Similar experiments with OsMYBS2 and OsMYBS3 showed that the two proteins did not interact homologously or heterologously (data not shown).
To confirm that only OsMYBS1 interacts homologously with itself and the three OsMYBSs do not interact with each other, the effector and reporter plasmids were reconstructed for expression in yeast. Although the background level of reporter gene expression in the presence of GBD-OsMYBS1 alone was too high (Figure 7B) to detect the interaction between GBD-OsMYBS1 and GAD-OsMYBSs, similar results were obtained in the yeast two-hybrid experiments (data not shown). These results suggest that only OsMYBS1 forms a homodimer under these experimental conditions.
DISCUSSION
Three Novel 1R MYBs
In the present study, we isolated and characterized three OsMYBS genes that encode MYBs with single DNA binding domains. In vitro gel mobility shift assay and in vivo transfection experiments in yeast and plant cells revealed that the three OsMYBSs are DNA binding proteins that interact specifically with the TATCCA element present in α-amylase gene promoters and that these proteins play essential roles in both GA- and sugar-regulated α-amylase gene expression.
The amino acid sequences of the 1R domains in OsMYBSs show equally similar homology with either the R2 or the R3 repeat of the animal and plant R2R3 MYBs. It has been proposed that a single original MYB repeat was replicated to give rise to two- and three-repeat MYBs (Lipsick, 1996). Therefore, R2 and R3 may have derived from duplication of one ancestor repeat, and the 1R may have evolved independently from the ancestor repeat as well. Because in c-Myb, both R2 and R3 are necessary for DNA binding and the putative base-contacting residues in the third helix of 1R is different from that of R2R3, it is likely that the 1R MYB binds DNA in a different manner.
Alignment of the MYB domains of all plant 1R MYBs shows that some of these domains contain the highly conserved SHAQK(Y/F)F motif in the third predicted α-helix (Figure 1). Because the third α-helix of MYBs has been shown to be involved in the sequence specificity of DNA binding (Gabrielsen et al., 1991), this family of MYBs containing 1R may recognize a DNA sequence motif different from those recognized by other MYBs. The exact consensus sequences recognized by the 1R MYBs have not been studied extensively. However, among the 1R MYBs containing the SHAQK(Y/F)F motif, only OsMYBS and StMYB1 recognize the TATCC core sequence. The 1R domains of OsMYBS and StMYB1 are highly conserved (Figure 1), suggesting that other amino acid residues immediately upstream of the SHAQK(Y/F)F motif and within the predicted third α-helix may contribute to the recognition of specific DNA sequence elements.
In a previous study, StMYB1, which was isolated from potato, activated the transcription of the CaMV35S promoter in tobacco protoplast transient expression assays and bound specifically to a DNA motif spanning positions −73 to −48 of the CaMV35S promoter in a gel mobility shift assay (Baranowskij et al., 1994). A random binding site selection assay indicated that StMYB1 has the highest binding affinity in vitro toward a GGATA motif (Baranowskij et al., 1994). We noted that the complementary sequence of 5′-GGATA-3′ is 5′-TATCC-3′. There is a TATCC(T) sequence located at −55 to −50 of the CaMV35S promoter, and the TATCC(T) sequence very likely is the target binding sequence of StMYB1. Interestingly, although OsMYBSs and StMYB1 have high-affinity binding for the same DNA sequence (except for OsMYBS3), OsMYBS1 and OsMYBS2 share very low homology with StMYB1 at regions outside of the 1R domain and its immediate vicinity. StMYB1 is expressed ubiquitously in potato, and its precise biological function is not known.
OsMYBSs Have Different Sequence Binding Preferences and Differential DNA Binding Affinities and Promoter Transactivation Abilities in Yeast and Plant Cells
Our gel mobility shift assays demonstrated that the three OsMYBSs possess specific binding activity for the TATCCA sequence in vitro. However, competition with mutant oligonucleotides revealed that OsMYBSs have different binding preferences for the TATCCA sequence. The three OsMYBSs can bind to either of the TATCCA repeats, because M2 and M3, with mutations at either repeat, still are able to compete out the binding (Figure 5B, lanes 5 and 6). Sequence CCA seems to be more important for OsMYBS1 binding, because M4 and M6, with mutations at TAT in one or both of these repeats, can compete for the binding better than M5, which has a mutation at CCA (Figure 5B, compare lanes 7 and 9 with lane 8).
By contrast, sequence TAT seems to be more important for OsMYBS2 binding, because M5 and M6, with mutations at CCA in one or both of these repeats, can compete for binding better than M4, which has a mutation at TAT (Figure 5C, compare lanes 8 and 9 with lane 7). Sequence CCA also seems to be more important for OsMYBS3 binding, because the binding behavior of OsMYBS3 for the TATCCA sequence is similar to that of OsMYBS1 (Figure 5D).
The sizes of the E. coli–expressed recombinant OsMYBSs are consistent with the sizes of OsMYBSs predicted from their nucleotide sequences, with OsMYBS3 (318 amino acids) > OsMYBS1 (306 amino acids) > OsMYBS2 (276 amino acids) (data not shown). The gel mobility shift assay showed that each of the three proteins formed protein complexes with DNA fragment F2 (Figure 4) or the wild type (Figure 5) that contained the TATCCA element. Interestingly, the size of the proteins complexes was not proportional to the size of OsMYBS. OsMYBS1 formed the largest, OsMYBS2 formed the second largest, and OsMYBS3 formed the smallest protein complex. We found that OsMYBS1 is able to form a homodimer (Figure 11), which may contribute to the formation of an OsMYBS1-DNA complex larger than the OsMYBS2- and OsMYBS3-DNA complexes.
Analysis in yeast revealed that the TATCCA element enhanced Cyc1 minimal promoter activity fivefold in the absence of OsMYBS (GAD control; compare Figure 6B, column 1, with Figure 6D, column 1), suggesting that yeast might contain OsMYBS homologs that recognize the TATCCA element. The GAD-OsMYBS2 fusion protein led to higher LacZ expression than the GAD-OsMYBS1 fusion protein in yeast (Figures 6D and 6F), suggesting that OsMYBS2 has higher binding affinity for the TATCCA element than OsMYBS1. The binding of GAD-OsMYBS1 to the 6xTATCCA-Cyc1 chimeric promoter was abolished (Figure 6F), indicating that the stable binding of OsMYBS1 to the TATCCA element may require additional sequences flanking the TATCCA element.
Interestingly, the GAD-OsMYBS3 fusion protein repressed the transcription of the 8x(TATCCA+F)-Cyc1 promoter in yeast (Figure 6D). It is possible that OsMYBS3 also bound to the TATCCA element but interfered with the transactivation ability of GAD. Another possibility is that GAD-OsMYBS3 competed with the yeast endogenous OsMYBS homolog for the TATCCA target sequence and caused a negative effect on promoter transcription.
The three OsMYBSs possess structural features characteristic of transcription activators. Protein sequences rich in acidic amino acids and Pro have been shown to act as transcription activation domains in both animal and plant MYBs (Mermod et al., 1989; Williams and Tjian, 1991; Schindler et al., 1992). The C terminus of OsMYBS1 is Pro rich (20 of 107 amino acids, or 19%) but has very few acidic amino acids. The C terminus of OsMYBS2 contains less Pro (12 of 172 amino acids, or 7%) but has more acidic amino acids (17 of 172 amino acids, or 10%) compared with OsMYBS1. OsMYBS3 is rich in both Pro (19 of 166 amino acids, or 11%) and acidic amino acids (22 of 166 amino acids, or 13%). In the present study, we show that OsMYBS1 and OsMYBS2 activate the transcription of the promoter containing SRS or the TATCCA element in yeast (Figure 7) and in barley aleurone cells (Figure 8).
In barley aleurone cells, under both repressed (in the presence of Glc) and derepressed (in the absence of Glc) conditions, OsMYBS1 induced higher luciferase expression driven by the SRS-CaMV35S or the 6x(TATCCA+F)-CaMV35S chimeric promoter than OsMYBS2 (Figures 8B and 9B). These results could be attributable to a higher binding affinity of OsMYBS1 to the SRS or to the TATCCA element compared with OsMYBS2, or perhaps OsMYBS1 had higher transactivation ability to the two promoters than OsMYBS2. In yeast, OsMYBS2 had higher binding affinity to the TATCCA element than OsMYBS1 (Figure 6), but OsMYBS1 showed higher promoter transactivation ability than OsMYBS2 (Figure 7).
Together, these findings demonstrate that OsMYBS1 had much higher transactivation ability than OsMYBS2 to activate a promoter containing the TATCCA element. Although the αAmy3 promoter contains two tandem repeats, whereas other α-amylase gene promoters contain only one repeat, of the TATCCA element (Yu, 1999a), OsMYBS might transactivate other α-amylase gene promoters as well. This notion is supported by our recent observation that OsMYBS1 transactivated the promoter of another rice α-amylase gene (αAmy8), which contains only one TATCCA element, in rice embryos under repressed conditions in a transient expression assay (P.-W. Chen and S.-M. Yu, unpublished results).
OsMYBS3 did not significantly transactivate the promoter containing SRS or the TATCCA element in barley aleurone cells under repressed conditions (Figures 8B and 9B). Furthermore, OsMYBS3 repressed the transcription of these promoters even under derepressed conditions. OsMYBS3 also repressed transcription in yeast cells (Figure 6D). All of these findings suggest that OsMYBS3 behaves like a transcriptional repressor in both yeast and barley aleurone cells.
OsMYBSs Act Independently of or Cooperatively with Other Protein Factors for Promoter Transactivation
MYB, with one repeat, may bind to DNA as a dimer, as has been proposed (Jin and Martin, 1999), although this has not been proved experimentally. In the present study, we showed that OsMYBS1 is capable of forming a homodimer in vivo. It is not clear why OsMYBS2 and OsMYBS3 do not form homodimers by themselves and why the three OsMYBSs do not form heterodimers with each other. The differential ability of OsMYBS in dimerization may lead to differential transactivation ability.
Both OsMYBS1 and OsMYBS2 transactivated the promoter containing an SRS in barley aleurone cells under repressed conditions (Figure 8B); however, only OsMYBS1 transactivated the promoter containing the TATCCA element with adjacent flanking sequences (TATCCA+F) under repressed conditions (Figure 9B). Furthermore, OsMYBS2 repressed the transcription of the promoter containing TATCCA+F even under derepressed conditions in a manner similar to OsMYBS3. These results suggest that additional sequences within SRS might be required for OsMYBS2 to function as a weak activator.
GARC contains the GARE sequence and three to four additional cis-acting promoter elements, including the TATCCA element, that are necessary for the GA activation of gene expression (Gubler and Jacobsen, 1992; Lanahan et al., 1992; Cercós et al., 1999). In the present study, a consensus GARE sequence was not found in the 1.1-kb rice αAmy3 promoter or in SRS, and the αAmy3 promoter–Luc or SRS–CaMV35S minimal promoter–Luc chimeric gene did not respond to GA in barley aleurone cells (data not shown). The 331-bp barley Amy32b promoter contains a GARE sequence but does not contain a complete SRS-like sequence except for a putative G-box and the TATCCA element. The Amy32b promoter–Luc chimeric gene did not respond to sugar in barley aleurone cells (data not shown).
It appears that a complete sugar response complex or GARC sequence may determine whether an α-amylase gene promoter has the potential to respond to sugar or GA. OsMYBS1 and OsMYBS2 act as activators, whereas OsMYBS3 acts as a repressor, in sugar starvation–induced α-amylase gene expression; however, they all cooperate with HvMYBGa in high-level, GA-induced α-amylase gene expression. These studies suggest that OsMYBSs are multifunctional depending on the target promoter sequences and their interactions with other protein factors in response to different signals. The three OsMYBS genes are expressed in various tissues of the rice plant, indicating that they may play certain, not yet identified, physiological roles in these tissues.
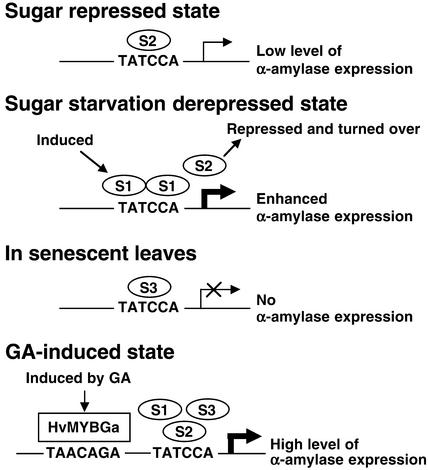
Role of OsMYBS in Sugar Starvation– and GA-Induced Gene Expression
The three OsMYBSs share a very similar structure in their DNA binding domains; hence, it is not surprising that they all bind to the same promoter sequence. However, the N and C termini of the three OsMYBSs are less homologous. Thus, questions are raised regarding whether these proteins perform similar or different biological functions and which of the proteins plays an essential role in the sugar and hormone regulation of α-amylase gene expression. Based on the differential binding affinities, transactivation abilities, and expression patterns of OsMYBSs, we suggest the model presented in Figure 12 to explain the biological relevance of our observations with respect to sugar- and GA-mediated α-amylase gene transcription.
Figure 12.
Proposed Roles of OsMYBSs in the Sugar and Hormone Regulation of α-Amylase Gene Expression in Cereals.
S1, S2, and S3 indicate OsMYBS1, OsMYBS2, and OsMYBS3, respectively.
The accumulation of both OsMYBS1 and OsMYBS3 mRNA in rice suspension cells and barley aleurone cells is suppressed by sugar (Figures 3B and 3C). By contrast, the accumulation of OsMYBS2 mRNA in these tissues is enhanced by sugar. For α-amylase genes subject to sugar regulation, OsMYBS2 is expressed and binds well to the TATCCA element when sugar is present. Although OsMYBS2 binds well to the TATCCA element, it has low transactivation ability; thus, only a low level of α-amylase expression is observed. However, under sugar starvation conditions, the expression of OsMYBS1 is induced but the expression of OsMYBS2 is suppressed. Thus, OsMYBS1 eventually replaces OsMYBS2 as the factor bound to the TATCCA element during sugar starvation.
Although OsMYBS1 has lower binding affinity for the TATCCA element, it leads to enhanced expression of α-amylase genes as a result of its high transactivation ability. As a repressor, whether OsMYBS3 plays a negative role in the sugar regulation of α-amylase gene expression is not clear. OsMYBS3 is most abundant in senescent leaves, in which sugars are depleted as a result of their transport to other tissues. OsMYBS3 may prevent α-amylase genes from being induced by the low sugar level in senescent leaves, which would be a wasteful process in dying cells.
For α-amylase genes subject to GA regulation, a GARE sequence is located just upstream of the TATCCA element. It has been shown that both the GARE sequence and the TATCCA element are necessary for GA-induced α-amylase gene expression in the aleurone layers. The GA-induced transcription factor HvMYBGa is able to bind to GARE and transactivate α-amylase gene expression. The transactivation activity of HvMYBGa is enhanced by OsMYBSs that bind to the neighboring TATCCA element. It is conceivable that interactions between HvMYBGa and OsMYBSs would stabilize the promoter-HvMYBGa-OsMYBS complex and that this could be part of the GA-responsive pathway, leading to the high-level expression of α-amylase genes.
Abscisic acid is an antagonist of GA that inhibits the expression of α-amylases in aleurone cells (Lovegrove and Hooley, 2000). The accumulation of OsMYBS1 and OsMYBS3 mRNA was suppressed by GA but unchanged by abscisic acid, whereas the accumulation of OsMYBS2 mRNA was suppressed by abscisic acid but unchanged by GA (Figure 3C, compare lane 1 with lanes 3 and 4). Additionally, OsMYBS1 and OsMYBS3 suppressed but OsMYBS2 maintained Amy32b promoter activity in the presence of GA, and cooperation between HvMYBGa and OsMYBS2 enhanced Amy32b promoter activity to a level higher than cooperation with OsMYBS1 and OsMYBS3 in the absence of GA (Figure 10). These results indicate that OsMYBS2 could be the dominant OsMYBS that interacts with HvMYBGa in response to GA in the aleurone layers (Figure 12).
In conclusion, the present study shows that three structurally related OsMYBSs share overlapping but nonidentical functions through the same target site in α-amylase gene promoters. It also demonstrates that common transcription factors are involved in the sugar and hormone regulation of α-amylase gene expression in cereals. OsMYBSs may play multiple roles through combinatorial interactions with other transcription factors in the generation of specific gene expression patterns. Our findings have opened up new avenues for investigating the molecular mechanisms of GA and sugar action in gene expression. Future functional studies using gene knockouts or silenced mutants and structure-function analysis on OsMYBS–DNA and OsMYBS–HvMYBGa interactions will help us better understand the biological function of this particular group of MYBs in plants.
METHODS
Rice Cell Culture
A suspension cell culture of rice (Oryza sativa cv Tainan 5) was established as described previously (Yu et al., 1991). Cells were subcultured every 7 days by transferring ∼0.5 mL of cells into 25 mL of fresh liquid Murashige and Skoog (1962) medium containing 3% Suc in a 125-mL flask. Cells were cultured on a reciprocal shaker at 120 rpm and incubated at 26°C in the dark.
Plasmids
Plasmid αAmy8-C carries a 1.4-kb rice α-amylase cDNA insert in pBluescript KS+ (Stratagene) (Yu et al., 1992). Plasmid pcRAc1.3 contains a 1.4-kb rice actin gene (Act1) cDNA insert in pBluescript II-KS+ (McElroy et al., 1990). Plasmid pRY18 carries a 3.8-kb DNA fragment that contains a rice genomic rDNA cluster, including the 3′ portion of 18S rRNA gene, the complete 5.8S rRNA gene, and the 5′ portion of the 25S rRNA gene in pUC13 (Sano and Sano, 1990).
Plasmid pGAMyb contains the HvMYBGa cDNA fused between the maize ubiquitin (Ubi) promoter and the nopaline synthase (Nos) terminator (Cercós et al., 1999). Plasmid pAHC18 contains the luciferase (Luc) cDNA fused between the Ubi promoter and the Nos terminator (Bruce et al., 1989). Plasmid pAmy32b-GUS contains the 331-bp promoter region, the entire 5′ untranslated sequence, and the first intron of Amy32b fused to the β-glucuronidase (GUS) coding sequence and the 3′ untranslated region of Amy32b (Gómez-Cadenas et al., 1999).
Protein Gel Blot Screening of a cDNA Library
Rice suspension cells were cultured in Suc-containing medium for 5 days and transferred to Suc-containing or Suc-free medium for 4 h. Cells were collected, and total RNA was purified. Poly(A)+ RNA was purified further using an oligo(dT) cellulose spin column (5 Prime → 3 Prime, Boulder, CO). The poly(A)+ RNA isolated from Suc-free cells was used to construct a cDNA library in λGEM-2 vector (Promega). Escherichia coli strain Y1090 (Stratagene) served as the bacterial host.
For screening and plaque purification, the 416-bp DNA fragment containing eight tandem repeats of the −133 to −82 region of the αAmy3 promoter was excised with PstI and XhoI from p3Luc.44 (Lu et al., 1998) and used as a probe for cDNA library screening. The library was screened according to the method described by Singh et al. (1988). Positive phage plaques were isolated, and phage DNA was purified for further characterization.
Plasmid Construction
OsMYBS1 and OsMYBS2 cDNAs were excised from λGEM-2 with SalI and NotI and inserted into the same sites in pBluescript II-KS+ (Stratagene) to generate pBS-S1 and pBS-S2, respectively. There is one SalI site within the OsMYBS3 cDNA; therefore, OsMYBS3 cDNA was excised from λGEM-2 with EcoRI and NotI and inserted into the same sites in pBluescript to generate pBS-S3. pAHC18 was digested with BamHI to remove the Luc cDNA insert and then was blunt ended. OsMYBS1 and OsMYBS2 were excised from pBS-S1 and pBS-S2, respectively, with SalI and NotI and blunt ended. OsMYBS3 was excised from pBS-S3 with EcoRI and NotI and blunt ended. The OsMYBS cDNAs then were ligated individually with the truncated pAHC18 to generate a Ubi-OsMYBS-Nos fusion gene.
Genomic DNA Gel Blot Analysis
Genomic DNA was isolated from rice calli according to the method of Sheu et al. (1996). Ten micrograms of genomic DNA was digested with restriction enzymes, fractionated on a 0.8% agarose gel, and transferred to a nylon membrane (Micron Separations Inc., Westborough, MA). Hybridization was performed at 42°C using 32P random primer–labeled OsMYBS gene-specific DNA as a probe. pBS-S1 was digested with SacII, and the 613-bp DNA fragment containing the 220-bp coding region and the 393-bp 3′ untranslated region of OsMYBS1 cDNA served as OsMYBS1-specific DNA. pBS-S2 was digested with SacI, and the 334-bp DNA fragment containing the 241-bp coding region and the 93-bp 3′ untranslated region of OsMYBS2 cDNA served as OsMYBS2-specific DNA. pBS-S3 was digested with SacI, and the 599-bp DNA fragment containing the 332-bp coding region and the 267-bp 3′ untranslated region of OsMYBS3 served as OsMYBS3-specific DNA.
RNA Gel Blot Analysis
Total RNA was purified from rice suspension cells using Trizol reagent (Gibco BRL). α-32P-labeled DNA probes were prepared, and RNA gel blot analysis was performed as described (Sheu et al., 1996). In cases in which the membrane blot was hybridized sequentially with various probes, the membrane was stripped and rehybridized as described (Sheu et al., 1994). Plasmid DNAs of αAmy8-C and pcRAc1.3 were digested with EcoRI. αAmy3 and αAmy8 gene-specific DNAs were prepared as described by Sheu et al. (1996). These insert DNAs were isolated individually, labeled with α-32P, and used as probes. A DNA fragment containing 25S, 18S, and 5.8S rDNAs was excised from pRY18 using BamHI, labeled with α-32P, and used as a probe to equalize RNA loading.
Expression and Purification of Recombinant Proteins
OsMYBS was digested with SalI and NotI and ligated into the same sites in pET28b(+) (Novagen, Madison, WI) to generate pET-S1, pET-S2, and pET-S3. The three plasmids were transferred into E. coli strain BL21(DE3), and OsMYBS1, OsMYBS2, and OsMYBS3 were expressed. Purification of the OsMYBSs was performed according to the instructions provided by Novagen. The protein concentration was determined with the Bradford reagent (Bio-Rad).
Gel Mobility Shift Assay
The gel mobility shift assay was performed essentially as described previously (Lu et al., 1998), except that the DNA–protein binding reaction was performed by incubation of 0.02 ng of 32P-labeled oligonucleotide with 0.2 μg of recombinant OsMYBS purified from E. coli and 1 μg of poly(dI-dC) in a total volume of 20 μL of solution. A DNA fragment containing the TATA box was prepared as described previously (Lu et al., 1998).
Construction of the Yeast Reporter Strains
The DNA fragment containing eight tandem repeats of −133 to −82 of the αAmy3 promoter was excised from p3Luc.44 (Lu et al., 1998) with EcoRI and XhoI, blunt ended, and inserted into the SmaI site in pLacZi (Clontech, Palo Alto, CA) to generate a 8x(TATCCA+F)–Cyc1 minimal promoter–LacZ fusion gene. The two complementary oligonucleotides, 5′-AATTCTATCCATATCCATATCCATATCCATATCCATATCCAC-3′ and 5′-GTGGATATGGATATGGATATGGATATGGATATGGATAG-3′, were annealed and inserted into the EcoRI and SmaI sites of pLacZi to generate a 6xTATCCA–Cyc1 minimal promoter–LacZ fusion gene. These plasmids were linearized with NcoI and introduced into the genome of yeast strain YM4271, according to the Matchmaker One-Hybrid system protocol (Clontech), to generate the yeast reporter strains.
Yeast One-Hybrid System for OsMYBS Binding Activity Assay
For construction of the GAD-OsMYBS fusion gene, the full-length OsMYBS cDNAs were amplified by PCR using pBS-S1, pBS-S2, and pBS-S3 as the DNA templates and oligonucleotides OsMYBS1-5′ (5′-AAACTCGAGAATGACCTCCCAGGCGGCGA-3′; XhoI site underlined) and OsMYBS1-3′ (5′-ATCGAATTCTCATTGGTGCATCTTGGCCGGA-3′; EcoRI site underlined) as primers for OsMYBS1, oligonucleotides OsMYBS2-5′ (5′-AAACTCGAGAATGCCCAACCT-CACCTCCA-3′; XhoI site underlined) and OsMYBS2-3′ (5′-AGCGAATTCTTATATAAAACTGGTGAA-3′; EcoRI site underlined) as primers for OsMYBS2, and oligonucleotides OsMYBS3-5′ (5′-AAA-CTCGAGTATGACGAGGCGGTGCTCGCA-3′; XhoI site underlined) and OsMYBS3-3′ (5′-ATCGAATTCTCATGCCTGTGCCCTTGT-3′; EcoRI site underlined) as primers for OsMYBS3.
The GAD sequence was amplified by PCR using pGAD-424 (Clontech) as the DNA template and oligonucleotides GAD-5′ (5′-CCAGAATTCTGCAAAGATGGATAAA-3′; EcoRI site underlined) and GAD-3′ (5′-CCACTCGAGCTCTCTTTTTTTGGGT-3′; XhoI site underlined) as the primers. All of the PCR products were digested with XhoI and EcoRI. The OsMYBS cDNAs were ligated individually with the GAD sequence in the XhoI site and inserted into the EcoRI site of pBluescript to generate pGAD-OsMYBS. pGAD-424 was digested with MluI and EcoRI to remove the 3′ portion of the GAD domain.
The GAD 3′ portion–OsMYBS fusion genes were excised from pGAD-OsMYBS with MluI and EcoRI and inserted into the MluI and EcoRI sites of pGAD-424 to generate pGAD–OsMYBS–424, which contained the ADH1 promoter–GAD–OsMYBS fusion gene. These plasmids were introduced individually into the yeast reporter strain, which hosted the 8x(TATCCA+F)–Cyc1 minimal promoter–LacZ fusion gene or the 6xTATCCA–Cyc1 minimal promoter–LacZ fusion gene, according to the Matchmaker One-Hybrid system protocol (Clontech). β-Galactosidase activity in yeast was assayed.
Yeast One-Hybrid System for OsMYBS Transactivation Assay
To construct the GBD-OsMYBS fusion gene, the GBD sequence was amplified by PCR using pGBT9 (Clontech) as the DNA template and oligonucleotides GBD-5′ (5′-CCAGAATTCAGATGAAGCTACTGTCT-3′; EcoRI site underlined) and GBD-3′ (5′-CCACTCGAGTTCGAT-ACAGTCAACTGT-3′; XhoI site underlined) as primers. Because there is one XhoI cutting site within the GBD sequence, the PCR product was digested with EcoRI first and then partially digested with XhoI. The resulting DNA fragment was ligated with OsMYBS cDNA and inserted into the EcoRI site of pBluescript to generate pGBD-OsMYBS. pGAD-424 was digested with HindIII to remove the GAD sequence and blunt ended.
The GBD-OsMYBS fusion genes were excised from pGBD-OsMYBS with EcoRI and HindIII, blunt ended, and ligated with the truncated pGAD-424 to generate pGBD–OsMYBS–424, which contained the ADH1 promoter–GBD–OsMYBS fusion gene. These plasmids were introduced into yeast strain CG-1945, which hosted the 3xUAS–Cyc1 minimal promoter–LacZ and GAL1 promoter–HIS3 reporter constructs (Clontech). 3xUAS is three tandem repeats of a synthetic UASG 17-mer consensus sequence that can be recognized by GBD. β-Galactosidase activity in yeast or growth of yeast cells on selective medium was assayed.
β-Galactosidase Activity and Yeast Cell Growth Assays
β-Galactosidase activity in transformed yeast cells was quantified using o-nitrophenyl β-galactopyranoside as the substrate, and growth of transformed cells were assayed in a medium containing 5 mM 3-aminotriazole but no His according to the Yeast Protocols Handbook (Clontech).
Barley Aleurone Tissue Transient Expression Assay
Particle bombardment of barley (Hordeum vulgare) aleurone tissue and transient assay of GUS were performed essentially as described by Lanahan et al. (1992) and Cercós et al. (1999). Bombarded barley embryoless half-seeds were incubated in a buffer (20 mM each CaCl2 and sodium succinate, pH 5.0) containing or lacking Glc or GA3 for 20 h, and luciferase or GUS activity was determined. All of the bombardments were repeated at least four times.
Acknowledgments
We thank Rodolfo Zentella, Jose Casaretto, and Lin-Tze Yu for technical assistance and Douglas Platt for help in the preparation of the manuscript. This work was supported by grants from Academia Sinica, the National Science Council (NSC-90-2311-B-001-008), and the Biomedical Research Foundation of the Republic of China to S.-M.Y. and by U.S. National Science Foundation Grant IBN-9983126 to T.-h.D.H.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.001735.
References
- Baranowskij, N., Frohberg, C., Prat, S., and Willmitzer, L. (1994). A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 13, 5383–5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, E.L., and Grotewold, E. (1999). Newly discovered plant c-myb-like genes rewrite the evolution of the plant myb gene family. Plant Physiol. 121, 21–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, W.B., Christensen, A.H., Klein, T., Fromm, M., and Quail, P.H. (1989). Photoregulation of a phytochrome gene promoter from oat transferred into rice by particle bombardment. Proc. Natl. Acad. Sci. USA 86, 9692–9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cercós, M., Gómez-Cadenas, A., and Ho, T.-H.D. (1999). Hormonal regulation of a cysteine proteinase gene, EPB-1, in barley aleurone layers: Cis- and trans-acting elements involved in the co-ordinated gene expression regulated by gibberellins and abscisic acid. Plant J. 19, 107–118. [DOI] [PubMed] [Google Scholar]
- Chan, M.-T., Chao, Y.-C., and Yu, S.-M. (1994). Novel gene expression system for plant cells based on induction of α-amylase promoter by carbohydrate starvation. J. Biol. Chem. 269, 17635–17641. [PubMed] [Google Scholar]
- Chan, M.-T., and Yu, S.-M. (1998. a). The 3′ untranslated region of a rice α-amylase gene mediates sugar-dependent abundance of mRNA. Plant J. 15, 685–696. [DOI] [PubMed] [Google Scholar]
- Chan, M.-T., and Yu, S.-M. (1998. b). The 3′ untranslated region of a rice α-amylase gene functions as a sugar-dependent mRNA stability determinant. Proc. Natl. Acad. Sci. USA 95, 6543–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldbrügge, M., Sprenger, M., Hahlbrock, K., and Weisshaar, B. (1997). PcMYB1, a novel plant protein containing a DNA-binding domain with one MYB repeat, interacts in vivo with a light-regulatory promoter unit. Plant J. 11, 1079–1093. [DOI] [PubMed] [Google Scholar]
- Frampton, J., Gibson, T.J., Ness, S.A., Doderiein, G., and Graf, T. (1991). Proposed structure for the DNA-binding domain of the Myb oncoprotein based on model building and mutational analysis. Protein Eng. 4, 891–901. [DOI] [PubMed] [Google Scholar]
- Gabrielsen, O.S., Sentenac, A., and Fromageot, P. (1991). Specific DNA binding by c-Myb: Evidence for a double helix-turn-helix-related motif. Science 253, 1140–1143. [DOI] [PubMed] [Google Scholar]
- Gómez-Cadenas, A., Verhey, S.D., Holappa, L.D., Shen, Q., Ho, T.-H.D., and Walker-Simmons, M.K. (1999). An abscisic acid-induced protein kinase, PKABA1, mediates abscisic acid-suppressed gene expression in barley aleurone layers. Proc. Natl. Acad. Sci. USA 96, 1767–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf, T. (1992). Myb: A transcriptional activator linking proliferation and differentiation in hematopoietic cells. Curr. Opin. Genet. Dev. 2, 249–255. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., Drummond, B.J., Bowen, B., and Peterson, T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76, 543–553. [DOI] [PubMed] [Google Scholar]
- Gubler, F., and Jacobsen, J.V. (1992). Gibberellin-responsive elements in the promoter of a barley high-pI α-amylase gene. Plant Cell 4, 1435–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler, F., Kalla, R., Roberts, J.K., and Jacobsen, J.V. (1995). Gibberellin-regulated expression of a myb gene in barley aleurone cells: Evidence for Myb transactivation of a high-pI α-amylase gene promoter. Plant Cell 7, 1879–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., and Martin, C. (1999). Multifunctionality and diversity within the plant MYB-gene family. Plant Mol. Biol. 41, 577–585. [DOI] [PubMed] [Google Scholar]
- Katzen, A.L., Kornberg, T.B., and Bishop, J.M. (1985). Isolation of the proto-oncogene c-myb from D. melanogaster. Cell 41, 449–456. [DOI] [PubMed] [Google Scholar]
- Kirik, V., and Bäumlein, H. (1996). A novel leaf-specific myb-related protein with a single binding repeat. Gene 183, 109–113. [DOI] [PubMed] [Google Scholar]
- Koch, K.E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 509–540. [DOI] [PubMed] [Google Scholar]
- Lanahan, M.B., Ho, T.-H.D., Rogers, S.W., and Rogers, J.C. (1992). A gibberellic response complex in cereal α-amylase gene promoters. Plant Cell 4, 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsick, J.S. (1996). One billion years of Myb. Oncogene 13, 223–235. [PubMed] [Google Scholar]
- Lovegrove, A., and Hooley, R. (2000). Gibberellin and abscisic acid signalling in aleurone. Trends Plant Sci. 5, 102–110. [DOI] [PubMed] [Google Scholar]
- Lu, C.-A., Lim, E.-K., and Yu, S.-M. (1998). Sugar response sequence in the promoter of a rice α-amylase gene serves as a transcriptional enhancer. J. Biol. Chem. 273, 10120–10131. [DOI] [PubMed] [Google Scholar]
- Majello, B., Kenyon, L.C., and Dalla-Favera, R. (1986). Human c-myb protooncogene: Nucleotide sequence of cDNA and organization of the genomic locus. Proc. Natl. Acad. Sci. USA 83, 9636–9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, C., and Paz-Ares, J. (1997). MYB transcription factors in plants. Trends Genet. 13, 67–73. [DOI] [PubMed] [Google Scholar]
- McElroy, D., Rothenberg, M., Reece, K.S., and Wu, R. (1990). Characterization of the rice (Oryza sativa) actin gene family. Plant Mol. Biol. 15, 257–268. [DOI] [PubMed] [Google Scholar]
- Mermod, N., O'Neill, E.A., Kelly, T.J., and Tjian, R. (1989). The proline-rich transcriptional activator of CTF/NF-1 is distinct from the replication and DNA binding domain. Cell 58, 741–753. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. [Google Scholar]
- Ogata, K., Hojo, H., Aimoto, S., Nakai, T., Nakamura, H., Sarai, A., Ishii, S., and Nishimura, Y. (1992). Solution structure of a DNA-binding unit of Myb: A helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc. Natl. Acad. Sci. USA 89, 6428–6432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata, K., Morikawa, S., Nakamura, H., Sekikawa, A., Inoue, T., Kanai, H., Sarai, A., Ishii, S., and Nishimura, Y. (1994). Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79, 639–648. [DOI] [PubMed] [Google Scholar]
- Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P.A., and Saedler, H. (1987). The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 6, 3553–3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perata, P., Matsukura, C., Vernieri, P., and Yamaguchi, J. (1997). Sugar repression of a gibberellin-dependent signalling pathway in barley embryos. Plant Cell 9, 2197–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakura, H., Kanei-Ishii, C., Nagase, T., Nakagoshi, H., Gonda, T.J., and Ishii, S. (1989). Delineation of three functional domains of the transcriptional activator encoded by the c-myb proto-oncogene. Proc. Natl. Acad. Sci. USA 86, 5758–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, Y., and Sano, R. (1990). Variation of the intergenic spacer region of ribosomal DNA in cultivated and wild rice species. Genome 33, 209–218. [Google Scholar]
- Schindler, U., Terzaghi, W., Beckmann, H., Kadesch, T., and Cashmore, A.R. (1992). DNA binding site preferences and transcriptional activation properties of Arabidopsis transcription factor GBF-1. EMBO J. 11, 1275–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu, J.-J., Jan, S.-P., Lee, H.-T., and Yu, S.-M. (1994). Control of transcription and mRNA turnover as mechanisms of metabolic repression of α-amylase gene expression. Plant J. 5, 655–664. [Google Scholar]
- Sheu, J.-J., Yu, T.-S., Tong, W.-F., and Yu, S.-M. (1996). Carbohydrate starvation stimulates differential expression of rice α-amylase genes that is modulated through complicated transcriptional and posttranscriptional processes. J. Biol. Chem. 271, 26998–27004. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., Yamaguchi-Shinozaki, K., Urao, T., and Koizumi, M. (1992). Nucleotide sequence of a gene from Arabidopsis thaliana encoding a myb homologue. Plant Mol. Biol. 19, 493–499. [DOI] [PubMed] [Google Scholar]
- Singh, H., LeBowitz, J.H., Baldwin, A.S., Jr., and Sharp, P.A. (1988). Molecular cloning an enhancer binding protein: Isolation by screening of an expression library with a recognition site DNA. Cell 52, 415–423. [DOI] [PubMed] [Google Scholar]
- Solano, R., Fuertes, A., Sánchez, L., Valencia, A., and Paz-Ares, J. (1997). A single residue substitution causes a switch from the dual DNA binding specificity of plant transcription factor MYB.Ph3 to the animal c-MYB specificity. J. Biol. Chem. 272, 2889–2895. [DOI] [PubMed] [Google Scholar]
- Solano, R., Nieto, C., Avila, J., Cañas, L., Diaz, I., and Paz-Ares, J. (1995). Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB.Ph3) from Petunia hybrida. EMBO J. 14, 1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke, R., Werber, M., and Weisshaar, B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4, 447–456. [DOI] [PubMed] [Google Scholar]
- Wang, Z.-Y., Kenigsbuch, D., Sun, L., Harel, E., Ong, M.S., and Tobin, E.M. (1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston, K. (1999). MYB proteins in life, death and differentiation. Curr. Opin. Genet. Dev. 8, 76–81. [DOI] [PubMed] [Google Scholar]
- Williams, T., and Tjian, R. (1991). Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev. 5, 670–682. [DOI] [PubMed] [Google Scholar]
- Yu, S.-M. (1999a). Regulation of α-amylase gene expression. In Molecular Biology of Rice, K. Shimamoto, ed (Tokyo: Springer-Verlag), pp. 161–178.
- Yu, S.-M. (1999. b). Cellular and genetic responses of plants to sugar starvation. Plant Physiol. 121, 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, S.-M., Kuo, Y.H., Sheu, G., Sheu, Y.-J., and Liu, L.-F. (1991). Metabolic derepression of α-amylase gene expression in suspension-cultured cells of rice. J. Biol. Chem. 266, 21131–21137. [PubMed] [Google Scholar]
- Yu, S.-M., Lee, Y.-C., Fang, S.-C., Chan, M.-T., Hwa, S.-F., and Liu, L.-F. (1996). Sugars act as signal molecules and osmotica to regulate the expression of α-amylase genes and metabolic activities in germinating cereal grains. Plant Mol. Biol. 30, 1277–1289. [DOI] [PubMed] [Google Scholar]
- Yu, S.-M., Tzou, W.-S., Lo, W.-S., Kuo, Y.-H., Lee, H.-T., and Wu, R. (1992). Regulation of α-amylase-encoding gene expression in germinating seeds and cultured cells of rice. Gene 122, 247–253. [DOI] [PubMed] [Google Scholar]