Abstract
Previous rotamer libraries showed little significant clustering for asparagine χ2 or glutamine χ3 values, but none of those studies corrected amide orientations or omitted disordered side chains. The current survey used 240 proteins at ≤1.7 Å resolution with <50% homology and <30 clashes per thousand atoms (atomic overlap ≥0.4 Å). All H atoms were added and optimized, and amide orientation was flipped by 180° if required by H bonding or atomic clashes. A side chain was included only if its amide orientation was clearly determined and if no atom had a B factor ≥40, alternate conformation, or severe clash; that selection process yielded 1,490 Asn and 863 Gln side chains. Clear clustering was observed for Asn χ2 and Gln χ3 (except when Gln χ2 is trans). For Gln, five major and four minor rotamers cover 87% of examples. For Asn, there are seven backbone-independent rotamers covering 94% of examples plus rotamers specified for strictly α-helical, β, and left-handed (+φ) Asn. Although the strongest influence on χ angles is avoidance of atomic clashes (especially with the NH2 hydrogens), some Asn or Gln rotamers are influenced by favorable van der Waals contacts and others by specific local H-bond patterns.
The most important variables for protein conformation are the φ, ψ, and ω angles of the backbone, which describe overall tertiary structure. However, the side-chain χ1, χ2, χ3 … dihedral angles constitute the other half of the conformational specification, determining how the parts of the protein fit together and how the functional groups of the side chains can interact with other molecules. Originally, each χ-angle distribution was studied separately, but since Ponder and Richards (1), most treatments have been organized around “rotamers,” or separate populated clusters in the n-dimensional space of χ values for a given amino acid type. Libraries of discrete side chain rotamers are widely used for homology modeling, protein redesign, Monte Carlo calculations, and crystallographic electron-density map fitting. The usefulness of the rotamer concept depends on two conditions: (i) the clusters are sharp and separated and (ii) the χ distributions are not simply independent of one another. Both conditions hold for most side-chain types, but Asn and Gln show notably poor clustering.
Asn and Gln χ1 and Gln χ2 angles are well behaved, with optima near the three staggered values. The major aspect at issue in the analysis of Asn/Gln conformations is the outermost dihedral angle (χ2 for Asn and χ3 for Gln). When the side-chain amide N and O atoms cannot be distinguished, that angle is uncertain by 180°. It involves the rotation of a planar amide relative to a tetrahedral group and has less distinct preferences than a rotation between methyl or methylene groups. The Asn/Gln amide χ angles are often omitted from side-chain conformer analyses and are treated differently each time they are included. Different studies have presumed 180° symmetry (2–4); have used 0°, +, t, and − classes (1); have used <180° and >180° classes (5); have used bins centered around 60°, 0°, and −60° (6); have energy-minimized the structures before cluster analysis (7); or have used the lack of clear clusters to argue that the rotamer concept is not very useful (8). Not surprisingly, the rotamers defined by using these disparate approaches have little in common. Nevertheless, one thing all of these studies agreed on is that Asn χ2 and Gln χ3 distributions were relatively broad and featureless.
The present work, in contrast, benefits from a number of noise reduction strategies not tried before in this context: (i) removing the most error-prone side chains with a B-factor cutoff (surprisingly, done only once previously; see ref. 9), (ii) removing residues with alternate conformations, (iii) removing residues with severe atomic clashes, and (iv) most importantly, correcting side-chain amide orientations misassigned by 180°, which has been studied before (10, 11) but not applied to rotamer libraries.
Recently, we developed a small-probe dot method for analyzing molecular contacts including all explicit H atoms (12) and applied this method to determine the orientation of Asn and Gln side-chain amides (ref. 13; in that study of high-resolution structures, ≈20% of the amides needed to be flipped, whereas another 15% were indeterminate and probably mobile). We observed that a substantial fraction of the serious NH2 clashes that force 180° amide flips are with other atoms in the same residue, implying that the side-chain conformation is actually disallowed. Because such cases occur even in very accurate protein structures, they have been included in rotamer lists and hence perpetuated in later structure determinations. The Asn or Gln rotamers with a trans amide χ angle in the libraries of Ponder and Richards (1), o version 6.2.2 (14), and Tuffery et al. (7) have serious internal clashes both at their average conformation and for a range on either side, as exemplified in Fig. 1. At the other extreme, Dunbrack and Cohen (6) assign twofold-symmetric rotamers for Asn and Gln, which is unnecessarily conservative. In the present work, we flip amides that clearly are shown to be wrong by clashes and/or H bonding, and we omit the indeterminate amides along with the high B-factor side chains. The resulting data demonstrate clear, useful patterns in the χ angle distributions for Asn and Gln, and so a new set of rotamers is defined.
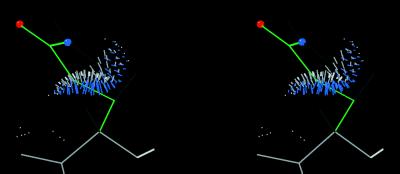
Figure 1.
Small-probe dots showing the internal contacts of an ideal-geometry Gln residue in the t + t rotamer (ref. 1; tp180° in our nomenclature). There are two bad clashes of Hɛ with Hα and/or Hβ at the mean angle (as shown) and for some distance on either side, but this is relieved if the side-chain amide is flipped by 180°.
METHODS
The set of 240 protein structures used here was chosen from the Protein Data Bank (15) as of August, 1998. All are crystal structures at 1.7 Å or better resolution and have a “clashscore” <30 [number of clashes or atomic overlaps ≥0.4 Å per thousand atoms (12)], giving priority to high resolution and wild-type sequence. For similar structures or chains with ≥50% sequence similarity only one structure was used; with <50% similarity, even the identical side chains usually differ in either rotamer or environment. Listed U2 (atomic displacement) values were converted to the more common B (temperature) factor by using the relationship B = 8π2 × U2.
The overall clashscore of a structure and the existence of a clash for a given residue were determined as in ref. 12. The program probe calculates contact dots wherever two atoms (including H) are within 0.5 Å of van der Waals contact and assigns a numerical score; the dot lists are displayed in mage (16). Correction of side-chain amide orientations (“flips”) was done by using the automated version of reduce (13). The program adds all H atoms and then combinatorially optimizes the rotations of movable hydrogens and the flips of Asn/Gln/His side chains using both H bonds and clashes. Each Asn/Gln amide orientation is assigned a state of “Keep” (61% of Asn and 53% of Gln), “Flip” (17% of Asn and 18% of Gln), “Clash” (if the amide shows an atomic overlap ≥0.4 Å in both orientations, as was observed in 3% of both Asn and Gln), or “X” for indeterminate (with a score difference <0.5 between the two flip states, as seen in 16% of Asn and 25% of Gln). These percentages differ slightly from those in ref. 13 because here we omitted the hand reassignment, which tends to remove residues from the indeterminate category.
The Asn/Gln rotamers were analyzed from these corrected coordinate files by using conformational angles from our own program dang, secondary-structure assignments of helical residues (“H”, but not in the first three “H” of a run) and β residues (“E”) from procheck (17), amide flip assignment (K, F, C, or X) and covalent modification flag from reduce, and the existence of an atomic clash or a local H bond from probe. Residues were omitted if any atom had a B factor ≥40 (12), either “a” or “b” alternate conformations, a severe atomic clash, or a covalent modification. Indeterminate and doubly clashing amides also were omitted.
We use the mode (or highest peak position) to define each rotamer because mean χ values can lie in a disallowed region between clusters and because means are quite sensitive to the boundaries of the bins inside which examples are counted. The mode is determined by placing a Gaussian mask (with an integral of 1.0 and a width at half-height of 4° for two-dimensional or 8° for three-dimensional data sets) at each data point and summing the masks. For counting occurrence frequency, bins are defined to include most but usually not all of the points in a cluster, with a minimum half-width of 30°. For Asn, rotamers sometimes were separated on the basis of H bonding.
Ideal-geometry (18) side chains were generated for theoretical analysis with hydrogens added by reduce. Their internal contact dots were displayed in mage in an interactive mode where the dots are updated by probe as bonds are rotated.
There is no consensus on rotamer nomenclature in the previous literature. For χ1 and for Gln χ2, concise symbols are desirable because those angles conform well to the usual staggered values. The terminology of g+, t, and g− would be suitable, except that within the field of side-chain conformation there are two competing traditions that use these same terms with exactly opposite meanings. One tradition (6, 9, 19) uses g− for −60° and g+ for +60°, as is standard in the chemistry community (20). The other tradition (3, 5, 8, 22) uses g+ for −60° and g− for +60° and refers to the international standards document (21), which changed the definition of dihedral angles (from trans = 0° to cis = 0°) but did not actually mention g+ or g−. To adopt the same-sign convention without adding to the confusion, we expand the common usage of “t” for trans to include “m” for minus and “p” for plus. The terminal amide χ angles sometimes show two distinct clusters within 120° and are seldom centered at staggered or 90° values. Therefore, for amides it seems necessary to list an actual angle value, rounded to the nearest 10°. Any intractably broad amide χ distributions are indicated by underscores: e.g., 0° . For backbone-dependent rotamers we prefix a α, β, or L (for instance αm−20°).
The Asn/Gln rotamer lists (both as tables and as o data files), the list of 240 Protein Data Bank files, plus the programs reduce, probe, prekin, mage, dang, and supporting scripts are all available from either the anonymous FTP site or the World Wide Web site at http://kinemage.biochem.duke.edu.
RESULTS
Improvement of χ Distributions.
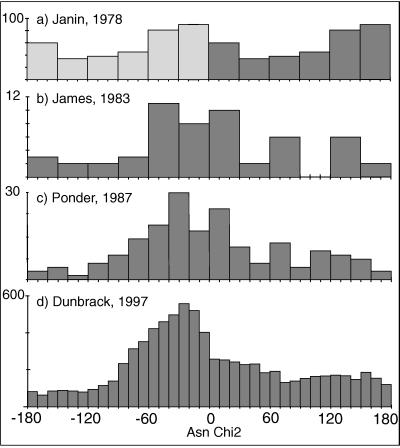
The use of a B-factor cutoff and the correction of amide-flip orientations are the most important ways in which our analysis differs from previous rotamer compilations, and we find that both changes significantly improve the clustering. Fig. 2 shows a historical progression of Asn χ2 distributions published between 1978 and 1997. Their most obvious features are the gradual improvement and the disappointing lack of clear and separate clusters. In Fig. 2a (modified from ref. 3), although the main peak shows asymmetry, its height is only twice the lowest value. Fig. 2b (modified from ref. 22), created with fewer but more accurate structures, shows that the peak near 0° is much larger than the one near 180°. Ponder and Richards (1) used data from 19 refined proteins to give χ distributions (e.g., Fig. 2c) and defined 6 Asn rotamers but omitted Gln χ3 when χ2 was trans. Schrauber et al. (8) did not give new rotamers for Asn because “the χ2 distribution is virtually uniform.” Kuszewski et al. (23) used potential-of-mean-force analysis to convert their χ distributions into relative energies; for Gln χ3 they show a nearly flat energy profile, stating that “the χ3 angle of Gln… is populated everywhere equally. ” Tuffery et al. (7) chose Asn rotamers “so that they sample the conformational space, since… χ2 can adopt any angular value.”
Figure 2.
Historical progression of published Asn χ2 distributions (number of observed occurrences in each range of χ2 value). (a) Figure modified from Janin et al. (3) uses 19 mostly unrefined proteins at ≤2.5 Å resolution: symmetrized data from 0° to 180° for Asn+Asp. (b) Figure modified from James and Sielecki (22) uses five proteases refined at ≤1.8 Å resolution: Asn χ2 from 0° to 360°. (c) Asn χ2 data from the 19 proteins used by Ponder and Richards (1), all but one of which were refined at ≤1.8 Å. (d) Data from the Web site http://www.fccc.edu/research/labs/dunbrack as of October 1998: data updated from the 518 proteins at ≤2.0 Å resolution in Dunbrack and Cohen (6). All examples reproduced with permission: a, S. Wodak; b; M. James and Academic Press; c, F. M. Richards; d, R. L. Dunbrack.
Most recently, Dunbrack and Cohen (6) overcame the problem of sample size by using 518 proteins at ≤2 Å resolution, which resulted in a smooth and reproducible overall Asn χ2 distribution (Fig. 2d). Separating the χ2 distribution by the three major χ1 values helps further because the peaks do not occur in the same places; however, all χ2 angles are populated over the full range. Not trusting the experimental amide orientations, their analysis combined values into the range from −90° to +90° with three equal bins; the resulting rotamers usually do not represent population clusters, and the mean angles are simply near the bin centers, although relative frequencies show the skew of the peak shape.
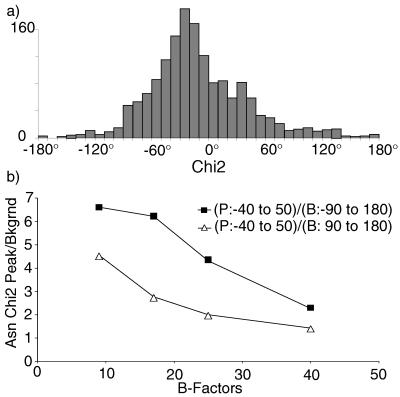
The Asn χ2 distribution for the raw data from our 240 files is indistinguishable from the Dunbrack and Cohen distribution shown in Fig. 2d. Fig. 3a shows our final, sharper distribution after B-factor, clash, and amide-flip correction, with few χ2 values near 180°. For the first time, there are unpopulated regions for the amide χ angles. The peak is still relatively broad and irregular, but when separated by χ1 and backbone conformation it resolves to several well-defined peaks.
Figure 3.
(a) Asn χ2 distribution from our 240-protein data set after all corrections. (b) Plot of peak/background ratio vs. B-factor range for the raw Asn χ2 values.
To obtain independent evidence for the validity of this shift of χ2 values away from 180°, we plotted the ratio of occurrence near 0° to occurrence in regions around 180° versus B-factor range for the uncorrected raw data. Fig. 3b shows that both ratios rise steeply and monotonically as B decreases and reliability improves, indicating that indeed there should be many fewer χ2 values near 180°.
Gln Side-Chain Conformations.
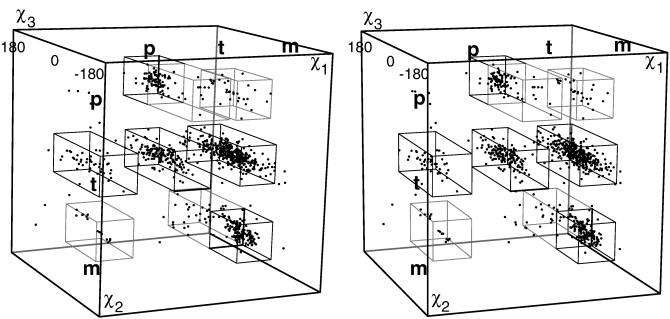
For Gln χ1 and χ2, the dihedral angles cluster tightly near the three possible staggered positions p, t, and m, as seen in the three-dimensional scatterplot in Fig. 4. However, the χ1–χ2 frequencies are not just the product of independent one-dimensional χ1 and χ2 distributions, either for peak positions or for relative frequencies. For instance, the ratio of χ2 m/p frequencies reverses from 5:1 (if χ1 is m) to 1:13 (if χ1 is t), with some peak positions offset by 20–30°. Part of such interdependence between successive χ angles can be explained by the fact that successive gauche dihedrals work well if they spiral in the same direction but produce unfavorable 1–5 contacts if they change direction (4, 24). The overall result is that, of the nine possible staggered positions for Gln χ1 and χ2, five show strong, well occupied clusters, another two (mp and pm) occur for only 2–3% of the examples, and two (pp and tm) are essentially forbidden (<1%).
Figure 4.
Three-dimensional scatterplot (in stereo) of χ1 vs. χ2 vs. χ3 values for corrected Gln with B <40, showing clusters at each rotamer. Boxes outline the ranges given in Table 1 and used for counting rotamer frequency.
In Fig. 4 the spread of points is broader in the χ3 dimension (even after all of the data corrections), but real clustering is visible in a distinctive pattern diagnostic of amide dihedral angles. Because χ3 is defined as the dihedral angle to the Oɛ atom (much smaller than the NH2 group), the cis region near χ3 = 0° is always more favorable than near 180°, so there are very few points near the front or back of the box. For the mm points (Lower Right) there is a major rotamer cluster around χ3 = −40°, whereas positive points are sparse and extend farther from zero. For the tp points (Upper layer, Middle), the large cluster is near χ3 = +40° and the negative side is sparse. That diagonal asymmetry arises from a clash to Hα, which occurs on opposite sides of 0° for χ2 m vs. for χ2 p.
The χ2 trans layer (Fig. 4 Middle) is populated over a wide range of χ3. For most of the region near χ3 = 180°, a Gln with ideal bond lengths and angles (18) clashes badly (e.g., Fig. 1); because the few points near χ3 trans in Fig. 4 have distorted bond angles, we do not define a rotamer in that region. However, a broad range of χ3 values between −90° and +90° is almost uniformly well populated, because the amide has only relatively weak interactions with Hγ and Hβ atoms. This conformational flexibility is inconvenient for modeling algorithms but surely is important to Gln’s role as a highly exposed polar residue interacting with solvent. Gln frequently has high B factors (29% of Gln in our starting data set had B ≥ 40 vs. 16% of Asn), and Gln is the most common uncharged polar residue on the exposed side of α-helices.
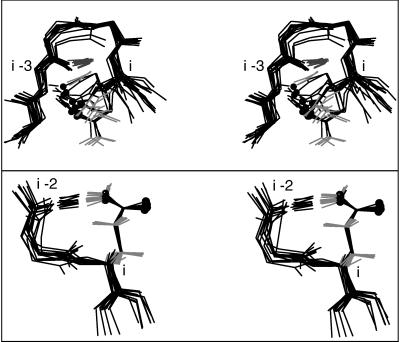
This flexibility of Gln also allows many possibilities for protein H bonding. Gln Oɛ can form a hydrogen bond with the backbone NH of residue i−3 (25) either around a tight turn or in the first turn of an α-helix; the latter pattern forms part of the helix N-terminal “capping box” (26). Our data set includes 32 cap-box Glns with i−3 H-bonds to the N-cap NH (e.g., 1AWD Q66), 31 of which have conformations near mt −30° ; the single N-cap+3 Gln with another rotamer (1ABA Q81) can approach from a different direction because the N-cap is a Gly. The data set includes 14 Gln i−3 H bonds around tight turns; 8 of them are mt−30°, but tt and tp also work in this arrangement. All 14 are superimposed in Fig. 5 Upper, showing that the Gln makes a pseudo antiparallel-β structure continuing past the tight-turn H bond. Both cap-box and turn-extension i−3 H bonds also are favorable for Glu.
Figure 5.
Superimposed examples, in stereo, of (Upper) Gln side- chain-to-backbone i−3 H bonds around a tight turn; (Bottom) Gln i−2 H bonds in pseudo-turns, all in the mt−30° rotamer.
Gln residues can form a pseudo-turn with an Nɛ to backbone CO (i−2) H bond. The 13 examples are shown in Fig. 5 Lower, with very tightly clustered conformations in the favorable mm−40° rotamer. Also, a Gln Oɛ can form a hydrogen bond to its own backbone NH, although the geometry is not ideal; such cases make up much of the mp0° and pm0° minor rotamers, and about 1/3 of them are in the first turn of α-helix.
Table 1 lists the five major rotamers that together account for 78% of the glutamine examples, plus four minor rotamers with <3% occurrence frequency. In strict α-helix, the distribution is similar except for the absence of χ1 p, so helical rotamers are not listed separately for Gln.
Table 1.
Glutamine rotamers
| No. | % | χ1,° | Range | χ2,° | Range | χ3,° | Range | |
|---|---|---|---|---|---|---|---|---|
| Major | ||||||||
| mt-30° | 304 | 35.2 | −67 | −35, −95 | 177 | −150, 150 | −25 | −90, 90 |
| tt 0° | 140 | 16.2 | −174 | −145, 155 | 173 | −150, 150 | −5 | −90, 90 |
| mm-40° | 126 | 14.6 | −66 | −35, −95 | −60 | −30, −90 | −40 | −90, 0 |
| tp60° | 77 | 8.9 | −175 | −150, 150 | 64 | 35, 95 | 60 | 0, 90 |
| pt 20° | 37 | 4.3 | 64 | 35, 95 | 180 | −150, 150 | 20 | −90, 90 |
| Minor | ||||||||
| mp0° | 25 | 2.9 | −60 | 85 | 0 | |||
| mm100° | 22 | 2.6 | −65 | −65 | 100 | |||
| pm0° | 15 | 1.7 | 75 | −75 | 0 | |||
| tp-100° | 14 | 1.6 | 180 | 65 | −100 | |||
| 760 | 88.0 |
Asn Side-Chain Conformations.
Asn exhibits the greatest backbone dependence of any amino acid for χ2 (5, 6) because it is short and asymmetrically polar. Therefore, we divide Asn residues into strictly α-helical, left-handed (0° < φ < 175°), β-sheet, and other. Fig. 6a plots χ1 vs. χ2 for the helical and the left-handed Asn side chains; contours give a smoothed representation of the distribution of points. There are sharp, clearly defined clusters as well as forbidden regions.
Figure 6.
(a) Plot of Asn χ1 vs. χ2 values for the 166 left-handed examples (red triangles and contours) and for the 310 strictly α-helical examples (×s and black contours). Contours representing the Gaussian smoothed distributions are shown in regions of clustering and stray points elsewhere to indicate the remaining scatter in the data. Note that χ1p is forbidden for left-handed as well as helical Asn. The two large α peaks for χ1m represent two mutually exclusive ways in which Asn interacts with residue i−4 on an α-helix. (b) Asn-99 of 1UDC with χ2 = −81°, for which Nδ forms H bonds to the i−4 backbone CO (shown by the overlapping dots). (c) Asn-12 of 1AB1 with χ2 = −21°, in which the amide makes broad van der Waals contact with the i−4 CO.
One unusual feature of Asn is that it is much more likely than any other non-Gly residue to be left-handed, probably because its ability to mimic a peptide makes it more nearly symmetric around the Cα atom (27). The 166 left-handed (+φ) Asn residues that compose 11% of this data set (compared with 4% for Asp and even less for other amino acids) have φ, ψ values that show a very tight distribution in the Lα/L310 region along a diagonal from +55°, +50° to +70°, +10°. As seen in Fig. 6a, left-handed Asns are quite constrained and have only two rotamers (Lm−30° and Lt30°), located close to the helical rotamers but are significantly offset. Neither L rotamer makes an H bond to the adjacent peptides. χ1 p is forbidden for left-handed Asn, because there is no room for the NH2 in that position, whereas Oδ would interact unfavorably with both peptide oxygens.
Although Asn is disfavored in strict α-helix, there are 310 in the data set. Three well-clustered major rotamers (see Fig. 6a) account for 87% of the Asns: αm−20° (65% of α Asn), αm−80°, and αt60°. Both αm rotamers interact with the i−4 CO of the preceding helix turn [αm−80° by H-bonding (Fig. 6b) and αm−20° by van der Waals contact (Fig. 6c)]. Not only are the two peaks well separated in χ1–χ2 values, but also, a given α-helical backbone usually is compatible with only one of the two possibilities. H-bonded αm−80° Asns usually are regular enough to qualify as α-helix, but they often involve distortion: either the Asn has 310 φ, ψ values or the turn between i−4 and i is slightly unwound; this is the same configuration described as a type of helix C-cap (28), and is indeed more common near the C-cap, but it also occurs in helix interiors. In either case the normal helical H bond of CO (i−4) also is present. The strongly preferred αm−20° rotamer not only makes good contact with the preceding turn, it also has the relationship with its own backbone that is most preferred for Asn in general (with Oδ lying against the backbone N). The αt60° rotamer is interesting because it has a tightly clustered peak but makes only weak local interactions. However, for about 2/3 of the αt60° examples, there is a crystallographically identified water making H bonds to both Asn NH2 and the i−3 CO.
The sparse trails of additional points in Fig. 6a, for which we assign two minor rotamers, represent permissible but not favored conformations. Their local interactions are weak and uncommon (e.g., a contact with Hα near m100° and a water bridging Oδ and CO near t40°), and they are stabilized by a variety of long-range interactions. As usual on strict α-helix, χ1 p is forbidden for Asn because it would sterically intersect the preceding turn.
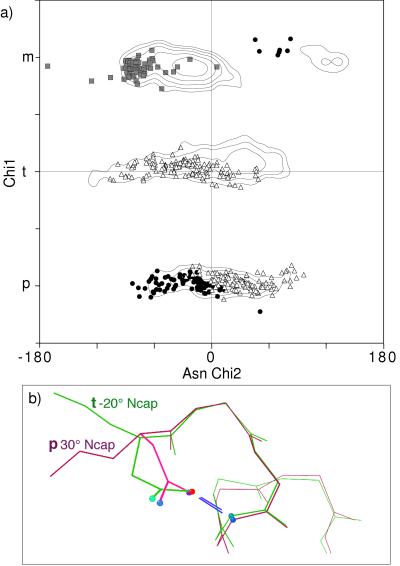
Asn is rare in the central portions of β-sheets but fairly frequent on edge strands, in β-bulges, or at the ends of β-strands. The χ1–χ2 distribution for β Asn is similar to that for all Asn but with enough changes in peak position or frequency that separate β rotamers are defined. For example, β Asn show no NH(i) H bonds, and thus no pm examples.
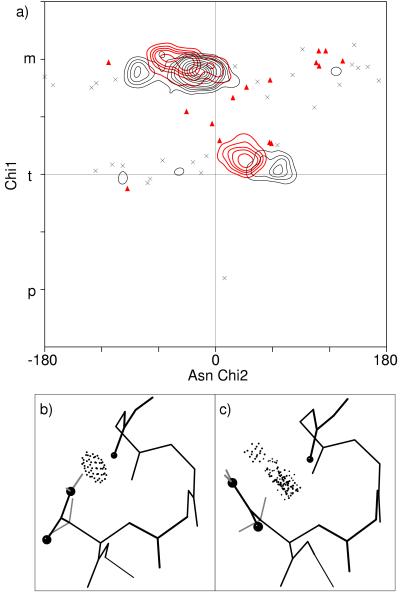
For the numerous Asn in turns, loops, and transitions, the distribution is largely dominated by the local side-chain-to-backbone H-bond patterns plotted in Fig. 7a. Overall, Asn has about twice as high a χ1 p percentage as most amino acids because the minor clash with backbone can be compensated by local H bonds. Local H-bond patterns common for Asn are NH(i) to Oδ and NH(i+2 or i+3) to Oδ (ref. 25 and our data). The χ1–χ2 distributions for i+2 and i+3 are indistinguishable and many examples have both, so they are plotted and discussed together. Both i and i+2/i+3 patterns occur in two quite different rotamers, which angle their approach to the NH from different sides, as seen for the N-cap examples in Fig. 7b. The two forms have been described specifically for helix N-caps (29), but they are general for all pseudo-turns with i+2 and/or i+3 H bonds: either a p30° rotamer with β φ,ψ or a t−20° rotamer (χ2 often near −60°) with poly(Pro) φ,ψ.
Figure 7.
a) Asn χ1 vs. χ2 points for the commonly occurring classes of local side-chain-backbone H bonds: i−4 (gray squares), i (•), and i+2 or i+3 (▵). Contours (at 2, 4, 8, 16, and 32) represent the smoothed distribution for all Asn. (b) Superimposed examples of the two different Asn conformations that make H-bonded helix N-caps. In pink is Asn-22 of 256B with β φ, ψ values and a p30° rotamer, and in green is Asn-153 of 1IXH with poly(Pro) φ,ψ values and a t−20° rotamer.
The second dominant influence on χ1–χ2 patterns for both general and “other” Asn is simply the favored orientations for the amide group relative to the adjacent peptides. The top peak in the all-Asn distribution is a tight cluster (as indicated in Fig. 7a) whose high local concentration is reflected in a height 3.5 times that of the next highest peak of the smoothed contours. That top peak has χ1 m, putting Cγ between the backbone Hα and NH; near χ2 −20°, the side-chain amide lies against the preceding peptide N with good van der Waals contacts. For t30° the same sort of contacts happen in relation to the following carbonyl carbon.
Table 2 lists a set of seven backbone-independent rotamers that can be used for the general case of Asn side chains. For strict α, for β-sheet, and for left-handed Asn, individual sets of major and minor rotamers are defined.
Table 2.
Asparagine rotamers
| No. | % | χ1,° | Range | χ2,° | Range | |
|---|---|---|---|---|---|---|
| Backbone-independent: | ||||||
| m-80° | 118 | 7.9 | −71 | −40, −100 | −76 | −100, −60 |
| m-20° | 582 | 39.0 | −71 | −40, −100 | −23 | −60, 10 |
| m120° | 58 | 3.9 | −64 | −40, −100 | 132 | 60, 160 |
| t30° | 235 | 15.7 | −168 | −140, 160 | 31 | 0, 80 |
| t-20° | 179 | 12.0 | −174 | −140, 160 | −20 | −120, 0 |
| p-10° | 103 | 6.9 | 63 | 30, 90 | −13 | −90, 0 |
| p30° | 132 | 8.8 | 64 | 30, 90 | 34 | 0, 90 |
| Major, backbone-dependent: | ||||||
| αm-20° | 204 | 65.8 | −72 | −45, −105 | −17 | −60, 10 |
| αm-80° | 26 | 8.4 | −71 | −45, −105 | −81 | −100, −60 |
| αt60° | 40 | 12.9 | −175 | −140, 160 | 64 | 30, 80 |
| βm-50° | 73 | 35.8 | −66 | −35, −95 | −49 | −90, 0 |
| βt 10° | 77 | 37.7 | −179 | −150, 150 | 11 | −90, 90 |
| Lm-30° | 91 | 54.8 | −65 | −40, −100 | −30 | −70, 10 |
| Lt30° | 58 | 34.9 | −166 | −130, 170 | 32 | 0, 60 |
| Minor, backbone-dependent: | ||||||
| αm100° | 9 | 2.9 | −70 | 120 | ||
| αt-60° | 10 | 3.2 | −175 | −60 | ||
| βm100° | 7 | 3.4 | −65 | 120 | ||
| βp60° | 17 | 8.3 | 65 | 60 | ||
DISCUSSION
Asn and Gln χ distributions are herein shown to have discrete and sometimes quite sharp clusters suitable for defining useful side-chain rotamers. The growing database is still improving rotamer statistics, but application of the corrections and filters used here also makes a large difference comparable to the improvement obtained in the 1980s by restriction to structures refined at high resolution. The unfortunate cycle, in which incorrectly oriented side-chain amides influence rotamer libraries which influence further structures, can be broken by using both H bonding and explicit-H atomic clashes to correct those orientations and by omitting the ambiguous ones. Furthermore, B-factor cutoffs make as much difference as resolution and homology criteria and are even easier to apply—we feel they should be standard practice in any statistical analysis of conformations.
Most side-chain rotamers are dominated by the negative effects of avoiding atomic clashes. However, well defined rotamer clusters also can be produced by favorable interactions: the sharp peak at m−20°, the most common Asn rotamer, reflects good contact of the amide with the peptide N, whereas the tightly clustered αt60° Asn rotamer is caused by a bridging water between Nδ and CO (i−3).
Unusual χ angles can be enforced by structural strain or compensated by other interactions: for example, an eclipsed χ1 angle can apparently be tolerated for the sake of four or five H bonds. Less extreme cases of this sort make the distinction between minor rotamers and nonrotamers an unclear one. Both for modeling calculations and for fitting experimental structures, the well populated rotamers should always be tried first; then rare or even “forbidden” conformations can be considered, but only when there is good evidence for them.
Acknowledgments
We thank Hope C. Taylor and Brent K. Presley for preparing ideal-geometry side chains. This work was supported by National Institutes of Health Research Grant GM-15000, by use of the Duke Comprehensive Cancer Center Shared Resource for Macromolecular Graphics, and by an educational leave for J.M.W. from Glaxo Wellcome.
References
- 1.Ponder J W, Richards F M. J Mol Biol. 1987;193:775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- 2.Chandrasekaran R, Ramachandran G N. Int J Prot Res. 1970;2:223–233. [PubMed] [Google Scholar]
- 3.Janin J, Wodak S, Levitt M, Maigret B. J Mol Biol. 1978;125:357–386. doi: 10.1016/0022-2836(78)90408-4. [DOI] [PubMed] [Google Scholar]
- 4.Bhat T N, Sasisekharan V, Vijayan M. Int J Pept Prot Res. 1979;13:170–184. doi: 10.1111/j.1399-3011.1979.tb01866.x. [DOI] [PubMed] [Google Scholar]
- 5.McGregor M J, Islam S A, Sternberg M J E. J Mol Biol. 1987;198:295–310. doi: 10.1016/0022-2836(87)90314-7. [DOI] [PubMed] [Google Scholar]
- 6.Dunbrack R L, Jr, Cohen F E. Protein Sci. 1997;6:1661–1681. doi: 10.1002/pro.5560060807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuffery P, Etchebest C, Hazout S. Protein Eng. 1997;10:361–372. doi: 10.1093/protein/10.4.361. [DOI] [PubMed] [Google Scholar]
- 8.Schrauber H, Eisenhaber F, Argos P. J Mol Biol. 1993;230:592–612. doi: 10.1006/jmbi.1993.1172. [DOI] [PubMed] [Google Scholar]
- 9.Kuszewski J, Gronenborn A M, Clore G M. J Magn Reson. 1997;125:171–177. doi: 10.1006/jmre.1997.1116. [DOI] [PubMed] [Google Scholar]
- 10.McDonald I K, Thornton J M. Protein Eng. 1994;8:217–224. doi: 10.1093/protein/8.3.217. [DOI] [PubMed] [Google Scholar]
- 11.Hooft R W W, Sander C, Vriend G. Proteins Struct Funct Genet. 1996;26:363–376. doi: 10.1002/(SICI)1097-0134(199612)26:4<363::AID-PROT1>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 12.Word, J. M., Lovell, S. C., LaBean, T. H., Taylor, H. C., Zalis, M. E., Presley, B. K., Richardson, J. S. & Richardson, D. C. (1999) J. Mol. Biol., in press. [DOI] [PubMed]
- 13.Word, J. M., Lovell, S. C., Richardson, J. S. & Richardson, D. C. (1999) J. Mol. Biol., in press. [DOI] [PubMed]
- 14.Jones T A, Zou J-Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein F C, Koetzle T F, Williams G J B, Meyer E F, Brice M D, Rodgers J R, Kennard O, Shimanouchi T, Tasumi M. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 16.Richardson D C, Richardson J S. Protein Sci. 1992;1:3–9. doi: 10.1002/pro.5560010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 18.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 19.Benedetti E, Morelli G, Nemethy G, Scheraga H A. Int J Pept Protein Res. 1983;22:1–15. doi: 10.1111/j.1399-3011.1983.tb02062.x. [DOI] [PubMed] [Google Scholar]
- 20.Eliel E L, Wilen S H. Stereochemistry of Organic Compounds. New York: Wiley; 1994. [Google Scholar]
- 21.IUPAC-IUB Commission on Biochemical Nomenclature. J Mol Biol. 1970;52:1–17. [PubMed] [Google Scholar]
- 22.James M N G, Sielecki A R. J Mol Biol. 1983;163:299–361. doi: 10.1016/0022-2836(83)90008-6. [DOI] [PubMed] [Google Scholar]
- 23.Kuszewski J, Gronenborn A M, Clore G M. Protein Sci. 1996;5:1067–1080. doi: 10.1002/pro.5560050609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunbrack R L, Jr, Karplus M. Nat Struct Biol. 1994;1:334–340. doi: 10.1038/nsb0594-334. [DOI] [PubMed] [Google Scholar]
- 25.Baker E N, Hubbard R E. Prog Biophys Mol Biol. 1984;44:97–179. doi: 10.1016/0079-6107(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 26.Harper E T, Rose G D. Biochemistry. 1993;32:7605–7609. doi: 10.1021/bi00081a001. [DOI] [PubMed] [Google Scholar]
- 27.Richardson D C, Richardson J S. In: Prediction of Protein Structure and the Principles of Protein Conformation. Fasman G D, editor. New York: Plenum; 1989. pp. 1–98. [Google Scholar]
- 28.Kumar S, Bansal M. Proteins Struct Funct Genet. 1998;31:460–476. doi: 10.1002/(sici)1097-0134(19980601)31:4<460::aid-prot12>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 29.Doig A J, MacArthur M W, Stapley B J, Thornton J M. Protein Sci. 1997;6:147–155. doi: 10.1002/pro.5560060117. [DOI] [PMC free article] [PubMed] [Google Scholar]