Abstract
To document genomic changes during long periods of storage, we analyzed Salmonella enterica serovar Typhimurium LT7, a mutator strain that was previously reported to have higher rates of mutations compared to other serovar Typhimurium strains such as LT2. Upon plating directly from sealed agar stabs that had been stocked at room temperature for up to four decades, many auxotrophic mutants derived from LT7 gave rise to colonies of different sizes. Restreaking from single colonies consistently yielded colonies of diverse sizes even when we repeated single-colony isolation nine times. Colonies from the first plating had diverse genomic changes among and even within individual vials, including translocations, inversions, duplications, and point mutations, which were detected by rare-cutting endonuclease analysis with pulsed-field gel electrophoresis. Interestingly, even though the colony size kept diversifying, all descendents of the same single colonies from the first plating had the same sets of detected genomic changes. We did not detect any colony size or genome structure diversification in serovar Typhimurium LT7 stocked at −70°C or in serovar Typhimurium LT2 stocked either at −70°C or at room temperature. These results suggest that, although colony size diversification occurred during rapid growth, all detected genomic changes took place during the storage at room temperature and were carried over to their descendents without further changes during rapid growth in rich medium. We constructed a genomic cleavage map on the LT7 strain that had been stocked at −70°C and located all of the detected genomic changes on the map. We speculated on the significance of mutators for survival and evolution under environmentally stressed conditions.
Comparative studies of bacteria have demonstrated the high level of conservation of the basic genome structure, i.e., the physical arrangement of DNA segments on the genome, in evolution. The earliest known example comes from the comparison of Salmonella enterica serovar Typhimurium and Escherichia coli, bacteria of two genera that have diverged for over 100 million years (6, 8, 39) but still have very similar genome structures (19). Specifically, they have similar genome sizes, similar sets of genes, and similar gene orders on the genome. Bacteria with closer phylogenetic relationships have even greater similarity. For instance, wild-type strains of serovar Typhimurium isolated from different sources have an indistinguishable genome structure as measured by I-CeuI cleavage patterns (25).
Genomic diversification does occur among cells of the same clone in culture and presumably also in nature, including duplication and inversion of genomic segments, at frequencies as high as 10−5 to 10−3 (3, 13). However, most of these types of genomic changes are selected against in nature and therefore cannot be easily detected. In order to capture such genomic changes for the elucidation of the basic rules in genomic diversification during evolution, we examined serovar Typhimurium cultures that had been stocked for several decades in sealed agar stabs at room temperature, a condition allowing slow growth for some time until the nutrients become exhausted. This archival system of serovar Typhimurium cultures provides a unique chance to detect genetic and genomic changes. Assuming that the originally uniform medium would become distinct in different parts of the stab with the growth of the bacteria and their competition with one another, we would expect to see survivors with unique sets of spontaneous genomic changes selected in different parts of the stab in adaptation to the local specific ecological conditions. We aim at detecting the hypothesized genomic changes and documenting the genomic diversification in this system. We have a collection of spontaneous mutants of serovar Typhimurium (43), including those derived from strain LT2 (16) and those from strain LT7 (15, 16, 35). When a program of developing a chromosomal map of serovar Typhimurium was initiated a half century ago, it was recognized that auxotrophic mutants selected from the LT2 strain had low rates of reversion to prototrophy. However, auxotrophs selected from LT7 had high reversion rates, making it difficult to distinguish recombinants from revertants upon transduction with phage P22. Therefore, the resulting chromosomal map was constructed with LT2, not LT7.
The LT7 strain was initially designated a phenotypic mutator and later shown to be due to a mut defect (15, 35). This genotype would indicate a possibility that populations within the LT7 culture might have greater diversity than those within the LT2 culture. In a preliminary parallel study with LT2 and LT7 mutants, we found great uniformity with LT2 mutants; however, the archival LT7 mutants (in stabs at room temperature) showed much diversity in both colony morphology and genome structure. In contrast, the wild-type strain of LT7 that had been stocked at −70°C did not show variation in colony morphology or genome structure. Diversification of genome structure of the archived LT7 mutants included inversions, translocations, duplications, and point mutations. Interestingly, these genomic changes obviously occurred during archiving in the past four decades, and no further changes had been observed when the bacteria were grown in rich medium for up to nine successive single-colony isolations. We now report the detected genomic changes of serovar Typhimurium LT7 during the diversification process and discuss their possible roles in evolution.
MATERIALS AND METHODS
Bacterial strains.
Serovar Typhimurium wild-type strains LT2 (SGSC1412) and LT7 (SGSC1417) were from the set of strains isolated by Lilleengen (16) in the 1940s as representative strains of phage types LT1 through LT22. These strains were analyzed by Zinder and Lederberg (43) and provided by J. Lederberg to the Salmonella Genetic Stock Center (SGSC) as lyophils; they are now maintained in lyophils and in 15% glycerol at −70°C. The serovar Typhimurium mutants were originally isolated in the laboratory of M. Demersec in the 1950s and, since the 1960s, have been stored in the laboratory of A. Eisenstark at room temperature in sealed agar stab cultures, 0.5 ml per agar stab vial. Over 20,000 stab cultures were collected for genetic mapping. Preliminary experiments showed that genome structure of most of the LT2-derived mutants archived at room temperature or stored in 15% glycerol at −70°C was very stable, whereas many of the LT7-derived mutants showed great diversity in genome structure as well as in colony morphology. Therefore, we concentrated on some randomly chosen strains of the LT7-derived mutants for the present study. The strains we used in the present study were: 8111 (met-115), 8117 (met-121), 8608 (pro-64), 8618 (pro-74), 9052 (ser-85), and 9059 (ser-92), with LT2 (SGSC1412) and LT7 (SGSC1417), which had been stocked at −70°C, being included for comparisons. All strains and their derivatives (see Results) are available from the SGSC (www.ucalgary.ca/∼kesander). Tn10 insertion mutants of serovar Typhimurium LT2 were obtained from numerous sources and are described elsewhere (2, 42); they were used for mapping the LT7 genome.
Transfer of Tn10 insertions through bacteriophage P22-mediated transduction.
A large number of Tn10 insertions into genes with known functions have been mapped on the genome of serovar Typhimurium LT2 (21). We transferred Tn10 insertions from serovar Typhimurium LT2 to serovar Typhimurium LT7 SGSC1417 by bacteriophage P22-mediated transduction to locate the same genes through homologous recombination. We made P22 lysates from a selected set of Tn10 insertion mutants of serovar Typhimurium LT2 by growing a 3-ml overnight culture in Luria-Bertani (LB) broth of these selected Tn10 mutants and inoculating these cultures with phage P22 at a multiplicity of infection of 1:100, followed by coincubation for 6 h. After destruction of living bacterial cells by two drops of chloroform and removal of the cell debris by centrifugation, the lysates, at 1011 PFU/ml, were ready for use in the transduction. For transferring the Tn10 insertions to serovar Typhimurium LT7, we spread 100 μl of an overnight culture of serovar Typhimurium LT7 and 20 μl of lysate onto an LB plate containing tetracycline. A colony was picked up and restreaked on another tetracycline plate for single-colony isolation. One colony from the second tetracycline plate was used for phenotype tests and mapping.
Enzymes and chemicals.
I-CeuI, AvrII, and SpeI were purchased from New England BioLabs; XbaI and proteinase K were from Boehringer Mannheim. [32P]dCTP was from New England Nuclear. Most other chemicals were from the Sigma Chemical Co.
PFGE methods and genomic mapping.
Preparation of intact genomic DNA, endonuclease cleavage of DNA in agarose blocks, and separation of the DNA fragments by pulsed-field gel electrophoresis (PFGE) were as described previously (22, 29). PFGE was performed with the Bio-Rad CHEF Mapper or Bio-Rad CHEF DRII electrophoresis system. For PFGE, we normally use three cycles of conditions: the first for general separation at 30 s ramping to 90 s for 16 h at 6 V/cm and buffer temperature of 12°C, the second for expanding areas with closely packed small bands at 3 s ramping to 6 s, and the third for expanding areas with closely packed large bands at pulsing times based on the sizes of the bands. The total run times for the second and third cycles were usually 6 to 12 h, depending on the extent of the separation. Most runs were carried out at 120° angle. For very crowded areas of bands, a 150° angle was used. For determining the sizes of DNA fragments on the PFGE gel, we most often used only λ ladder (New England Biolabs) as the size marker, but in many cases we also used bacterial genomic DNA cleaved with an endonuclease as markers. Among the ones we often used was serovar Typhimurium LT2 or serovar Typhi Ty2 DNA cleaved with XbaI, AvrII, or SpeI; sizes of these fragments had been determined previously (21, 24). Genomic mapping methods with I-CeuI were described (19) and further optimized (25). The techniques of double cleavage and end labeling were also described previously (21).
Long-range PCR procedure.
For the preparation of templates, genomic DNA was extracted with the Promega Wizard genomic DNA purification kit (catalog no. A1120). We used the following 16 primers: primer 1, GGAACGTTGAAGACGACGAC; primer 2, GCAAGCTGCTTCCTGTTACC; primer 3, TCAATGCTGGAAAAGTCTTGC; primer 4, AACTGGTTCCTGGCAAAGTG; primer 5, CAGGCGCTCAGTAGTTGTTG; primer 6, CCCGTTTTACAGCGTTATGG; primer 7, AACCGAATGCAGGGATAGTC; primer 8, AAATGTCGGGACAAAAGTGC; primer 9, CATTTGGCTGCAAAACAGC; primer 10, CTGGGCGAATTCGATGATAC; primer 11, TTGGGAAACCGTATCCATTG; primer 12, AGACGGACATCGCCAATAAC; primer 13, GCGGTGGTTAGAGAAAGCAC; primer 14, CTATTTCGCCGGGAAGAATC; primer 15, ATGGGTATCATTGCGCTTTC; and primer 16, GCCGATGTCGTTAATCTGC. The locations of all of the primers except primers 1 and 2, which amplify a 0.5-kb segment of genomic DNA as a positive control of the conditions for the experiments, are shown on the map of LT7 (see Fig. 7B). The lower mix included 2 μl of deoxynucleoside triphosphates (10 mM), 4 μM forward primer (5 μl), 4 μM reverse primer (5 μl), and template (10 μl). After the mixture was heated at 90°C for 30 s, the upper mix (10× PCR buffer, 5 μl; 25 mM MgCl2, 3 μl; Tap polymerase, 0.25 μl; double-distilled H2O, 19.75 μl) was added. PCR was carried out for 30 cycles with denaturation at 96°C for 1 min, annealing at 57°C for 1 min, and extension at 72°C for 10 min. The PCR products were analyzed by gel electrophoresis with 1% agarose. We used a procedure similar to that described by Helm and Maloy (12).
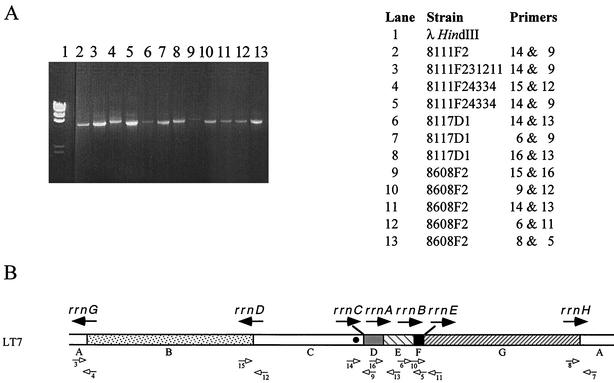
FIG. 7.
Long-range PCR to confirm the genomic changes revealed by PFGE. (A) PCR products on an agarose gel, with the templates and primers shown on the right; (B) locations and orientations of the primers shown on the genome of LT7. HindIII-cleaved λ DNA was used as the DNA fragment size marker; negative controls of PCR products are not shown in this figure.
RESULTS
Diverse colony morphology of the archived serovar Typhimurium LT7 mutants.
Replicate vials for each of the serovar Typhimurium mutants have been archived. One vial from each mutant strain was opened, and the content was smashed into 3 ml of LB broth. An aliquot of 100 μl of such suspension was spread onto an LB plate and incubated at 37°C. The rest of the suspension was incubated by shaking at 37°C overnight before being given an SGSC number and frozen at −70°C in 15% glycerol. On inspection of the plates the next day, we found that mutants derived from serovar Typhimurium LT2, as well as reference strains of LT2 (SGSC1412) and LT7 (SGSC1417), produced uniform colonies, whereas colonies of mutants derived from serovar Typhimurium LT7 were very diverse. We scored the colonies with the letters A, B, C, D, E, and F, with the letters representing colony sizes (in diameter) as follows: A, >4 mm; B, 2 to 4 mm; C, 1 to 2 mm; D, 0.5 to 1 mm; E, 0.3 to 0.5 mm; and F, <0.3 mm. We then tried to “purify” the colonies by streaking individual colonies out onto another LB plate. However, the purification was not possible, because colonies from the single colonies showed diversity again. For example, from strain 8111, we obtained six colonies of different sizes, A, B, C, D, E, and F on the first plating, as defined above. When we picked up a single colony from 8111A and streaked it onto another LB plate, we obtained colonies with a whole range of sizes from >4 mm to <0.3 mm, which we designated 1, 2, 3, 4, 5 and 6, instead of A, B, C, D, E and F, respectively. For instance, 8111F231213 means: from a tiny colony of strain 8111 (8111F), we obtained a normal-sized colony (in addition to the other five sizes), 8111F2. From 8111F2, we in turn obtained a small colony (8111F23), a large colony (8111F231), a normal-sized colony (8111F2312), a large colony (8111F23121), and a small colony (8111F231213), all in addition to the other five sizes. In every replating, we obtained colonies of all sizes. The colony morphology diversification continued to occur and did not stop even at the ninth plating when we stopped the restreaking. Such colony size diversification was not observed with the LT2 lineages stocked at room temperature or −70°C or the LT7 wild-type strain that had been frozen at −70°C.
We suspected that there might be genomic changes among the bacteria with diverse colony sizes, which may be revealed through genomic analyses. For that, we needed first to make a genome map for LT7 and then locate the possible genomic changes on the map.
Endonuclease cleavages of serovar Typhimurium LT7 genomic DNA and the use of Tn10.
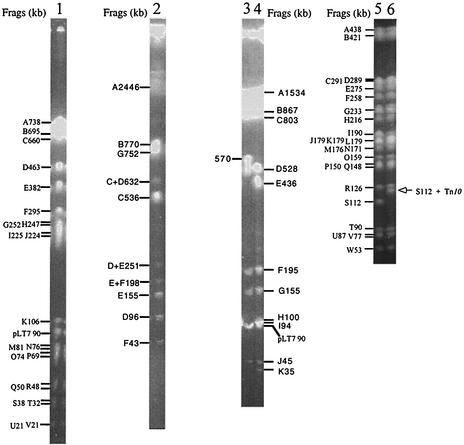
The wild-type serovar Typhimurium LT7, SGSC1417, which had been stocked at −70°C and did not show diversification in colony size or genome structure (see below), was used to construct a genome map. Cleavages of genomic DNA of serovar Typhimurium LT7 with XbaI, I-CeuI, AvrII, and SpeI generated 25, 7, 13, and 35 fragments, respectively (Fig. 1). Taking advantage of the XbaI and AvrII cleavage sites within the Tn10 DNA sequence, we located a number of genes on the LT7 genome as landmarks through the Tn10 insertions transferred from LT2. Most of the Tn10 insertions were located at homologous sites in the genome of LT7. In the case of a Tn10 insertion mutant, an original XbaI fragment was usually missing because of the extra XbaI site in the Tn10. In the meantime, two new XbaI fragments would appear, adding up to the size of the missing XbaI fragment plus 9.3 kb, the size of Tn10; see the illustration in Fig. 3 in reference 22. AvrII works in a similar manner, except that it has two cleavage sites, one each in the IS10 sequences of Tn10 at the left and right ends. Therefore, an AvrII fragment will be split into three: two of the bacterial DNA (summing up to the wild-type fragment plus 2 kb, 1 kb each of the IS10 sequence from the end to the AvrII site) and one of the inner part of Tn10 between the two AvrII sites (ca. 7 kb). Unlike XbaI or AvrII, SpeI does not cut the Tn10 sequence. However, the SpeI fragments that have Tn10 insertions can be recognized by size increase (9.3 kb; see Fig. 1, lane 6). This feature is sometimes very advantageous for reliably assigning a Tn10 to a certain genomic location by the increased size of an SpeI fragment, whereas in the case of XbaI or AvrII, a Tn10 inserted a few kilobases to an end of a fragment may be difficult to locate on the genome.
FIG. 1.
PFGE patterns of genomic DNA of the wild serovar Typhimurium LT7 strain stored at −70°C cleaved with the endonucleases XbaI, AvrII, I-CeuI, and SpeI. Lanes: 1, XbaI cleavage of LT7 (3 small fragments—W [6.5 kb], X [6.4 kb], and Y [1 kb]—had run out of the gel); 2, I-CeuI cleavage of LT7; 3 and 4, AvrII cleavages of LT2 and LT7, respectively, for a comparison; two small fragments of LT7 had run out of the gel, including L (4 kb) and M (2 kb); 5, SpeI cleavage of LT7 (12 small fragments, X through II, ranging from 49 to 7 kb, had run out of the gel); 6, SpeI cleavage of LT7 serB::Tn10, showing the size shift of S112 with the Tn10 insertion (it is now 112 + 9, i.e., 121 kb).
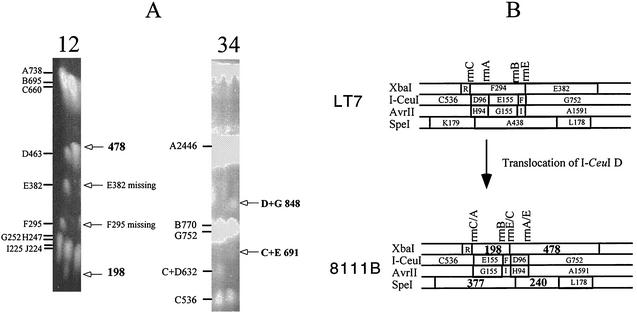
FIG. 3.
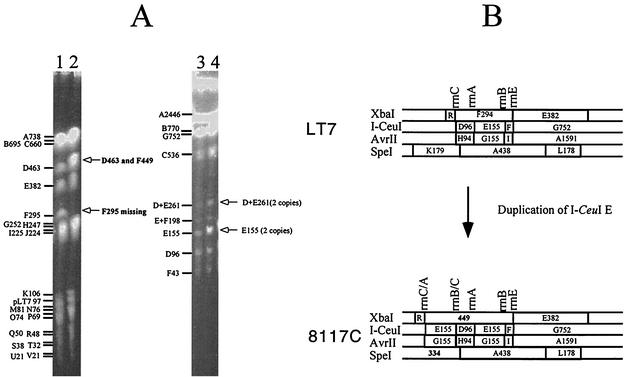
Genomic translocation exemplified by strain 8111B. (A) PFGE gels of LT7 and 8111B. Lanes: 1, LT7 DNA cleaved by XbaI, with the fragments and their sizes indicated on the left of the PFGE gel; 2, 8111B DNA cleaved by XbaI, with deviations of the cleavage pattern from that of LT7 indicated on the right of the PFGE gel; 3, LT7 DNA cleaved by I-CeuI, with the fragments and their sizes indicated on the left of the PFGE gel; and 4, 8111B DNA cleaved by I-CeuI, with deviations of the cleavage pattern from that of LT7 indicated on the right of the PFGE gel. (B) Local comparison of LT7 and 8111B showing the translocation of I-CeuI D, which resulted in three hybrid rrn operons, disappearance of two XbaI fragments (F and E) and two SpeI fragments (K and A [not shown on the PFGE picture]), and appearance of two new fragments each from XbaI (198 and 478 kb) and SpeI (377 and 240 kb) digestions.
Genome map of serovar Typhimurium LT7.
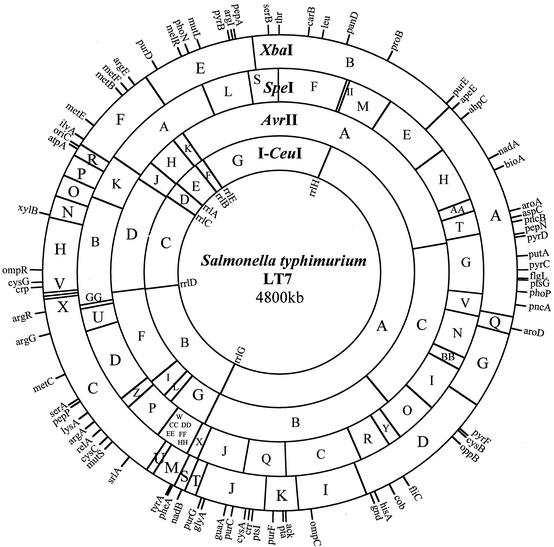
Analysis of the cleavage and Tn10 data led to the map of the LT7 genome, which is 4,800 kb in size (Fig. 2), 57 kb smaller than LT2 (33). The decrease in genome size in LT7 relative to LT2 is essentially because of the presence in LT2, but not in LT7, of Fels1 prophage (33) that contains nanH, the gene encoding sialidase (14). nanH is acquired by LT2 but not by other serovar Typhimurium strains and is located at 1,002 kb from thr. The location for oriC (at 3,918 kb from thr) was inferred from E. coli K-12 by its distances to uncA (atpA) and to rrlC. Overall, the LT7 map is very similar to that of LT2 (21), but there are differences that can distinguish LT7 from LT2, such as the AvrII sites at 2,935 kb, 2,970 kb, and 3,070 kb from thr (between genes srlA and argA), which are present in LT7 but not in LT2.
FIG. 2.
Genome map of serovar Typhimurium LT7. The seven rrn operons are shown inside the map.
Translocations.
On the LT7 map, we then located the genomic changes in the LT7 mutants, among which translocations were the most frequent events detected. All translocations we located had obviously occurred by homologous recombination between rRNA genes and therefore were easily revealed by I-CeuI analysis (29). Figure 3 shows translocation of a segment between rrnC and rrnA, i.e., I-CeuI fragment D, which was between I-CeuI C and I-CeuI E but now is inserted into a new location between I-CeuI F and I-CeuI G, generating three hybrid rrn operons.
Duplications.
I-CeuI E is duplicated in several lines of the 8117 derivatives, including 8117C, 8117D, 8117E, and 8117F, and other strains (8615A, etc.). I-CeuI F is duplicated in 8618E1. These duplications also obviously occurred through homologous recombination between rrn operons (Fig. 4).
FIG. 4.
Genomic duplication exemplified by strain 8117C. (A) PFGE gels of LT7 and 8117C. Lanes: 1, LT7 DNA cleaved by XbaI; 2, 8117C DNA cleaved by XbaI; 3, LT7 DNA cleaved by I-CeuI; and 4, 8117C DNA cleaved by I-CeuI. (B) Local comparison of LT7 and 8117C showing the duplication of I-CeuI E, which resulted in two hybrid rrn operons, the disappearance of the XbaI fragment F and SpeI fragment K (not shown on the PFGE picture), and the appearance of one new fragment each from XbaI (449 kb) and SpeI (334 kb) digestions.
Inversions.
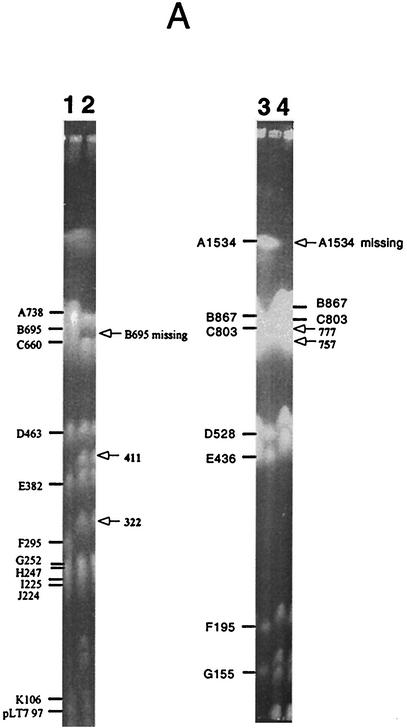
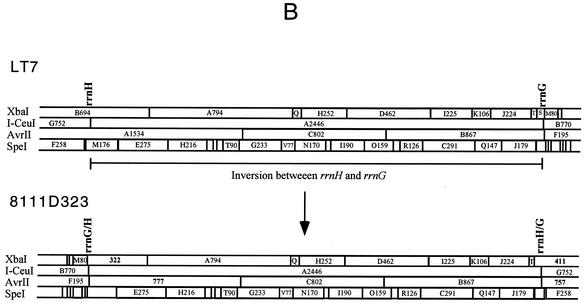
Inversions were detected in several strains as the result of recombination between rrnH and rrnG. Inversions resulting from recombination between rrn operons can be revealed by analyses with XbaI and AvrII or by long-range PCR (See below) but not by I-CeuI (Fig. 5).
FIG. 5.
Genomic inversion exemplified by strain 8111D323. (A) PFGE gels of LT7 and 8111D323. Lanes: 1, LT7 DNA cleaved by XbaI; 2, 8111D323 DNA cleaved by XbaI; 3, LT7 DNA cleaved by AvrII; and 4, 8111D323 DNA cleaved by AvrII. (B) Local comparison of LT7 and 8111D323 showing the inversion of I-CeuI A, which resulted in two hybrid rrn operons, the disappearance of XbaI B and AvrII A, and appearance of two new fragments each from XbaI (411 and 322 kb) and AvrII (777 and 757 kb) digestions.
Point mutations.
Point mutations were inferred here by the gain or loss of endonuclease cleavage sites (Fig. 6). They were inferred only after all other possible causes that might result in the disappearance of the original endonuclease cleavage sites or the appearance of new endonuclease cleavage sites had been ruled out. In the present study, disappearance of the endonuclease cleavage sites caused by translocations, duplications, or inversions of genomic DNA segments can be easily and unambiguously distinguished as described above.
FIG. 6.
Point mutations as inferred from the creation or loss of endonuclease cleavage sites. (A) XbaI-cleaved genomic DNA of LT7 (lane 1) and 9059A (lane 2); (B) AvrII-cleaved genomic DNA of LT7 (lane 1) and 8608D (lane 2); (C) I-CeuI-cleaved genomic DNA of LT7 (lane 1) and 8111F231213 (lane 2). For these three strains, genomic translocations, duplications, and inversions had been ruled out.
Confirmation of genomic rearrangements by long-range PCR.
Although most of the genomic changes reported above could be detected very efficiently by PFGE, we wanted to confirm some of them by other methods to solve any potential ambiguities, especially inversions between rrnH and rrnG and those between rrnD and rrnC. Using long-range PCR, we confirmed all of the translocations, duplications, and inversions that had been revealed by PFGE analysis. Figure 7 shows some examples out of a series of well-controlled experiments. In each case, if a PCR product was obtained with the pair of primers that would amplify the rrn operon in LT7 (Fig. 7B), the neighboring relationship between the two I-CeuI fragments flanking that particular rrn operon would be confirmed to be the same as in LT7, such as strains 8111F2, 8111F231211, and 8111F24334 for I-CeuI C and D (primers 14 and 9), 8111F24334 for I-CeuI B and C (primers 15 and 12), 8117D1 for I-CeuI D and E (primers 16 and 13), and 8618D1 for I-CeuI A and B (primers 3 and 4 [data not shown in Fig. 7]). Otherwise, the PCR products would clearly reveal a rearrangement, such as 8117D1 for I-CeuI C and E (primers 14 and 13) and for I-CeuI E and D (primers 6 and 9), and all combinations of 8608F2 (Fig. 7). Note that, with 8117D1, both primer pair 6 and 9 (Fig. 7, lane 7) and primer pair 16 and 13 (Fig. 7, lane 8) yielded positive results, a situation that would only be possible if there are two I-CeuI E fragments, and they sandwich I-CeuI D (i.e., ---E-D-E---); in Fig. 4, 8117C illustrates a similar situation.
Diversity of the global structure of the genome among the serovar Typhimurium LT7 mutants.
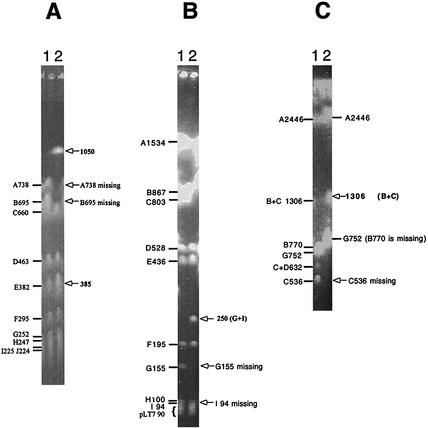
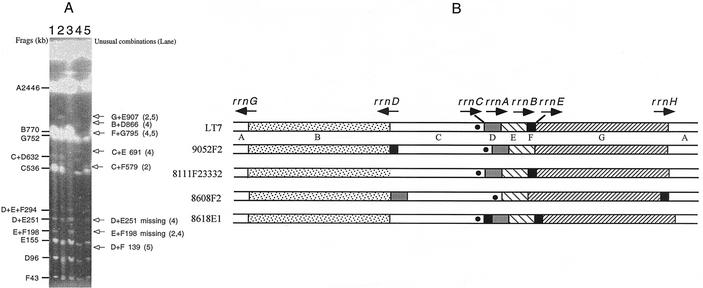
Translocations, duplications, inversions, and point mutations all contributed to diversification of genome structure in the serovar Typhimurium LT7 mutants. Although many of the genomic changes reported above seemed to be a single event in a given strain, we could not rule out the possibility that they were the “net” result of multiple changes. For example, the translocations may be the final result of a number of inversions. A special case with strain 8608F2 clearly demonstrates that multiple genomic changes could occur in a given strain (Fig. 8). In 8608F2, multiple translocations resulted in an extraordinary genome structure. Figure 8 also presents some other examples of the diversifying genomes among the serovar Typhimurium LT7 mutants, including the I-CeuI F translocation in 9052F2, the point mutation in rrnD of 8111F23332, and I-CeuI F duplication in 8618E1. Note that in spite of the dramatic genomic changes, the genomic location of oriC changed not much.
FIG. 8.
Diverse genome structures among representative serovar Typhimurium LT7 mutants. (A) PFGE patterns of I-CeuI-cleaved genomic DNA of representative serovar Typhimurium LT7 mutants; (B) I-CeuI maps of these strains based on the data from panel A. In lane 3, the loss of the I-CeuI cleavage site in rrlD resulted in the disappearance of I-CeuI C; the disappearance of I-CeuI B and the appearance of I-CeuI B+C (1306 kb) are not obvious on this picture but have been confirmed in other experiments (data not shown). These maps, although constructed based initially on only I-CeuI data, were all confirmed by long-range PCR. The solid circle in I-CeuI C indicates the position of oriC.
Genomic diversification during archiving.
Each of the auxotrophic cultures was initially inoculated in quintuplicate from a single colony. Cells from “sibling” vials in the collection had been tested and shown to exhibit genomic diversity indistinguishable from the diversity observed from cells in random vials (data not shown), suggesting that genomic diversification did not exist prior to the archiving but rather occurred during storage. We did not design our experiments to test the debate whether genes can mutate in the absence of chromosomal replication, since we assumed that cells continued to divide while stored in vials, by using the trace nutrients in the medium or materials from the dead cells of their siblings. Representative data summarized in Table 1 strongly suggest that genomic diversification had occurred during archiving in the sealed agar stabs at room temperature in the past four decades. For example, strain 8111 gave rise to colonies of different sizes, ranging from >4 mm to <0.3 mm (A through F). Since these colonies were formed on the first plating, they should have come from cells that had been in the stab. They might each have their own set of genomic changes, since they might represent different lineages descending from the same ancestor 8111. This assumption was substantiated by the findings that each of the primary colonies gave rise to descendents with identical genomic features. As shown in Table 1, 8111A had no genomic change detected, 8111B had I-CeuI D translocation, 8111E lost an AvrII site presumably by a point mutation(s), and the 8111F lineage lost an I-CeuI site. The 8111D lineage gives a good example showing that each lineage had maintained the particular genome structure during the fast growth in the laboratory for at least nine replatings no matter how colony morphology had diversified during the replating process. For instance, all derivatives from 8111D, i.e., 8111D321, 8111D322, 8111D323, 8111D324, etc., had the same genomic features as 8111D: I-CeuI F translocation and I-CeuI A inversion (Fig. 5 and Table 1). All lineages, including 8111F and derivatives, 8117C and derivatives, and those that are not reported here, had the same situation. So far we have not found a single case in which a descendant had a different set of genomic features from that of the original colony of the first plating, implying that all genomic changes documented in the present study occurred during archiving in the stabs at room temperature and that the genome structures determined in the present study are stable, no matter how different they may appear compared with the wild-type LT7.
TABLE 1.
Summary of genomic changes on representative strains of S. enterica serovar Typhimurium
| Straina | Genomic change(s) |
|---|---|
| LT2 | No change |
| LT7 | No change |
| 8111A | No change |
| 8111B | I-Ceu D translocation (Fig. 3) |
| 8111D | I-CeuI F translocation and I-CeuI A inversion (Fig. 5) |
| 8111D321 | I-CeuI F translocation and I-CeuI A inversion |
| 8111D322 | I-CeuI F translocation and I-CeuI A inversion |
| 8111D323 | I-CeuI F translocation and I-CeuI A inversion |
| 8111D324 | I-CeuI F translocation and I-CeuI A inversion |
| 8111D443 | I-CeuI F translocation and I-CeuI A inversion |
| 8111E | Point mutation: AvrII GH join |
| 8111F2 | Point mutation: loss of I-CeuI site between B and C |
| 8111F23111 | Point mutation: loss of I-CeuI site between B and C |
| 8111F23112 | Point mutation: loss of I-CeuI site between B and C |
| 8111F231211 | Point mutation: loss of I-CeuI site between B and C |
| 8111F231213 | Point mutation: loss of I-CeuI site between B and C (Fig. 6) |
| 8111F23332 | Point mutation: loss of I-CeuI site between B and C (Fig. 8) |
| 8111F24334 | Point mutation: loss of I-CeuI site between B and C |
| 8117A | No change |
| 8117B | No change |
| 8117C | I-CeuI E duplicated (Fig. 4) |
| 8117C31111 | I-CeuI E duplicated |
| 8117C31112 | I-CeuI E duplicated |
| 8117C31121 | I-CeuI E duplicated |
| 8117C31124 | I-CeuI E duplicated |
| 8117D1 | I-CeuI E duplicated (Fig. 7, 14 and 13 positive, 6 and 9 positive, and 16 and 13 positive) |
| 8608A | No change |
| 8608B | No change |
| 8608C | No change |
| 8608D | Point mutation: loss of AvrII site between G and I (Fig. 6) |
| 8608E | No change |
| 8608F2 | I-CeuI D translocation (Fig. 7, 15 and 16 positive, 9 and 12 positive, and 14 and 13 positive); I-CeuI F translocation (Fig. 7, 6 and 11 positive and 8 and 5 positive); see also Fig. 8 for I-CeuI D and F translocations |
| 8618A | No change |
| 8618B | No change |
| 8618E1 | Duplication of I-CeuI F (Fig. 8) |
| 9052F2 | I-CeuI F translocation (Fig. 8) |
| 9059A | Point mutation: loss of XbaI site between A and B and creation of an XbaI site resulting in a 380-kb fragment (Fig. 6) |
All strains from 8111A to the bottom of the table are derivatives of the wild type LT7; their genotypes are given in Materials and Methods.
DISCUSSION
In previous work, we observed high degree of conservation in physical structure of the bacterial genomes and pointed out the potential importance of this phenomenon in both theoretical and applied areas of study, such as in elucidating the mode of bacterial evolution and in bacterial phylogenetic studies (17, 19, 30). As the first step toward exploring the mechanisms behind such a high degree of genomic conservation, we needed to analyze the genomes of archival mutator strains of serovar Typhimurium, attempting to capture the genomic changes that would escape detection under natural conditions. In the present study, our physical mapping methods have proven efficient, accurate, and unambiguous.
The diversity of genome structure among the archived LT7 strains that we have revealed in the present study is striking, considering the high degree of conservation in genome structure between serovar Typhimurium LT2 and E. coli K-12. Among nearly 200 strains examined, we found translocations, duplications, inversions, and point mutations. We did not find genomic insertions, which could be accounted for by the fact that the archived bacteria were in pure culture and therefore did not have the chance to acquire exogenous DNA. We did not find large-scale (12 kb or larger) deletions either, but we do not rule out the possibility that small deletions that are beyond the resolution of our mapping methods (ca. 12 kb) may have escaped our detection. We also considered the possible roles of IS200 in the genomic diversification. IS200 has six copies in serovar Typhimurium (44) and 25 copies in S. enterica serovar Typhi (40), so it may be a hot spot for homologous recombination. In serovar Typhi, an inversion around the replication terminus is mediated by a pair of IS200 (1). However, none of the rearrangements of the LT7 mutants detected in the present study was mediated by IS200. All copies of IS200 in serovar Typhimurium (6 copies) and in serovar Typhi (25 copies) are located in intergenic regions and are usually fairly stable in the genome, causing no genotypic or phenotypic changes. So we do not expect a major role of IS200 in the genomic diversification of LT7.
Translocations were frequent and always involved one of the three small I-CeuI fragments—D, E, and F—in the present study. They could be the results of deletion and reinsertion, but it is more likely that they are the final products of multiple homologous recombination events because inversions seem to be more frequent among Salmonella spp. (28). Duplications also involve one of the three small I-CeuI fragments. In the present study, I-CeuI E and F are duplicated in 8117C and 8618E1, respectively; I-CeuI D is seen duplicated in other Salmonella strains (S.-L.L, unpublished data). Duplications may double the amount of certain gene products, which might be needed by the bacteria for adaptation.
Inversions often occur among enteric bacteria, presumably as a compensation mechanism to restore a physical balance of the chromosome when there is a major insertion or deletion on the genome (23, 26). In this work, however, inversion occurred when there was no detectable insertion or deletion. Probably, after archival storage for several decades under nonnatural conditions, a large number of genes have lost functions, but products of some other genes might be in greater demand and therefore have somehow obtained closer proximity to the origin of DNA replication (oriC) by inversion to reach a higher gene dosage.
Point mutations were inferred from the gain or loss of endonuclease cleavage sites. The number of nucleotides used in the cleavage sites by the four endonucleases, including 6 times 25 for XbaI, 6 times 13 for AvrII, 6 times 35 for SpeI, and 26 times 7 for I-CeuI, totals 620 bp, and the genome is 4,800 kb. Therefore, one base change in the cleavage sequences for the four endonucleases represents 7.7-kb base changes in the whole genome, although this number seems to be too large to be true and needs to be verified at least by sample sequencing. More startling, however, is the loss of an I-CeuI site in the 8111F derivatives (8111F and its descendants), including 8111F231213 (Fig. 6). I-CeuI is an intron-encoded endonuclease (10, 31, 32), which cleaves DNA within bacterial rrl genes and thus determines the copy number and genomic distribution of rrl genes (19). The I-CeuI cleavage site is highly conserved in most bacteria; a limited degeneracy is known, such as in Mycobacterium tuberculosis (4), but no recent mutation has been reported. If one base in the I-CeuI cleavage site (26 bp) represents 110 bp in the whole 23S rRNA gene (2.9 kb), frequent mutations may make 23S rRNA sequence of little value in phylogenetic studies, which is obviously not the case. We need to find out which base is changed in the 8111F strains that made one of the I-CeuI sites no longer cleavable and whether this 23S rRNA molecule is still functional with the mutation in the sequence.
One interesting question is where the genomic diversification events may have occurred. Based on our results, we speculate that slow growth in the stab during archiving at room temperature may have selected for some mutations that enhanced survival in the stab, which in turn led to genomic diversification, with the special genome structure that adapts best to the local ecological settings becoming fixed, although we do not completely rule out the possibility that some diversification events might have occurred in the first burst of luxuriant growth (40 years ago) before the period of stationary phase. In fact, it seems likely that cells were not in stationary phase in the stab but were growing very slowly, using as energy the bodies of their siblings. Initial heavy growth generated a large population that died over time and released nutrients that could be used by the surviving cells to support slow growth; ongoing death exceeded growth and the population was dying off very slowly over four decades. If growth (by partial replacement of dead cells) was occurring in the stabs, it would be hard to know whether the high frequency of mutant cells in an old stab reflects an increased mutation rate or differential reproduction of rare mutant types originally introduced into the stab at very low frequency. It is more likely that the observed increase in mutant frequency reflected selection rather than mutagenesis.
What guarantees the stability of the physical structure of the bacterial genome during evolution? Mechanisms responsible for such high degree of genomic conservation are still not fully understood. However, the methyl-directed mismatch repair (MMR) system may be a key factor, as the main genetic difference between the two commonly used serovar Typhimurium strains, LT7 and LT2, is that LT7 is a mutator with defects in mutL but LT2 is not and does not show genomic diversification (7). MMR systems repair DNA replication errors on newly synthesized DNA strands to guarantee the precise replication of the genomic DNA, whereby they inhibit recombination between nonidentical DNA sequences and safeguard genetic stability (11, 37). Genes involved in MMR, including mutH, mutL, mutS, etc., have been identified and characterized for their individual roles (5, 38, 41), with the MMR system in E. coli being the most extensively studied (36, 37). Cells having defects in MMR are mutators and have been reported for elevated spontaneous mutation rates (34, 35, 45). In the present study, LT7 as a mutator provided us with excellent chances to analyze genomic diversification; we are now sequencing genes in the MMR system, including mutL, for representative strains to investigate the relationships between the MMR defects and genomic diversification and to gain further insights into the molecular basis of genomic diversification. Whether or not MMR is involved in genomic diversification, two issues remain to be discussed and further explored. The first is how bacterial genome structure could be conserved if diversification took place all of the time, as in the case of the LT7 archival cultures. Working with stationary-phase cultures of E. coli, Finkel and Kolter found that populations of surviving cells were highly dynamic even after many months of incubation, and the diversification proceeded along different paths, resulting in the coexistence of multiple mutant forms (9). Many lines of evidence from researchers working with natural populations also indicate that genomic changes do occur frequently among bacteria, but most of them are lethal. As a result, only a very small proportion of the bacterial population undergoing genomic changes may survive under the specific natural selection pressure. Most genomic changes may be of small scale and may not drastically change the overall structure of the genome, as demonstrated by the distinct genomic differences and at the same time highly conserved overall genome structure of most of the more than 2,500 Salmonella species (18, 20, 21, 25, 28). Some lineages of bacteria of the same phylogenetic group, however, may have different genome structures, usually when they have large genomic insertions that may have disrupted the original genomic balance (23, 26, 27). Once a new balance is reached after genomic rearrangements, a new genome structure, very different as it may be from the majority of the bacteria in the same phylogenetic group, will be established and conserved (23). Combining our findings in previous and the current studies, we believe that the conservation of physical structure of bacterial chromosomes is guaranteed jointly by the requirement of a physical balance and by a genomic function that prohibits replication errors, such as the MMR system, although we need to experimentally evaluate the roles of MMR in the conservation of the bacterial genome structure.
The second issue is about the significance of genome diversification in bacterial evolution and, more specifically, speciation. In a model of bacterial speciation, the Adopt-Adapt Model (17, 30), we hypothesize that bacteria speciate by acquisition of exogenous DNA (the “adopt” step) and the ensuing adaptive genomic rearrangements (the “adapt” step). In fact, the bacterial speciation process involves a special kind of genomic diversification that ultimately leads to a new bacterial species. Both steps of the speciation process, adopt and adapt, would require the genome to be more tolerant to mismatches, i.e., for accepting the incoming DNA and for rearranging the chromosome. Further work is needed to establish whether or how the MMR system contributes to the genomic diversification events, as reported here, and to bacterial speciation.
Acknowledgments
We thank Barney Truong, Suneetha Alokam, and Sushma Kothapalli for technical assistance.
The present study was supported by an operating grant from the Medical Research Council of Canada (MRC grant GOP-38106) to S.-L.L., an operating grant from the Natural Sciences and Engineering Research Council of Canada (NSERC; grant 216912-00) to S.-L.L., an operating grant from the Canadian Institutes for Health Research (CIHR; grant MOP-47817) to S.-L.L., an NSERC operating grant and a grant from the National Institutes of Health (AI34829) to K.E.S., a CIHR operating grant to R.N.J., and Cancer Research Center and University of Missouri Research Board grants to A.E. S.L.L. was also supported by the Alberta Cancer Board. W.-Q.L. is a summer student supported by Alberta Heritage Foundation for Medical Research (June-August 2001 and May-August 2002).
REFERENCES
- 1.Alokam, S., S. L. Liu, K. Said, and K. E. Sanderson. Inversions over the terminus region in Salmonella and E. coli: IS200s as the sites of homologous recombination inverting the chromosome of Salmonella enterica serovar Typhi. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 2.Altman, E., J. R. Roth, A. Hessel, and K. E. Sanderson. 1996. Transposons in current use in genetic analysis in Salmonella, p. 2613-2626. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 3.Anderson, R. P., and J. R. Roth. 1978. Tandem genetic duplications in Salmonella typhimurium: amplification of the histidine operon. J. Mol. Biol. 126:53-71. [DOI] [PubMed] [Google Scholar]
- 4.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 5.Cox, E. C. 1976. Bacterial mutator genes and the control of spontaneous mutation. Annu. Rev. Genet. 10:135-156. [DOI] [PubMed] [Google Scholar]
- 6.Doolittle, R. F., D. F. Feng, S. Tsang, G. Cho, and E. Little. 1996. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science 271:470-477. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, K., I. Linetsky, C. Hueser, and A. Eisenstark. 2001. Genetic variability among archival cultures of Salmonella typhimurium. FEMS Microbiol. Lett. 199:215-219. [DOI] [PubMed] [Google Scholar]
- 8.Feng, D. F., G. Cho, and R. F. Doolittle. 1997. Determining divergence times with a protein clock: update and reevaluation. Proc. Natl. Acad. Sci. USA 94:13028-13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkel, S. E., and R. Kolter. 1999. Evolution of microbial diversity during prolonged starvation. Proc. Natl. Acad. Sci. USA 96:4023-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gauthier, A., M. Turmel, and C. Lemieux. 1991. A group I intron in the chloroplast large subunit rRNA gene of Chlamydomonas eugametos encodes a double-strand endonuclease that cleaves the homing site of this intron. Curr. Genet. 19:43-47. [DOI] [PubMed] [Google Scholar]
- 11.Harfe, B. D., and S. Jinks-Robertson. 2000. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 34:359-399. [DOI] [PubMed] [Google Scholar]
- 12.Helm, R. A., and S. Maloy. 2001. Rapid approach to determine rrn arrangement in Salmonella serovars. Appl. Environ. Microbiol. 67:3295-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hill, C. W., and B. W. Harnish. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. USA 78:7069-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoyer, L. L., A. C. Hamilton, S. M. Steenbergen, and E. R. Vimr. 1992. Cloning, sequencing, and distribution of the Salmonella typhimurium LT2 sialidase gene, nanH, provides evidence for interspecies gene transfer. Mol. Microbiol. 6:873-884. [DOI] [PubMed] [Google Scholar]
- 15.Kirchner, C. E., and M. J. Rudden. 1966. Location of a mutator gene in Salmonella typhimurium by cotransduction. J. Bacteriol. 92:1453-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lilleengen, K. 1948. Typing Salmonella by means of bacteriophage. Acta Pathol. Microbiol. Scand. 77:11-125. [DOI] [PubMed] [Google Scholar]
- 17.Liu, G. R., A. Rahn, W. Q. Liu, K. E. Sanderson, R. N. Johnston, and S. L. Liu. 2002. The evolving genome of Salmonella pullorum. J. Bacteriology 184:2626-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, S.-L., A. Hessel, H.-Y. M. Cheng, and K. E. Sanderson. 1994. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella paratyphi B. J. Bacteriol. 176:1014-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, S.-L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-CeuI, an intron-encoded endonuclease, specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, S.-L., A. Hessel, and K. E. Sanderson. 1993. The Xba I-Bln I-Ceu I genomic cleavage map of Salmonella enteritidis shows an inversion relative to Salmonella typhimurium LT2. Mol. Microbiol. 10:655-664. [DOI] [PubMed] [Google Scholar]
- 21.Liu, S.-L., A. Hessel, and K. E. Sanderson. 1993. The XbaI-BlnI-CeuI genomic cleavage map of Salmonella typhimurium LT2 determined by double digestion, end-labeling, and pulsed-field gel electrophoresis. J. Bacteriol. 175:4104-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, S.-L., and K. E. Sanderson. 1992. A physical map of the Salmonella typhimurium LT2 genome made by using XbaI analysis. J. Bacteriol. 174:1662-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, S.-L., and K. E. Sanderson. 1995. The chromosome of Salmonella paratyphi A is inverted by recombination between rrnH and rrnG. J. Bacteriol. 177:6585-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, S.-L., and K. E. Sanderson. 1995. The genomic cleavage map of Salmonella typhi Ty2. J. Bacteriol. 177:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, S.-L., and K. E. Sanderson. 1995. I-CeuI reveals conservation of the genome of independent strains of Salmonella typhimurium. J. Bacteriol. 177:3355-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, S.-L., and K. E. Sanderson. 1995. Rearrangements in the genome of the bacterium Salmonella typhi. Proc. Natl. Acad. Sci. USA 92:1018-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, S.-L., and K. E. Sanderson. 1996. Highly plastic chromosomal organization in Salmonella typhi. Proc. Natl. Acad. Sci. USA 93:10303-10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, S.-L., and K. E. Sanderson. 1998. Homologous recombination between rrn operons rearranges the chromosome in host-specialized species of Salmonella. FEMS Microbiol. Lett. 164:275-281. [DOI] [PubMed] [Google Scholar]
- 29.Liu, S.-L., and K. E. Sanderson. 1998. Physical analysis of the Salmonella typhimurium genome, p. 371-381. In P. H. Williams, J. Ketley, and G. Salmond (ed.), Methods in microbiology. Academic Press, Inc., New York, N.Y.
- 30.Liu, S.-L., A. B. Schryvers, K. E. Sanderson, and R. N. Johnston. 1999. Bacterial phylogenetic clusters revealed by genome structure. J. Bacteriol. 181:6747-6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall, P., T. B. Davis, and C. Lemieux. 1994. The I-CeuI endonuclease: purification and potential role in the evolution of Chlamydomonas group I introns. Eur. J. Bacteriol. 220:855-859. [DOI] [PubMed] [Google Scholar]
- 32.Marshall, P., and C. Lemieux. 1991. Cleavage pattern of the homing endonuclease encoded by the fifth intron in the chloroplast large subunit rRNA-encoding gene of Chlamydomonas eugametos. Gene 104:241-245. [DOI] [PubMed] [Google Scholar]
- 33.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 34.Miller, J. H. 1996. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu. Rev. Microbiol. 50:625-643. [DOI] [PubMed] [Google Scholar]
- 35.Miyake, T. 1960. Mutator factor in Salmonella typhimurium. Genetics 45:11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modrich, P. 1989. Methyl-directed DNA mismatch correction. J. Biol. Chem. 264:6597-6600. [PubMed] [Google Scholar]
- 37.Modrich, P. 1991. Mechanisms and biological effects of mismatch repair. Annu. Rev. Genet. 25:229-253. [DOI] [PubMed] [Google Scholar]
- 38.Nevers, P., and H. C. Spatz. 1975. Escherichia coli mutants uvrD and uvrE deficient in gene conversion of lambda-heteroduplexes. Mol. Gen. Genet. 139:233-243. [DOI] [PubMed] [Google Scholar]
- 39.Ochman, H., and A. C. Wilson. 1987. Evolutionary history of enteric bacteria, p. 1649-1654. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 40.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 41.Rydberg, B. 1978. Bromouracil mutagenesis and mismatch repair in mutator strains of Escherichia coli. Mutat. Res. 52:11-24. [DOI] [PubMed] [Google Scholar]
- 42.Sanderson, K. E., A. Hessel, S.-L. Liu, and K. E. Rudd. 1996. The genetic map of Salmonella typhimurium, edition VIII, p. 1903-1999. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 43.Sanderson, K. E., A. Hessel, and B. A. D. Stocker. 1996. Strains of Salmonella typhimurium and other Salmonella species used in genetic analysis, p. 2496-2503. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington DC.
- 44.Sanderson, K. E., P. Sciore, S. L. Liu, and A. Hessel. 1993. Location of IS200 on the genomic cleavage map of Salmonella typhimurium LT2. J. Bacteriol. 175:7624-7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Treffers, H. P., V. Spinelli, and N. O. Belser. 1954. A factor (or mutator gene) influencing mutation rates in Escherichia coli. Proc. Natl. Acad. Sci. USA 40:1064-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]