Abstract
The orf245 gene is located immediately upstream of, and divergently transcribed from, the replication initiation gene, rep, of the Staphylococcus aureus multiresistance plasmid pSK1, and related genes have been found in association with a range of evolutionarily distinct replication genes on plasmids from various gram-positive genera. orf245 has been shown previously to extend the segregational stability of a pSK1 minireplicon. Here we describe an investigation into the basis of orf245-mediated stabilization. orf245 was not found to influence transcription of pSK1 rep, indicating that it is not directly involved in plasmid replication. This was confirmed by demonstrating that orf245 is able to enhance the segregational stability of heterologous theta- and rolling-circle-replicating replicons, suggesting that it encodes a plasmid maintenance function. Evidence inconsistent with postsegregational killing and multimer resolution mechanisms was obtained; however, the intergenic region upstream of orf245 was found to mediate orf245-dependent incompatibility, as would be expected if it encodes a cis-acting centromere-like site. Taken together, these findings implicate active partitioning as the probable basis of the activity of orf245, which is therefore redesignated par. Since it is unrelated to any gene known to play a role in plasmid segregation, it seems likely that pSK1 par potentially represents the prototype of a novel class of active partitioning systems that are distinguished by their capacity to enhance plasmid segregational stability via a single protein-encoding gene.
Clinical isolates of Staphylococcus aureus and coagulase-negative staphylococci commonly possess one or more plasmids on which antimicrobial resistance determinants are frequently encoded. These plasmids range from small rolling-circle (RC) plasmids that may carry a single resistance determinant and are multicopy to large multiresistance and conjugative plasmids that are generally 15 to 60 kb in size and maintained at a low copy number. Based on structural and functional properties, the staphylococcal multiresistance plasmids have been divided into three groups: the β-lactamase-heavy-metal resistance plasmids, the pSK41-like conjugative plasmids, and the pSK1 family (14, 15, 43). The β-lactamase-heavy-metal resistance plasmids have been detected in isolates from the 1940s and typically carry the transposon Tn552 or a derivative thereof, encoding the blaZ determinant, which confers penicillin resistance, and the cad, mer, and/or asa operons, encoding resistance to cadmium, mercury, and arsenate, respectively. The pSK41-like conjugative plasmids, which were first detected in the mid-1970s, usually carry a Tn4001-like element encoding the bifunctional aminoglycoside resistance gene aacA-aphD and often contain cointegrated remnants of small plasmids encoding genes such as aadD, mediating resistance to the aminoglycosides neomycin and kanamycin; smr, conferring antiseptic and disinfectant resistance; and dfrA, encoding resistance to trimethoprim. The 28.4-kb plasmid pSK1 is the prototype of a family of plasmids isolated from epidemic S. aureus strains in Australia and the United Kingdom since 1980. Members of the pSK1 family typically confer resistance to antiseptics and disinfectants, encoded by qacA, and may also carry Tn4001, a Tn552-like transposon, and/or a cointegrated remnant of a dfrA-carrying plasmid previously designated Tn4003.
Replication initiation genes of representatives from the three families of staphylococcal multiresistance plasmids have been shown to be related to one another, and, on the basis of size and experimental and sequence data, they are believed to utilize a theta-mode replication system (14, 20, 49). Based on amino acid sequence similarity, these plasmids can be placed in a family of gram-positive theta replicons, which include the β-lactamase-heavy-metal resistance family plasmids pSX267 and pI9789::Tn552, the conjugative plasmid pSK41, and pSK1 from Staphylococcus; pAD1, pCF10, and pPD1 from Enterococcus; pLJ1, pSAK1, and pLH1 from Lactobacillus; and pLS32 from Bacillus natto (14). In pLS32, replication has been shown to be independent of host-encoded DNA polymerase I, and the origin of replication (ori) has been localized to a region of iterons within the rep gene (54).
Low-copy-number plasmids require stabilization mechanisms to prevent their loss at cell division (58). Such mechanisms include plasmid multimer resolution, postsegregational killing, and active partitioning systems, all of which have been identified in both gram-negative and gram-positive bacteria (18, 22, 28, 53, 56). In pSK1, orf245 has been shown to enhance segregational stability of the pSK1 replicon while not being required for replication (14). Similar to orf256 from pI9789::Tn552, with which it shares nucleotide sequence similarity, orf245 is located upstream of, and transcribed divergently from, the rep gene (14). Genes encoding proteins homologous to the deduced orf245 product have been identified in a range of gram-positive genera, including Staphylococcus, Streptococcus, Lactococcus, Lactobacillus, Clostridium, and Tetragenococcus, on plasmids carrying six apparently evolutionarily distinct types of replication genes (14). Here, we demonstrate that orf245 is able to extend the maintenance of heterologous replicons, and we provide evidence supporting partitioning as its mechanism of plasmid stabilization. We have consequently renamed orf245 as par and will henceforth use this designation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and primers.
The S. aureus and Escherichia coli strains and plasmids and oligonucleotides used in this study are listed in Table 1. All strains were cultured at 37°C in Luria-Bertani (LB) medium (46) containing, where appropriate, ampicillin (100 μg/ml), chloramphenicol (10 to 25 μg/ml), gentamicin (20 μg/ml), trimethoprim (100 μg/ml), neomycin (15 μg/ml), erythromycin (10 to 20 μg/ml), or tetracycline (5 μg/ml). The B2 and NYE media of Schenk and Laddaga (47) were used for electroporated cells. PCR utilizing primers 3HindIII and 1353EcoRI (Table 1) was performed to generate a 1.3-kb fragment from pSK1 carrying par, the par-rep intergenic region, and 47 bp of rep, which contains the sequence corresponding to nucleotide positions 3 to 1353 of the pSK1 replication region sequence (GenBank accession no. AF203376). This was cloned into pRB394, producing pSK5316 (Table 1), with the predicted promoter for rep placed upstream of the ribosome binding site of the chloramphenicol acetyltransferase (CAT) reporter gene (cat) to produce a rep-cat transcriptional fusion. A similar construct, but lacking par, was made by first using primers 842HindIII and 1353EcoRI to generate a 0.5-kb PCR product which contains the sequence corresponding to nucleotide positions 842 to 1353 of the pSK1 replication region sequence (GenBank accession no. AF203376). This fragment, carrying the par-rep intergenic region and 47 bp of rep, was cloned into the same vector to produce pSK5322 (Table 1), with the predicted promoter for pSK1 rep placed upstream of the cat gene ribosome binding site.
TABLE 1.
Bacterial strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Genotype, description, or sequence | Reference or source |
|---|---|---|
| E. coli DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Bethesda Research Laboratories |
| S. aureus | ||
| RN4220 | Derivative of NCTC8325-4 that accepts foreign DNA | 42 |
| SK982 | Rifampcin and novobiocin-resistant laboratory strain | 33 |
| Plasmids | ||
| pAM401 | E. coli-E. faecalis shuttle vector with pACYC184 and pIP501 replicons | 59 |
| pRB394 | E. coli-B. subtilis promoter shuttle vector with pBR322 and pUB110 replicons containing a promoterless cat gene | 8 |
| pSK1 | S. aureus multiresistance plasmid conferring trimethoprim, gentamicin, and multidrug resistance | 45 |
| pSK4828 | pSK1 HindIII-BamHI PCR product of 2.3 kb containing par and rep cloned into pUC118 | 1 |
| pSK4829 | pSK1 HindIII-BamHI PCR product of 2.3 kb containing par and rep cloned into pWE180 | 14 |
| pSK5316 | pSK1 Prep-cat fusion, par in cis, HindIII-EcoRI PCR product of 1.3 kb cloned into pRB394 | This study |
| pSK5322 | pSK1 Prep-cat fusion, HindIII-EcoRI PCR product of 0.5 kb cloned into pRB394 | This study |
| pSK5334 | Site-directed mutation in par gene carried by pSK5316 | This study |
| pSK5335 | Site-directed mutation in par gene carried by pSK4829 | This study |
| pSK5378 | pSK1 BamHI par PCR product of 1.3 kb cloned into pAM401 | This study |
| pSK6110 | pSK1 BamHI PCR product of 204 bp containing region upstream of par cloned into pAM401 | This study |
| pSK6111 | Site-directed mutation in par gene carried by pSK5378 | This study |
| pT181 | 4.45-kb S. aureus plasmid containing tetA(K), Tcr | 29 |
| Primersa | ||
| 3BamHI | 5′-GCGGGATCCCCCTAGATAATTCTTCTG-3′b | |
| 3HindIII | 5′-GCGAAGCTTCCCTAGATAATTCTTCTGATAATTTAG-3′ | |
| 818 | 5′-GCATTATTAACAACAGTTTGTTTAG-3′ | |
| 842 | 5′-GTTACATTCAATTCATCAGCAACCAT-3′ | |
| 842HindIII | 5′-GCGAAGCTTGTTACATTCAATTCATCAGCAACC-3′ | |
| 844EcoRI | 5′TACATTCAATTCATCAGCAAGAATTCTTCAAGTTTACATATCATCCACCCTTAG-3′ | |
| 880BamHI | 5′-CGCGGATCCCATATCATCCACCCTTAGGCACC-3′ | |
| 897EcoRI | 5′-CTAAGGGTGGATGATATGTAAACTTGAAGAATTCTTGCTGATGAATTGAATGTA-3′ | |
| 1028 | 5′-GTTGTTTTAAGGTCATAACATGATTCTG-3′ | |
| 1084BamHI | 5′-CCGGGATCCGGTGTATATAAAGCGTTTTAAGTAC-3′ | |
| 1282 | 5′-CTTAGAGTTGAGGTAATAAAACAAT-3′ | |
| 1353BamHI | 5′-GCGGGATCCCGTAATGTTTCGAATTCACTTGC-3′ | |
| 1353EcoRI | 5′-CGTAATGTTTCGAATTCACTTGCAG-3′ |
The number of each primer refers to the 5′-most base in each instance that corresponds to a nucleotide in the previously published sequence of the pSK1 replication region (GenBank accession no. AF203376).
As indicated in the primer name, restriction endonuclease recognition sites incorporated into primers are underlined. Nucleotide base substitutions to produce a mutant par are in boldface.
For stability studies, the par gene and par-rep intergenic region were amplified from pSK1 by PCR with primers 3BamHI and 1353BamHI and cloned into pAM401 to generate pSK5378 (Table 1), which contains the sequence corresponding to nucleotide positions 3 to 1353 of the pSK1 replication region sequence (GenBank accession no. AF203376). Identical mutations were created in par in pSK4829, pSK5316, and pSK5378 by PCR-based site directed mutagenesis with primers 844EcoRI (corresponding to par antisense nucleotides) and 897EcoRI (corresponding to par sense nucleotides), producing pSK5335, pSK5334, and pSK6111, respectively (Table 1). This resulted in the insertion of two stop codons (TAA and TGA) at the second and fourth codons of par. In the same mutagenesis reaction, an EcoRI site between the fifth and seventh codons was also created to facilitate screening of mutant transformants. To localize the par-related incompatibility determinant, a 204-bp fragment from the par-rep intergenic region was cloned into pAM401 by inserting a PCR product amplified by primers 880BamHI and1084BamHI, to produce pSK6110 (Table 1), which contains the sequence corresponding to nucleotide positions 880 to 1084 of the pSK1 replication region sequence (GenBank accession no. AF203376). All clones produced from the products of PCR were sequenced to confirm that no errors had been incorporated during amplification.
DNA manipulations.
Plasmid DNA was isolated from E. coli by using the Quantum Prep miniprep kit (Bio-Rad) according to the manufacturer's instructions. S. aureus DNA isolations were performed as described previously (32). All restriction endonuclease digestion and ligation reactions were performed according to the manufacturers' instructions. DNA cloning and transformation of E. coli were performed by standard techniques (46). PCR was performed with Pfu (Stratagene) or Pfx (Gibco BRL) enzyme, according to the manufacturers' instructions, in an MJ Research PTC-100 with Hot Bonnet. Primers were synthesized with a Beckman Oligo 1000 DNA synthesizer or supplied by Sigma-Aldrich. Electroporation of S. aureus (47) was performed with a Bio-Rad Gene Pulser with pulse controller. Plasmid copy numbers were determined by densitometric analysis of the amount of DNA isolated from S. aureus RN4220 harboring both the plasmid of interest and pT181, a plasmid with a copy number of 22 per cell (41), as previously described (55).
DNA sequencing and sequence analysis.
Automated cycle sequencing was performed by the Sydney University and Prince Alfred Macromolecular Analysis Centre or the Australian Genome Research Facility. Manual sequencing was performed to obtain sequence ladders for transcript mapping by using a Sequitherm sequencing kit (Epicentre Technologies). Sequence data were stored and assembled with the program SEQUENCHER (Gene Codes Corp.). Coiled-coil motifs in amino acid sequences were identified by using the MultiCoil program (60) (http://nightingale.lcs.mit.edu/cgi-bin/multicoil).
Transcript mapping by primer extension.
Total cellular RNA was isolated from S. aureus strains as previously described (30). Transcript mapping was performed essentially as described by Ausubel et al. (3). For determination of the rep transcription start point (TSP), primers 1353EcoRI and 1282 (Table 1) were used, the former complementary to sequences within the rep structural gene and the latter complementary to sequences 25 bp upstream of the rep start codon. The par TSPs were determined by using primers 818 and 842, complementary to sequences located within the par structural gene, and primer 1028, complementary to sequences located 145 bp upstream of the par start codon. Primers were end labeled with [γ-32P]ATP (Bresatec), mixed with total RNA, denatured by heating at 80°C for 3 min, and then hybridized at 42°C for 90 min before being extended by the addition of deoxynucleoside triphosphates and avian leukemia virus reverse transcriptase (Promega). The primer extension products were loaded on a 6% polyacrylamide gel and electrophoresed alongside dideoxy sequencing ladders generated with the same primers and DNA template (pSK4828 [Table 1]). The gels were then dried and autoradiographed.
Determination of promoter activity and CAT assays.
CAT assays were performed as previously described (24). Overnight cultures of S. aureus RN4220 harboring rep promoter fusion constructs pSK5316, pSK5322, and pSK5334 and the vector pRB394 were diluted 1:100 and grown with selection to an optical density at 600 nm of 0.5. Cells from 2-ml cultures were harvested by centrifugation and resuspended in WL buffer (10 mM EDTA, 25 mM Tris, pH 8.0) containing 50 μg of lysostaphin per sample. Cultures were incubated for 30 min at 37°C, and then the cell lysates were centrifuged (16,000 × g) for 30 min at 4°C and the supernatant was held on ice. CAT assays were performed on 50-μl samples with acetyl coenzyme A (Sigma) by the spectrophotometric method (48). CAT activity is expressed as nanomoles of chloramphenicol acetylated per minute per milligram of protein. Experiments were performed on three separate days. The total protein concentration was estimated by the method of Bradford (6), using bovine serum albumin (Pierce) as a standard.
Assay of plasmid segregational stability.
One hundred microliters of an overnight culture of the strain to be assayed, grown in medium selective for the plasmid, was used to inoculate 10 ml of LB medium and grown with selection for 4 h. Dilutions of this culture were prepared in saline, and a viable count was performed with nonselective LB agar plates. This culture was used to inoculate 10 ml of LB (10−2 dilution, or 10−4 for more stable plasmids), and after overnight growth without selection, the culture was diluted, counted, and subcultured as before; this process was repeated until approximately 50 or 100 generations of growth was achieved. Fifty to 100 colonies from viable-count plates were patched onto media with and without selection for the plasmid so that the proportion of the population retaining the resistance phenotype conferred by the plasmid could be quantitated. DNA was isolated from selected colonies to confirm the absence or presence of the relevant plasmid.
Growth assays.
Overnight cultures were diluted 1:100 in 10 ml of LB medium with selection for the plasmids, grown for 4 h, and then diluted to 105 CFU/ml in 100 ml of LB medium. A viable count was performed with nonselective and selective LB agar plates to determine the CFU per milliliter and the proportion of the population carrying the resistance phenotype conferred by the plasmid. This was repeated at hourly intervals for 6 h, representing approximately 12 generations. Experiments were performed on three separate days.
Incompatibility determination.
Plasmid incompatibility was determined by measuring retention of the resident plasmid pSK4829 or pSK5335 (Table 1) in S. aureus RN4220 over 25 to 30 generations after transformation of the incoming plasmid, pSK6110 or its parent pAM401. One hundred microliters of an overnight culture of the strain to be assayed, grown in medium selective for both resident and incoming plasmids, was used to inoculate 10 ml of LB medium and grown with selection for both plasmids for 4 h. The culture was then diluted in saline, and a viable count was performed with nonselective LB agar plates. One hundred microliters of this culture was used to inoculate 10 ml of LB, and after overnight growth with selection for the incoming plasmid only, the culture was diluted, counted, and subcultured as before; this process was repeated for approximately 25 to 30 generations. Fifty colonies from viable-count plates were patched onto media with and without selection for the resident plasmid so that the proportion of the population retaining the resistance phenotype conferred by this plasmid could be quantitated. DNA isolations were subsequently performed on representative isolates to the confirm presence or absence of plasmids. Experiments were performed in triplicate. A significant difference (P < 0.05) in retention of the resident plasmids between the strains carrying the introduced plasmids pAM401 and pSK6110 was taken to indicate incompatibility.
Statistical analysis.
Statistical analysis was carried out with Statview 5.0.1 (SAS Institute Inc.). Differences between groups were evaluated by Fisher's protected least-significant-difference test after analysis of variance and by repeated-measures analysis where appropriate. A significant difference was defined as a P value of <0.05.
RESULTS
Identification of promoter elements and TSPs of rep and par.
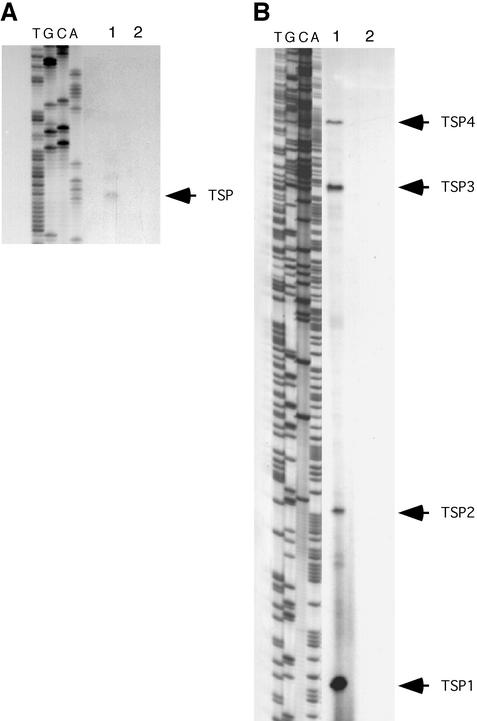
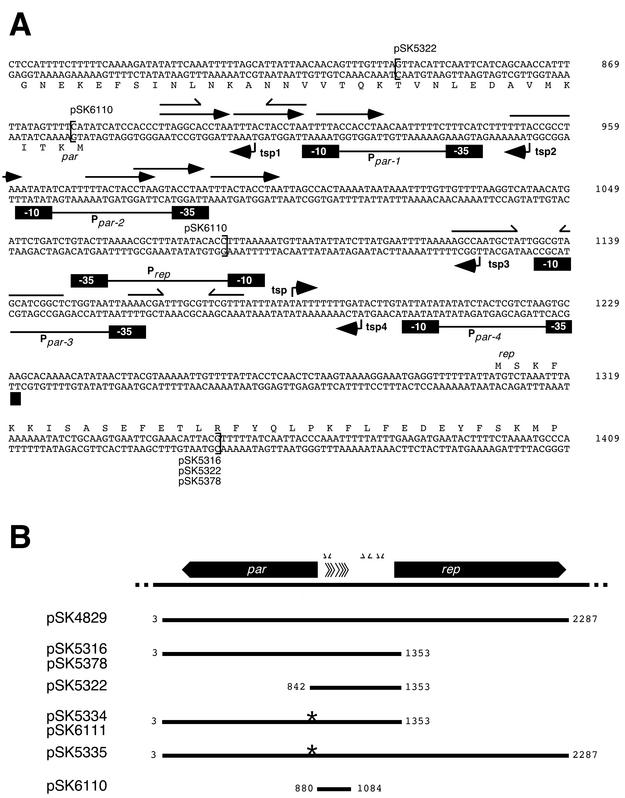
To facilitate studies aimed at establishing the nature of par and the mechanistic basis of its activity, the TSPs and putative promoter elements for the replication initiation gene, rep, and the par gene were identified by primer extension analysis. With RNA isolated from S. aureus strain SK982 harboring pSK1, transcript mapping analysis of rep revealed one faint extension product, corresponding to a thymine nucleotide at position 1185, 122 bp upstream of the rep start codon (Fig. 1A and 2A). Examination of the sequence upstream confirmed the presence of an appropriately positioned candidate promoter, Prep, consisting of −35 and −10 sequences (TGGTAA and TTTATT, respectively) separated by 19 bp (Fig. 2A). The position of the rep TSP shown in Fig. 1A, determined by using primer 1282, was confirmed in a separate primer extension experiment which utilized primer 1353EcoRI (data not shown). The putative rep promoter region overlaps a 6-bp inverted repeat (IR) and is adjacent to an 11-bp IR (Fig. 2A), suggesting that it may be subject to regulatory control.
FIG. 1.
Transcript mapping by primer extension, showing par and rep transcription initiation. Autoradiographs show primer extension products from pSK1, fractionated on 6% polyacrylamide-7 M urea sequencing gels. Total RNA was isolated from log-phase cultures of SK982 harboring pSK1 (grown with trimethoprim and gentamicin) (lanes 1) and from the negative control SK982 (lanes 2). Sequence ladders (lanes T, G, C, and A), derived from pSK4828 template, were coelectrophoresed to enable determination of TSPs. Primers 818 and 1282 (Table 1) were used to generate sequence ladders and transcript mapping products for rep (A) and par (B), respectively. Arrows indicate identified TSPs.
FIG. 2.
(A) Organization of the rep and par promoter regions in pSK1. The sequence corresponds to that obtained from the replication region of pSK1. The numbers at the end of each line indicate the position in the pSK1 sequence corresponding to GenBank accession no. AF203376. The short labeled arrows indicate TSPs identified in this study. The boxes represent −10 and −35 sequences of the putative promoters Ppar-1 to Ppar-4 and Prep. DR sequences are indicated by arrows, and IRs are shown by half arrows. Brackets indicate the boundaries of cloned fragments in the indicated plasmids (Table 1). The leftward extents of pSK5316 and pSK5378 correspond to nucleotide 3 of the GenBank entry. The deduced amino acid sequences for the gene products, Rep and Par, are given adjacent to their coding strands. (B) Schematic diagram of plasmid constructs employed in this study. The par and rep genes from the pSK1 replication region are shown by arrowed boxes; the arrows indicate the direction of transcription. DRs and IRs are indicated by arrowheads and half arrows, respectively. The lines indicate the extents of the pSK1 replication regions cloned into recombinant plasmids; numbers at the ends indicate the nucleotide position in the pSK1 sequence. Plasmid names are given on the left. The asterisks indicate the locations of stop codons inserted into the par coding sequence by site-directed mutagenesis.
Primer extension analysis of par, utilizing RNA isolated from S. aureus strain SK982 harboring pSK1, facilitated the detection of four extension products, corresponding to three thymine nucleotides and an adenine nucleotide at positions 909, 953, 1125, and 1196, respectively (Fig. 1B and 2A). Primer 818, complementary to sequence located within the par structural gene, was utilized to generate the extension products shown in Fig. 1B. The positions of TSP1, TSP2, and TSP3 were subsequently confirmed in separate primer extension studies utilizing primer 842, and the position of TSP4 was confirmed by using primer 1028, which is complementary to sequence 145 bp upstream of the par start codon (data not shown). Examination of the sequence upstream of TSP1 to -4 confirmed the presence of appropriately positioned candidate promoters, Ppar-1 to Ppar-4 (Fig. 2A). Expression of par therefore appears to be initiated from multiple promoters; however, the possibility that some of the extension products detected actually represent processed forms of a longer transcript cannot be excluded. For instance, the extension products from putative promoters Ppar-2 and Ppar-3, both of which represent a suboptimal match to the promoter consensus sequence (23, 37), may represent processed forms of message from Ppar-4 (Fig. 1B and 2A).
Effect of par on rep transcription.
It was previously hypothesized that the enhanced segregational stability determined for a minireplicon carrying the pSK1 rep gene and par, in comparison to a derivative lacking par, may reflect an effect of the par product on transcription of the divergently transcribed rep gene (14). To determine whether par enhances the stability of pSK1 through an effect on transcription of the rep gene, a DNA segment from pSK1, containing the rep promoter region and the entire par gene, was cloned upstream of a promoterless cat reporter gene in pRB394, to produce pSK5316 (Fig. 2B). For comparison, an equivalent DNA fragment containing the rep promoter but lacking par was similarly cloned into pRB394, to produce pSK5322 (Fig. 2B). Additionally, in order to exclude the possibility of an effect of insert size on rep transcription and to confirm that any effect was indeed mediated by the par open reading frame (ORF), a mutation was created in the par gene in pSK5316 by PCR-based site-directed mutagenesis. The mutation resulted in the insertion of two stop codons (TAA and TGA) at the second and fourth codons of par, thereby producing the par nonsense mutant pSK5334 (Fig. 2B). Following electroporation of pSK5316, pSK5322, pSK5334, and pRB394 into S. aureus RN4220, the level of CAT activity was determined for cells harboring the various constructs. As shown in Table 2, the CAT activity measured for the rep promoter in cells harboring pSK5316, which carries par, did not differ significantly from that determined for cells which contained pSK5322 or pSK5334, which lack a functional par gene. These data indicate that par does not affect transcription of the pSK1 rep gene and therefore rule out such an effect as the basis of par-mediated enhancement of pSK1 minireplicon segregational stability.
TABLE 2.
Effect of par on Prep transcription
| Plasmid | rep promoter | par | CAT activitya (mean ± SD) |
|---|---|---|---|
| Nil | Absent | Absent | 2.52 ± 1.46 |
| pRB394 | Absent | Absent | 3.48 ± 1.61 |
| pSK5316 | Present | Present | 10.87 ± 4.25 |
| pSK5322 | Present | Absent | 10.92 ± 4.63 |
| pSK5334 | Present | Mutant | 11.17 ± 1.26 |
Mean values from three independent cultures are presented. CAT activity is expressed as nanomoles of chloramphenicol acetylated per minute per milligram of protein.
Stabilization effect of par on segregation of theta and RC replicons.
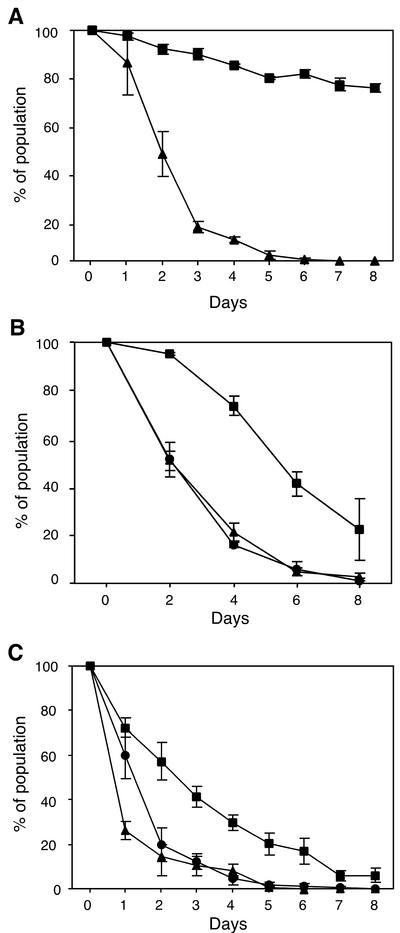
To confirm that par-mediated stabilization (14) is indeed mediated by the par ORF, rather than by any other element within the par sequence, a mutation equivalent to that of pSK5334 described above was introduced into the par gene of the pSK1 minireplicon pSK4829 to produce pSK5335 (Fig. 2B), carrying a par nonsense mutation. The segregational stabilities of pSK4829 and pSK5335 were assayed in S. aureus RN4220 in the absence of selection during 8 days of subculturing, corresponding to approximately 50 generations. After 8 days, pSK4829 was found to be maintained by 75% of the cell population (Fig. 3A). In contrast, pSK5335 showed significantly lower (P < 0.0001) segregational stability, being entirely lost from the bacterial population by day 6, representing approximately 40 generations (Fig. 3A), thereby confirming that the pSK1 par ORF mediates enhanced segregational stability of its cognate replicon.
FIG. 3.
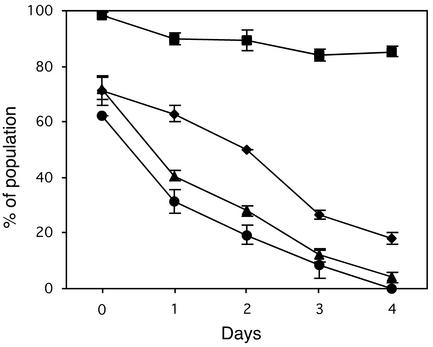
Segregational stability of plasmids in the presence and absence of par, in cis. Each data point is the mean from three experiments. The error bars represent standard errors. (A) Effect of par on segregational stability of pSK1 minireplicons. Cells of S. aureus RN4220 containing pSK4829 (▪) (carrying functional par) or pSK5335 (▴) (carrying nonfunctional par) were examined for stable maintenance of plasmids by determination of the proportion of the cell population which remained erythromycin resistant during growth in nonselective medium. Eight days of growth represents approximately 50 generations. (B) Effect of par on pIP501 theta replicon stability.Cells of S. aureus RN4220 harboring pAM401 (•) (vector only), pSK5378 (▪) (carrying functional par), or pSK6111 (▴) (carrying nonfunctional par) were examined for stable maintenance of plasmids by determination of the proportion of the cell population which remained chloramphenicol resistant during growth in nonselective medium. Eight days of growth represents approximately 100 generations. (C) Effect of par on pUB110 RC replicon stability. Cells of S. aureus RN4220 harboring pRB394 (•) (vector only), pSK5316 (▪) (carrying functional par), and pSK5334 (▴) (carrying nonfunctional par) were examined for stable maintenance of plasmids by determination of the proportion of the cell population which remained neomycin resistant during growth in nonselective medium. Eight days of growth represents approximately 50 generations.
To demonstrate that par-mediated stabilization is not facilitated via an effect on its cognate replication system, the ability of pSK1 par to stabilize an unrelated theta-replicating plasmid was investigated. A DNA fragment containing the par gene and upstream intergenic region from pSK1 was cloned into pAM401, an E. coli-Enterococcus faecalis shuttle vector which replicates via a theta mechanism mediated by a pIP501-derived replication region (59), to produce pSK5378 (Fig. 2B). For comparison, a par nonsense mutation was introduced into pSK5378, as described for pSK5335 above, to produce pSK6111 (Fig. 2B). pSK5378, pSK6111, and pAM401 were introduced into S. aureus RN4220, and plasmid stability assays were undertaken (Fig. 3B). After 4 days of growth, representing 50 generations, pSK5378 was retained by 74% of the cells in the population, whereas pSK6111 and pAM401 were retained by just 21 and 16%, respectively. Thus, the presence of a functional par gene significantly enhanced (P < 0.05) the segregational stability of the heterologous pIP501-based theta replicon. To confirm that the par phenotype is independent of any effect on replication, the relative copy numbers of pSK5378 and pSK6111, carrying wild-type and mutant par, respectively, were determined as described by Weaver et al. (55). Using pT181 as an internal standard with a copy number of 22 (41), copy numbers of 7.92 ± 0.52 and 7.56 ± 0.45 were obtained for pSK5378 and pSK6111, respectively, which are not significantly different (P > 0.05).
The ability of pSK1 par to stabilize an RC-replicating replicon was also investigated. The E. coli-Bacillus subtilis shuttle vector pRB394 carries the replication region from the small staphylococcal RC plasmid pUB110 (34). The segregational stabilities of pSK5316, carrying par; pSK5334, carrying a par nonsense mutation; and the vector pRB394 were determined in S. aureus RN4220. As shown in Fig. 3C, the presence of par in cis to rep of pUB110 in pSK5316 significantly enhanced the stability of the RC replicon (P < 0.0002) in an S. aureus population grown without selection for the plasmid, relative to pSK5334 and its parent pRB394. Together these results demonstrate that, in addition to stabilizing its cognate replicon, pSK1 par can extend the segregational stability of unrelated theta- and RC-replicating plasmids.
Mechanism of par-mediated segregational stability.
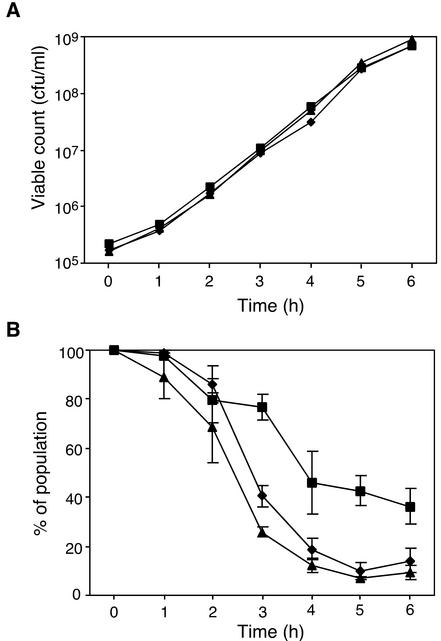
When carried by a segregationally unstable plasmid, postsegregational killing typically mediates a significant reduction in host growth rate as a consequence of its effect on plasmid-free segregants (27, 40). To investigate whether pSK1 par facilitates enhanced segregational stability via a postsegregational killing mechanism, we measured and compared the growth of cells which carried derivatives of the highly unstable RC pUB110 rep-based plasmid pRB394 (Fig. 3C) (8). Figure 4A shows the relative growth of S. aureus RN4220 cells variously carrying pSK5316, which carries par; pSK5322, which lacks par; and pSK5334, which carries a par nonsense mutation, in media without plasmid selection. Over a 6-h period, during which plasmid loss was observed for a major proportion of all three cell populations (Fig. 4B), no significant difference in growth rate between the strains was observed (P > 0.05) (Fig. 4A). Significantly, in the case of S. aureus RN4220 carrying the par+ plasmid pSK5316, more than 60% of the cell population had lost the plasmid at the conclusion of the experiment (Fig. 4B), with no concomitant reduction in growth rate (Fig. 4A). Since a postsegregational killing mechanism would be expected to have a negative effect on growth under such conditions, these data suggest that pSK1 par acts via another mechanism.
FIG. 4.
Relative growth (A) and plasmid maintenance (B) of S. aureus strains with plasmids carrying pUB110 rep in the presence or absence of par. Four-hour cultures were subcultured in LB medium and grown for 6 h, which represents approximately 12 generations. Each data point is the mean from three experiments. The error bars represent standard errors. Since there is no significant difference between strains in growth rate, error bars are omitted for clarity in panel A. Data for the following strains are shown: S. aureus RN4220 harboring pSK5316 (▪) (pUB110 replicon with par in cis), pSK5322 (♦) (pUB110 replicon lacking par), and pSK5334 (▴) (pUB110 replicon with nonfunctional par in cis).
A second mechanism of plasmid stabilization, multimer resolution, was investigated as the possible mechanistic basis of pSK1 par-mediated plasmid stabilization. DNA was isolated from S. aureus RN4220 cells harboring plasmids carrying functional and mutant par genes (pSK5378 and pSK6111, respectively) and was fractionated by agarose gel electrophoresis to allow visualization of plasmid monomers and multimers. No significant difference between the numbers or intensities of the various plasmid forms was observed (data not shown), indicating that the presence of par in pSK5378 does not result in a reduction in the quantity of plasmid multimers and suggesting that par does not function as a multimer resolution system. Consistent with this notion, pSK1 carries another gene, sin, that encodes a resolvase that belongs to the recombinase superfamily (44).
par-mediated incompatibility.
Partition-mediated incompatibility occurs when two plasmids introduced into the same cell carry identical centromere-like sites, which are indistinguishable from each other during the partitioning process. Under the replicon-pairing model, this results in the formation of mixed pairs of plasmids prior to cell division, random assortment of which ultimately results in cell lines arising that carry only one of the two plasmids (2). To gain support for the notion that pSK1 par mediates active partitioning, the ability of par to confer cis-acting incompatibility on otherwise compatible replicons was tested. Furthermore, since partitioning sites have been shown to be incompatibility determinants in other partitioning systems (2), identification of the region conferring par-mediated incompatibility would likely delimit the binding site of the par product Par in the partitioning process. The plasmids pSK4829 and pAM401 replicate via unrelated replicons (pSK1 and pIP501, respectively) and are therefore compatible and useful in the determination of incompatibility properties of sequences that are cloned into them. A 204-bp DNA fragment from pSK1, containing the region immediately upstream of par, was cloned into pAM401, to produce pSK6110 (Fig. 2B). This plasmid was then introduced by electroporation into S. aureus RN4220 harboring the pSK1 par+ minireplicon pSK4829. As a comparison, pAM401 was similarly transformed into the same S. aureus strain. Following daily subculturing (1:100 dilution) in L broth selective for the incoming plasmid, cultures were grown for approximately 25 to 30 generations and erythromycin resistance, encoded by the pSK1 minireplicon, was used to indicate retention of pSK4829.
A significant difference (P < 0.05) in retention of the resident plasmid, pSK4829, between the strains carrying the introduced plasmids pAM401 and pSK6110 was taken to indicate incompatibility. As shown in Fig. 5, introduction of pAM401 yielded a pSK4829 retention rate of 85% after 25 to 30 generations. However, when pAM401 carried the 204-bp BamHI fragment immediately upstream of the par gene (pSK6110), the pSK4829 retention rate was significantly lower, 18% (P < 0.05) (Fig. 5). This indicates that the 204-bp fragment, in the presence of par, is able to mediate incompatibility when carried by two unrelated replicons that are otherwise compatible. To establish that the destabilization effect was indeed due to par-mediated incompatibility rather than rep-mediated incompatibility, the same stability assays were carried out with pSK5335, a pSK1 minireplicon carrying a par nonsense mutation. No significant difference in retention of pSK5335 was detected in the presence of coresident pAM401 or pSK6110 (P > 0.05) (Fig. 5), confirming that replication of the pSK1 minireplicon is not affected by the 204-bp fragment carried by pSK6110 and therefore that this fragment does not confer rep-mediated incompatibility. The 204-bp region cloned in pSK6110, which contains seven 12-bp direct repeats (DRs) and one 7 bp IR (Fig. 2) therefore probably encompasses the binding site of the Par protein in the partitioning of the pSK1 plasmid. The most likely explanation for the observed incompatibility is that this fragment carries a par cis-acting site, although we cannot exclude the possibility that it might be due to a regulatory effect on par expression emanating from within this fragment.
FIG. 5.
par-mediated incompatibility. Cells of S. aureus RN4220 harboring pSK4829 (pSK1 minireplicon that contains par) plus pAM401 (pIP501 minireplicon) (▪), pSK4829 plus pSK6110 (pIP501 minireplicon carrying 204-bp region upstream of par) (♦), pSK5335 (pSK1 minireplicon with par nonsense mutation) plus pAM401 (•), or pSK5335 plus pSK6110 (▴) were examined for stable maintenance of pSK1 minireplicons by determination of the proportion of the cell population which remained erythromycin resistant during growth in medium selective for pAM401 or its derivative. Four days of growth represents 25 to 30 generations. For details, see Materials and Methods. Each data point is the mean from three experiments. The error bars represent standard errors.
Prediction of a coiled-coil domain.
Across the entire family of proteins showing homology to the deduced par product, amino acid sequence conservation is restricted to two segments, an N-terminal region comprising the first half of a helix-turn-helix (HTH) domain and a second segment, towards the C terminus, of unknown structure (14). The HTH DNA-binding motif identified at the N terminus of Par extends from amino acid residue 3 to 24 and is located at an equivalent position in the homologous proteins (14). Amino acid sequence conservation is confined to the first α-helix (14), which usually associates nonspecifically with DNA so as to provide a rigid underlying framework that supports the second, so-called recognition helix (7). The recognition helix has a role in binding to specific DNA sequences (52), so it is not surprising that this helix is not conserved across the Par homologs. HTH motifs have also been identified in other partitioning proteins including F SopB and P1 ParB (13).
In an effort to determine whether the region of homology towards the C terminus also represented a common structural motif, the probability of Par forming a coiled coil was determined by using the MultiCoil program (60). The coiled-coil structure, which consists of two to five α-helices wrapped around each other, is associated with the formation of protein oligomers and has been observed in a broad range of proteins, including transcription factors and proteins involved in the cytoskeleton of eukaryotic cells (9). The probability of coiled-coil formation in Par was shown to be very high in the region from amino acid residue 132 to 155 (probability of >0.9), with a section within this region (amino acid residues 141 to 155) predicted to form a coiled-coil dimer (probability of >0.8) (data not shown). Analysis of all full-length Par homologs similarly revealed a moderate to high probability of coiled-coil formation at a position corresponding to the conserved region in each case (data not shown), suggesting an important role for this part of the protein. Consistent with the notion that Par forms a coiled coil, preliminary protein purification revealed that Par exhibits aberrant migration during sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). His-tagged Par protein migrates with an apparent molecular weight of approximately 42,000, which is 50% greater than its predicted molecular weight of 28,800 (14). Such unusual migration has been reported for other proteins with a coiled-coil motif (10).
DISCUSSION
The results of this study demonstrate that pSK1 par mediates enhanced plasmid segregational stability independently of any effect on plasmid replication. Additionally, active partitioning is implicated as the probable mechanism of par action. To date, relatively little information is available on plasmid stabilization mechanisms in staphylococci other than the putative multimer resolution systems res and sin, identified in pSK41 (4) and in pSK1 and pI9789::Tn552 (44), respectively. The putative active partitioning mechanism reported here represents the first partitioning system identified in staphylococci. Notably, par homologs have been found not only on other theta-replicating plasmids but also on the RC plasmids pSSU1, pLH2, and pVA380-1 (14). To our knowledge, partitioning systems have not previously been recognized on RC plasmids. The results described here demonstrated that the pSK1 par system was able to extend the segregational stability of a heterologous RC replicon. The carriage of this type of partitioning system by some RC plasmids may be facilitated by its compact nature, since the RC replication strategy imposes restraints on plasmid size.
All plasmid-encoded partitioning systems characterized to date share a number of common features: two trans-acting proteins, encoded by genes in an autoregulated operon and with ATPase and DNA-binding activities, and a cis-acting centromere-like site (19, 35). In all known cases the upstream gene within the partitioning operon encodes an ATPase that is essential for the segregation of DNA (19). Partitioning systems characterized to date can be divided into two families based on the type of ATPase they encode. Type I partitioning loci encode Walker-type ATPases, whereas type II partitioning systems encode ATPases from the actin-hsp70 superfamily (19). As the ATPase activities of R1 ParM, P1 ParA, and F SopA are believed to be too low to release sufficient energy for the active movement of plasmids within the cell, it has been proposed that the role of ATPase in partitioning is as a molecular switch (5, 25). It has been suggested that the activities of P1 ParA are controlled by an ATP-ADP molecular switch; the ADP-bound form of ParA binds to the promoter region in its role as a repressor of the par operon, and the ATP-bound form interacts with ParB in the partition complex at parS, thereby participating in partitioning (5, 16). Type I loci are further divided into two subgroups: type Ia, in which the centromere-like site is located downstream of the par operon, represented by the par/sop gene family, and type Ib, represented by par of pTAR, which comprises partitioning systems with a centromere-like site contained within the par promoter region. Type Ib ATPases also lack an N-terminal DNA-binding domain, which is present in type Ia systems. The replicon-pairing model of DNA segregation, based largely on studies of par from plasmid R1 (26) and the par/sop family from prophage P1 and F plasmid, respectively (39, 50, 51), is currently favored by most groups and involves the pairing of plasmids, as a result of the partitioning proteins binding to the centromere-like site, followed by separation of the paired complexes towards the cell poles at cell division (58). Analyses involving direct visualization of plasmids within prokaryotic cells by using cytological techniques have suggested the existence of a mitotic spindle-like apparatus (35). Indeed, evidence has recently been provided for a prokaryotic analog of the eukaryotic mitotic spindle encoded by plasmid R1, where the ParM ATPase has been found to form actin-like filaments which generate the force required for the movement of plasmids to opposite cell poles (36). It is conceivable that the coiled-coil domain of pSK1 Par forms a structural element in the staphylococcal cell analogous to that reported for both coiled-coil-containing proteins in the cytoskeleton of eukaryotic cells and R1 ParM actin-like filaments in E. coli cells (9, 36). Interestingly, a coiled-coil domain is present in MukB, a protein involved in chromosome and plasmid partitioning in E. coli (57).
The pSK1 par system differs significantly from the active partitioning systems described above. Specifically, there is no observable homology with previously described partitioning systems. The par-containing sequence cloned into the heterologous plasmids (Fig. 2B) delimits the extent of a 1,350-bp fragment from nucleotide position 3 to 1353 (GenBank accession no. AF203376) (14). Only one significant ORF is evident. Thus, only a single protein-encoding gene appears to be required. Furthermore, it seems likely that partitioning is achieved in the absence of ATPase activity, since no ATP-binding motif is evident within either the deduced Par product or the deduced six-frame translations of the additional upstream or downstream sequences in the cloned fragment. ParB/SopB proteins from plasmids P1 and F have been shown to bind to their respective centromere-like sites as dimers (12, 31, 38, 50). The MultiCoil prediction of a coiled-coil dimer domain suggests that Par might also undergo dimerization. Like other proteins which contain coiled coils, Par was found to migrate aberrantly upon sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10). Since the apparent molecular weight was approximately 50% greater than that predicted for Par, it seems likely that the species detected represents a dimer linked by coiled-coil domains. Recent studies have provided evidence that coiled-coil domains can act as molecular switches (9). While hydrophobic interactions are the main driving force for coiled-coil stability, ionic interactions also contribute and have been shown to modulate the assembly and stability of coiled coils in a pH-dependent manner in macrophage scavenger receptor, influenza virus hemagglutinin, and the Fos leucine zipper dimer (9). pH-dependent electrostatic repulsions are believed to destabilize the coiled-coil structure, thereby facilitating conformational change, which, in the case of macrophage scavenger receptor, allows the release of bound ligands (9). Thus, in addition to mediating dimerization, the coiled coil in Par may form the basis of a molecular switch, analogous to the ATPase activity identified in other partitioning systems.
A 204-bp fragment upstream of par was shown to mediate incompatibility, raising the possibility that this region contains the centromere-like site. In R1 and pTAR, the centromere-like site consists of 10 11-bp DRs and 12 7-bp DRs, respectively, located upstream of the partitioning genes (11, 17). In the class of par systems typified by P1 and F, this function is performed by multiple repeat sequences located downstream of the par genes (21). In pSK1, a series of seven 12-bp DRs and one IR of 7 bp represent possible candidate binding sites involved in centromere-like functions (Fig. 2).
Partitioning loci previously identified on plasmids of gram-positive origin belong to the type 1b class. The results presented here suggest that the pSK1 par locus is distinct from all known classes of partitioning systems and therefore potentially represents the prototype of a new class of partitioning systems, type III.
Acknowledgments
We thank Keith Weaver for providing plasmid pAM401.
This work was supported by project grant 153816 from the National Health and Medical Research Council (Australia). A.E.S. was the recipient of an Australian Postgraduate Award.
REFERENCES
- 1.Apisiridej, S. 1997. Ph.D. thesis. University of Sydney, Sydney, Australia.
- 2.Austin, S., and K. Nordstrom. 1990. Partition-mediated incompatibility of bacterial plasmids. Cell 60:351-354. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. S. Smith, and K. Struhl. 1990. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 4.Berg, T., N. Firth, S. Apisiridej, A. Hettiaratchi, A. Leelaporn, and R. A. Skurray. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative plasmids. J. Bacteriol. 180:4350-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouet, J. Y., and B. E. Funnell. 1999. P1 ParA interacts with the P1 partition complex at parS and an ATP-ADP switch controls ParA activities. EMBO J. 18:1415-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Brennan, R. G., and B. W. Matthews. 1989. The helix-turn-helix binding motif. J. Biol. Chem. 264:1903-1906. [PubMed] [Google Scholar]
- 8.Bruckner, R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122:187-192. [DOI] [PubMed] [Google Scholar]
- 9.Burkhard, P., S. V. Strelkov, and J. Stetefeld. 2001. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11:82-88. [DOI] [PubMed] [Google Scholar]
- 10.Cope, L. D., E. R. Lafontaine, C. A. Slaughter, C. A. Hasemann, Jr., C. Aebi, F. W. Henderson, G. H. McCracken, Jr., and E. J. Hansen. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 181:4026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dam, M., and K. Gerdes. 1994. Partitioning of plasmid R1. Ten direct repeats flanking the parA promoter constitute a centromere-like partition site parC, that expresses incompatibility. J. Mol. Biol. 236:1289-1298. [DOI] [PubMed] [Google Scholar]
- 12.Davis, M. A., and S. J. Austin. 1988. Recognition of the P1 plasmid centromere analog involves binding of the ParB protein and is modified by a specific host factor. EMBO J. 7:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Firth, N., S. Apisiridej, T. Berg, B. A. O'Rourke, S. Curnock, K. G. H. Dyke, and R. A. Skurray. 2000. Replication of staphylococcal multiresistance plasmids. J. Bacteriol. 182:2170-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Firth, N., and R. A. Skurray. 2000. The Staphylococcus genetics: accessory elements and genetic exchange, p. 326-338. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 16.Fung, E., J. Y. Bouet, and B. E. Funnell. 2001. Probing the ATP-binding site of P1 ParA: partition and repression have different requirements for ATP binding and hydrolysis. EMBO J. 20:4901-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallie, D. R., and C. I. Kado. 1987. Agrobacterium tumefaciens pTAR parA promoter region involved in autoregulation, incompatibility and plasmid partitioning. J. Mol. Biol. 193:465-478. [DOI] [PubMed] [Google Scholar]
- 18.Gerdes, K., S. Ayora, I. Canosa, R. Ceglowski, R. Diaz-Orejas, T. Franch, A. P. Gultyaev, R. B. Jensen, I. Kobayshi, C. Macpherson, D. Summers, C. M. Thomas, and U. Zielenkiewicz. 2000. Plasmid maintenance systems, p. 49-75. In C. M. Thomas (ed.), The horizontal gene pool. Hardwood Academic Publishers, Amsterdam, The Netherlands.
- 19.Gerdes, K., J. Moller-Jensen, and R. B. Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 20.Gering, M., F. Götz, and R. Bruckner. 1996. Sequence and analysis of the replication region of the Staphylococcus xylosus plasmid pSX267. Gene 182:117-122. [DOI] [PubMed] [Google Scholar]
- 21.Gordon, G. S., and A. Wright. 2000. DNA segregation in bacteria. Annu. Rev. Microbiol. 54:681-708. [DOI] [PubMed] [Google Scholar]
- 22.Greenfield, T. J., T. Franch, K. Gerdes, and K. E. Weaver. 2001. Antisense RNA regulation of the par post-segregational killing system: structural analysis and mechanism of binding of the antisense RNA, RNAII and its target, RNAI. Mol. Microbiol. 42:527-537. [DOI] [PubMed] [Google Scholar]
- 23.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson, M. C., and G. C. Stewart. 1986. Differential utilization of Staphylococcus aureus promoter sequences by Escherichia coli and Bacillus subtilis. Gene 48:93-100. [DOI] [PubMed] [Google Scholar]
- 25.Jensen, R. B., and K. Gerdes. 1997. Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR-parC complex. J. Mol. Biol. 269:505-513. [DOI] [PubMed] [Google Scholar]
- 26.Jensen, R. B., and K. Gerdes. 1999. Mechanism of DNA segregation in prokaryotes: ParM partitioning protein of plasmid R1 co-localizes with its replicon during the cell cycle. EMBO J. 18:4076-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen, R. B., E. Grohmann, H. Schwab, R. Diaz-Orejas, and K. Gerdes. 1995. Comparison of ccd of F, parDE of RP4, and parD of R1 using a novel conditional replication control system of plasmid R1. Mol. Microbiol. 17:211-220. [DOI] [PubMed] [Google Scholar]
- 28.Kearney, K., G. F. Fitzgerald, and J. F. Seegers. 2000. Identification and characterization of an active plasmid partition mechanism for the novel Lactococcus lactis plasmid pCI2000. J. Bacteriol. 182:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan, S. A., and R. P. Novick. 1983. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid 10:251-259. [DOI] [PubMed] [Google Scholar]
- 30.Leelaporn, A., N. Firth, M. E. Byrne, E. Roper, and R. A. Skurray. 1994. Possible role of insertion sequence IS257 in dissemination and expression of high- and low-level trimethoprim resistance in staphylococci. Antimicrob. Agents Chemother. 38:2238-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lobocka, M., and M. Yarmolinsky. 1996. P1 plasmid partition: a mutational analysis of ParB. J. Mol. Biol. 259:366-382. [DOI] [PubMed] [Google Scholar]
- 32.Lyon, B. R., J. W. May, and R. A. Skurray. 1983. Analysis of plasmids in nosocomial strains of multiple-antibiotic-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 23:817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lyon, B. R., J. W. May, and R. A. Skurray. 1984. Tn4001: a gentamicin and kanamycin resistance transposon in Staphylococcus aureus. Mol. Gen. Genet. 193:554-556. [DOI] [PubMed] [Google Scholar]
- 34.McKenzie, T., T. Hoshino, T. Tanaka, and N. Sueoka. 1986. The nucleotide sequence of pUB110: some salient features in relation to replication and its regulation. Plasmid 15:93-104. [DOI] [PubMed] [Google Scholar]
- 35.Moller-Jensen, J., R. B. Jensen, and K. Gerdes. 2000. Plasmid and chromosome segregation in prokaryotes. Trends Microbiol. 8:313-320. [DOI] [PubMed] [Google Scholar]
- 36.Moller-Jensen, J., R. B. Jensen, J. Lowe, and K. Gerdes. 2002. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 21:3119-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moran, C. P., N. Lang, S. F. J. LeGrice, G. Lee, M. Stephens, A. L. Sonenshein, J. Pero, and R. Losick. 1982. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol. Gen. Genet. 186:339-346. [DOI] [PubMed] [Google Scholar]
- 38.Mori, H., Y. Mori, C. Ichinose, H. Niki, T. Ogura, A. Kato, and S. Hiraga. 1989. Purification and characterization of SopA and SopB proteins essential for F plasmid partitioning. J. Biol. Chem. 264:15535-15541. [PubMed] [Google Scholar]
- 39.Niki, H., and S. Hiraga. 1997. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell 90:951-957. [DOI] [PubMed] [Google Scholar]
- 40.Nordstrom, K., and S. J. Austin. 1989. Mechanisms that contribute to the stable segregation of plasmids. Annu. Rev. Genet. 23:37-69. [DOI] [PubMed] [Google Scholar]
- 41.Novick, R. P. 1989. Staphylococcal plasmids and their replication. Annu. Rev. Microbiol. 43:537-565. [DOI] [PubMed] [Google Scholar]
- 42.Novick, R. P. 1990. The Staphylococcus as a molecular genetic system, p. 1-37. In R. P. Novick (ed.), Molecular biology of the staphylococci. VCH, New York, N.Y.
- 43.Paulsen, I. T., N. Firth, and R. A. Skurray. 1997. Resistance to antimicrobial agents other than β-lactams, p. 175-212. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 44.Paulsen, I. T., M. T. Gillespie, T. G. Littlejohn, O. Hanvivatvong, S. J. Rowland, K. G. Dyke, and R. A. Skurray. 1994. Characterisation of sin, a potential recombinase-encoding gene from Staphylococcus aureus. Gene 141:109-114. [DOI] [PubMed] [Google Scholar]
- 45.Rouch, D. A., L. J. Messerotti, L. S. Loo, C. A. Jackson, and R. A. Skurray. 1989. Trimethoprim resistance transposon Tn4003 from Staphylococcus aureus encodes genes for a dihydrofolate reductase and thymidylate synthetase flanked by three copies of IS257. Mol. Microbiol. 3:161-175. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 47.Schenk, S., and R. A. Laddaga. 1992. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73:133-138. [DOI] [PubMed] [Google Scholar]
- 48.Shaw, W. V. 1975. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737-755. [DOI] [PubMed] [Google Scholar]
- 49.Sheehy, R. J., and R. P. Novick. 1975. Studies on plasmid replication. V. Replicative intermediates. J. Mol. Biol. 93:237-253. [DOI] [PubMed] [Google Scholar]
- 50.Surtees, J. A., and B. E. Funnell. 1999. P1 ParB domain structure includes two independent multimerization domains. J. Bacteriol. 181:5898-5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Surtees, J. A., and B. E. Funnell. 2001. The DNA binding domains of P1 ParB and the architecture of the P1 plasmid partition complex. J. Biol. Chem. 276:12385-12394. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki, M., N. Yagi, and M. Gerstein. 1995. DNA recognition and superstructure formation by helix-turn-helix proteins. Protein Eng. 8:329-338. [DOI] [PubMed] [Google Scholar]
- 53.Swinfield, T. J., L. Janniere, S. D. Ehrlich, and N. P. Minton. 1991. Characterization of a region of the Enterococcus faecalis plasmid pAMβ1 which enhances the segregational stability of pAMβ1-derived cloning vectors in Bacillus subtilis. Plasmid 26:209-221. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka, T., and M. Ogura. 1998. A novel Bacillus natto plasmid pLS32 capable of replication in Bacillus subtilis. FEBS Lett. 422:243-246. [DOI] [PubMed] [Google Scholar]
- 55.Weaver, K. E., D. B. Clewell, and F. An. 1993. Identification, characterization, and nucleotide sequence of a region of Enterococcus faecalis pheromone-responsive plasmid pAD1 capable of autonomous replication. J. Bacteriol. 175:1900-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weaver, K. E., K. D. Jensen, A. Colwell, and S. I. Sriram. 1996. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol. Microbiol. 20:53-63. [DOI] [PubMed] [Google Scholar]
- 57.Weitao, T., S. Dasgupta, and K. Nordstrom. 2000. Role of the mukB gene in chromosome and plasmid partition in Escherichia coli. Mol. Microbiol. 38:392-400. [DOI] [PubMed] [Google Scholar]
- 58.Williams, D. R., and C. M. Thomas. 1992. Active partitioning of bacterial plasmids. J. Gen. Microbiol. 138:1-16. [DOI] [PubMed] [Google Scholar]
- 59.Wirth, R., F. Y. An, and D. B. Clewell. 1986. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J. Bacteriol. 165:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolf, E., P. S. Kim, and B. Berger. 1997. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 6:1179-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]