Abstract
We have investigated the conditions required for polar localization of the CheZ phosphatase by using a CheZ-green fluorescent protein fusion protein that, when expressed from a single gene in the chromosome, restored chemotaxis to a ΔcheZ strain. Localization was observed in wild-type, ΔcheZ, ΔcheYZ, and ΔcheRB cells but not in cells with cheA, cheW, or all chemoreceptor genes except aer deleted. Cells making only CheA-short (CheAS) or CheA lacking the P2 domain also retained normal localization, whereas cells producing only CheA-long or CheA missing the P1 and P2 domains did not. We conclude that CheZ localization requires the truncated C-terminal portion of the P1 domain present in CheAS. Missense mutations targeting residues 83 through 120 of CheZ also abolished localization. Two of these mutations do not disrupt chemotaxis, indicating that they specifically prevent interaction with CheAS while leaving other activities of CheZ intact.
CheZ is a phosphatase that accelerates the removal of the intrinsically labile phosphoryl group from CheY-P (19, 28). Zhao et al. (30) recently determined the crystal structure of the CheZ dimer complexed with two CheY monomers containing the phosphoryl analog BeF3−. CheZ can also be isolated in a complex with CheA-short (CheAS) (26), a short form of CheA produced from an internal translation initiation site at codon 98 of cheA (9). This complex was first identified in coimmunoprecipitation experiments, but it can also form with purified proteins. In experiments done in vitro at 4°C to slow the spontaneous dephosphorylation of CheY-P, the CheZ/CheAS complex showed a 2.3-fold-higher phosphatase activity than free CheZ (26). Binding of CheAS to CheZ may be inhibited by CheW, as indicated by the decreased level of coimmunoprecipitation when CheW is overexpressed (27). CheAS lacks the phosphoryl-accepting His-48 residue of CheA-long (CheAL), but it is catalytically active and can phosphorylate CheAL in trans (29).
Using CheZ fused to yellow fluorescent protein (1), Sourjik and Berg (23) found that CheZ localizes to the subpolar chemoreceptor clusters identified by Maddock and Shapiro (12). We had been using CheZ fused to green fluorescent protein (GFP) (1) to study the distribution of CheZ in cells and, after learning of Sourjik and Berg's work, asked whether CheAS is needed for this localization and what part of CheZ is required.
Construction of a CheZ-GFP chimera.
A cheZ-gfp gene fusion was created by PCR with primers encoding a 7-amino-acid flexible linker (GGSSAAG). The fusion gene was cloned into the pBAD18 vector, and the resulting plasmid, pBJC101 (Table 1), was shown to enable the ΔcheZ strain RP1616 to form wild-type chemotactic swarms in tryptone semisolid agar containing 0.002% arabinose. The fusion gene was subsequently cloned into the vector plasmid pCJ30 to facilitate its insertion into the chromosome by using the λInCh system (5). The resulting pBJC104 plasmid was also able to restore chemotactic swarming to strain RP1616, even in the absence of the inducer IPTG (isopropyl-β-d-thiogalactopyranoside). The cheZ-gfp gene was inserted from plasmid pBC104 into the chromosome of strain RP1616. The BC200 strain created in this way made wild-type chemotactic swarms when transcription of cheZ-gfp was induced with 1 mM IPTG. In the absence of IPTG, little or no swarming was observed.
TABLE 1.
Strains, plasmids, and phage
| Strain, plasmid, or phage | Genotype or phenotype | Comments | Reference or source |
|---|---|---|---|
| Strains | |||
| RP437 | thr-1(Am) leuB6 his-4 metF159(Am) eda-50 rpsL1356 thi-1 ara-14 mtl-1 xyl-5 tonA31 tsx-78 lacY1 F− | 18 | |
| AJW536 | RP437 cheA(M98L) zig::Tn10, polA(Ts) | 14 | |
| RP1616 | RP437 ΔcheZ6725 | J. S. Parkinson | |
| RP9535 | RP437 ΔcheA1643 eda+ | 16 | |
| RP1078 | RP437 Δ(cheW-tap)2217 | 18 | |
| RP2867 | RP437 Δ(tap-cheB)224 eda+ | 18 | |
| RP5231 | RP437 Δ(cheY-cheZ)4213 eda+ | J. S. Parkinson | |
| RP1515 | RP437 cheA169(Am) Δlac-169 eda+ | 22 | |
| RP1516 | RP437 cheA157(Am) Δlac-169 eda+ | 22 | |
| UU1118 | RP437 cheA(Δ7-247) eda+ | 7 | |
| UU1121 | RP437 cheA(Δ150-247) eda+ | J. S. Parkinson | |
| VB13 | RP437 Δtsr7021 Δ(tar-tap)5201 trg::Tn10 thr+eda+ | 28 | |
| DHB6521 | SM551 (λInCh1 lysogen) | 5 | |
| BC200 | RP1616 Δ(λatt-lom)::bla lacIq ptac-cheZ-gfp | pBJC104 into RP1616 via λInCh1 | This study |
| BC201 | RP437 Δ(λatt-lom)::bla lacIq ptac-cheZ-gfp | pBJC104 into RP437 via λInCh1 | This study |
| BC203 | VB13 Δ(λatt-lom)::bla lacIq ptac-cheZ-gfp | pBJC104 into VB13 via λInCh1 | This study |
| BC206 | RP9535 Δ(λatt-lom)::bla lacIq ptac-cheZ-gfp | pBJC104 into RP9535 via λInCh1 | This study |
| BC207 | RP1078 Δ(λatt-lom)::bla lacIq ptac-cheZ-gfp | pBJC104 into RP1078 via λInCh1 | This study |
| BC208 | RP5231 Δ(λatt-lom)::bla lacIq ptac-cheZ-gfp | pBJC104 into RP5231 via λInCh1 | This study |
| BC209 | AJW536 Δ(λatt-lom)::bla lacIq ptac-cheZ-gfp | pBJC104 into AJW536 via λInCh1 | This study |
| BC210 | UU1121 Δ(λatt-lom)::bla lacIq ptac-cheZ-gfp | pBJC104 into UU1121 via λInCh1 | This study |
| BC211 | UU1118 Δ(λatt-lom)::bla lacIq ptac-cheZ-gfp | pBJC104 into UU1118 via λInCh1 | This study |
| BC212 | RP1515 Δ(λatt-lom)::bla lacIq ptac-cheZ-gfp | pBJC104 into RP1515 via λInCh1 | This study |
| BC213 | RP1516 Δ(λatt-lom)::bla lacIq ptac-cheZ-gfp | pBJC104 into RP1516 via λInCh1 | This study |
| BC214 | RP2867 Δ(λatt-lom)::bla lacIq ptac-cheZ-gfp | pBJC104 into RP2867 via λInCh1 | This study |
| Plasmids and phage | |||
| pAG3 | ptac cheA(1-149) Ampr | Expresses CheA P1 domain | 7 |
| pBAD18 | araC+ Ampr | paraBAD expression vector | 8 |
| pCJ30 | lacIq Ampr | ptac expression vector | 2 |
| pPM2 | gfp mut-2 | Expresses GFP Mut2 | 6 |
| pBJC100 | cheZ Ampr | cheZ in pBAD18 | This study |
| pBJC101 | paraBAD cheZ-gfp Ampr | cheZ-gfp in pBAD18 | This study |
| pBJC102 | gfp mut-2 Ampr | gfp mut 2 in pBAD18 | This study |
| pBJC104 | ptac cheZ-gfp Ampr | cheZ-gfp in pCJ30 | This study |
| λInCh1 | Kanr CI857 | λInCh for pBR-derived plasmids | 5 |
Extracts from cells containing plasmid pBJC104 or the chromosomal insertion of cheZ-gfp contained a protein of the size expected (∼54 kDa) for CheZ-GFP, whether the immunoblots were developed with anti-CheZ or anti-GFP antibody. Only a small amount of normal-length CheZ was detected. CheZ expressed from plasmid pBJC100 also supported the best swarming in strain RP1616 at 0.002% arabinose. Since the amounts of CheZ and CheZ-GFP, estimated from immunoblots, appeared to be about the same under these conditions of induction, we concluded that CheZ-GFP is functional and responsible for the observed complementation of ΔcheZ.
Subcellular localization of CheZ-GFP.
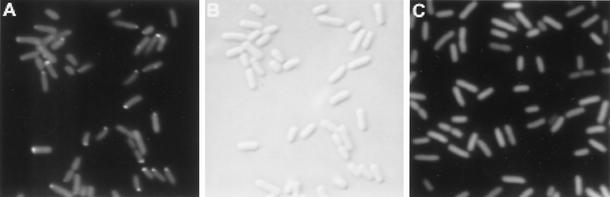
Subcellular localization of GFP fluorescence was examined in strains containing the cheZ-gfp gene in single copy on the chromosome. In strain BC200 (ΔcheZ), CheZ-GFP localized to patches as was previously observed by Sourjik and Berg (23) for plasmid-encoded CheZ-yellow fluorescent protein. All cells exhibited diffuse cytoplasmic fluorescence, but bright, localized patches of fluorescence were seen in 85% of the cells, primarily near the poles but also laterally in 15 to 20% of the cells (Fig. 1A and B). This pattern was similar to that seen by immunofluorescence (12) with antibody to CheA or the serine chemoreceptor Tsr. Thus, these patches are likely to represent CheZ-GFP associated with clusters containing chemoreceptors, CheA and CheW. Similar patterns of fluorescence were observed in cells of strain RP1616 carrying plasmid pBJC101.
FIG. 1.
CheZ-GFP localizes to patches. (A) Fluorescence micrograph of BC200 cells grown to late exponential phase at 32°C in tryptone broth (15) containing 1 mM IPTG. The cells exhibit uniform background fluorescence, presumably due to CheZ-GFP dimers in the cytoplasm. Localized bright patches of fluorescence are visible in most cells. (B) Differential interference contrast photomicrograph of the same cells as in panel A. Comparison of the two images reveals that nearly all cells fluoresce with the same intensity, indicating that their CheZ-GFP contents are similar. (C) Fluorescence micrograph of cells of strain BC206 (ΔcheA) grown as described for panel A. Note that the level of background fluorescence is the same but that no intense patches of fluorescence are visible. Cells were observed at a magnification of ×1,575 with a Zeiss Axioplan 2, and the images were captured with a Hamamatsu C5810 charge-coupled device camera. The peak excitation wavelength was 484 nm, and emitted light was detected at 510 to 530 nm.
Essentially identical numbers and distributions of patches of fluorescence were seen cells of strain BC214 (Δtap-cheB) and BC208 (Δche-cheZ), but no patches were detectable in cells of strain BC206 (ΔcheA) (Fig. 1C), strain BC207 (ΔcheW-tap) or strain BC203 (Δtar-tap Δtsr trg::Tn10). These results mirror the strain dependence for formation of receptor clusters (11, 12). Immunogold labeling of thin sections of cells that was carried out with anti-CheZ antibody indicated that the wild-type CheZ protein also localizes in clusters near the cell poles (J. R. Maddock, personal communication).
Localization of CheZ-GFP to receptor patches requires CheAS but not CheAL.
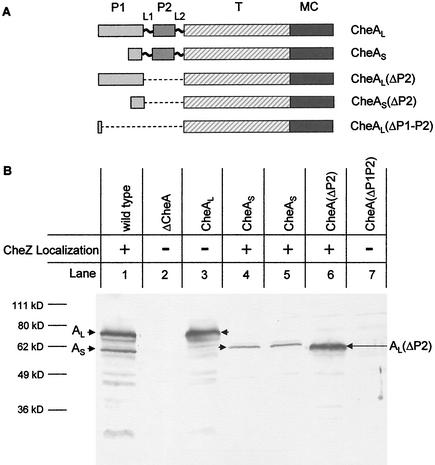
We next examined the dependence of CheZ-GFP patching on CheAS and CheAL. Strain BC209 produces only CheAL because ATG codon 98 of cheA, the CheAS start codon (9), has been changed to CTG. The M98L version of CheAL has about 70% of the kinase activity of wild-type CheA, and a strain producing M98L CheAL makes swarms with 70% the diameter of wild-type swarms (21). Strains BC212 and BC213 contain amber mutations in the cheA sequence between the start codon of CheAL and codon 98, so that both strains produce only CheAS. A schematic of the CheA polypeptides produced by these strains is shown in Fig. 2A. The ratio of the intensities of bands of CheAL versus CheAS detected on immunoblots prepared with our CheA antiserum (22) is 2.3:1 in extracts from strain BC200 (Fig. 2B). The ratio of the intensities of the CheAL band in extracts of strain BC209 and of the CheAS band in extracts from strains BC212 and BC213 is about the same. Thus, each form of CheA exists in a normal amount in the absence of the other. We note that because most of the highly antigenic P1 domain is missing from CheAS, our immunoblots may underestimate its amount. Thus, the actual CheAL/CheAS ratio in strain BC200 may be closer to the 1:1 value previously reported for late exponential-phase, highly motile Escherichia coli cells (27).
FIG. 2.
Cellular levels of various forms of the E. coli CheA protein. (A) Schematic representation of CheAL, CheAS, CheAL(ΔP2), CheAS(ΔP2), and CheA(ΔP1-P2). P1, P2, T, and MC represent the phosphorylation, CheY-binding, dimerization and catalytic, and CheW/receptor input domains, respectively. (B) Immunoblot with polyclonal CheA antiserum (24). Extracts were prepared from 1 ml of cells grown to an optical density at 590 nm of 0.8 and resuspended in 100 μl of sodium dodecyl sulfate loading dye. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% gel). Lanes: 1, BC201; 2, BC206; 3, BC209; 4, BC212; 5, BC213; 6, BC210; 7, BC211. The CheA protein species produced by each strain is indicated above the lane, and the corresponding band is indicated with an arrow.
Cells of strain BC209 contained no detectable bright patches and showed only the diffuse cytoplasmic fluorescence found in cells of strain BC206 (ΔcheA) (Fig. 1C). Cells of the CheAS-only strains BC212 and BC213 localized CheZ-GFP to patches just like strain BC200 (Fig. 1A). These data suggested that CheZ interacts with CheAS at the patch in vivo and is required for the pattern of CheZ localization observed in wild-type cells. It has previously been shown that CheAL-only and CheAS-only cells form receptor patches with equal facility (21).
The N-terminal region of CheAS may bind CheZ.
Strain BC210 (cheAΔP2) expresses a CheA protein missing the P2 domain, which binds CheY (16, 25). Its swarming ability is only slightly decreased from that of its wild-type parental strain RP437. Strain BC211 [cheAΔ(P1-P2)] expresses a CheA protein in which a cheA-internal deletion removes most of the P1 and P2 domains, and it does not form chemotactic swarms. Chromosomally encoded CheZ-GFP had a patchy distribution in cells of strain BC210 but not in cells of strain BC211. Expression of the P1 domain (25) from plasmid pAG3 enabled strain BC211 to form chemotactic swarms with about half the diameter of those made by strain RP437 but did not restore CheZ-GFP localization in cells of this strain. Although our polyclonal CheA antiserum did not visualize the CheAΔ(P1-P2) protein in immunoblots, it must be present at some level for complementation to occur. A protein of the size expected for CheALΔP2 was detected with this antiserum in an extract from strain BC210 (Fig. 2B), but no band was seen at the position expected for CheASΔP2.
The inability to detect CheAΔ(P1-P2) and CheASΔP2 with our polyclonal antiserum may represent the absence of epitopes recognized by the antiserum, since 23 of 27 monoclonal antibodies raised against full-length CheA target P1 and P2 (J. S. Parkinson, personal communication). Those data suggest that P1 and P2 are the most antigenic portions of CheA and that even polyclonal antisera against intact CheA are deficient in recognizing the remainder of the protein. We obtained monoclonal serum directed against the P4 (catalytic) domain of E. coli CheA (C. O'Connor and P. Matsumura, unpublished data), but it failed to visualize any form of CheA, including either full-length CheAL or CheAS, on immunoblots.
Mutations within a specific region of cheZ eliminate polar localization of CheZ-GFP.
To identify which part(s) of CheZ is responsible for localization, we introduced a selection of previously identified cheZ missense mutations (3, 4, 20) scattered throughout the gene into plasmid pBJC101. Of these 17 mutations, the ones causing the L90S and F117S substitutions completely eliminated polar localization of CheZ-GFP, and cells expressing these proteins failed to form patches, looking just like cells of the ΔcheA strain BC206 (Fig. 1C). Error-prone PCR mutagenesis (31) generated a mutant CheZ-GFP protein containing the W94R substitution that also did not localize to receptor patches.
A summary of the swarming behavior and localization patterns supported by these mutant proteins is given in Table 2. Note that the T25P, L28P, A87V, and A87G mutant proteins showed an intermediate level of patch formation; many cells lacked visible patches, but a significant minority of cells showed essentially normal patterns of CheZ-GFP localization. Except for A87V, which completely eliminated swarming, these substitutions caused partial defects in chemotactic swarm formation. Immunoblot analyses with anti-CheZ antibody indicated that most of the mutant CheZ-GFP proteins were present in normal amounts, regardless of their ability to function in chemotaxis or localize to patches. The exceptions were the T25P and L28P proteins, which were found at ∼50% of normal levels based on relative band intensities on immunoblots.
TABLE 2.
Effect of cheZ mutations on CheZ-GFP localization and chemotactic swarming
| Mutation source | Mutation | Localizationa | Swarm phenotypeb | Reference |
|---|---|---|---|---|
| Random mutagenesis | W94R | − | − | This study |
| Previously published | T25P | + | + | 20 |
| L28P | + | + | 20 | |
| D50G | ++ | + | 20 | |
| A65V | ++ | − | 4 | |
| M83T | ++ | − | 4 | |
| A87G | + | + | 4 | |
| A87V | + | − | 4 | |
| L90S | − | − | 4 | |
| F117S | − | − | 4 | |
| F141I | ++ | − | 3 | |
| D143G | ++ | − | 4 | |
| T145M | ++ | − | 3 | |
| I149T | ++ | − | 20 | |
| E158G | ++ | + | 20 | |
| N182Y | ++ | − | 20 | |
| G188E | ++ | − | 4 | |
| V205E | ++ | − | 4 | |
| Site-directed | W94S | − | − | This study |
| mutagenesis of | W97S | − | + | This study |
| amphipathic | F98S | − | ++ | This study |
| helices | I102S | ++ | ++ | This study |
| L104S | ++ | ++ | This study | |
| A107S | ++ | + | This study | |
| L110S | − | − | This study | |
| V111S | − | − | This study | |
| T114A | − | − | This study | |
| L118S | − | − | This study | |
| V121S | − | − | This study |
++, wild-type localization of CheZ-GFP; +, weaker localization or localization in fewer cells; −, no localization.
++, wild-type swarm in aspartate-minimal motility agar; +, swarm with a smaller diameter and less-distinct chemotactic rings; −, no swarming, as with the ΔcheZ parent strain.
Mutations that disrupt CheZ-GFP localization alter residues on the hydrophobic faces of two amphipathic helices.
A computer-generated secondary-structure prediction for CheZ indicated that the region of CheZ targeted by localization-defective (Loc−) mutations forms two α helices separated by a short loop. This prediction has been confirmed by the recently published structure of CheZ (30). Helical-wheel projections revealed that each helix should be amphipathic and that Ala-87, Leu-90, Trp-94, and Phe-117 are located on hydrophobic faces of the helices. Since Loc− mutations affecting these residues exchange polar residues for nonpolar ones, we hypothesized that hydrophobic interactions involving these residues could be important for CheZ function.
Almost all of the residues on the hydrophobic faces of the predicted helices were replaced with Ser by site-directed mutagenesis. The only exceptions were Met-83, which we had already tested as the previously identified M83T mutation (4), and Thr-114, which was replaced with Ala. Each mutant protein was expressed from plasmid pBJC104 in strain RP1616, and its chemotaxis and CheZ-GFP localization phenotype were analyzed (Table 2). Ser replacements in the region from L110 to V121 and the T114A substitution conferred a Che− Loc− phenotype, whereas the I102S, L104S, and A107S substitutions had little effect on either chemotaxis or localization. The W94S replacement imposed the same Che− Loc− phenotype as W94R. Cells expressing the W97S or F98S version of CheZ-GFP had a Che+ Loc− phenotype, implying that the overall conformation and function of these mutant proteins were not significantly compromised. In the absence of the inducer IPTG, each mutant protein was produced at physiologically normal levels in about the same amount as wild-type CheZ-GFP.
Proposed CheAS-CheZ interaction sites are conserved in enteric bacteria.
The deduced amino acid sequences corresponding to the predicted helix-turn-helix hairpin of E. coli CheZ were compared for the 14 gram-negative proteobacteria for which CheZ sequences were available in the Entrez database. Figure 3A presents the alignment of nine of these sequences, selected to avoid redundancy and to represent at least one member of each genus. Residues 95 to 98 are conserved as D(D/E)WF in all of the enteric species, which are the only bacteria known to express CheAS (14). Nonenteric γ-proteobacteria, including Vibrio and Pseudomonas, do not display this motif. However, there is substantial sequence conservation at other positions in this region, with the notable exception of the CheZ proteins from the two Xanthomonas species, which lack the entire region encompassing the apical helix-turn-helix hairpin. When the sequences of CheA from these same species are compared (Fig. 3B), it is clear that sequences corresponding to the putative N terminus of CheAS are also conserved in the enteric bacteria.
FIG. 3.
Alignment of amino acid sequences of the CheZ apical loop region (30) and the C-terminal portion of the P1 domain of CheA (17). (A) Alignment of CheZ sequences from representatives of each bacterial genus for which a putative cheZ gene has been sequenced. Asterisks indicate the conserved D(D/E)WF motif at residues 95 to 98 of E. coli CheZ. The number in parentheses indicates the residue number for the first position in the sequence shown. Residues highlighted in dark gray are identical in all of the aligned sequences except that of X. campestri. Residues highlighted in light gray are identical within the enteric or the nonenteric bacteria. Boldface type indicates positions at which residues are chemically conserved within the enteric or nonenteric bacteria. The sequence listed as Salmonella enterica is for serovar Typhimurium. In addition to the species and serovars shown, comparisons were made with the cheZ sequences from Salmonella enterica serovar Typhi, Yersinia enterocolitica, Vibrio parahaemolyticus, Pseudomonas syringae, and Xanthomonas axonopodis. Those sequences did not differ significantly from those shown for their congeners. (B) Alignment of the available amino acid sequences for the C-terminal portion of the P1 domain of CheA from the species whose CheZ sequences are shown in panel A. All sequences were obtained from the Entrez database (http://www.ncbi.nlm.nih.gov/PubMed/) and aligned by using the AlignX program of the Vector NTI Suite molecular biology software package.
Conclusion.
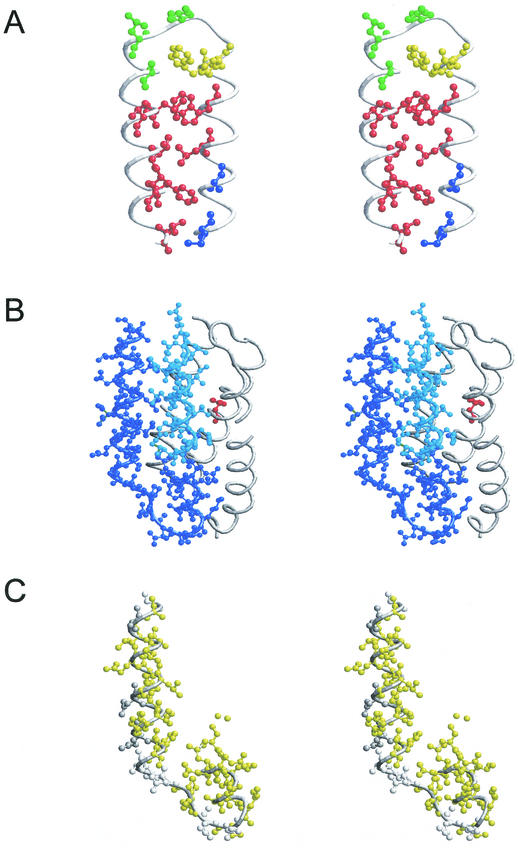
The results presented here establish that CheZ localization to the receptor patch occurs only when CheAS is present. The two residues of CheZ most clearly implicated in interaction with CheAS are Phe-97 and Trp-98. These two aromatic residues, especially Phe-97, are solvent exposed in the apical hairpin loop of the CheZ crystal structure (Fig. 4A) and might be expected to have an energetically favorable interaction with a hydrophobic partner. Mutations causing a Che− Loc− phenotype replace hydrophobic residues at the interhelix packing surface of the hairpin (Fig. 4A) or residues located at the subunit interface of the CheZ homodimer (30). These substitutions may destabilize the hairpin or interfere with CheZ dimerization, respectively.
FIG.4.
Stereo views of the crystal structures for the apical loop region of CheZ (28) and the P1 domain of CheA (15). (A) Apical loop of E. coli CheZ. Residues 83 to 121 are shown. Substitutions at the residues in blue displayed a Che− Loc+ phenotype. Substitutions at the residues in red produced a Che− Loc− phenotype. Substitutions at the residues in yellow resulted in a Che+ Loc− phenotype. Residues shown in green were not affected in either chemotaxis or localization by substitution at these positions. (B) Salmonella CheA P1 domain. Residues in dark blue are in α-helices 4 and 5 and are present CheAS. Residues in cyan are in the N-terminal portion of α-helix 4 missing in CheAS and make hydrophobic contacts with residues in α-helix 5 of full-length P1. The site of phosphorylation (His-48) is indicated in red. (C) Conserved residues in the remnant P1 domain of CheAS. Yellow residues are conserved among all enteric species. Those indicated in dark gray are chemically conserved among enteric species, whereas those shown in light gray are variable.
The N-terminal sequence of CheAS corresponds to the C-terminal portion of the P1 domain. The crystal structure of the Salmonella P1 domain (17) is shown in Fig. 4B. It contains five α-helices, and Met-98, the first residue of CheAS, resides in the middle of the fourth helix, which extends to Lys-106. Residues Ala-113 through Ala-130 comprise the fifth helix. Based on this structure, the N terminus of CheAS is predicted to be an amphipathic helix of 8 residues followed by a turn and an amphipathic helix of 18 residues. In CheAS, the hydrophobic residues Leu-123 and Leu-126 should be exposed to solvent and available to interact with CheZ. Furthermore, purified CheZ binds to a ′P1-P2 fragment derived from CheAS but not to a P1-P2 fragment derived from CheAL (L. Kott and R. M. Weis, personal communication), and CheZ can be coprecipitated with an N-terminal fragment of CheAS containing its first 42 residues fused to GST (O'Connor and Matsumura, personal communication).
The absence of CheAS does not diminish chemotaxis in the assays that have been employed (19). However, within the enteric bacteria the selective conservation of amino acid sequences in the regions of CheAS and CheZ that we propose to interact (Fig. 3) constitutes a clear example of an evolutionary trace as defined by Lichtarge and colleagues (10, 13). When combined with knowledge of the concentrated distribution of a set of conserved residues on the surfaces of both of two potentially interacting proteins, such “evolutionarily privileged clusters” are strong predictors of protein-protein binding sites. The central premise of evolutionary trace analysis is that evolution has already done the experiment of mutation and functional analysis to identify important residues. The ability to disrupt CheZ localization through two otherwise benign substitutions of residues in a sequence that is conserved in the CheZ proteins of enteric bacteria (14) implies that the CheZ-CheAS interaction confers a selective advantage under some environmental conditions.
Acknowledgments
Larry Griffing mentored B.J.C in fluorescence microscopy. Michael Eisenbach, Ruth Silversmith, and Viktor Sourjik commented on an early draft of the manuscript. Sandy Parkinson and Alan Wolfe supplied strains, and Phil Matsumura supplied monoclonal antibody directed against the P4 domain of CheA. Janine Maddock graciously communicated unpublished data on CheZ subcellular localization determined by immunogold labeling. Laila Kott and Bob Weis, and Phil Matsumura and Chris O'Connor, kindly shared unpublished information about binding of CheZ to the N terminus of CheAS in vitro.
Financial support was provided by Public Health Service grants GM37369 to M.D.M. and GM52583 to R.C.S.
REFERENCES
- 1.Baumann, C. T., C. S. Lim, and G. L. Hager. 1998. Simultaneous visualization of the yellow and green forms of the green fluorescent protein in living cells. J. Histochem. Cytochem. 46:1073-1076. [DOI] [PubMed] [Google Scholar]
- 2.Bibikov, S. I., R. Biran, K. E. Rudd, and J. S. Parkinson. 1997. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 179:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blat, Y., and M. Eisenbach. 1996. Mutants with defective phosphatase activity show no phosphorylation-dependent oligomerization of CheZ. J. Biol. Chem. 271:1232-1236. [DOI] [PubMed] [Google Scholar]
- 4.Boesch, K. C., R. E. Silverman, and R. B. Bourret. 2000. Isolation and characterization of nonchemotactic CheZ mutants of Escherichia coli. J. Bacteriol. 182:3544-3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, D., D. S. Weiss, J. C. Chen, and J. Beckwith. 2000. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J. Bacteriol. 182:842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 7.Garzon, A., and J. S. Parkinson. 1996. Chemotactic signaling by the P1 phosphorylation domain liberated from the CheA histidine kinase of Escherichia coli. J. Bacteriol. 178:6752-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guzman, L., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kofoid, E. C., and J. S. Parkinson. 1991. Tandem translation starts in the cheA locus of Escherichia coli. J. Bacteriol. 173:2116-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lichtarge, O., and M. E. Sowa. 2002. Evolutionary predictions of binding surfaces and interactions. Curr. Opin. Struct. Biol. 12:21-27. [DOI] [PubMed] [Google Scholar]
- 11.Lybarger, S. R., and J. R. Maddock. 1999. Clustering of the chemoreceptor complex in Escherichia coli is independent of the methyltransferase CheR and the methylesterase CheB. J. Bacteriol. 181:5527-5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in Escherichia coli. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 13.Madubushi, S., H. Yao, H., M. Marsh, M., D. M. Kristensen, A. Philippi, M. E. Sowa, and O. Lichtarge. 2002. Structural clusters of evolutionary trace residues are statistically significant and common in proteins. J. Mol. Bio. 316:139-154. [DOI] [PubMed] [Google Scholar]
- 14.McNamara, B. P., and A. J. Wolfe. 1997. Coexpression of the long and short forms of CheA, the chemotaxis histidine kinase, by members of the family Enterobacteriaceae. J. Bacteriol. 179:1813-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Morrison, T. B., and J. S. Parkinson. 1994. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of Escherichia coli. Proc. Natl. Acad. Sci. USA 91:5485-5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mourey, L., S. Da Re, J. D. Pedelacq, T. Tolstykh, C. Faurie, V. Guillet, J. B. Stock, and J.-P. Samama. 2001. Crystal structure of the CheA histidine phosphotransfer domain that mediates response regulator phosphorylation in bacterial chemotaxis. J. Biol. Chem. 276:31074-31082. [DOI] [PubMed] [Google Scholar]
- 18.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanatinia, H., E. C. Kofoid, T. B. Morrison, and J. S. Parkinson. 1995. The smaller of two overlapping cheA gene products is not essential for chemotaxis in Escherichia coli. J. Bacteriol. 177:2713-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanna, M. G., and M. I. Simon. 1996. In vivo and in vitro characterization of Escherichia coli protein CheZ gain- and loss-of function mutants. J. Bacteriol. 178:6275-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skidmore, J. M., D. D. Ellefson, B. P. McNamara, M. M. P. Couto, A. J. Wolfe, and J. R. Maddock. 2000. Polar clustering of the chemoreceptor complex in Escherichia coli occurs in the absence of complete CheA function. J. Bacteriol. 182:967-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith, R. A., and J. S. Parkinson. 1980. Overlapping genes at the cheA locus of Escherichia coli. Proc. Natl. Acad. Sci. USA 77:5370-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sourjik, V., and H. C. Berg. 2000. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol. Microbiol. 37:740-751. [DOI] [PubMed] [Google Scholar]
- 24.Stewart, R. C., R. VanBruggen, D. D. Ellefson, and A. J. Wolfe. 1998. TNP-ATP and TNP-ADP as probes of the nucleotide binding site of CheA, the histidine protein kinase in the chemotaxis signal transduction pathway of Escherichia coli. Biochemistry 37:12269-12279. [DOI] [PubMed] [Google Scholar]
- 25.Swanson, R. V., S. C. Schuster, and M. I. Simon. 1993. Expression of CheA fragments which define domains encoding kinase, phosphotransfer, and CheY binding activities. Biochemistry 32:7623-7629. [DOI] [PubMed] [Google Scholar]
- 26.Wang, H., and P. Matsumura. 1996. Characterization of the CheAS/CheZ complex: a specific interaction resulting in enhanced dephosphorylating activity on CheY-phosphate. Mol. Microbiol. 19:695-703. [DOI] [PubMed] [Google Scholar]
- 27.Wang, H., and P. Matsumura. 1997. Phosphorylating and dephosphorylating complexes in bacterial chemotaxis. J. Bacteriol. 179:287-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward, S. M., A. Delgado, R. P. Gunslaus, and M. D. Manson. 2002. A NarX-Tar chimera mediates repellent taxis to nitrate and nitrite. Mol. Microbiol. 44:709-719. [DOI] [PubMed] [Google Scholar]
- 29.Wolfe, A. J., B. P. McNamara, and R. C. Stewart. 1994. The short form of CheA couples chemoreception to CheA phosphorylation. J. Bacteriol. 176:4483-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao, R., E. J. Collins, R. B. Bourret, and R. E. Silversmith. 2002. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat. Struct. Biol. 9:570-575. [DOI] [PubMed] [Google Scholar]
- 31.Zhou, Y., X. Zhang, and R. H. Ebright. 1991. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 19:6052.. [DOI] [PMC free article] [PubMed] [Google Scholar]