Abstract
The prevalence of the Moraxella catarrhalis immunoglobulin D (IgD)-binding outer membrane protein MID and its gene was determined in 91 clinical isolates and in 7 culture collection strains. Eighty-four percent of the clinical Moraxella strains expressed MID-dependent IgD binding. The mid gene was detected in all strains as revealed by homology of the signal peptide sequence and a conserved area in the 3′ end of the gene. When MID proteins from five different strains were compared, an identity of 65.3 to 85.0% and a similarity of 71.2 to 89.1% were detected. Gene analyses showed several amino acid repeat motifs in the open reading frames, and MID could be called a putative autotransport protein. Interestingly, homopolymeric {polyguanine [poly(G)]} tracts were detected at the 5′ ends within the open reading frames. By flow cytometry, using human IgD and fluorescein isothiocyanate-conjugated anti-IgD polyclonal antibodies, most strains showed two peaks: one high- and one low-intensity peak. All isolates expressing high levels of MID had 1, 2, or 3 triplets of G's in their poly(G) tracts, while strains not expressing MID had 4, 7, 8, or 10 G’s in their poly(G) tracts or point mutations causing a putative preterminated translation. Northern blot analysis revealed that the mid gene was regulated at the transcriptional level. Experiments with nonclumping variants of M. catarrhalis proved that bacteria lost their MID expression by removing a G in their poly(G) tracts. Moraxella strains isolated from the nasopharynx or from blood and sputum specimens expressed MID at approximately the same frequency. In addition, no variation was observed between strains of different geographical origins (Australia, Europe, Japan, or the United States). MID and the mid gene were found solely in M. catarrhalis, whereas related Neisseria and Moraxella species did not express MID. Taken together, MID appears to be a conserved protein that can be found in essentially all M. catarrhalis strains. Furthermore, MID is governed by poly(G) tracts when bacteria undergo phase variation.
Moraxella catarrhalis is a gram-negative unencapsulated diplococcus that during the last two decades has increasingly gained respect as a respiratory pathogen (9, 26). Although M. catarrhalis can be found as a commensal organism in many healthy children, it is the third most frequent bacterium isolated in patients with otitis media. It has also been associated with sinusitis, acute laryngitis, suppurative keratitis, and in rare cases, septicemia. In adults with predisposing conditions, e.g., chronic obstructive pulmonary disease, M. catarrhalis causes lower respiratory tract disease. The pathogenesis of M. catarrhalis infection is not completely understood, but recently an increasing number of virulence factors involved in adhesion and colonization have been determined.
Several different M. catarrhalis outer membrane proteins (OMPs) have been isolated and more or less characterized (33). The best-defined group of moraxella OMPs is the ubiquitous surface protein A (UspA) family consisting of UspA1, UspA2 (HMWP), and UspA2H (1, 13, 27). Interestingly, the UspA proteins together with Yersinia YadA belong to a novel class of adhesins consisting of a tripartite organization with an N-terminal oval head domain and a putative coiled-coil rod terminated by a C-terminal membrane anchor domain (24). UspA1 and UspA2H are responsible for adherence to epithelial cells in vitro as revealed in transformation experiments using a nonadhesive Haemophilus influenzae (29). uspA1 gene regulation depends upon phase variation, and differences in the lengths of the homopolymeric {polyguanine [poly(G)]} tracts have been defined upstream of the open reading frame (ORF) (30). Antibodies against UspA1 and UspA2 protect mice from infection in a pulmonary clearance animal model (21), suggesting that the UspA proteins might be promising vaccine candidates. Furthermore, antibodies with bactericidal activity against UspA1 and UspA2 are already detected in children less than 1 year old (11).
Another M. catarrhalis OMP is protein CD that shares sequence homology with porins and has been suggested to promote adhesion to nasal and inner ear mucins (36, 43). M. catarrhalis outer membrane protein B (CopB) is involved in the acquisition of iron from human transferrin and lactoferrin, and it has been demonstrated that CopB expression is regulated by iron (2). In parallel to the UspA proteins, antibodies raised against protein CD (37) and CopB (20) also are protective in the pulmonary clearance mouse model. Like CopB, the lactoferrin-binding proteins (Lbp) and the transferrin-binding proteins (Tbp) are involved in iron metabolism (for reviews, see references 26 and 33). The sequences of these OMP receptors have homology to those from other bacterial species including Escherichia coli (8, 38), and these OMP receptors are immunogenic as revealed by analysis of patient sera (53).
We have recently discovered and characterized a novel high-molecular-weight OMP of M. catarrhalis displaying a strong affinity for both soluble and surface-bound human immunoglobulin D (IgD) (14). The protein has been designated MID (for Moraxella immunoglobulin D-binding protein) and has an apparent molecular mass of 200 kDa as estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). MID comprises 2,139 amino acid residues, and the sequence has no similarity to other Ig-binding proteins. Recombinantly expressed MID binds IgD and translocates to the outer membrane of E. coli. MID binds two purified IgD myeloma proteins, four IgD myeloma sera, and finally one IgD standard serum. In contrast, MID does not attract IgG, IgM, IgA, or IgE myeloma proteins. Fluorescein isothiocyanate (FITC)-conjugated MID specifically binds to the IgD B-cell receptor on human CD19+ B cells as revealed by flow cytometry experiments. Finally, MID proteins conjugated to Sepharose beads are mitogenic for human purified B lymphocytes and comparable with proliferation induced by whole formalin-treated M. catarrhalis (19).
To identify the IgD-binding region, the MID protein was digested with proteases (41). In addition, a series of truncated fragments of MID were manufactured and expressed in E. coli, followed by analysis for IgD binding in Western and dot blots. The smallest fragment with essentially preserved IgD binding contained 238 amino acid residues (MID962-1200). Ultracentrifugation experiments and gel electrophoresis revealed that native MID962-1200 was a tetramer. Interestingly, tetrameric MID962-1200 attracted IgD ≥20-fold efficiently than the monomeric form. Thus, a tetrameric structure of MID962-1200 was crucial for optimal IgD-binding capacity.
The goals of this study were to define the frequency of the mid gene, compare the genomic organization and regulation, and determine the IgD-binding activity of M. catarrhalis in a defined population of clinical isolates originating from various countries and anatomical sites. The mid gene is detected in all strains investigated. MID protein expression is found to a different extent in all strains analyzed, and the predominant expression and consequent IgD-binding activity are detected irrespective of the site of isolation. The expression of MID is related to phase variation, and sequence analyses of the 5′ end of the genes reveal that mid is regulated by poly(G) tracts located within the ORF.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Clinical M. catarrhalis isolates and type strains in addition to related species were included as indicated in Table 1. Clinical isolates were from the Department of Medical Microbiology, University Hospital Malmö, Lund University, Malmö, Sweden, and the State Serum Institute, Copenhagen, Denmark (12). Type strains were from the Culture Collection University of Gothenburg (CCUG; Department of Clinical Bacteriology, Sahlgrenska Hospital, Gothenburg, Sweden), the National Collection of Type Cultures (NCTC; Central Public Health Laboratory, London, United Kingdom), or the American Type Culture Collection (ATCC; Manassas, Va.). All bacterial strains were grown on solid medium (1.1% IsoVitaleX, 7.8% human blood, 0.9% proteose peptone) or in nutrient broth (NB) (Oxoid, Basingstoke, Hampshire, United Kingdom). To study phase variation, selected M. catarrhalis strains were cultured in 10 ml of NB without shaking. One milliliter of the supernatant (without clumping bacteria) was transferred to a new tube containing 9 ml of NB. This procedure was repeated six times in 4 weeks.
TABLE 1.
M. catarrhalis strains and related species analyzed in this study
| M. catarrhalis strain or related species | Geographic origin | No. of strains isolated from:
|
Total no. | ||
|---|---|---|---|---|---|
| Nasopharynx | Sputum | Blood | |||
| M. catarrhalis | |||||
| Clinical isolates | Australiaa | 6 | 6 | ||
| Belgiuma | 6 | 6 | |||
| Denmarka | 9 | 6 | 15 | ||
| Finlandb | 2 | 2 | |||
| Hollanda | 6 | 6 | |||
| Hungaryb | 10 | 10 | |||
| Japana | 6 | 6 | |||
| Spaina | 3 | 3 | |||
| Swedenc | 24 | 24 | |||
| UKa,d | 6 | 6 | |||
| Type strains | USa,e | 7 | 7 | ||
| ATCC 8176 | 1 | ||||
| ATCC 8193 | 1 | ||||
| CCUG 11766 | 1 | ||||
| CCUG 353T | 1 | ||||
| CCUG 3292 | 1 | ||||
| CCUG 8539 | 1 | ||||
| NCTC 4103 | 1 | ||||
| Total no. | 36 | 49 | 6 | 98 | |
| Related species | |||||
| Acinetobacter baumannii (ATCC 9955) | |||||
| A. baumannii (ATCC 9956) | |||||
| Moraxella bovis (ATCC 10900T) | |||||
| M. caviae (CCUG 354T) | |||||
| M. cuniculi (CCUG 2496) | |||||
| Moraxella nonliquefaciens (28010/89)c | |||||
| Moraxella osloensis (969165)c | |||||
| M. osloensis (ATCC 10973) | |||||
| M. osloensis (ATCC 12429) | |||||
| M. ovis (CCUG 355T) | |||||
| Moraxella phenylpyruvica (23716/81)c | |||||
| N. gonorrheae (18143)c | |||||
| N. meningitidis serotype Ac | |||||
| N. meningitidis serotype Bc | |||||
| N. meningitidis serotype Cc | |||||
| Neisseria mucosa (10774)c | |||||
| Neisseria pharyngisc | |||||
| Neisseria siccac | |||||
| Neisseria subflava (26509/77)c | |||||
| Oligella ureolytica (99585/92)c | |||||
| O. ureolytica (965072)c | |||||
Strains described in detail in reference 12.
Clinical isolates from Finland and Hungary were supplied by M. Lönnrot (Department of Clinical Microbiology, Tampere University Hospital, Tampere) and E. Nagy (Department of Clinical Microbiology, Albert Szent-Györgyi Medical University, Szeged) respectively.
Isolated at Department of Medical Microbiology, Malmö, Sweden.
UK, United Kingdom.
US, United States.
Flow cytometry analysis and antibodies.
IgD binding to whole bacterial cells was analyzed using purified human myeloma protein IgD(κ) (IgD with κ light chains) (The Bindingsite, Birmingham, United Kingdom) and FITC-conjugated rabbit anti-human IgD polyclonal antibodies (pAb) (Dakopatts, Glostrup, Denmark). MID protein expression was determined with a specific rabbit anti-MID962-1200 antiserum. Rabbits were immunized intramuscularly with 200 μg of the purified recombinant MID962-1200 fragment (41) emulsified in complete Freund's adjuvant (Difco, Heidelberg, Germany) and given booster doses with the same amount of protein in incomplete Freund's adjuvant on days 18 and 36. Blood was drawn 2 to 3 weeks later. For flow cytometry analyses, bacteria from liquid cultures were washed twice with phosphate-buffered saline (PBS) containing 2% bovine serum albumin and incubated with 0.5 μg of IgD(κ) per ml or rabbit anti-MID962-1200 pAb in a final volume of 100 μl of PBS containing 2% bovine serum albumin at 4°C for 1 h. After the bacteria were washed, they were incubated with FITC-conjugated anti-IgD pAb or FITC-conjugated swine anti-rabbit pAb for 30 min at 4°C. After two more washes, the bacteria were analyzed by flow cytometry (EPICS XL-MCL flow cytometer; Coulter, Hialeah, Fla.). A single-cell population was gated in the forward scatter/side scatter histogram excluding aggregating bacteria. To monitor nonspecific binding, the FITC-conjugated human anti-IgD pAb or FITC-conjugated swine anti-rabbit pAb were added separately as negative controls for each strain analyzed.
OMP preparations.
Bacteria in stationary or logarithmic phase were washed twice with 50 mM Tris-HCl buffer, pH 8.0. The pellet was resuspended in Tris-HCl buffer containing 3% Empigen and protease inhibitors (Complete mini; Roche, Bromma, Sweden). OMPs were extracted by rotating the mixture at 37°C for 2 h. The bacterial cells, stripped of their outer membranes, were centrifuged at 23,000 × g for 20 min at 4°C. The supernatants were collected and centrifuged once more. The resulting fractions containing the OMPs were stored in aliquots at −20°C. Protein concentrations were determined by the bicinchoninic acid protein assay (Pierce, Rockford, Ill.).
Dot blot assays.
Protein preparations were diluted (250 to 0.06 ng) in Tris-buffered saline (TBS) and applied to membranes (Immobilon-P; Millipore Corporation, Bedford, Mass.) using a dot blot apparatus (Schleicher & Schuell, Dessel, Germany). Filters were blocked in TBS with 0.05% Tween 20 containing 5% milk powder for 1 h at room temperature. Thereafter, incubation with human IgD(κ) was done at 4°C overnight. After extensive washing of the membranes with TBS containing 0.05% Tween 20, membranes were incubated with a horseradish peroxidase-conjugated goat anti-human IgD antiserum (Biosources, Camarillo, Calif.) for 45 min at room temperature. After several washings, filters were developed using enhanced chemiluminescence detection reagents (Amersham Pharmacia Biotech, Uppsala, Sweden). Resulting chemiluminescence was analyzed by a Fluor-S Multimager (Bio-Rad, Sundbyberg, Sweden).
Gel electrophoresis and detection of proteins on membranes (Western blotting).
SDS-PAGE (10% polyacrylamide) was run as described before (14). Gels were stained with Coomassie brilliant blue R-250 (Bio-Rad). In parallel, the proteins were electrophoretically transferred (30 V overnight) from a gel to an Immobilon-P membrane. After transfer, the membrane was blocked with 5% milk powder in PBS with 0.1% Tween 20 (PBS-Tween) for 2 h. After four washes with PBS-Tween, the membrane was incubated with purified IgD myeloma protein for 1 h. Horseradish peroxidase-conjugated goat anti-human IgD, diluted 1:1,000, was added after four washes and incubated for 45 min. The Ig proteins were diluted in PBS-Tween containing 2% milk powder, and the incubated membranes were kept at room temperature. Finally, the membrane was washed with PBS-Tween four times. Development was performed with enhanced chemiluminescence Western blotting detection reagents and visualized with a Fluor-S Multimager.
Southern blot analysis.
Genomic DNA was extracted from M. catarrhalis, digested to completion with SspI, run on a 0.7% agarose gel, and blotted onto a nylon membrane (Hybond N+; Amersham Pharmacia Biotech). One SspI restriction enzyme site exists at position 911 in the M. catarrhalis Bc5 mid gene (14). A radioactively labeled probe was generated by PCR, resulting in a 932-bp fragment of the 3′ end of the Bc5 mid gene. The primers 5′-ATGTCAACGATGGCAATCAAGAGCC-3′ and 5′-CCCCAAGCTTAAAGTGAAAACCTGCACCAACTGCTGCC-3′ were used. Labeling was performed by adding Redivue [α-33P]dCTP (Amersham Pharmacia Biotech) directly to the PCR mixture. Filters were hybridized at 60°C in a mixture containing 0.25 M phosphate buffer (pH 7.2), 7% SDS, and 1 mM EDTA (pH 8). After overnight hybridization, the filters were washed twice for 5 min at 60°C in 0.1 M phosphate buffer (pH 7.2) containing 2% SDS. Resulting filters were exposed to intensifying screens and analyzed by a Fluor-S Multimager (Bio-Rad).
DNA isolation, primers, and PCR conditions.
DNA was extracted using a genomic DNA preparation kit (Qiagen, Hilden, Germany). The full-length mid gene was amplified by PCR (Expand Long Template PCR System; Roche) using the specific primers 5′-GCTTGAATGACGATCCCAATCATCAG-3′ and 5′-GCGAGGATCCGCTAAAAAGTGAAAACCTGCAC-3′ at a final concentration of 3 μM (14). A standard protocol with MgCl2 at a final concentration of 2.25 mM was used. The PCR protocol was as follows: 10 cycles of PCR, with 1 cycle consisting of denaturation at 94°C for 2 min, denaturation at 94°C for 10 s, annealing at 53°C for 45 s, and extension at 68°C for 6 min. Thereafter, 10 cycles were run, with 1 cycle consisting of 10 s at 94°C, 45 s at 53°C, and 8 min at 68°C. Finally, the DNA was subjected to 10 cycles, with 1 cycle consisting of 10 s at 94°C, 45 s at 53°C, and 10 min at 68°C. Primers for detecting the poly(G) tracts and the sequences encoding the signal peptides were 5′-GCTTGAATGACGATCCCAATCATCAG-3′ and 5′-GCTAGTGCTCCATAATTGATAGCTTGTGC-3′. In these reactions, a standard PCR protocol was used. Resulting PCR products were sequenced by gene walking and capillary electrophoresis on a Beckman CEQ 2000 and a dye-terminator cycle sequencing kit (CEQ DTCS kit; BeckmanCoulter, Stockholm, Sweden). The resulting DNA sequences were edited and aligned using PHRED (CodonCode, Dedham, Mass.) and SEQUENCHER (MedProbe, Oslo, Norway). To analyze the frequency of the mid gene in clinical isolates, the primers 5′-ATGTCAACGATGGCAATCAAGAGCC-3′ and 5′-CCCCAAGCTTAAAGTGAAAACCTGCACCAACTGCTGCC-3′ detecting a conserved area in the 3′ end of the gene were used in addition to the specific primers detecting the sequence encoding the signal peptide as described above.
RNA isolation, Northern blotting, and RT-PCR.
Total RNA was isolated from logarithmic-phase bacterial culture using the RNAWIZ reagent (Ambion, Austin, Tex.). Bacteria were harvested by freezing 100-μl samples of log-phase culture in liquid nitrogen. The RNA preparations were treated with DNase (Atlas Pure Total RNA Labeling System; Clontech, Palo Alto, Calif.). For Northern analysis, RNA (10 μg) was run on denaturing (formaldehyde) agarose gels and transferred to Hybond N+ membranes (Amersham Pharmacia, Uppsala, Sweden). RNA was fixed by treatment with 0.05 M NaOH for 5 min. The probe used in the Southern blot analysis was used for hybridizations. The membrane was incubated overnight at 42°C, washed twice with 1× SSC-0.1% SDS (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature, and washed twice with 0.1× SSC-0.1% SDS at 55°C. Thereafter, the membrane was exposed to intensifying screens and analyzed by a Personal Molecular Imager FX. Ready-To-Go RT-PCR beads (Amersham Pharmacia Biotech) were used for reverse transcription-PCR (RT-PCR). For first-strand synthesis, the primer mix pd(N)6 was used, while subsequent amplification of a mid-specific cDNA fragment of 456 bp was done using the primers 5′-GCAACAGGTACGGATCCAGGCTTTGCTG-3′ and 5′-CCCCAAGCTTAAAGTGAAAACCTGCACCAACTGCTGCC-3′. To exclude the possibility of DNA contamination, negative controls (with inactivated reverse transcriptase) were included for all RNA preparations analyzed.
RAPD.
Genomic DNA was randomly amplified using the primers 5′-ACGGTGCCTG-3′ (A70-3; Sigma-Genosys, Pampisford, Cambridge, United Kingdom) and 5′-ATGTAAGCTCCTGGGGATTCAC-3′ (ERIC1-R) (49). PCRs were run using the Expand High Fidelity PCR System (Roche). PCR was performed as follows: an initial denaturation step at 95°C for 1 min, followed by four cycles of PCR, with one cycle consisting of denaturation at 94°C for 45 s, annealing at 30°C for 120 s, and elongation at 72°C for 60 s. Thereafter, 26 cycles of PCR was performed, with 1 cycle consisting of 5 s at 94°C, 30 s at 36°C, and 30 s at 72°C, followed by a final step of 10 min at 72°C. The randomly amplified polymorphic DNA (RAPD) products were analyzed on a 1% agarose gel and subsequently scanned using a GelDoc system (Bio-Rad).
PFGE.
Bacteria were grown overnight on solid medium and resuspended in PBS. Thereafter, bacteria were mixed with 10 mM Tris (pH 8.0), 1.0 M NaCl, and 1% low-melting-point agarose. The mixture was poured into a Bio-Rad mold as recommended by the manufacturer. Once solidified, the agarose disks were treated with lysis buffer (6 mM Tris [pH 8.0], 1 M NaCl, 100 mM EDTA [pH 8.0], 0.2% Na deoxycholate, 0.5% Na laurylsarcosine, 0.5% Brij 58 [Fluka, Buchs, Switzerland]) supplemented with RNase A (50 μg/ml) and lysozyme at 37°C overnight. The gel disks were then washed three times at room temperature in TE (Tris-EDTA) buffer (30 min for each wash) and treated with proteinase K (1 mg/ml) in ES buffer (0.5 M EDTA [pH 9.0], 34 mM sarcosyl) at 56°C overnight. The gel disks were washed as described above, and the DNA was digested overnight with restriction enzyme SpeI. After the digestion, the agarose-DNA disks were rinsed in electrophoresis buffer and pulsed-field gel electrophoresis (PFGE) was performed using a CHEF Mapper (Bio-Rad) (51).
Nucleotide sequence analysis and software for prediction of DNA structures.
Signal peptide prediction analysis was performed using the SignalP version 1.1 software (http://www.cbs.dtu.dk/services/SignalP/) (40). Protein sequences were aligned using the GeneJockey Clustal II software package (Biosoft, Cambridge, United Kingdom). In addition, sequences were subjected to a statistical analysis of protein sequences (SAPS; http://www.ch.embnet.org/software/SAPS_form.html) (6). Homologies and identities were calculated using the software Needle (39) in The European Molecular Biology Open Software Suite (EMBOSS; http: //www.hgmp.mrc.ac.uk/Software/EMBOSS/) (44). To predict coiled-coil structures, the MacStripe 2.0b1 software (www.yourk.ac.uk/depts/biol/units/coils /coilcoil/html) was used (32).
RESULTS
Characterization of MID protein in clinical isolates; most M. catarrhalis strains bind IgD and express MID.
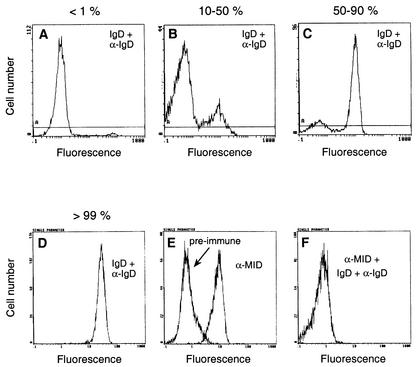
To analyze the IgD-binding capacity and MID expression in an array of M. catarrhalis isolates, we developed a direct binding flow cytometry assay consisting of human IgD(κ) and a FITC-conjugated secondary antibody directed against IgD. In initial experiments, bacteria collected at different time points were analyzed. No difference in the IgD binding or MID expression of logarithmically growing or stationary-phase bacteria was observed, suggesting that the MID expression of M. catarrhalis was not dependent on the growth phase. Stationary-phase bacteria were thus used in all further analyses. Most strains characteristically gave two peaks, one low- and one high-intensity peak in our assay. However, the relative proportion of the two peaks varied. In Fig. 1A to D, our assay is exemplified by strains where the high-intensity peak comprises <1, 10 to 50, 50 to 90, and >99% of analyzed bacterial cells.
FIG. 1.
Flow cytometry profiles demonstrating four different isolates of M. catarrhalis displaying various levels of MID protein expression. (A to D) Bacterial populations with <1%, 10 to 50%, 50 to 90%, and >99% of the bacteria analyzed in high-intensity peaks are shown as representative examples. IgD binding is also shown. (E) Fluorescence with anti-MID pAb and (F) an experiment with IgD binding blocked by anti-MID pAb. Bacteria were grown to stationary phase and incubated with human myeloma IgD(κ) or rabbit anti-MID pAb (α-MID) on ice. After 1 h and washes, FITC-conjugated rabbit anti-human IgD pAb (α-IgD) or swine anti-rabbit pAb was added for an additional 30 min, followed by additional washes and subsequent flow cytometry analysis. In panel E, “pre-immune” indicates fluorescence of bacteria incubated with a rabbit preimmune serum and then with the FITC-conjugated swine anti-rabbit pAb.
To confirm the MID-dependent IgD binding, bacteria were preincubated with the anti-MID pAb, and then IgD and the FITC-conjugated anti-IgD pAb were added. The anti-MID antiserum significantly reduced the IgD binding (Fig. 1E and F). All isolates examined (n = 10) showed the same pattern of inhibition. These experiments also suggested that there is only one OMP attracting IgD in M. catarrhalis.
To investigate whether the source of the isolate was important for MID protein expression, M. catarrhalis strains originating from sputum, nasopharynx, and blood were included in our study. All strains were clinical isolates that came from strain collections. Therefore, several past subcultures of the various strains could not be excluded. When MID expression of the bacteria was analyzed by flow cytometry using human IgD(κ) and FITC-conjugated anti-human IgD pAb, there was a high degree of variability in the IgD-binding pattern measured for the different isolates (Table 2). Of 91 clinical isolates analyzed, 15 (16%) showed a very small or no (<1%) high-intensity peak, while 34 (37%) of isolates contained more than 99% high-intensity IgD-binding bacteria (Table 2). However, the MID expression patterns of the strains isolated from the different sources did not differ (Table 2). Moreover, strains obtained from different laboratories worldwide (Table 1) showed similar variations in MID expression, suggesting that MID is a conserved OMP of M. catarrhalis. In contrast, no IgD binding (i.e., MID expression) was detected in strains closely related to M. catarrhalis (Table 1), i.e., the mean fluorescence intensity of these isolates was equal to the background level.
TABLE 2.
Frequency of MID protein expression in clinical isolates of M. catarrhalisa
| Isolate source | No. of isolates with the following % of bacteria in high-intensity peakb
|
Total no. of isolates | |||||
|---|---|---|---|---|---|---|---|
| <1 | 1-10 | 11-50 | 50-90 | 90-99 | >99 | ||
| Sputum | 8 | 3 | 3 | 7 | 13 | 15 | 49 |
| Nasopharynx | 4 | 4 | 2 | 8 | 2 | 16 | 36 |
| Blood | 3 | 0 | 0 | 0 | 0 | 3 | 6 |
| Total no. | 15 | 7 | 5 | 15 | 15 | 34 | 91 |
MID expression was defined by flow cytometry. Bacteria were incubated with a human IgD(κ) myeloma protein followed by incubation with a secondary FITC-conjugated pAb and subsequent flow cytometry analyses.
Number of isolates with the indicated percentage of IgD-binding bacteria in high-intensity peak (percentage of total).
To further examine the moraxella-dependent specific IgD binding and MID protein expression, OMP preparations followed by dot blotting or SDS-PAGE and Western blotting were performed. MID appeared as two bands on stained gels, one band representing a multimeric complex with a molecular mass of more than 1,000 kDa and one band representing a monomer with an approximate molecular mass of 200 kDa (14, 41). Both MID bands were readily detected in the OMP fraction isolated from high- and intermediate-MID-expressing strains, i.e., >1% high-intensity peak in flow cytometry analysis, whereas the two bands could not be detected by SDS-PAGE and Western blotting in strains expressing low levels of MID (<1% high-intensity peak). MID or any specific IgD-binding activity was not detected in the related Moraxella species M. caviae, M. cuniculi, or M. ovis. Thus, IgD-binding and protein levels of MID corresponded with the flow cytometry results.
The mid gene is distributed in all M. catarrhalis strains.
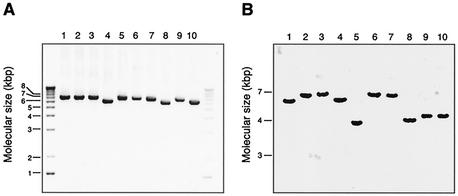
To investigate whether the mid gene existed in all M. catarrhalis strains, primers were chosen on the basis of a conserved area upstream of the ORF and a conserved area downstream including the stop codon sequence (14). The mid gene was detected in all 91 clinical isolates and 7 type strains analyzed, and the genomic mid DNA was approximately 6,000 to 7,000 bp long. The PCR products obtained from 10 strains are shown in Fig. 2A. The existence of the mid gene was further verified by Southern blotting using a probe containing a sequence selected from the 3′ end of the gene. Southern blot experiments revealed that the moraxella strains contained only one mid gene (Fig. 2B). The mid gene was not found in the moraxella (neisseria)-related species in Table 1 (not shown).
FIG. 2.
M. catarrhalis contains only one gene encoding the MID protein. (A) PCR analysis demonstrating mid genes ranging from 6,000 to 7,000 bp. (B) Southern blot of SspI-digested genomic DNA. Our model strains Bc5, BBH17, Perez112, RH1, and RH4 were compared to five randomly selected clinical isolates. Genomic DNA from M. catarrhalis was prepared. PCR was run with a primer pair annealing to a sequence upstream of the ORF and a conserved area downstream including the stop codon sequence. In the Southern blot experiment, DNA was digested with SspI, which cleaves the mid gene once, and subjected to Southern blot analysis using a 33P-labeled probe originating from the 3′ end of the gene.
Sequence analysis of the mid gene: amino acid repeat motifs and autotransporter homologies are found in the mid genes from selected strains.
In addition to MID protein from our reference strain Bc5 (14), the MID sequences obtained from four other clinical isolates were analyzed in detail. Comparison of the signal peptide sequences by the method of Nielsen et al. (40) revealed that the most common predicted signal peptidase cleavage site was located at positions 67 and 68 and was preceded by the amino acid residues alanine, tyrosine, and alanine.
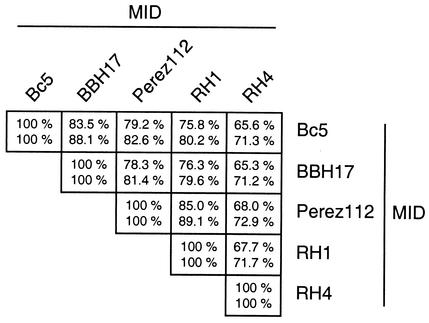
For an overview of the similarity and identity between different mid genes, the sequences of the five ORF MID proteins were analyzed (39). For four strains, the degrees of identity and similarity were ≥75.8 and ≥79.6%, respectively (Fig. 3). In contrast, slightly lower values for identity and similarity, ≥65.3 and ≥71.2%, respectively, were obtained for the fifth isolate (RH4).
FIG. 3.
Comparison of full-length MID amino acid sequences from five different M. catarrhalis strains. The degrees of identity and similarity between the various MID sequences were calculated using the software Needle (39).
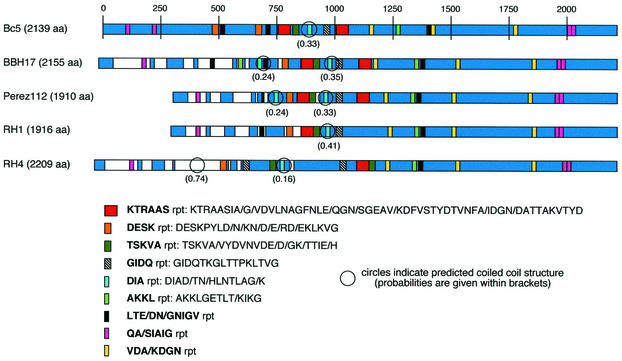
The full-length MID ORFs were also aligned and compared to the sequence from strain Bc5 as shown in Fig. 4. In conformity with the similarity and identity data (Fig. 3), homology existed between the various strains. The hypervariable regions (shown in white) were located in the N-terminal regions. OMPs often contain conserved amino acid stretches that are repeated several times in the same protein. We subjected our MID sequences to similarity analyses and found approximately nine conserved boxes spanning between 6 and 48 amino acids (Fig. 4). The amino acid consensus sequence KTRAAS that occurred adjacent to the TSKVA repeat was found in all sequences and was repeated once in three of the five sequences studied in detail. Other long consensus sequences that occurred in all genes were the DESK and GIDQ repeats. It has been suggested that the OMPs in the UspA family of M. catarrhalis form coiled coils (13). When the MID amino acid sequence was analyzed for coiled-coil structures (32), one or two coiled-coil stretches (probability of 0.16 to 0.74) were localized in the middle third of the molecules. Finally, a careful analysis of MID revealed several sequence homologies that offer support for MID being an autotransporter protein (22). In conclusion, MID consists of several amino acid repeats, contains a few coiled coils, and is finally suggested to be classified as a putative autotransporter protein.
FIG. 4.
Schematic representation of homology between M. catarrhalis Bc5 and four other strains. Repetitive amino acid sequence motifs are also shown. The entire ORF is represented by a solid blue line for strain Bc5. Homology of the other strains with Bc5 is indicated in blue, whereas dissimilarities with the amino acid sequence of Bc5 are in white. Repetitive amino acid motifs are indicated with colored boxes. The circled sequences show the areas with a higher probability of forming coiled-coil structures (32). Abbreviations: aa, amino acids; rpt, repetitive sequence.
mid gene expression is controlled by a poly(G) tract.
A poly(G) tract is found downstream of the ATG start codon in the mid gene (14) and would be a feasible regulatory sequence as has been shown for the uspA set of genes for example (30). Since M. catarrhalis isolates express MID to different degrees as judged by flow cytometry (Fig. 1 and Table 2), we analyzed the poly(G) tracts in strains expressing high (>99% high-intensity peak) and low (<1% high-intensity peak) levels of MID. Table 3 summarizes the results obtained for 23 different strains. Interestingly, all isolates with high MID expression (n = 13) possessed 1, 2, or 3 triplets of G residues. In contrast, bacteria with mid genes containing 4, 7, 8, or 10 G's did not express MID. In five additional mid genes with 1 or 2 triplets of G's, point mutations had occurred after the predicted signal peptidase cleavage site, causing a putative preterminated translation.
TABLE 3.
Characterization of the MID poly(G) tract in 23 M. catarrhalis strainsa
| No. of G residues | No. of isolates with the following level of expression of MID:
|
|
|---|---|---|
| High | Low | |
| 3 | 2 | 1b |
| 4 | 1 | |
| 6 | 4 | 4b |
| 7 | 1 | |
| 8 | 2 | |
| 9 | 7 | |
| 10 | 1 | |
Bacteria were incubated with a human IgD(κ) myeloma protein followed by incubation with a secondary FITC-conjugated pAb and analysis by flow cytometry.
Point mutations after the predicted signal peptide cleavage site were observed in these strains.
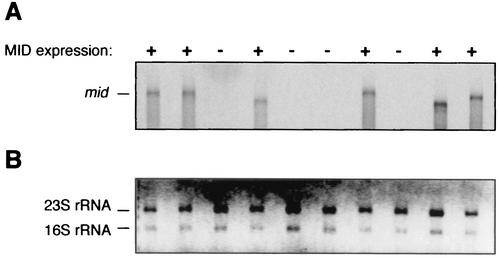
To examine whether mid was regulated at the transcriptional level, total RNA from high- and low-MID-expressing Moraxella strains was isolated and subjected to Northern (RNA) analysis. Using a conserved sequence derived from the 3′ end of the gene as a probe, we found that strains with 4, 7, 8, or 10 G’s in their poly(G) tracts were devoid of mid transcripts. In contrast, high-MID-expressing strains (with 1, 2, or 3 triplets of G’s) displayed high concentrations of mid RNA (Fig. 5). Since the cutoff for Northern blots is relatively high, a sensitive RT-PCR was also performed. To rule out DNA contamination, RNA was treated with DNase before RT was performed. mid gene transcripts were detected in both high- and low-expressing strains by RT-PCR (data not shown). Taken together, MID expression is suggested to be regulated by poly(G) tracts localized within the ORF and is thus governed at the RNA molecular level.
FIG. 5.
High-MID-expressing M. catarrhalis strains express mid mRNA, whereas low-MID-expressing strains are mid mRNA deficient. (A) Northern blot hybridized with a radioactive mid cDNA probe and (B) the filter with ethidium bromide-stained RNA after blotting. The positions of rRNAs are indicated. Six moraxella strains with high-intensity peaks >99% (MID expression, +) representing 1, 2, or 3 triplets of G’s and four strains with high-intensity peaks <1% (MID expression, −) representing 4, 7, 8, or 10 G’s in their poly(G) tracts are shown. Total RNA was isolated and loaded onto a denaturing formaldehyde gel. After separation, RNA was blotted to a nylon filter that was subsequently hybridized with a 33P-labeled DNA probe originating from the 3′ end of the mid gene.
mid gene regulation is related to phase variation.
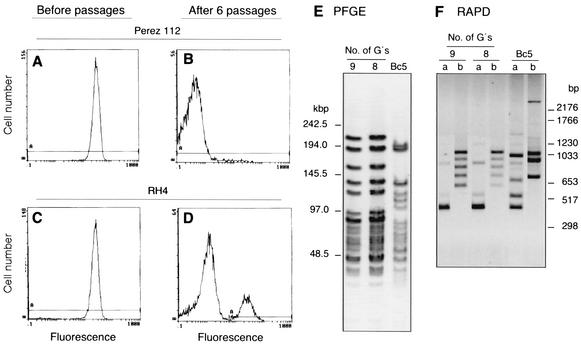
A characteristic feature of M. catarrhalis, which is related to phase variation, is the bacterium's ability to clump, i.e., autoagglutinate, when it grows in broth (28). To relate the number of G's in the poly(G) tracts to mid gene expression, our highly MID-expressing strains Perez112 and RH4 were incubated in NB without shaking for six passages in 4 weeks. In contrast to the clumping wild-type Perez112, the nonclumping variant found in the supernatant barely expressed MID as revealed by flow cytometry analysis (Fig. 6A and B). Before passages, the M. catarrhalis RH4 strain showed a single peak of high (>99%) IgD-binding bacteria (Fig. 6C). After six passages, an additional low two peaks were observed in the nonclumping fraction (Fig. 6D). Similar patterns were observed when six other strains were investigated. Interestingly, when the number of G's were analyzed, five of six strains lost a G after the six passages, resulting in considerably decreased MID expression. To ensure that we analyzed bacteria originating from the same isolates, PFGE and RAPD typing were performed in parallel with sequencing of the poly(G) tracts. As shown with the Perez112 strain, the same genetic profile was found in the wild type and the nonclumping variants with poly(G) tracts containing 9 and 8 G's, respectively (Fig. 6E and F).
FIG. 6.
MID protein expression is shut down during phase variation. IgD binding of M. catarrhalis strains Perez112 (A and B) and RH4 (C and D) is shown before (clumping variants [A and C]) and after (nonclumping variants [B and D]) six passages. The genetic profiles of Perez112 before (9 G's) and after (8 G's) the six passages obtained by PFGE (E) and RAPD (F) were identical. M. catarrahlis strains were cultured in NB medium without shaking. A small fraction of the supernatant (without clumping bacteria) was transferred to a new tube six times in 4 weeks. IgD binding was analyzed by flow cytometry as described in the legend to Fig. 1. The numbers of G's in the poly(G) tracts were determined by DNA sequencing. Details on PFGE and RAPD are given in Materials and Methods. In panel F, the letters a and b indicate the primers A70-3 and ERIC1-R, respectively. The M. catarrhalis Bc5 strain was included as a control in panels E and F.
DISCUSSION
In this study, the mid gene can be detected in all 91 clinical M. catarrhalis isolates from different continents. Flow cytometry analysis was found to be a convenient method for screening MID protein expression in a large number of isolates. It was shown that the degree of MID expression varied without relation to the anatomical site or geographical origin of the isolates. It must be stressed, however, that the bacteria we tested were not freshly isolated but collection strains. Despite the fact that not all strains expressed high levels of mature MID protein, the mid gene was present in all isolates tested. This is consistent with the results of a recent study by Lafontaine et al. (30) demonstrating that expression of M. catarrhalis UspA1 exhibits phase variation. This phenomenon points to the importance of examining the phenotype by protein analyses in contrast to characterization of the genotype only.
It has been known for many years that bacteria randomly undergo phase variation (23). However, depending on the specific environmental growth conditions, genes can also be turned on or off through sophisticated molecular switches. To date, the majority of known phase variants are reversible, and the random occurrence results in phenotypically heterogenous populations. Henderson et al. (23) have divided phase variation into three major groups: (i) variation by genomic rearrangement, (ii) variation by slipped-strand mispairing, or (iii) variation mediated by differential methylation. One of the best-characterized reversible phase variations is the regulation of type 1 fimbriae in E. coli (35). The type 1 fimbria operon is governed by inversion of a chromosomal region (314 bp) located upstream of the gene. Slipped-strand mispairing occurs in short sequence repeats or in variable numbers of tandem repeats, resulting in pretermination of translation or abolished transcription. Examples are poly(C) or poly(G) tracts represented in the opc and hpuA genes, respectively, of N. meningitidis (31, 45) and tandem repeats in the hif gene cluster in H. influenzae (34).
Aggregation of M. catarrhalis is a feature that by definition is related to phase variation (28). Indeed, when we examined the ORFs of several isolates and related the IgD-binding capacity to the number of G's in the characteristic poly(G) tracts, we found that MID-expressing isolates contained 1, 2, or 3 triplets of G's in their poly(G) tracts, keeping the ORF intact. In contrast, most low-MID-expressing isolates harbored 4, 7, 8, or 10 G's, causing preterminated translation (Table 3). A similar gene regulation has been observed for the uspA1 gene (30). However, a distinct feature of the mid gene poly(G) tract is that it is located within the ORF and therefore might cause a translational frameshift compared to the uspA1 gene that contains the poly(G) tract upstream of the predicted start codon ATG. Northern blot analysis revealed, however, that mid mRNA did not exist in low-MID-expressing strains with 4, 7, 8, or 10 G's, suggesting that like uspA1, mid is regulated at the transcriptional level. However, when we used a sensitive RT-PCR, mid transcripts could also be detected in the low-MID-expressing strains. Therefore, two different functions can be related to the poly(G) tract. (i) The mid gene is controlled at the DNA level by transcriptional regulation factors binding to the poly(G) tract. (ii) The mid gene is controlled at the mRNA level; the stability (or half-life) of the mid mRNA is dependent on the poly(G) tract. However, since low-MID-expressing isolates could also contain a minor MID-expressing population (Fig. 1A and Table 2), a few mid transcripts originating from the low-MID-expressing population may have been detected by the RT-PCR analysis. Other bacterial proteins, for example, Neisseria meningitidis and Neisseria gonorrhoeae hemoglobin receptors (HpuA) are regulated within the ORF, resulting in frameshift mutations (10, 31). In the case of the N. gonorrhoeae HpuA protein, the “off-phase” variants contained 9 G's, whereas the “on-phase” variants contained 10 G's. In contrast, 10 G's were found in the N. meningitidis hpuA gene in isolates expressing the gene and 11 G's were detected in the silent variants.
Another interesting feature of the MID protein is that the signal peptide together with the C-terminal ends share similarities with autotransporter proteins (reference 22 and references therein). Autotransporters are often related to the virulence of gram-negative bacteria. When the mid gene product was analyzed in detail, repetitive amino acid motifs were detected (Fig. 4). Similar patterns have been observed in the M. catarrhalis UspA1 and UspA2 proteins (13). The middle part of the MID molecule, which is downstream of and adjacent to the hypervariable domain, was significantly conserved among the five strains analyzed, suggesting that this part of the molecule has an important function that remains to be determined. Indeed, results of our recent experiments show that IgD binds to this part of MID (41). Furthermore, our functional studies reveal that MID is also an adhesin and hemagglutinin (A. Forsgren, M. Karamehmedovic, and K. Riesbeck, submitted for publication). A specific structure that is responsible for this function was also defined.
The degree of identity for MID proteins from four different strains that we sequenced was ≥75.8%, and for the MID gene product from a fifth strain, it was ≥65.3%. Sasaki et al. (46) sequenced a 200-kDa protein. That sequence shows only 53.5% identity with the first mid gene that we sequenced (14). However, the first 60 amino acids of the MID proteins shown in Fig. 4 in this paper have 95% identity with those of the 200-kDa protein that Sasaki et al. sequenced. In addition, there is an internal region where, in a segment containing 200 amino acids, there is 95% identity between MID and the 200-kDa protein of Sasaki et al. Finally, the last 400 amino acid residues of the MID protein have 97% identity with the last 400 amino acids of the 200-kDa protein of Sasaki et al. In view of these striking levels of identity, MID could be identical to the 200-kDa protein of Sasaki et al.
After this paper was submitted, Pearson et al. (42) published a study in which the gene encoding the 200-kDa protein (designated Hag) of an M. catarrhalis strain was inactivated by insertional mutagenesis. The isogenic hag mutant was unable to agglutinate human erythrocytes and lost its ability to autoagglutinate (but was still attached at wild-type levels to several human epithelial cell lines). The hag mutation also eliminated the ability of this mutant strain to bind human IgD. Thus, the Hag protein described by Pearsson (42) is most likely identical to the MID protein that we described (14, 19, 41; Forsgren et al., submitted).
Several gram-positive bacteria (e.g., Staphylococcus aureus and group A streptococcus) have been demonstrated to bind Igs (17, 18, 54). In contrast, nonimmune Ig binding to gram-negative bacteria is rare (50, 52). Interestingly, both M. catarrhalis and H. influenzae have a strong affinity for soluble human IgD (15). It was shown several years ago that IgD binding parallels a similar interaction with surface-bound IgD on B lymphocytes, a fact that explains the strong mitogenic effects on human lymphocytes by the two gram-negative bacteria (7, 16).
Interestingly, The MID protein conjugated to Sepharose beads activates purified lymphocytes to the same degree as formalin-killed whole bacteria (19). To obtain the maximal proliferative response, the T-helper (Th) cytokines IL-2 and IL-4 are required, whereas the Th2-specific cytokines IL-4 and IL-10 are needed for optimal IgA and IgG production. Protein D from H. influenzae is another IgD-binding OMP that we have characterized, and it has been shown to be an important pathogenicity factor (3, 25). However, protein D does not bind to the majority of IgD myelomas tested, and it has been proposed that encapsulated H. influenzae serotype b expresses an additional IgD receptor (47).
Why does M. catarrhalis harbor the IgD-binding protein MID? It is well-known that high concentrations of soluble IgD are present in the middle ear and nasopharynx (5, 48). IgD has thus been hypothesized to play a role in the local immune response. It has been suggested that the bacteria may benefit from the secreted IgD molecules and perhaps use the IgD as nutrients or as a protective coat. On the other hand, if the immune system adapts to the environmental bacterial threat rapidly, coating bacteria with soluble IgD perhaps decreases the bacterium's virulence. In that case, it is possible that the local polyclonal IgD production occurs in order to neutralize the bacteria. When M. catarrhalis (or H. influenzae) is coated with IgD, the bacterium would not activate B cells via the IgD B-cell surface receptor. Although this is a speculative hypothesis, this strategy would reduce cellular activation and consequently decrease a proinflammatory cytokine production and local inflammation that otherwise would damage the host tissue.
In conclusion, in this paper we have characterized the distribution of the MID protein in several clinical isolates. MID was a conserved M. catarrhalis OMP and was regulated by the number of guanosine nucleotides in characteristic poly(G) tracts located within the ORF. The mature gene product was present to a variable degree in all the strains investigated. Recently, M. catarrhalis has been acknowledged as a respiratory pathogen, and there is a strong need for a vaccine. The frequency of β-lactamase-resistant M. catarrhalis (4, 26) has also intensified the research for a suitable OMP to be included in a future vaccine against moraxella (33). Since MID has been proven to be conserved among the strains of the bacterial species and interact with the host immune system due to its unique IgD-binding features, the full-length protein or parts thereof would be an important candidate for vaccine development.
Acknowledgments
This work was supported in part by grants from the Alfred Österlund Foundation, the Anna and Edwin Berger Foundation, the Crafoord Foundation, the Greta and Johan Kock Foundation, the IngaBritt and Arne Lundberg Foundation, the Magnus Bergvall Foundation, the Swedish Medical Research Council, the Swedish Society of Medicine, The T.H.C. Bergh Foundation, the Åke Wiberg foundation, and the Cancer Foundation at the University Hospital in Malmö, Sweden.
We thank Marta Brant and Maria Sterner for excellent technical assistance.
REFERENCES
- 1.Aebi, C., E. R. Lafontaine, L. D. Cope, J. L. Latimer, S. L. Lumbley, G. H. McCracken, Jr., and E. J. Hansen. 1998. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis 035E. Infect. Immun. 66:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi, C., B. Stone, M. Beucher, L. D. Cope, I. Maciver, S. E. Thomas, G. H. McCracken, Jr., P. F. Sparling, and E. J. Hansen. 1996. Expression of the CopB outer membrane protein by Moraxella catarrhalis is regulated by iron and affects iron acquisition from transferrin and lactoferrin. Infect. Immun. 64:2024-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahrén, I. L., H. Janson, A. Forsgren, and K. Riesbeck. 2001. Protein D expression promotes the adherence and internalization of non-typeable Haemophilus influenzae into human monocytic cells. Microb. Pathog. 3:151-158. [DOI] [PubMed] [Google Scholar]
- 4.Bootsma, H. J., P. C. Aerts, G. Posthuma, T. Harmsen, J. Verhoef, H. van Dijk, and F. R. Mooi. 1999. Moraxella (Branhamella) catarrhalis BRO beta-lactamase: a lipoprotein of gram-positive origin? J. Bacteriol. 181:5090-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandtzaeg, P., S. T. Gjeruldsen, F. Korsrud, K. Baklien, P. Berdal, and J. Ek. 1979. The human secretory immune system shows striking heterogeneity with regard to involvement of J chain-positive IgD immunocytes. J. Immunol. 122:503-510. [PubMed] [Google Scholar]
- 6.Brendel, V., P. Bucher, I. Nourbakhsh, B. E. Blaisdell, and S. Karlin. 1992. Methods and algorithms for statistical analysis of protein sequences. Proc. Natl. Acad. Sci. USA 89:2002-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvert, J. E., and A. Calogeres. 1986. Characteristics of human B cells responsive to the T-independent mitogen Branhamella catarrhalis. Immunology 58:37-41. [PMC free article] [PubMed] [Google Scholar]
- 8.Campagnari, A. A., T. F. Ducey, and C. A. Rebmann. 1996. Outer membrane protein B1, an iron-repressible protein conserved in the outer membrane of Moraxella (Branhamella) catarrhalis, binds human transferrin. Infect. Immun. 64:3920-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catlin, B. W. 1990. Branhamella catarrhalis: an organism gaining respect as a pathogen. Clin. Microbiol. Rev. 3:293-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, C. J., C. Elkins, and P. F. Sparling. 1998. Phase variation of hemoglobin utilization in Neisseria gonorrhoeae. Infect. Immun. 66:987-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, D., V. Barniak, K. R. VanDerMeid, and J. C. McMichael. 1999. The levels and bactericidal capacity of antibodies directed against the UspA1 and UspA2 outer membrane proteins of Moraxella (Branhamella) catarrhalis in adults and children. Infect. Immun. 67:1310-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christensen, J. J., J. Ursing, and B. Bruun. 1994. Genotypic and phenotypic relatedness of 80 strains of Branhamella catarrhalis of worldwide origin. FEMS Microbiol. Lett. 119:155-159. [DOI] [PubMed] [Google Scholar]
- 13.Cope, L. D., E. R. Lafontaine, C. A. Slaughter, C. A. Hasemann, Jr., C. Aebi, F. W. Henderson, G. H. McCracken, Jr., and E. J. Hansen. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 181:4026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsgren, A., M. Brant, A. Möllenkvist, A. Muyombwe, H. Janson, N. Woin, and K. Riesbeck. 2001. Isolation and characterization of a novel IgD-binding protein from Moraxella catarrhalis. J. Immunol. 167:2112-2120. [DOI] [PubMed] [Google Scholar]
- 15.Forsgren, A., and A. Grubb. 1979. Many bacterial species bind human IgD. J. Immunol. 122:1468-1472. [PubMed] [Google Scholar]
- 16.Forsgren, A., A. Penta, S. F. Schlossman, and T. F. Tedder. 1988. Branhamella catarrhalis activates human B lymphocytes following interactions with surface IgD and class I major histocompatibility complex antigens. Cell. Immunol. 112:78-88. [DOI] [PubMed] [Google Scholar]
- 17.Forsgren, A., and J. Sjöqvist. 1966. Protein A from Staphylococcus aureus. Pseudo-immune reaction with human gamma-globulin. J. Immunol. 97:822-827. [PubMed] [Google Scholar]
- 18.Frick, I. M., M. Wikström, S. Forsen, T. Drakenberg, H. Gomi, U. Sjöbring, and L. Björck. 1992. Convergent evolution among immunoglobulin G-binding bacterial proteins. Proc. Natl. Acad. Sci. USA 89:8532-8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gjörloff Wingren, A., R. Hadzic, A. Forsgren, and K. Riesbeck. 2002. A novel IgD-binding bacterial protein from Moraxella catarrhalis induces human B lymphocyte activation and isotype switching in the presence of Th2 cytokines. J. Immunol. 168:5582-5588. [DOI] [PubMed] [Google Scholar]
- 20.Helminen, M. E., I. Maciver, M. Paris, J. L. Latimer, S. L. Lumbley, L. D. Cope, G. H. McCracken, Jr., and E. J. Hansen. 1993. A mutation affecting expression of a major outer membrane protein of Moraxella catarrhalis alters serum resistance and survival in vivo. J. Infect. Dis. 168:1194-1201. [DOI] [PubMed] [Google Scholar]
- 21.Helminen, M. E., I. Maciver, J. L. Latimer, J. Klesney-Tait, L. D. Cope, M. Paris, G. H. McCracken, Jr., and E. J. Hansen. 1994. A large, antigenically conserved protein on the surface of Moraxella catarrhalis is a target for protective antibodies. J. Infect. Dis. 170:867-872. [DOI] [PubMed] [Google Scholar]
- 22.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 23.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches-the ON and OFF of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 24.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janson, H., B. Carlén, A. Cervin, A. Forsgren, A. Björk-Magnusdottir, S. Lindberg, and T. Runer. 1999. Effects on the ciliated epithelium of protein D-producing and -nonproducing nontypeable Haemophilus influenzae in nasopharyngeal tissue cultures. J. Infect. Dis. 180:737-746. [DOI] [PubMed] [Google Scholar]
- 26.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2:547-559. [DOI] [PubMed] [Google Scholar]
- 27.Klingman, K. L., and T. F. Murphy. 1994. Purification and characterization of a high-molecular-weight outer membrane protein of Moraxella (Branhamella) catarrhalis. Infect. Immun. 62:1150-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyd, J. M., A. W. Cripps, and T. F. Murphy. 1998. Outer-membrane antigen expression by Moraxella (Branhamella) catarrhalis influences pulmonary clearance. J. Med. Microbiol. 47:159-168. [DOI] [PubMed] [Google Scholar]
- 29.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lafontaine, E. R., N. J. Wagner, and E. J. Hansen. 2001. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol. 183:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis, L. A., M. Gipson, K. Hartman, T. Ownbey, J. Vaughn, and D. W. Dyer. 1999. Phase variation of HpuAB and HmbR, two distinct haemoglobin receptors of Neisseria meningitidis DNM2. Mol. Microbiol. 32:977-989. [DOI] [PubMed] [Google Scholar]
- 32.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 33.McMichael, J. C. 2000. Vaccines for Moraxella catarrhalis. Vaccine 8(Suppl. 1):S101-S107. [DOI] [PubMed]
- 34.Mhlanga-Mutangadura, T., G. Morlin, A. L. Smith, A. Eisenstark, and M. Golomb. 1998. Evolution of the major pilus gene cluster of Haemophilus influenzae. J. Bacteriol. 180:4693-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mol, O., and B. Oudega. 1996. Molecular and structural aspects of fimbriae biosynthesis and assembly in Escherichia coli. FEMS Microbiol. Rev. 19:25-52. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, T. F., C. Kirkham, and A. J. Lesse. 1993. The major heat-modifiable outer membrane protein CD is highly conserved among strains of Branhamella catarrhalis. Mol. Microbiol. 10:87-97. [DOI] [PubMed] [Google Scholar]
- 37.Murphy, T. F., J. M. Kyd, A. John, C. Kirkham, and A. W. Cripps. 1998. Enhancement of pulmonary clearance of Moraxella (Branhamella) catarrhalis following immunization with outer membrane protein CD in a mouse model. J. Infect. Dis. 178:1667-1675. [DOI] [PubMed] [Google Scholar]
- 38.Myers, L. E., Y. P. Yang, R. P. Du, Q. Wang, R. E. Harkness, A. B. Schryvers, M. H. Klein, and S. M. Loosmore. 1998. The transferrin binding protein B of Moraxella catarrhalis elicits bactericidal antibodies and is a potential vaccine antigen. Infect. Immun. 66:4183-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Needleman, S. B., and C. D. Wunsch. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443-453. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 41.Nordström, T., A. Forsgren, and K. Riesbeck. 2002. The immunoglobulin D-binding part of the outer membrane protein MID from Moraxella catarrhalis comprises 238 amino acids and a tetrameric structure. J. Biol. Chem. 277:34692-34699. [DOI] [PubMed] [Google Scholar]
- 42.Pearson, M. M., E. R. Lafontaine, N. J. Wagner, J. W. St. Geme III, and E. J. Hansen. 2002. A hag mutant of Moraxella catarrhalis strain O35E is deficient in hemagglutination, autoagglutination, and immunoglobulin D-binding activities. Infect. Immun. 70:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reddy, M. S., T. F. Murphy, H. S. Faden, and J. M. Bernstein. 1997. Middle ear mucin glycoprotein: purification and interaction with nontypable Haemophilus influenzae and Moraxella catarrhalis. Otolaryngol. Head Neck Surg. 116:175-180. [DOI] [PubMed] [Google Scholar]
- 44.Rice, P., I. Longden, and A. Bleasby. 2000. “EMBOSS: The European Molecular Biology Open Software Suite.” Trends Genet. 16:276-277. [DOI] [PubMed] [Google Scholar]
- 45.Sarkari, J., N. Pandit, E. R. Moxon, and M. Achtman. 1994. Variable expression of the Opc outer membrane protein in Neisseria meningitidis is caused by size variation of a promoter containing poly-cytidine. Mol. Microbiol. 13:207-217. [DOI] [PubMed] [Google Scholar]
- 46.Sasaki, K., R. E. Harkness, and M. H. Klein. September 1998. Nucleic acids encoding high molecular weight major outer membrane protein of Moraxella. U.S. patent 5808024.
- 47.Sasaki, K., and R. S. Munson, Jr. 1993. Protein D of Haemophilus influenzae is not a universal immunoglobulin D-binding protein. Infect. Immun. 61:3026-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sørensen, C. H., and P. L. Larsen. 1988. IgD in nasopharyngeal secretions and tonsils from otitis-prone children. Clin. Exp. Immunol. 73:149-154. [PMC free article] [PubMed] [Google Scholar]
- 49.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 25:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Widders, P. R., L. A. Dorrance, M. Yarnell, and L. B. Corbeil. 1989. Immunoglobulin-binding activity among pathogenic and carrier isolates of Haemophilus somnus. Infect. Immun. 57:639-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolf, B., M. Kools-Sijmons, C. Verduin, L. C. Rey, A. Gama, J. Roord, J. Verhoef, and A. van Belkum. 2000. Genetic diversity among strains of Moraxella catarrhalis cultured from the nasopharynx of young and healthy Brazilian, Angolan, and Dutch children. Eur. J. Clin. Microbiol. Infect. Dis. 19:759-764. [DOI] [PubMed] [Google Scholar]
- 52.Yarnall, M., P. R. Widders, and L. B. Corbeil. 1988. Isolation and characterization of Fc receptors from Haemophilus somnus. Scand. J. Immunol. 28:129-137. [DOI] [PubMed] [Google Scholar]
- 53.Yu, R. H., R. A. Bonnah, S. Ainsworth, and A. B. Schryvers. 1999. Analysis of the immunological responses to transferrin and lactoferrin receptor proteins from Moraxella catarrhalis. Infect. Immun. 67:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, L., K. Jacobsson, J. Vasi, M. Lindberg, and L. Frykberg. 1998. A second IgG-binding protein in Staphylococcus aureus. Microbiology 144:985-991. [DOI] [PubMed] [Google Scholar]