Abstract
When soluble extracts of the extreme acidothermophilic archaeon Sulfolobus solfataricus were incubated with [γ-32P]ATP, several proteins were radiolabeled. One of the more prominent of these, which migrated with a mass of ∼46 kDa on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), was purified by column chromatography and SDS-PAGE and subjected to amino acid sequence analysis via both the Edman technique and mass spectroscopy. The best match to the partial sequence obtained was the potential polypeptide product of open reading frame sso0417, whose DNA-derived amino acid sequence displayed many features reminiscent of the 2,3-diphosphoglycerate-independent phosphoglycerate (PGA) mutases [iPGMs]. Open reading frame sso0417 was therefore cloned, and its protein product was expressed in Escherichia coli. Assays of its catalytic capabilities revealed that the protein was a moderately effective PGA mutase that also exhibited low levels of phosphohydrolase activity. PGA mutase activity was dependent upon the presence of divalent metal ions such as Co2+ or Mn2+. The recombinant protein underwent autophosphorylation when incubated with either [γ-32P]ATP or [γ-32P]GTP. The site of phosphorylation was identified as Ser59, which corresponds to the catalytically essential serine residue in bacterial and eucaryal iPGMs. The phosphoenzyme intermediate behaved in a chemically and kinetically competent manner. Incubation of the 32P-labeled phosphoenzyme with 3-PGA resulted in the disappearance of radioactive phosphate and the concomitant appearance of 32P-labeled PGA at rates comparable to those measured in steady-state assays of PGA mutase activity.
Interest in the members of the Archaea has grown steadily over the last two decades, stimulated to a large degree by their extremophilic lifestyles and distinct phylogenetic status. Within archaeal proteomes resides a wealth of information concerning unique metabolic pathways for extracting energy from sulfur, producing methane from CO2, etc., as well as biochemical and biophysical mechanisms for sustaining life processes under conditions of temperature, pH, and salinity hostile to “conventional” organisms (reviewed in references 1, 12, 23, and 37). Archaeal genomes combine unique, bacterial, and eucaryal features in a complex puzzle whose deconvolution will provide important insights into the nature and order of the evolutionary process (reviewed in references 13, 14, and 17). However, our current library of information concerning fundamental metabolic, sensory, regulatory, and other processes in the Archaea remains lacking in both breadth and depth.
In a previous study, we observed that when extracts of the hyperthermophilic archaeon Sulfolobus solfataricus were incubated with [γ-32P]ATP, numerous polypeptides incorporated radiolabeled phosphate (41). One of the more visually prominent of these was a phosphoserine-containing polypeptide with a mass of ∼46 kDa. Using the recently released genome sequence of S. solfataricus P2 (38), we have been able to link this phosphoprotein to the open reading frame (ORF) that encodes it. We describe here the identification of the ∼46-kDa phosphoprotein as an enzyme with phosphoglycerate (PGA) mutase activity that is encoded by ORF sso0417.
MATERIALS AND METHODS
Materials.
[γ-32P]ATP, [α-32P]ATP, and [γ-32P]GTP were purchased from NEN Research Products (Boston, Mass.). Matrix Gel Blue A and Matrix Gel Blue B were purchased from Amicon Corp. (Danvers, Mass.). Hydroxyapatite was from Bio-Rad (Richmond, Calif.). Enolase was obtained from Sigma (St. Louis, Mo.). Oligonucleotide primers were from Life Technologies, Inc. (Gaithersburg, Md.). Genomic DNA from S. solfataricus was obtained from the American Type Culture Collection. The Gene Editor in vitro site-directed mutagenesis system and sequencing grade trypsin were from Promega (Madison, Wis.). EK Max enterokinase was from Invitrogen (San Diego, Calif.). The Jupiter C18 (10 μm/300 A) reversed-phase high-pressure liquid chromatography (HPLC) resin was from Phenomenex, Inc. (Torrance, Calif.). 3-PGA was puchased as the tricyclohexylammonium salt from Roche Diagnostics GmbH (Mannheim, Germany). General laboratory reagents, molecular biology supplies, all other sugars, and microbial culture media were obtained from Fisher (Pittsburgh, Pa.) or Sigma.
Standard procedures.
S. solfataricus P1 (ATCC 35091) was grown in continuous culture with vigorous aeration at 70°C in de Rosa's standard medium (11) with the level of yeast extract increased to 2 g/liter. Protein concentrations were determined as described by Bradford (3), using a premixed reagent and a standardized solution of bovine serum albumin from Pierce (Rockford, Ill.). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli (29), and gels were stained with Coomassie brilliant blue as described by Fairbanks et al. (15). 32P-labeled phosphoproteins were visualized in SDS-polyacrylamide gels by electronic autoradiography using a Packard (Meriden, Conn.) Instantimager.
Partial purification of the ∼46-kDa phosphoprotein.
Soluble extract was prepared from a 20-g (wet weight) sample of S. solfataricus and incubated with [γ-32P]ATP for 30 min as described by Solow et al. (41). The radiolabeled extract was then applied to a 3-by-50-cm column of DE-52 cellulose that had been equilibrated in 20 mM (N-morpholino)-ethanesulfonic acid (MES; pH 6.5) containing 50 mM NaCl and 0.5 mM EDTA. The column was washed with this same buffer and then developed with a 400-ml linear gradient of 50 to 500 mM NaCl in 20 mM MES (pH 6.5) containing 0.5 mM EDTA. Fractions, 2 ml each, were collected and analyzed by SDS-PAGE, followed by autoradiography. Those containing the ∼46-kDa phosphoprotein were pooled, dialyzed versus 4 liters of 20 mM MES (pH 6.5) containing 0.5 mM EDTA, and applied to a 2-by-15-cm column of Matrix Gel Blue A that had been equilibrated in the same buffer. The column was washed in equilibration buffer (5 volumes) and then developed with a 60-ml linear gradient of 0 to 500 mM NaCl in 20 mM MES (pH 6.5) containing 0.5 mM EDTA. Fractions (2 ml) were collected and analyzed by SDS-PAGE, followed by autoradiography.
Fractions containing the ∼46-kDa phosphoprotein were pooled, dialyzed versus 4 liters of 10 mM sodium phosphate (pH 6.8), and applied to a 1-by-8-cm column of hydroxyapatite that had been equilibrated in the same buffer. The column was washed with equilibration buffer and developed with a 60-ml linear gradient of 10 to 200 mM sodium phosphate (pH 6.8). Fractions were collected and analyzed as described above. Those containing the ∼46-kDa phosphoprotein were pooled and dialyzed versus 20 mM MES (pH 6.5) containing 0.5 mM EDTA. The dialyzed material was applied to a 1-by-8-cm column of Matrix Gel Blue B that had been equilibrated in dialysis buffer. The column was washed with 20 mM MES (pH 6.5) containing 0.5 mM EDTA. Bound proteins were eluted with 20 mM MES (pH 6.5) containing 0.5 mM EDTA and 500 mM NaCl.
Partial sequence analysis of the ∼46-kDa phosphoprotein.
For Edman sequencing, a sample of the ∼46-kDa phosphoprotein was treated with Staphylococcus aureus V8 protease, and the resulting peptides were resolved on a high-osmolarity SDS-polyacrylamide gel according to established procedures (31). The peptides were electroblotted onto an Immobilon-P membrane, and the most visually prominent peptide subjected to 20 cycles of Edman sequencing at the Protein Chemistry Facility of the W. Alton Jones Cell Science Center in Lake Placid, N.Y.
For sequence analysis by mass spectroscopy, a 4.2-μg portion of partially purified ∼46-kDa phosphoprotein was applied to a 10% (wt/vol) SDS-polyacrylamide gel. After electrophoresis, the gel was stained with Coomassie blue, and the section of the gel containing the ∼46-kDa phosphoprotein was excised and sent to the W. M. Keck Biomedical Mass Spectrometry Laboratory at the University of Virginia for analysis.
Cloning of sso0417 and expression of its recombinant protein product, rSso0417.
ORF sso0417 was amplified from genomic DNA from S. solfataricus and cloned into expression vector pRSET-C, which had been cut with EcoRI and BglII, by standard procedures (39). The primers used for PCR amplification were 5′-GAAGATCTCGGTGATTGGTTGAAGC-3′ (forward primer) and 5′-AAGGAATTCTCATGAACGTATTTCTCTG-3′ (reverse primer). The resulting fusion protein, rSso0417, which contained an N-terminal hexahistidine tag, was expressed in E. coli BL21(DE3)pLysS and purified by Ni2+-chelate chromatography according to established protocols (39).
Site-directed mutagenesis of Ser59.
Site-directed mutagenesis was performed by using the Promega Gene Editor in vitro site-directed mutagenesis system according to the manufacturer's instructions. For mutagenically altering Ser59 to Ala, the following primer was used: 5′-TAATTCCTGGGGCTGATACTTCACATC-3′. To alter Ser59 to Thr, we used following primer 5′-TTCCTGGGACTGATACTTCACATC-3′.
Spectrophotometric assay of PGA mutase activity.
PGA mutase activity was measured by the coupled enzyme assay procedure described in Grisolia and Carreras (21) with minor modifications. Under standard assay conditions, the activity of 40 μg of rSso0417 was measured in 1 ml of 25 mM HEPES (pH 7.5) containing 10 U of enolase, 15 mM 3-PGA, and the indicated divalent metal ion cofactors at a concentration of 5 mM. The assay temperature was 25°C. The coupling enzyme, enolase, converts the product of the PGA mutase reaction, 2-PGA, to phosphoenolpyruvate. The latter was measured by exploiting its UV absorbance at 240 nm, using an extinction coefficient of 1,750 M−1 cm−1.
Assay of phosphatase activity.
Phosphatase activity was measured by incubating 4 μg of rSso0417 for 30 min at 65°C in 50 μl of 25 mM HEPES (pH 7.5) containing either 5 mM MnCl2 or 5 mM CoCl2 and either 15 mM 3-PGA or 2.5 mM ATP as the substrate. Portions (50 μl) were then analyzed for phosphate content by the malachite green method (30).
Assay of kinase activity.
rSso0417 (2 μg) was incubated at 25°C in 50 μl of 25 mM HEPES (pH 7.5) containing 5 mM MgCl2, 5 mM MnCl2, 1 mM [γ-32P]ATP (specific radioactivity, 10 to 150 cpm/pmol), and one of the following sugars, each at a concentration of 15 mM: 3-PGA, 2-PGA, glyceric acid, glycerol, erythrose-4-phosphate, or ribulose-5-phosphate. Where indicated, enolase (0.5 U) was also included. The reaction was terminated by the addition of 1 μl of 500 mM EDTA (pH 8.0). Portions (1 μl) of the reaction mixture were then spotted onto 20-by-20-cm sheets of polyethyleneimine cellulose, and the adenine nucleotides in the mixture were separated by thin-layer chromatography by using 1 M LiCl as the mobile phase (34). The presence and quantity of the predicted products of a kinase reaction, [α-32P]ADP and [α-32P]AMP, were determined by electronic autoradiography. Unlabeled ATP, ADP, and AMP were applied to adjacent lanes as standards and visualized by exploiting their fluorescence under UV light.
Autophosphorylation of rSso0417.
rSso0417 was autophosphorylated with [32P]phosphate by incubating 0.4 mg of the purified recombinant protein at 35°C for 90 min in 0.5 ml of 45 mM Tris-HCl (pH 7.5) containing 5 mM morpholinepropanesulfonic acid (MOPS), 0.45 mM EDTA, 50 μM ATP, 70 μCi of [γ-32P]ATP, 2 mM dithiothreitol (DTT), 5 mM MgCl2, and 5 mM MnCl2. The autophosphorylated protein was then separated from ATP and other small molecules by gel filtration chromatography on a 1.5-by-20-cm column of Sephadex G-25 that had been equilibrated in and was developed with 50 mM Tris (pH 7.5) containing 50 mM NaCl. The eluted phosphoprotein was dialyzed versus 25 mM Tris (pH 7.5) containing 0.5 mM EDTA. This method typically yielded rSso0417 that was phosphorylated to a stoichiometry of ∼0.5 mol/mol. Autophosphorylation of rSso0417 “in gel” was performed with 20 μg of rSso0417 essentially as described by Lower et al. (32).
Phosphoamino acid analysis.
Acid hydrolysis of phosphorylated proteins and analysis of their phosphoamino acid content by thin-layer electrophoresis were performed by an adaptation of the method of Cooper et al. (10).
Effects of potential substrates and cofactors on the quantity of [32P]phosphate in rSso0417.
In order to assay the effect of potential substrates on the quantity of [32P]phosphate in rSso0417, 5-μg portions of rSso0417 that had been autophosphorylated as described above were incubated for 15 min at 65°C in 50 μl of 25 mM MOPS (pH 7.0) containing, unless otherwise indicated, 5 mM MgCl2, 5 mM MnCl2, and 0.1 mM concentrations of the indicated sugars. Incubations were terminated by adding 16 μl of SDS sample buffer lacking reducing agents, followed by heating at 100°C for 5 min. Portions (33 μl) of the heated mixture were then applied to a 10% (vol/vol) SDS-polyacrylamide gel. After electrophoresis, the protein-bound radioactivity was measured by electronic autoradiography using a Packard Instantimager. For assay of the effect of divalent metal ions, incubations were performed essentially as described above in the presence or absence of 0.1 mM 3-PGA. The incubation time was decreased to 5 min, and MgCl2 and MnCl2 were replaced by the indicated metal ions, each at a concentration of 0.1 mM. [32P]inorganic phosphate was extracted into organic solvents as a molybdic acid complex by the method of Martin and Doty (33).
Identification of [32P]PGA by paper chromatography.
Autophosphorylated rSso0417 (∼0.4 μg) that contained 3.6 pmol of [32P]phosphate was incubated with 15 mM 3-PGA for 5 min at 25°C in 50 μl of 25 mM HEPES (pH 7.5) containing 5 mM MnCl2. A portion of the incubation mixture (20 μl) was then spotted, 5 μl at a time, 2 cm from the bottom of a 20- by 24-cm (width by height) sheet of Whatman no. 1 filter paper. A second 20-μl portion was spotted coincident with 1 μl of 100 mM 2,3-diphosphoglycerate (2,3-diPGA) that was dissolved in 10 mM HEPES (pH 7.5). Other lanes contained PGA and the 2,3-diPGA standards alone. The filter paper was developed until the solvent front was 20 cm from the origin with solvent system I, and the positions of the PGA and diPGA standards were determined as described by Jacobs and Grisolia (25). Radiolabeled species were detected by electronic autoradiography.
Identification of the phosphorylation site on rSso0417 by mass spectroscopy.
rSso0417 (20 μg) was autophosphorylated as described above except that the ATP used was not radiolabeled. A 10-μg portion of the autophosphorylated protein was applied to an SDS-polyacrylamide gel, and the gel stained with Coomassie blue after electrophoresis. The gel was destained by incubating with several changes of 25 mM ammonium bicarbonate containing 50% (vol/vol) acetonitrile. The section of the gel containing the protein was excised, incubated in 50 mM ammonium bicarbonate (pH 8.0) containing 10 mM DTT, and then soaked in a solution of 55 mM iodoacetamide in 50 mM ammonium bicarbonate (pH 8.0). The gel slice was rinsed several times with ammonium bicarbonate (pH 8.0) and then with acetonitrile. After vacuum drying in a Speed-Vac, the gel slice was rehydrated for 15 min in a solution of 0.02 mg of TPCK (tosylphenylalanyl chloromethyl ketone)-treated trypsin/ml and then transferred to a tube containing 25 μl of 25 mM ammonium bicarbonate (pH 8.0). The mixture was incubated for 8 h at 37°C. Trifluoroacetic acid (5 μl of a 5% [vol/vol] solution) was added, and the mixture was immediately agitated with a Vortex mixer to terminate proteolysis. The supernatant liquid was removed and stored at −20°C until needed.
For liquid chromatography-mass spectrometry (LC-MS) and liquid chromatography-tandem mass spectrometry (LC-MS-MS), 1-μl portions of the peptide mixture were applied to a 75-μm capillary column packed with Jupiter C18 (10 μm/300 A) resin to a bed length of 10 cm. Prior to loading, the column was equilibrated in 0.5% (vol/vol) acetic acid (HOAc) at a flow rate of 50 μl/min using an LCPackings Ultimate HPLC pump. The column tip was gravity pulled, using a microtorch to eliminate the need for a frit. The column was developed with 0.5% (vol/vol) HOAc containing 5% (vol/vol) acetonitrile for 5 min, followed by a linear gradient of 5 to 80% (vol/vol) acetronitrile in 0.5% (vol/vol) HOAc for 40 min. A low dead volume stainless steel tee was used to split the eluant and divert a 250-nl/min portion to the mass spectrometer. Mass spectra were acquired on a ThermoFinnigan LCQ DecaXP quadrupole ion trap mass spectrometer. The electrospray voltage was 2.3 kV, the capillary temperature was 150°C, and the capillary voltage was 10 V. Three microscans (∼1.5 s each) were acquired for each spectrum recorded. The threshold for tandem mass spectrum acquisition was set at 5E6, and precursor ions were dynamically excluded for 5 min. Peptides were identified by using the SEQUEST algorithm searching an in-house FASTA-formated database for the S. solfataricus P2 genome, using a dynamic modification (80.0 atomic mass units [amu]) on S, T, and Y residues and a static modification (carbamidomethylation, 58.03 amu) on cysteine residues.
RESULTS
Partial amino acid sequence of the ∼46-kDa phosphoprotein of S. solfataricus.
When a soluble extract of the archaeon S. solfataricus was incubated with [γ-32P]ATP, numerous polypeptides were observed to incorporate radiolabeled phosphate. Among the more visually prominent of these was a polypeptide with an apparent mass of ∼46-kDa that contained [32P]phosphoserine (41). The large number of polypeptides present in the extract rendered the unambiguous identification of the Coomassie stained band corresponding to the ∼46-kDa phosphoprotein problematic. Therefore, the phosphoprotein was partially purified by column chromatography on DE-52 cellulose, Matrix Gel Blue A, hydroxyapatite, and Matrix Gel Blue B as described in Materials and Methods. The resulting protein mixture was sufficiently simplified in its polypeptide composition to permit the ∼46-kDa phosphoprotein to be identified on an SDS-polyacrylamide gel.
Two methods were employed in an effort to obtain amino acid sequence information from the ∼46-kDa phosphoprotein. First, a sample was subjected to partial proteolysis with S. aureus V8 protease. The resulting peptides were resolved from one another by SDS-PAGE on a high-osmolarity gel and transferred to a polyvinylidene difluoride membrane. After being stained, the most visually prominent polypeptide was sent to the Protein Chemistry Facility of the W. Alton Jones Cell Science Center in Lake Placid, N.Y., for Edman sequence analysis. Although the low yields of phenylthiohydantoin (PTH) amino acids obtained rendered their identification difficult, a partial sequence was obtained (Table 1). In order to obtain further sequence information, a sample of the ∼46-kDa phosphoprotein was sent to the W. M. Keck Biomedical Mass Spectrometry Laboratory at the University of Virginia for analysis by MS. Here, the protein was reduced, alkylated, and proteolyzed with trypsin, and the resulting peptides were separated by reversed-phase HPLC and sequenced by MS-MS using collisionally induced dissociation (CID) to induce peptide fragmentation. This analysis yielded complete or partial sequence information from 21 peptides (Table 1).
TABLE 1.
Partial sequence analysis of ∼46-kDa phosphoprotein
| Analysis | Peptide no. | Mass (Da) | Sequence |
|---|---|---|---|
| Edman sequencinga | ??qY?IL(l/p)FI(a/p)(f/d)G?K??p | ||
| ::::::::: | |||
| MKQYKILLIVADGLGDRP18 | |||
| Mass spectrometryb | 1 | 1,020.1 | QFEAXGAQR |
| :::::: | |||
| AFEALGAGA88 | |||
| 2 | 1,650.4 | ??FATVDN??TVVER | |
| :::::::: | |||
| GNFATVNNDLVVVDR113 | |||
| 3 | 772.2 | VAVV--- | |
| ::: | |||
| IAVV154 | |||
| 4 | 1,195.4 | WXPANXVXXR | |
| :::::::: | |||
| EKPANIVLLR229 | |||
| 5 | 1,244.6 | XNAAAV---K | |
| ::::: | |||
| LKAAAV251 | |||
| 6 | 1,267.8 | QTGGXD---xGK | |
| :::::: | |||
| ATGGIDTNYNAK285 | |||
| 7 | 1,654.0 | ??DVXNXNXXYSYR | |
| :::::::: | |||
| GLDVTNILLNYSNR408 | |||
| 8 | 865.0 | XQYTxaR | |
| 9 | 888.2 | XTASt--xK | |
| 10 | 1,044.6 | evVQEXXSK | |
| 11 | 1,054.0 | XYAX---K | |
| 12 | 1,059.4 | xGEX---qvK | |
| 13 | 1,062.8 | EAQYVDXPK | |
| 14 | 1,262.4 | QXEXEFQAQR | |
| 15 | 1,277.4 | ??qNSqp--- | |
| 16 | 1,494.0 | ??VADXV---EK | |
| 17 | 1,538.6 | WSNVSGV---GAR | |
| 18 | 1,722.7 | tpXEAVQEG---K | |
| 19 | 2,251.4 | XXXF---K | |
| 20 | 2,556.1 | ??NXYGS??DXA---K | |
| 21 | 2,744.8 | ??AYAP---WFeY--- |
The partially purified ∼46-kDa phosphoprotein was extracted from an SDS-polyacrylamide gel and digested with S. aureus V8 protease. The resulting peptides were separated by SDS-PAGE and transferred to a polyvinylidene difluoride membrane, and one of the more visually prominent bands was analyzed by the Edman method. Shown is the sequence obtained, and below it is the region of the protein product of ORF sso0417 with which its shares sequence similarity. Lowercase letters indicate tentative identifications, question marks indicate amino acid residues of undetermined identity, and dashes designate the presence of unknown numbers of unidentified amino acid residues. Parentheses enclose possible alternative assignments.
The partially purified ∼46-kDa phosphoprotein was extracted from an SDS-polyacrylamide gel, digested with trypsin, and analyzed by LC-MS and LC-MS-MS by standard procedures at the W. M. Keck Mass Spectrometry Laboratory at the University of Virginia. Shown are the masses of the peptides obtained, as well as the amino acid sequences deduced from their patterns of fragmentation. Below peptides 1 to 7 are shown the regions of the protein product of ORF sso0417 with which they share sequence similarity. “X” indicates the presence of I or L, which cannot be distinguished on the basis of mass.
The ∼46-kDa phosphoprotein is encoded by ORF sso0417.
When the partial sequence obtained via Edman analysis was searched against the genome of S. solfataricus, the best discernible match was to the N-terminal sequence of the predicted polypeptide product of ORF sso0417 (Table 1). The calculated molecular mass of the deduced protein, 45,179 Da, closely matched the mass empirically determined for the phosphoprotein by SDS-PAGE. Additional homology searches utilizing the 21 peptide sequences obtained by LC-MS-MS indicated that one-third, i.e., 7, displayed discernible similarity to portions of the predicted protein product of ORF sso0417 as well. However, none of the empirically determined sequences matched the potential product of ORF sso0417 in an exact, residue-for-residue basis, a potential consequence of errors and ambiguities in residue assignments arising from the low levels of sample available for these analyses.
In the face of the large number of peptides left unaccounted for, as well as the many discrepancies between the predicted protein sequence and those of the putative “matching” peptides, the proposition that the ∼46-kDa phosphoprotein was the product of ORF sso0417 was tested further. ORF sso0417 was cloned, and its protein product was expressed as a recombinant fusion protein in E. coli. rSso0417, which contained an N-terminal hexahistidine sequence, was purified to apparent homogeneity by using metal chelate chromatography. When incubated with [γ-32P]ATP, the protein became phosphorylated on serine to a stoichiometry of roughly 0.5 mol/mol, strongly suggesting that sso0417 did indeed encode the ∼46-kDa phosphoprotein observed in soluble extracts of S. solfataricus. Radiolabeling also was observed when [γ-32P]GTP was substituted for ATP. After SDS-PAGE, rSso0417 could be renatured and phosphorylated with [γ-32P]ATP as phosphoryl donor while still immobilized within the gel matrix, indicating that phosphate incorporation resulted from an autophosphorylation event (data not shown).
Autophosphorylation of recombinant Sso0417 takes place on Ser59 in vitro.
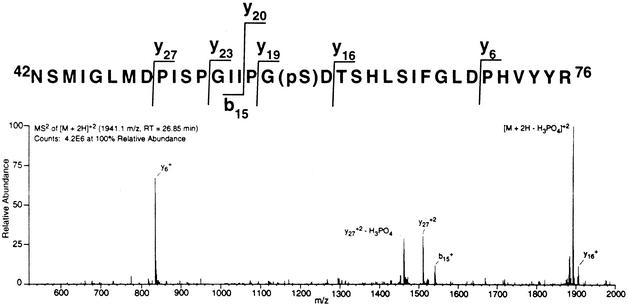
rSso0417 was incubated with ATP, and the autophosphorylated protein was isolated by SDS-PAGE. After incubation with trypsin, the resulting peptide mixture was analyzed by LC-MS-MS. Thanks in part to the large quantity of recombinant protein available for analysis, tryptic peptides encompassing 94% of the DNA-derived amino acid sequence encoded by ORF sso0417, including every predicted serine residue, were identified and sequenced. Computer analysis of the data obtained, by using parameters that accounted for the possible phosphorylation of all serine residues and neutral fragment (phosphate) loss, coupled with manual spectral inspection, indicated that only a single serine residue was phosphorylated. This serine residue was contained in a peptide spanning Asn42 through Arg76 (Fig. 2). Although the singly charged molecular ion [M + H+]1+ (3,881.4 amu) for this peptide was not detected, strong signals were obtained for three multiply charged ions: [M + 2H+]2+ (1,941.6 amu), [M + 3H+]+3 (1,294.8 amu), and [M + 4H+]4+ (971.4 amu).
FIG. 2.
Comparison of DNA-derived amino acid sequence of Sso0417 with archaeal, bacterial, and eukaryotic iPGMs. Shown is the DNA-derived amino acid sequence of the protein product of ORF sso0417 from S. solfataricus (Sso0417 [38], GenBank accession no. Q980A0) and, immediately below it, the sequence of iPGMs from the archaeon P. furiosus (Pfs iPGM [42]; GenBank accession no. AAL82083), the eukaryote T. brucei (Tbr iPGM [8]; GenBank accession no. CAB85498), and the bacterium B. stearothermophilus (Bst iPGM [6]; GenBank accession no. AF120091). The sequences were aligned by using the CLUSTALW program available from the European Bioinformatics Institute, with the C-terminal ca. 60 residues adjusted by eye. Amino acid identities between each of the established iPGMs and Sso0417 are indicated by colons, whereas nonidentical amino acids of very similar character are indicated by periods. Amino acids comprising subdomain A of iPGM from B. stearothermophilus (26) are underlined. Amino acids involved in metal ion binding in iPGM are marked by asterisks. Other amino acid residues that are conserved among the subdomain B regions of bacterial and eucaryal iPGMs are indicated by plus signs. The catalytically essential serine residue of the iPGM from B. stearothermohpilus is marked by “@.”
CID of the multiply charged ions was used to determine which of the five serines within the 35-residue peptide was phosphorylated. Figure 1 shows one of the patterns obtained when the doubly charged peptide ion [M + 2H+]2+ was fragmented by CID. The most abundant peak corresponds to peptide that has undergone neutral loss of phosphate [M + 2H+ − H3PO4]2+. Singly charged ions corresponding to fragments Ans42 to Ile56 (b15+) and Thr61 to Arg76 (y16+) are both visible, but not their potential phosphorylation products. On the other hand, doubly charged ions corresponding to the phosphorylated form of peptide fragment Pro50 to Arg76 (y272+), as well as the corresponding peptide fragment that had undergone neutral loss of phosphate (y272+ − H3PO4), were readily apparent. The only serine residue whose phosphorylation would be predicted to produce this fragmentation pattern, as well as the patterns that were obtained from the triply and quadruply charged ions (data not shown), was Ser59.
FIG. 1.
Identification of site of phosphorylation on rSso417 by mass spectrometry. rSso0417 was incubated with ATP, isolated by SDS-PAGE, and hydrolyzed into peptides by using trypsin. These peptides were then analyzed by LC-MS and LC-MS-MS as described in Materials and Methods. Shown is the sequence of the phosphate containing peptide, and below it the MS2 spectra of the [M + 2H+]2+ ion (average m/z = 1,941.6) used to determine the position of the serine residue phosphorylated (pS) with the major ions and the sites of collisional-induced dissociation that produced them indicated. The peak resulting from neutral loss of phosphate is designated [M + 2H+ − H3PO4]2+.
The assignment deduced from mass spectroscopic analysis was verified by using site-directed mutagenesis. Substitution of Ser59 with Ala produced a protein that failed to undergo autophosphorylation when incubated with [γ-32P]ATP, whereas substitution of Ser59 with Thr produced a protein that underwent autophosphorylation exclusively on threonine residues (data not shown).
rSso0417 exhibits PGA mutase activity.
Although the predicted protein product of ORF sso0417 was originally annotated as a potential phosphonopyruvate decarboxylase (38), homology searches indicated the existence of significant sequence homology with 2,3-diPGA-independent PGA mutases [iPGM] from Eucarya and Bacteria (Fig. 2). PGA mutases catalyze the net transfer of phosphate between the 3- and 2-hydroxyl groups of PGA (16, 27, 35). In addition to the iPGMs, a second, structurally distinct family of PGA mutases—the 2,3-diPGA-dependent PGA mutases [dPGMs]—are present in many members of the Bacteria and Eucarya.
The regions of sequence similarity between Sso0417 and prototypic iPGMs from Bacteria and Eucarya were not symmetrically distributed along the polypeptide chain. Rather, they were confined almost exclusively to domain A of the latter, which includes residues drawn from both the N- and C-terminal portions of the polypeptide. X-ray crystallographic analysis indicates that domain A contains the residues responsible for binding the two divalent metal ions that serve as catalytic cofactors for iPGMs, whereas domain B includes many of the residues responsible for binding the substrate or product, PGA (26). Domain A also contains a conserved serine residue that is essential for activity (26). It is generally believed that the catalytic mechanism of the iPGMs involves the formation of a phosphoenzyme intermediate (2, 4, 26, 27), with this serine residue constituting the most likely site of modification (9, 26). Intriguingly, not only does Sso0417 contain a plausible candidate for this conserved serine residue, it was the same residue that was autophosphorylated when rSso0417 was incubated with ATP in vitro, i.e., Ser59.
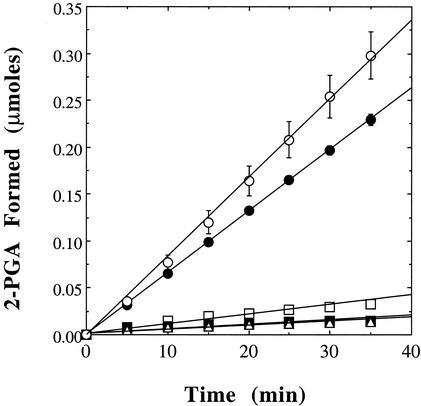
The presence in the DNA-derived amino acid sequence of Sso0417 of plausible candidates for each of the metal ion binding residues conserved among the iPGMs, as well as the catalytically essential serine residue, suggested that it might also be a phosphomutase. Therefore, rSso0417 was assayed for PGA mutase activity by using a coupled enzyme assay system that utilizes enolase to convert the reaction product, 2-PGA, to phosphoenolpyruvate, which absorbs UV light at 240 nm. As can be seen in Fig. 3, rSso0417 exhibited PGA mutase activity when incubated with the components of this coupled assay system. As has been observed for prototypic iPGMs (7, 9, 28), the PGA mutase activity of rSso0417 was dependent upon the presence of exogenous metal ions such as Co2+ or Mn2+. The rate of phosphoenolpyruvate production remained constant when the concentration of enolase was doubled, indicating that, under these conditions, the conversion of 3-PGA to 2-PGA by rSso0417 was rate limiting. Activity was maximal at around pH 7.5.
FIG. 3.
rSso0417 possesses PGA mutase activity. The PGA mutase activity of both rSso0417 (•) and a mutationally altered form in which Ser59 was altered to Thr (○) was measured spectrophotometrically by using a coupled assay system that measures the increase in absorbance at 240 nm that takes place when 2-PGA is converted to phosphoenolpyruvate by enolase. Also shown are the changes in absorbance observed in control assays in which either the potential PGM (□), enolase (▵), or 3-PGA (▪) was omitted. All assays were performed in triplicate, with error bars indicating the magnitude of the standard error. For further details, see Materials and Methods.
The measured rate of reaction in the presence of Co2+, i.e., 0.515 μmol/min/mg, was approximately three times that observed with Mn2+ (0.182 μmol/min/mg), which in turn was a more effective activator than Cd2+ (Table 2). Removal of the N-terminal fusion domain via proteolytic cleavage with enterokinase had no discernible effect on catalytic activity (data not shown). Substitution of the presumed catalytic Ser, Ser59, with Ala eliminated the PGA mutase activity of rSso0417, while its replacement by Thr restored catalytic function (Fig. 3). The addition of exogenous 2,3-diPGA had no discernible effect on enzyme activity.
TABLE 2.
Catalytic activity of rSso0417a
| Activity type (enzyme) | Substrate | Concn (mM) | Cofactor | Temp (°C) | Mean reaction rate (μmol/min/mg) ± SE |
|---|---|---|---|---|---|
| PGA mutase | 3-PGA | 15 | Mn2+ | 25 | 0.182 ± 0.045 |
| 3-PGA | 15 | Co2+ | 25 | 0.515 ± 0.032 | |
| 3-PGA | 15 | Cd2+ | 25 | 0.113 ± 0.018 | |
| 3-PGA | 15 | Mn2+ + Mg2+ | 25 | 0.165 ± 0.004 | |
| Phosphatase | 3-PGA | 15 | Mn2+ | 65 | 0.017 ± 0.001 |
| 3-PGA | 15 | Co2+ | 65 | 0.050 ± 0.002 | |
| ATP | 2.5 | Mn2+ | 65 | 0.018 ± 0.009 | |
| ATP | 2.5 | Co2+ | 65 | 0.067 ± 0.056 |
Shown are the results of steady-state analyses of the rates at which rSso0417, 40 μg per assay, catalyzed conversion of 3- to 2-PGA or the hydrolysis of 3-PGA and ATP. The temperatures of the assays of PGA mutase activity were dictated by the stability of the coupling enzyme, enolase. The temperature of the phosphohydrolase assays was raised to 65°C in order to increase reaction rates to more readily detectable levels. Shown are the results of triplicate determinations. All metal ions were present at a concentration of 5 mM. For further details, see Materials and Methods.
What is the fate of the enzyme-bound phosphoryl group?
The catalytic mechanism of the iPGMs remains a subject of controversy. The most widely accepted reaction sequence involves the transfer of a phosphoryl group from 3-PGA (or 2-PGA) to the enzyme's catalytically essential serine residue, forming glycerate and a phosphoenzyme intermediate, followed by the subsequent rephosphorylation of the tightly bound glycerate to form 2-PGA (or 3-PGA) (reviewed in reference 27). This double-displacement mechanism readily accounts for three observations concerning prototypic iPGMs. First, catalysis does not require an exogenous cofactor such as 2,3-diPGA. Second, phosphoryl transfer is intramolecular, i.e., the phosphate group bound to the carbon backbone of the product is the same one that was bound to the backbone of the corresponding substrate molecule (4, 19). Third, phosphotransfer occurred with net retention of configuration by the chiral phosphoryl group of 2-[(R)-16O,17O,18O]PGA in vitro (2). On the other hand, repeated attempts to detect the presumed intermediates in the iPGM reaction sequence, glycerate and the phosphoenzyme, have met with little success (4, 5, 9, 22, 24). Moreover, even when minute quantities (<0.05 mol/mol) of the presumed phosphoenzyme intermediate of an iPGM from Trypsanosoma brucei were detected, the rate at which the phosphoryl group turned over was much too low (t1/2 ∼ 40 s) to be considered kinetically competent (9).
If autophosphorylated rSso0417 corresponds to the postulated phosphoenzyme intermediate of prototypic iPGMs from the Bacteria and Eucarya, then incubation with its cognate reaction intermediate, glycerate, should result in completion of the catalytic cycle with the concomitant displacement of [32P]phosphate from the enzyme. However, even when rSso0417 that had been autophosphorylated with [γ-32P]ATP was incubated for a prolonged period (15 min) with excess glycerate, a significant portion of the [32P]phosphate remained bound to the enzyme (Table 3). In contrast, incubation under similar conditions with equal or lower concentrations of 3-PGA resulted in the near-complete disappearance of protein-bound [32P]phosphate. Several other phosphorylated sugars, including 2-PGA, β-glycerol phosphate, erythrose-4-phosphate, and ribulose-5-phosphate, also proved noticeably more effective than glycerate in their ability to chase protein-bound radioactivity from autophosphorylated rSso0417 (Table 3). However, none was as effective as 3-PGA. These observations suggested either that rSso0417 is not a PGA mutase or that the reaction sequence employed by the autophosphorylated protein differed from that of prototypic iPGMs.
TABLE 3.
Substrate specificity of rSso0417 PGA mutasea
| Potential substrate | [32P]Pi remaining (% control) at a substrate concn of:
|
|||||
|---|---|---|---|---|---|---|
| 0.01 mM | 0.02 mM | 0.04 mM | 0.1 mM | 1 mM | 10 mM | |
| 3-PGA | 17 | 17 | 15 | 11 | 9 | 9 |
| 2-PGA | - | - | - | 11 | 9 | 6 |
| Glyceric acid | - | - | - | 69 | 34 | 24 |
| β-Glycerolphosphate | 51 | - | - | 37 | - | - |
| Erythrose-4-phosphate | 91 | 78 | 59 | 23 | - | - |
| Ribulose-5-phosphate | 57 | 40 | 29 | 17 | - | - |
Potential substrates were assayed for their ability to cause the disappearance of bound [32P]phosphate from the putative PGA mutase. Recombinant Sso0417 PGA mutase was incubated with [γ-32P]ATP, and the [32P]phosphorylated protein was isolated by gel filtration chromatography. Portions (5 μg, each containing ∼0.5 mol of Pi/mol) were incubated for 15 min at 65°C in 50 μl of 25 mM MOPS (pH 7.0) containing 5 mM MgCl2, 5 mM MnCl2, and one of the potential substrates listed at the indicated concentrations. Incubation was terminated by the addition of SDS sample buffer lacking DTT, followed by heating at 100°C for 5 min, followed by SDS-PAGE. Shown is the proportion of 32P radioactivity remaining bound to the protein relative to controls to which no sugar was added. A total of 70% or more of the radioactivity remained after incubation with the following compounds, each at a concentration of 0.1 mM: ribose-1-phosphate, ribose-5-phosphate, xylose-1-phosphate, glucose, glucose-1-phosphate, glucose-6-phosphate, fructose, fructose-1-phosphate, fructose-6-phosphate, mannose-1-phosphate, mannose-6-phosphate, ATP, or ADP. -, Not determined.
We first asked whether rSso0417 was a phosphatase. iPGMs belong to the same enzyme superfamily as does alkaline phosphatase, a phosphohydrolase whose catalytic mechanism involves a phosphoserine intermediate (18), whereas a dPGM-like protein from Bacillus stearothermophilus was recently reported to be a broad-specificity phosphatase (36). As can be seen in Table 2, rSso0417 exhibited weak but detectable phosphohydrolase activity toward both 3-PGA and ATP. However, the rate at which it catalyzed their hydrolysis fell well below that at which it converted 3-PGA to 2-PGA. In addition, when autophosphorylated rSso0417 was incubated with 3-PGA, <20% of the 32P radioactivity that was displaced from the protein was recovered as inorganic phosphate, indicating that the majority was transferred to 3-PGA or some other solute and not water.
We next sought to determine whether Sso0417 was a kinase. Certainly, the protein's autophosphorylation in the presence of ATP constitutes strong circumstantial evidence for such a function. However, when rSso0417 was incubated with [α-32P]ATP and either 3-PGA or any one of a variety of other sugars, including glycerate, 2-PGA, glycerol, β-glycerol phosphate, erythrose-4-phosphate, or ribulose-5-phosphate, neither [α-32P]ADP nor [α-32P]AMP was produced in significant quantities. Specifically, under conditions in which rSso0417 catalyzed the production of ∼10 nmol of 2-PGA, <0.1 nmol of [α-32P]ADP or [α-32P]AMP was detected (data not shown)
The second predicted product of a potential PGA kinase reaction is 2,3-diPGA. However, analysis by paper chromatography—which separates monophosphorylated glycerates from 2,3-diPGA—revealed that very little if any 2,3-diPGA was formed when 32P-labeled rSso0417 was incubated with 3-PGA. Instead, the vast majority (≥85%) of the radiolabeled phosphate migrated as 2-PGA and/or 3-PGA, as if autophosphorylated rSso0417 catalyzed the interconversion of 3-PGA and 2-PGA (data not shown). Thus, under these circumstances autophosphorylated rSso0417 behaved more like a dPGM, with ATP serving as cofactor instead of 2,3-diPGA.
As with prototypic iPGMs, dPGMs utilize a double-displacement mechanism involving a phosphoenzyme intermediate (reviewed in reference 16). However, they must first be primed for catalysis via prior autophosphorylation of an active-site histidine residue by using the cofactor 2,3-diPGA as phosphodonor. Upon binding 3-PGA (or 2-PGA), the now catalytically competent phosphoenzyme transfers its phosphoryl group to the free hydroxyl group of the substrate to form 2,3-diPGA. This intermediate subsequently donates a phosphoryl group back to the active site histidine to produce 2-PGA (or 3-PGA) and regenerate the catalytically competent phosphoenzyme. Phosphoryl transfer by dPGMs is therefore intermolecular. The phosphate group that is present on the 2-PGA (or 3-PGA) produced by a dPGM originates from the free phosphoenzyme, which acquired it either during the preceding catalytic cycle or from the priming cofactor 2,3-diPGA.
We next investigated whether transfer of the enzyme-bound phosphoryl group to PGA took place with sufficient rapidity to account for at which rSso0417 catalyzed the conversion of 3-PGA to 2-PGA in the steady state. Using Mn2+ as the cofactor, the 32P radiolabel should disappear from rSso0417 with a half-life of ≤6 s under the conditions described in Table 2 if phosphotransfer was kinetically competent. In other words, ≤32% of the initial protein-bound [32P]phosphate should remain after a 10-s incubation in the presence of Mn2+ and 3-PGA. As can be seen from Table 4, the experimentally determined figure was ≤24%.
TABLE 4.
3-PGA induces the rapid disappearance of 32P radioactivity from autophosphorylated rSso0417a
| Incubation period | Addition
|
Amt of [32P]phosphate remaining
|
||
|---|---|---|---|---|
| 5 mM MnCl2 | 15 mM 3-PGA | Mean pmol ± SE | % Control | |
| 10 s | − | − | 3.6 ± 0.1 | 109 |
| 10 s | + | − | 3.7 ± 0.2 | 112 |
| 10 s | − | + | 3.3 ± 0.2 | 100 |
| 10 s | + | + | 0.8 ± 0.1 | 24 |
| 5 min | + | + | 0.4 ± 0.1 | 12 |
Portions, ∼0.4 μg each (7.5 pmol), of rSso0417 that had been autophosphorylated by using [γ-32P]ATP were incubated at 25°C in a volume of 50 μl of 25 mM HEPES (pH 7.5) containing the compounds listed at the indicated final concentrations. The incubation was terminated by the addition of 500 μl of ice-cold acetone, and the denatured protein was collected by centrifugation at 10,000 × g for 3 min. The superatant liquid was decanted, and the pellet was resuspended in 25 mM Tris (pH 7.5) containing 0.5 mM EDTA. Then, 30 μl of SDS-PAGE sample buffer was immediately added and the mixture was heated for 5 min at 100°C. The mixture was applied to an SDS-gel. After, electrophoresis, the amount of [32P]phosphate remaining bound to the protein was determined by electronic autoradiography. Shown are the results of triplicate analyses.
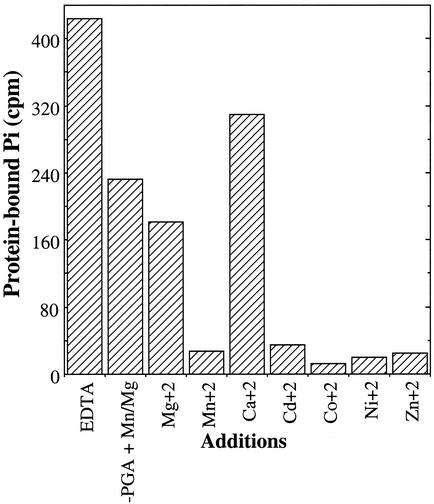
As was observed in steady-state assay of PGA mutase activity, Mn2+ and Cd2+ proved to be effective cofactors for the 3-PGA-induced disappearance of [32P]phosphate from autophosphorylated rSso0417, whereas Mg2+ proved relatively ineffective (Fig. 4). Assessing the role of Co2+ proved problematic, since this metal effectively supported the transfer of the enzyme-bound phosphoryl group to water when 3-PGA was not present. The same proved true for Ni2+ and Cu2+. Zn2+, on the other hand, facilitated substrate-dependent transfer much more readily than hydrolytic transfer to water, whereas Mg2+ had little discernible effect on phosphoenzyme hydrolysis.
FIG. 4.
Metal ion preference of rSso0417. rSso0417 was phosphorylated with [γ-32P]ATP to a stoichiometry of ∼0.5 mol/mol, purified, and then dialyzed against buffer containing 0.05 mM EDTA to remove any remaining metals. The phosphoenzyme was incubated with one of the metal ions listed, each at a concentration of 0.1 mM, for 5 min at a temprature of 65°C in the presence or absence of 3-PGA at 0.1 mM. The protein was then isolated by SDS-PAGE, and the amount of protein-bound [32P]phosphate determined by electronic autoradiography. The control represents enzyme that was incubated for 5 min at 65°C in the absence of both metals and 3-PGA. In prior experiments it was observed that incubation with 3-PGA in the absence of metals had no effect on enzyme-bound phosphate. For further details, see Materials and Methods.
DISCUSSION
Several lines of evidence indicate that the ∼46-kDa phosphoprotein previously detected in soluble extracts of S. solfataricus was the protein product of ORF sso0417. First, whereas the deduced protein product of ORF sso0417 represented an imperfect and incomplete match to the amino acid sequence data generated from the ∼46-kDa phosphoprotein, it plausibly accounted for a higher proportion of the sequence information obtained than any other component of the S. solfataricus genome. Second, the calculated mass of Sso0417, 45,179 Da, closely matched the estimated mass of the ∼46-kDa phosphoprotein from S. solfataricus extracts. Third, both proteins became phosphorylated when incubated with [γ-32P]ATP and did so on serine residues. The appearance of unaccounted-for peptides in the mass spectral analyses most likely reflected the presence of polypeptide contaminants in the sample. We suggest that the discrepancies noted between the empirically and genomically derived sequences for those peptides that displayed similarity to Sso0417 were the consequence of inaccuracies in amino acid assignments arising from the low levels of sample available for these analyses.
Although Sso0417 was originally annotated as a phosphonopyruvate decarboxylase (38), our own homology searches indicated that it contained plausible candidates for virtually all of the mechanistically critical residues found in bacterial and eucaryal iPGMs. The iPGMs, in turn, are part of a larger family of metal-requiring phosphotransferases and phosphohydrolases that includes the alkaline phosphatases (reviewed in references 18 and 27). Enzyme assays indicated that rSso0417 displayed measurable levels of both activities but was a markedly more efficient PGA mutase than phosphohydrolase. As has proven to be the case for established iPGMs, the PGA mutase activity of rSso0417 was dependent upon the presence of divalent metal ions such as Mn2+, Co2+, or Cd2+.
Portions of the amino acid sequence of this archaeal enzyme dramatically diverged from that of prototypic iPGMs from the Bacteria and Eucarya, specifically in the substrate-binding region, subdomain B. The specific activity of rSso0417, i.e., ∼0.52 μmol/min/mg when Co2+ was utilized as a cofactor, was also low relative to values reported for bacterial iPGMs. Even if one allows for the roughly 25-fold increase in activity that would take place if the assays been conducted at the growth temperature of S. solfataricus (∼75°C), the estimated specific activity of ∼16 μmol/min/mg still falls an order of magnitude below that determined for prototypic iPGMs from Bacillus megaterium (568 μmol/min/mg [40]) and Bacillus subtilis (621 μmol/min mg [43]). On the other hand, the estimated specific activity of rSso0417 falls near that reported for the iPGM from T. brucei (26 μmol/min mg [9]).
As the present study neared completion, two proteins with PGA mutase activity were characterized from the archaeons Pyrococcus furiosus and Methanococcus jannaschii (20, 42). Like Sso0417, the areas of similarity between these archaeal proteins and prototypical iPGMs from the Eucarya and Bacteria were restricted to domain A (Fig. 2). These similarities extended to the molecular masses of their component polypeptides (ca. 45 to 46 kDa) and their specific activities, an estimated 20 μmol/min mg at 90°C for the iPGM from P. furiosus and an estimated 80 μmol/min mg at 100°C for the iPGM from M. jannaschii (42). Although all three archaeal enzymes could utilize Mn2+, Co2+, or Zn2+ as cofactors in vitro, only the iPGMs from P. furiosus and M. jannaschii utilized Mg2+ as well.
The significance of Sso0417's most unusual property, the ability to autophosphorylate its catalytically essential serine residue using ATP as phosphoryl donor, remains cryptic. The greater efficiency with which the phosphoenzyme transferred its phosphoryl group to 3-PGA as opposed to glycerate is exactly the reciprocal of what would be predicted based on the presumed mechanism of catalysis for eucaryal and bacterial iPGMs. Such behavior mirrors, at least superficially, that of dPGMs in which 3- and 2-PGA are interconverted by a previously phosphorylated enzyme via the transient formation of 2,3-diPGA. However, this sequence of events may represent an adventitious and physiologically irrelevant side reaction peculiar to the phosphoenzyme generated in the laboratory. It must be remembered that the autophosphorylated enzyme utilized in these studies was produced by incubation with ATP, not the presumed substrate 3-PGA. Nor did catalysis by rSso0417 in the coupled assay system require the addition of ATP, as would be expected if this nucleotide triphosphate functioned as a priming cofactor in vivo.
On the other hand, archaeal iPGMs differ significantly in sequence from their better-characterized bacterial and eucaryal counterparts. It is not unreasonable to suggest that these differences extend to the level of catalytic mechanism as well. Autophosphorylated rSso0417 behaved in an apparently chemically and kinetically competent manner as a PGA mutase. Moreover, the differences between the mechanisms of dPGMs and prototypic iPGMs are relatively small, as it is the order rather than the chemical nature of the individual events that vary. Further studies are needed to definitively determine the sequence of steps that take place within the active sites of Sso0417 and other archaeal iPGMs and whether these differ from those of bacterial and eucaryal iPGMs. However, the ability to produce reagent quantities of the phosphoenzyme in vitro using ATP will provide a potent tool for examining the catalytic mechanism of this metabolically critical enzyme family.
Acknowledgments
This work was supported by grant MCB 0077484 from the National Science Foundation (P.J.K.) and an NSF and Alfred P. Sloan Foundation Postdoctoral Research Fellowship in Molecular Evolution (K.M.B.).
REFERENCES
- 1.Amend, J. P., and E. L. Shock. 2001. Energetics of overall metabolic reactions of hyperthemophilic Archaea and bacteria. FEMS Microbiol. Rev. 25:175-243. [DOI] [PubMed] [Google Scholar]
- 2.Blattler, W. A., and J. R. Knowles. 1980. Phosphoglycerate mutases: stereochemical course of phosphoryl group transfers catalyzed by cofactor-dependent enzyme from rabbit muscle and cofactor-independent enzyme from wheat germ. Biochemistry 19:738-743. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Breathnach, R., and J. R. Knowles. 1977. Phosphoglycerate mutase from wheat germ: studies with 18O-labeled substrate, investigations of phosphatase and phosphoryl transfer activities, and evidence for a phosphoryl-enzyme intermediate. Biochemistry 16:3054-3060. [DOI] [PubMed] [Google Scholar]
- 5.Britton, H. G., J. Carreras, and S. Grisolia. 1971. Mechanism of action of 2,3-diphosphoglycerate-independent phosphoglycerate mutase. Biochemistry 10:4522-4533. [DOI] [PubMed]
- 6.Chander, M., P. Setlow, E. Lamani, and M. J. Jedrzejas. 1999. Structural studies on a 2,3-diphosphoglycerate independent phosphoglycerate mutase from Bacillus stearothermophilus. J. Struct. Biol. 126:156-165. [DOI] [PubMed] [Google Scholar]
- 7.Chander, M., B. Setlow, and P. Setlow. 1998. The enzymatic activity of phosphoglycerate mutase from gram-positive endospore-forming bacteria requires Mn2+ and is pH sensitive. Can. J. Microbiol. 44:759-767. [DOI] [PubMed] [Google Scholar]
- 8.Chevalier, N., D. J. Rigden, J. Van Roy, F. R. Opperdoes, and P. A. M. Michels. 2000. Trypanosoma brucei contains a 2,3-bisphosphoglycerate independent phosphoglycerate mutase. Eur. J. Biochem. 267:1464-1472. [DOI] [PubMed] [Google Scholar]
- 9.Collet, J. F., V. Stroobant, and E. van Schaftingen. 2001. The 2,3-bisphosphoglycerate-independent phosphoglycerate mutase from Trypanosoma brucei: metal-ion dependency and phosphoenzyme formation. FEMS Microbiol. Lett. 204:39-44. [DOI] [PubMed] [Google Scholar]
- 10.Cooper, J. A., B. M. Sefton, and T. Hunter. 1983. Detection and quantification of phosphotyrosine in proteins. Methods Enzymol. 99:387-402. [DOI] [PubMed] [Google Scholar]
- 11.de Rosa, M. A., A. Gambacorta, and J. D. Bullock. 1975. Extremely thermophilic acidophilic bacteria convergent with Sulfolobus solfataricus. J. Gen. Microbiol. 86:156-164. [DOI] [PubMed] [Google Scholar]
- 12.Deppenmeier, U., T. Lienard, and G. Gottschalk. 1999. Novel reactions involved in energy conservation by methanogenic archaea. FEBS Lett. 457:291-297. [DOI] [PubMed] [Google Scholar]
- 13.Doolittle, W. F. 1999. Lateral genomics. Trends Cell Biol. 9:M5-M8. [PubMed] [Google Scholar]
- 14.Doolittle, R. F. 2000. Searching for the common ancestor. Res. Microbiol. 151:85-89. [DOI] [PubMed] [Google Scholar]
- 15.Fairbanks, G., T. L. Steck, and D. F. H. Wallach. 1971. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry 10:2606-2617. [DOI] [PubMed] [Google Scholar]
- 16.Fothergill-Gilmore, L. A., and H. C. Watson. 1989. The phosphoglycerate mutases. Adv. Enzymol. 62:227-313. [DOI] [PubMed] [Google Scholar]
- 17.Gaasterland, T. 1999. Archaeal genomics. Curr. Opin. Microbiol. 2:542-547. [DOI] [PubMed] [Google Scholar]
- 18.Galperin, M. Y., and M. J. Jadrzejas. 2001. Conserved core structure and active site residues of the alkaline phosphatase superfamily of enzymes. Proteins 45:318-324. [DOI] [PubMed] [Google Scholar]
- 19.Gatehouse, A., and J. R. Knowles. 1977. Phosphogylcerate mutase from wheat germ: studies with isotopically labeled 3-phospho-d-glycerates showing that the catalyzed reaction is intramolecular. Biochemistry 16:3045-3050. [DOI] [PubMed] [Google Scholar]
- 20.Graham, D. E., H. Xu, and R. H. White. 2002. A divergent archaeal member of the alkaline phosphatase binuclear metalloenzyme family has phosphoglycerate mutase activity. FEBS Lett. 517:190-194. [DOI] [PubMed] [Google Scholar]
- 21.Grisolia, S., and J. Carreras. 1975. Phosphoglycerate mutase from wheat germ (2,3-PGA-independent). Methods Enzymol. 42:429-435. [DOI] [PubMed] [Google Scholar]
- 22.Grisolia, S., B. K. Joyce, and M. Fernandez. 1961. Studies on the mechanism of action of phosphoglyceratemutases. Biochim. Biophys. Acta 50:81-89. [DOI] [PubMed] [Google Scholar]
- 23.Grogan, D. W. 2000. The question of DNA repair in hyperthermophilic archaea. Trends Microbiol. 8:180-185. [DOI] [PubMed] [Google Scholar]
- 24.Ito, N., and S. Grisolia. 1959. Phosphoglyceric acid mutase activity without added 2,3-diphosphoglycerate in preparations purified from wheat germ. J. Biol. Chem. 234:242-245. [PubMed] [Google Scholar]
- 25.Jacobs, R. J., and S. Grisolia. 1966. Phosphoryl intermediates formed with phosphoglycerate mutase: role and labilization of 2,3-diphosphoglycerate. J. Biol. Chem. 241:5926-5935. [PubMed] [Google Scholar]
- 26.Jedrzejas, M. J., M. Chander, P. Setlow, and G. Krishnasamy. 2000. Structure and mechanism of a novel phosphogylcerate mutase from Bacillus stearothermophilus. EMBO J. 19:1419-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jedrzejas, M. J., and P. Setlow. 2001. Comparison of the binuclear metalloenzymes diphosphoglycerate-independent phosphoglycerate mutase and alkaline phosphatase: their mechanism of catalysis via a phosphoserine intermediate. Chem. Rev. 101:607-618. [DOI] [PubMed] [Google Scholar]
- 28.Kuhn, N. J., B. Setlow, and P. Setlow. 1993. Manganese(II) activation of 3-phosphoglycerate mutase of Bacillus megaterium: pH-sensitive interconversion of active and inactive forms. Arch. Biochem. Biophys. 306:342-349. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Lanzetta, P. A., L. J. Alvarez, P. S. Reinach, and O. A. Candia. 1979. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100:95-97. [DOI] [PubMed] [Google Scholar]
- 31.Leng, J., A. J. Cameron, S. Buckel, and P. J. Kennelly. 1995. Isolation and cloning of a protein-serine/threonine phosphatase from an archaeon. J. Bacteriol. 177:6510-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lower, B. H., K. M. Bischoff, and P. J. Kennelly. 2000. The archaeon Sulfolobus solfataricus contains a membrane-associated protein kinase activity that preferentially phosphorylates threonine residues in vitro. J. Bacteriol. 182:3452-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin, J. B., and D. M. Doty. 1949. Determination of inorganic phosphate: modification of isobutyl alcohol procedure. Anal. Chem. 21:965-967. [Google Scholar]
- 34.Randerath, K., and E. Randerath. 1967. Thin-layer methods for nucleic acid derivatives. Methods Enzymol. 12(Pt. A):323-347. [Google Scholar]
- 35.Ray, W. J., Jr., and E. J. Peck, Jr. 1972. Phosphomutases, p. 407-477. In P. D. Boyer (ed.), The enzymes, vol. II. Academic Press, Inc., New York, N.Y.
- 36.Rigden, D. J., I. Bagyan, E. Lamani, P. Setlow, and M. J. Jedrzejas. 2001. A cofactor-dependent phosphoglycerate mutase from Bacillus stearothermophilus is actually a broad specificity phosphatase. Protein Sci. 10:1835-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scandurra, R., V. Consalvi, R. Chiaraluce, L. Politi, and P. C. Engel. 2000. Protein stability in extremophilic archaea. Front. Biosci. 5:D787-D795. [DOI] [PubMed] [Google Scholar]
- 38.She, Q., R. K. Singh, F. Confalonieri, Y. Zivanovic, G. Allard, M. J. Awayez, C. C.-Y. Chan-Weiher, G. Clausen, B. A. Curtis, A. De Moors, G. Erauso, C. Fletcher, P. M. K. Gordon, I. Heikamp-de Jong, A. C. Jeffries, C. J. Kozera, N. Medina, X. Peng, H. P. Thi-Ngoc, P. Redder, M. E. Schenk, C. Theriault, N. Tolstrup, R. L. Charlebois, W. F. Doolittle, M. Duguet, T. Gaasterland, R. A. Garrett, M. A. Ragan, C. W. Sensen, and J. Van der Oost. 2001. The complete genome of trhe crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA 98:7835-7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi, L., K. M. Bischoff, and P. J. Kennelly. 1999. The icfG gene cluster of Synechocystis sp. PCC6803 encodes an Rsb/Spo-like protein kinase, protein phosphatase, and two phosphoproteins. J. Bacteriol. 181:4761-4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh, R. P., and P. Setlow. 1979. Purification and properties of phosphoglycerate phosphomutase from spores and cells of Bacillus megaterium. J. Bacteriol. 137:1024-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solow, B., K. M. Bischoff, M. J. Zylka, and P. J. Kennelly. 1998. Archaeal phosphoproteins: identification of a hexosephosphate mutase and the α-subunit of succinyl-CoA synthetase in the extreme acidothermophile Sulfolobus solfataricus. Protein Sci. 7:105-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Oost, J., M. A. Huynen, and C. H. Verhees. 2002. Molecular characterization of phosphoglycerate mutase in archaea. FEMS Microbiol. Lett. 212:111-120. [DOI] [PubMed] [Google Scholar]
- 43.Watabe, K., and E. Freese. 1979. Purification and properties of the manganese-dependent phosphoglycerate mutase of Bacillus subtilis. J. Bacteriol. 137:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]