Abstract
Human and mouse cDNAs encoding a new β-1,3-N-acetylglucosaminyltransferase (β3GnT) have been isolated from fetal and newborn brain libraries. The human and mouse cDNAs included ORFs coding for predicted type II transmembrane polypeptides of 329 and 325 aa, respectively. The human and mouse β3GnT homologues shared 90% similarity. The β3GnT gene was widely expressed in human and mouse tissues, although differences in the transcript levels were visible, thus indicating possible tissue-specific regulation mechanisms. The β3GnT enzyme showed a marked preference for Gal(β1–4)Glc(NAc)-based acceptors, whereas no activity was detected on type 1 Gal(β1–3)GlcNAc and O-glycan core 1 Gal(β1–3)GalNAc acceptors. The new β3GnT enzyme was capable of both initiating and elongating poly-N-acetyllactosamine chains, which demonstrated its identity with the poly-N-acetyllactosamine synthase enzyme (E.C. 2.4.1.149), showed no similarity with the i antigen β3GnT enzyme described recently, and, strikingly, included several amino acid motifs in its protein that have been recently identified in β-1,3-galactosyltransferase enzymes. The comparison between the new UDP–GlcNAc:βGal β3GnT and the three UDP–Gal:βGlcNAc β-1,3-galactosyltransferases-I, -II, and -III reveals glycosyltransferases that share conserved sequence motifs though exhibiting inverted donor and acceptor specificities. This suggests that the conserved amino acid motifs likely represent residues required for the catalysis of the glycosidic (β1–3) linkage.
Glycosyltransferases are grouped in families defined by the type of carbohydrate donor transferred and the glycosidic linkage catalyzed (1). The relevance of this classification system is being confirmed at the genetic level as it is often found that genes encoding glycosyltransferases of the same family share similar primary and genomic structures (2–4). However, exceptions to this rule have been documented, as exemplified by β-1,6-N-acetylglucosaminyltransferase enzymes, which share little resemblance to each other (reviewed in ref. 5). Structurally related enzymes that transfer different donor molecules represent another kind of exception. For example, the blood group A and B enzymes differ by only four amino acids but catalyze either the α-1,3 transfer of Gal or GalNAc to galactosylated acceptors, respectively (6). A similar case has been reported with the identification of a β-1,4-N-acetylglucosaminyltransferase from Lymnaea stagnalis that is homologous to mammalian β-1,4-galactosyltransferases (7). However, although the donor molecules differ, these homologous glycosyltransferases still share common acceptor substrates. We now provide another exception to the list by presenting a glycosyltransferase that is structurally related to the family of β-1,3-galactosyltransferases (β3GalT) but uses distinct donor and acceptor substrates.
Similarity searches in the expressed sequence tag (EST) division of the GenBank database have revealed the existence of several ORFs encoding proteins that share conserved motifs with β3GalT enzymes (8–10). Among those ORFs, we have isolated a full-length cDNA that includes all of the conserved motifs identified in the four β3GalT enzymes characterized to date. Surprisingly, this cDNA was found to encode a UDP-GlcNAc:Gal(β1–4)Glc(NAc) β3GnT enzyme that was able to catalyze both the initiation and elongation of N-acetyllactosamine chains, thereby enabling the formation of poly-N-acetyllactosaminoglycans and showed no structural similarity to the i-antigen β3GnT (iGnT) enzyme reported by others (11).
MATERIALS AND METHODS
Isolation of Human and Mouse β3GnT cDNAs.
EST fragments similar to the mouse β3GalT-I, -II, and -III enzymes were retrieved from the EST division of GenBank by using the tblastx algorithm (12). The human and mouse β3GnT cDNAs were isolated from bacteriophage λ libraries of human fetal brain cDNA (CLONTECH) and mouse newborn brain cDNA (Stratagene) by using a 367-bp fragment derived from the AA150140 EST as a probe. The probing fragment was generated by using PCR with 50 ng of human T cell cDNA as template with the primers 5′-GCGACTACTACCTGCCCTACG-3′ and 5′-CTCCCTTCTCTGGCAAGCACT-3′ for 30 cycles at 95°C for 45 s, 58°C for 30 s, and 72°C for 45 s. Once purified and subcloned into the pBluescript vector (Stratagene), the cDNAs of interest were sequenced by primer walking by using the AmpliTaq FS Dye Terminator sequencing kit (Perkin-Elmer/Applied Biosystems).
Cloning and Expression of Recombinant Baculoviruses.
A EcoRI–XhoI 1.9-kbp fragment isolated from a partial human β3GnT cDNA lacking the transmembrane domain was subcloned into the FastBac-HTc vector (Life Technologies) linearized with EcoRI–XhoI. The mouse β3GnT gene was subcloned as a 1.0-kbp StuI–StuI fragment, representing the protein-coding region devoid of transmembrane domain, into the FastBac-HTc vector. Recombinant baculoviruses were generated by transposon-mediated recombination (13) following the recommendations of the manufacturer. Sf9 insect cells were infected at a multiplicity of 10 and further incubated at 27°C before assaying for β3GnT activity.
Glycosyltransferase Activity Assays.
All donor and acceptor substrates were from Sigma, except for Gal(β1–4)GlcNAc(β1-OpNP) (pNP = p-nitrophenyl), which was purchased from Toronto Research Chemicals (Downsview, ON, Canada) and the O(CH2)8CO2Me-derivatized (Mco) acceptors, which were kindly provided by Markus Streiff (Novartis, Basel, Switzerland). Sf9 cells (5 × 106) infected with wild-type or with recombinant β3GnT baculoviruses were lysed 72 hr postinfection in 1 ml of 2% Triton X-100 in phosphate-buffered saline on ice for 15 min. The cytoplasmic and solubilized membrane fractions were recovered by centrifugation at 300 × g for 5 min. β3GnT activity was assayed by using 10 μl of cell lysate in 50 μl of 50 mM cacodylate buffer (pH 7.0), 20 mM MnCl2, 5% Me2SO, 0.75 mM ATP, 0.5 mM UDP–GlcNAc including 5 × 104 cpm of UDP–[14C]GlcNAc (Amersham) and various acceptors (see Table 1). Galactosyltransferase and N-acetylgalactosaminyltransferase actvivity were assayed as above using UDP–Gal and UDP–GalNAc as donor molecules. Reaction products were quantified as described (9, 14).
Table 1.
Acceptor substrate specificity of the β3GnT enzyme
| Acceptors | pmol/min/mg prot added
|
|
|---|---|---|
| Sf9 mock* | β3GnT† | |
| UDP–GlcNAc‡ | ||
| Gal(β1–4)GlcNAc(β1–OpNP) | 6 | 1,478 |
| Gal(β1–4)Glc(β1–OBn) | 4 | 1,034 |
| Gal(β1–4)GlcNAc(octyl) | 5 | 284 |
| Gal(β1–3)GlcNAc(octyl) | 5 | 8 |
| Gal(β1–3)GalNAc(octyl) | 5 | 4 |
| Gal(α1–OpNP) | 4 | 89 |
| Gal(β1–OpNP) | 4 | 26 |
| GalNAc(α1–OBn) | 4 | 5 |
| GalNAc(β1–OBn) | 5 | 6 |
| GlcNAc(α1–OBn) | 4 | 4 |
| GlcNAc(β1–OBn) | 11 | 7 |
| GlcNAc(β1–3)GalNAc(α1–OBn) | 8 | 5 |
| Gal(β1–4)Glc | 13 | 502 |
| Gal(β1–4)GlcNAc | 14 | 714 |
| Lac-N-neo-tetraose§ | 14 | 748 |
| Lac-N-tetraose§ | 13 | 45 |
| UDP–Gal‡ | ||
| GalNAc(α1–OBn) | 170 | 190 |
| GalNAc(β1–OBn) | 15 | 18 |
| GlcNAc(α1–OBn) | 11 | 10 |
| GlcNAc(β1–OBn) | 23 | 22 |
| UDP–GalNAc‡ | ||
| Gal(α1–OpNP) | 3 | 5 |
| Gal(β1–OpNP) | 3 | 5 |
| GlcNAc(α1–OBn) | 3 | 3 |
| GlcNAc(β1–OBn) | 27 | 31 |
All acceptors were assayed at 5 mM except the (Mco)-derivatized acceptors, which were assayed at 2 mM.
Lysate of Sf9 cells infected with a wild-type baculovirus.
Lysate of Sf9 cells infected with a recombinant baculovirus expressing the human β3GnT enzyme.
Carbohydrate donor.
Lac-N-neo-tetraose, Gal(β1–4)GlcNAc(β1–3)Gal(β1–4)Glc; Lac-N-tetraose, Gal(β1–3)GlcNAc(β1–3)Gal(β1–4)Glc
β3GnT Gene Expression in Human and Mouse Tissues.
The β3GnT mRNAs were detected by using Northern blot analysis with commercially available multiple-tissue poly(A)+ RNA blots (CLONTECH). A SacI–PstI 668-bp fragment from the mouse β3GnT cDNA and a 367-bp region of the human β3GnT cDNA between nucleotides 616 and 983 were labeled with [α-32P]CTP (Hartmann Analytics, Braunschweig, Germany) by random priming and hybridized to the poly(A)+ RNA blots. Blots were washed in 0.1× SSC, 0.1% SDS heated to 55°C and exposed for 4 days between intensifying screens at −70°C.
Flow Cytometry.
A 1.7-kbp fragment including the human β3GnT cDNA was subcloned into the pcDNA3.1 plasmid (Invitrogen) opened with BamHI and XhoI. HeLa cells were transfected with 5 μg of pcDNA3.1-β3GnT plasmid by using LipofectAmine (Life Technologies). Three days later, cells were stained with Lycopersicon esculentum (tomato)-fluorescein isothiocyanate lectin (1 μg/ml; Vector Laboratories). Alternatively, transfected cells were stained with human anti-i antiserum as described (11). Fluorescence was analyzed on a FACScan flow cytometer and cellquest software (Becton Dickinson).
HPLC.
The carbohydrate-containing fraction of a 1-ml incubation using 10 mM Gal(β1–4)Glc[β1-OBn(benzyl)] as acceptor and 100 μl of β3GnT-expressing Sf9 cell lysate as enzyme source was prepurified through a Sep-Pak C18 cartridge and purified by using HPLC with a Lichrosorb-NH2 column (4.6 × 250 mm, Chrompack, Bergen op Zoom, The Netherlands) at a flow rate of 2 ml/min by using a Kratos SF 400 HPLC system (ABI Analytical, Kratos Division). The column was eluted isocratically with solvent A (aqueous 80% acetonitrile), followed by a linear gradient from 100% solvent A/0% solvent B (aqueous 20% acetonitrile) to 65% solvent A/35% solvent B in 18 min. The effluent was monitored at 257 nm by using a 757 absorbance detector (Applied Biosystems).
Methylation Analysis.
Permethylation was performed essentially as described by Ciucanu et al. (15). After hydrolysis with 2 M trifluoroacetic acid (2 hr, 120°C), the mixture of partially methylated monosaccharides was reduced with 0.5 M sodium borodeuteride (2 hr) and neutralized. Boric acid was removed, and the mixture was acetylated with acetic anhydride (3 hr, 120°C). The partially methylated alditol acetates were analyzed by GLC/electron-impact MS (16, 17) on a Fisons MD800/8060 system (electron energy = 70 eV) equipped with a CPSil5 fused silica capillary column (25 m × 0.32 mm, Chrompack) by using a temperature program of 130–240°C at 4°C/min.
Matrix-Assisted Laser Desorption Ionization (MALDI) Mass Spectrometry.
Positive-ion mode MALDI-TOF (time-of-flight) MS analysis of permethylated oligosaccharide was performed on a Voyager-DE (PerSeptive Biosystems, Framingham, MA) instrument operating at an accelerating voltage of 24 kV (grid voltage 92.5%, ion guide wire voltage 0.01%) and equipped with a VSL-337ND-N2 laser. The sample was dissolved in methanol (1 μg/μl) and subsequently mixed in the sample well with 2,5-dihydroxybenzoic acid (10 mg/ml in H2O) at a ratio of 1:2. Linear mass scans were recorded over 1,000 Da by using a pulse delay time of 90 ns. Recorded data were processed by using grams/386 software (v. 3.04, Galactic Industries, Salem NH).
NMR Spectroscopy.
Before analysis, the oligosaccharide sample was repeatedly exchanged in 2H2O (99.9 atom % 2H, Isotec) with intermediate lyophilization and finally dissolved in 450 μl of 2H2O (99.96 atom % 2H, Isotec). Resolution-enhanced 1H one-dimensional and two-dimensional NMR spectra were recorded on a Bruker DRX-600 instrument (NSR Center, Nijmegen University) at probe temperatures of 300 K. Chemical shifts (δ) were expressed in parts per million relative to internal acetate (δ 1.908; acetone, δ 2.225). HO2H signal suppression was achieved by applying a water-eliminated Fourier transform pulse sequence (18) in one-dimensional 1H experiments and by presaturation for 1 s in two-dimensional experiments. Two-dimensional total correlation spectroscopy (TOCSY) spectra were recorded by using MLEV-17 mixing sequences with effective spin-lock times between 20 and 100 ms. Two-dimensional rotating frame nuclear Overhauser effect spectroscopy (ROESY) spectra were recorded with a mixing time of 250 ms. The spin-lock field strength corresponded to a 90° pulse of about 120 μs. 1H one-dimensional and two-dimensional spectra were processed on Silicon Graphics IRIS workstations (Indigo 2 and O2) by using Bruker uxnmr software (Bijvoet Center, Department of NMR Spectroscopy).
RESULTS
While searching for sequences related to the β3GalT-I, -II, and -III enzymes (9) in the EST division of GenBank, we found several entries corresponding to distinct ORFs that shared sequence similarity with the β3GalT enzymes. The 399-bp human EST fragment AA150140, which included the β3GalT-motif EDVx[V/L]G, was therefore considered as a potential member of the β3GalT gene family. The AA150140 EST was part of the Hs.48730 cluster of the UniGene collection (19), which represents sequences obtained from uterus, colon, tonsil, and brain tissues. By using the AA150140 fragment as a probe, we have isolated full-length cDNAs from human and mouse brain cDNA libraries. The human and mouse cDNAs included each an ORF of 990 bp and 978 bp, which encoded proteins of 329 and 325 amino acids, respectively (Fig. 1). The encoded proteins were referred to as the human and mouse β3GnT. Noteworthy, the human and mouse ORFs had an elevated G/C content of 75% and 67%, respectively. Two especially G/C rich clusters, reaching 82% and 74%, were both found in the human and mouse cDNAs between nucleotide positions 1–350 and 500–600. The human and mouse β3GnT proteins shared 83% identity and 90% similarity, whereas the highest divergence was observed in the N-terminal 1/3 of the amino acid sequences. A Kyte–Doolittle hydropathy analysis (20) revealed a potential transmembrane domain of 19 amino acids located at the N terminus of both human and mouse isoforms, thereby supporting a type II transmembrane topology typical of most glycosyltransferase enzymes (21).
Figure 1.
Primary structure and deduced amino acid sequence of the human and mouse β3GnT cDNA. Amino acids identical in both species are in boldface. The predicted transmembrane region is shaded and the single potential N-glycosylation site (N-X-[S/T]) is underlined.
A comparison between the β3GnT studied here and the previously characterized β3GalT proteins revealed that, whereas the overall identity was only between 15 and 19%, various motifs were conserved (Fig. 2). In addition to the EDVx[V/L]G stretch noticed in the AA150140 EST, all of the motifs identified previously in the four β3GalT proteins were found at similar locations along the β3GnT amino acid sequence. By contrast, the cysteine residues of the β3GnT protein could not be aligned with those of β3GalT enzymes (Fig. 2).
Figure 2.
clustalw alignment of mouse β3GalT-I, -II, -III, and -IV and mouse β3GnT proteins. Conserved residues are shaded. The white arrows mark the positions of the cysteine residues conserved among β3GalT proteins. The black arrow shows the position of the cysteines conserved in the five proteins.
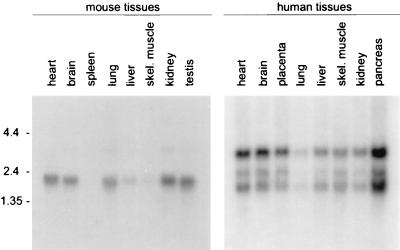
The β3GnT gene was expressed in all human tissues examined as determined by using Northern blot analysis (Fig. 3). Three transcripts were detected at 1.6, 2.4, and 3.3 kb, with the 3.3-kb mRNA producing the strongest hybridization signal. The intensity of the signal varied among the tissues examined, indicating differences in the level of β3GnT gene expression in human tissues. In the mouse, a major 2.2-kb transcript was detected in most tissues and a minor 3.7-kb mRNA was detected in lung and kidney tissues (Fig. 3). No hybridization signal was detected in spleen and moderate-to-weak signals were found in liver and skeletal muscle, respectively.
Figure 3.
Expression pattern of the β3GnT gene in adult human and mouse tissues as determined by using Northern blot analysis. Each lane represents about 2 μg of poly(A)+ RNA. At the left, the size of the RNA markers is indicated in kilobases.
The mouse and human β3GnT proteins were expressed in Sf9 insect cells as recombinant baculoviruses. The β3GnT enzyme was able to transfer GlcNAc to Gal(α1-OpNP), Gal(β1-OpNP) but much more efficiently to Gal(β1–4)Glc(NAc)-based acceptors (Table 1). No activity was detected toward type 1 Gal(β1–3)GlcNAc-, O-glycan core-1 Gal(β1–3)GalNAc-, and GlcNAc- and GalNAc-based acceptors (Table 1). The strict requirement for type 2 Gal(β1–4)Glc(NAc) acceptors indicated that this β3GnT protein was distinct from the core-3 β3GnT enzyme (EC 2.1.4.147) and the elongation core-1 β3GnT enzyme (EC 2.4.1.146). Although structurally related to the β3GnT protein, the previously characterized β3GalT-I to -III enzymes did not exhibit any N-acetylglucosaminyltransferase activity (9). The β3GnT enzyme showed an absolute requirement for the divalent cation Mn2+ and a pH optimum at 7.0 (data not shown). The Km values for the UDP–GlcNAc donor and for the Gal(β1–4)Glc(β1-OBn) acceptor were 0.2 mM and 6.6 mM, respectively. These values were in the range of those obtained with the β3GnT poly-N-acetyllactosamine extension enzyme purified from calf serum by Kawashima et al. (22). This similitude suggested that this β3GnT enzyme may indeed represent a poly-N-acetyllactosamine synthase. In fact, β3GnT catalyzed both the initiation and the elongation of poly-N-acetyllactosamine chains as it transferred GlcNAc to the tetrasaccharide acceptor Gal(β1–4)GlcNAc(β1–3)Gal(β1–4)Glc (lac-N-neo-tetraose) as efficiently as to a single N-acetyllactosamine unit (Table 1). By contrast, the β3GnT enzyme could not utilize the tetrasaccharide Gal(β1–3)GlcNAc(β1–3)Gal(β1–4)Glc (lac-N-tetraose) acceptor, confirming the adverse effect of the (β1–3) linkage between the terminal Gal and the penultimate GlcNAc onto the elongation activity. Also, the β3GnT enzyme did not exhibit significant Gal transferase and GalNAc transferase activity in comparison to the transferase activity measured with UDP–GlcNAc as donor (Table 1).
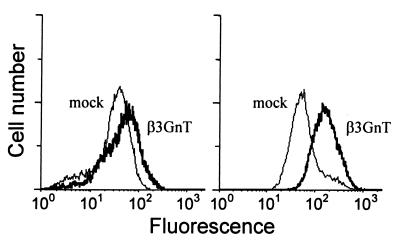
The relation between the β3GnT enzyme and synthesis of polylactosamine chains in vivo was investigated by analyzing the levels of i-antigen and tomato-lectin ligand on the surface of HeLa cells transiently transfected with an β3GnT-expression vector. The i-antigen represents repeated (β1–3)-linked N-acetyllactosamine units (23) and tomato lectin binds to N-acetylglucosamine oligomers (24). As shown by flow cytometry results, the overexpression of the β3GnT cDNA in HeLa resulted in a slight increased reactivity for tomato lectin and a more pronounced elevation of i-antigen on the cell surface (Fig. 4), thus indicating the identity of this β3GnT protein with the poly-N-acetyllactosamine synthase enzyme (EC 2.4.1.149).
Figure 4.
Flow cytometry analysis of HeLa cells. HeLa cells transfected with an empty pcDNA3.1 vector (mock) or with the β3GnT expression vector (β3GnT) were stained with tomato lectin (Left) or anti-i antiserum (Right).
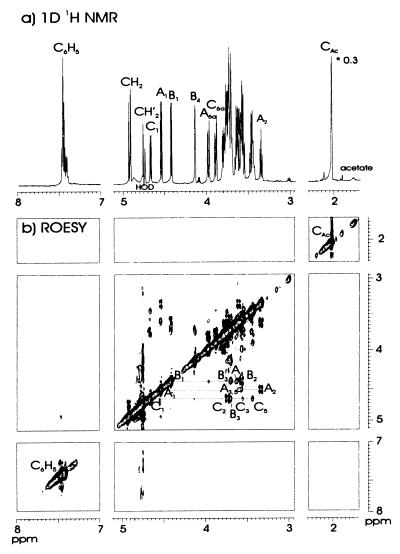
To prove the nature of the glycosidic linkage in the product formed by this novel glycosyltransferase, a large-scale incubation was performed with Gal(β1–4)Glc(β1-OBn) as acceptor. The isolated product from the HPLC trisaccharide region was analyzed along different routes. Its molecular mass as determined (after permethylation) by positive-ion mode MALDI TOF mass spectrometry indicated a trisaccharide with the brutoformula HexNAcHex2Bn (m/z 798, [M+Na]+). Methylation analysis revealed the presence of 4-substituted Glc, 3-substituted Gal, and terminal GlcNAc, demonstrating a (1–3)-linkage between GlcNAc and Gal. To confirm this conclusion, and to establish the anomeric configuration of the GlcNAc(1–3)Gal linkage, one-dimensional and two-dimensional 1H NMR experiments were performed. The most downfield signals in the one-dimensional 1H NMR spectrum (Fig. 5a) of the trisaccharide belonged to the benzyl group (δ 7.431, 4.930 (Jgem 11.6 Hz), 4.755 (Jgem 11.6 Hz). By means of two-dimensional TOCSY (20 and 100 ms) and ROESY experiments, almost all monosaccharide resonances in the one-dimensional spectrum could be assigned (Table 2), and the three anomeric doublets present at δ 4.673 (3J1, 2 8.6 Hz), 4.546 (3J1, 2 8.0 Hz), and 4.423 (3J1, 2 8.0 Hz) were attributed to β-GlcNAc, β-Glc, and β-Gal, respectively. The ROESY spectrum (200 ms, Fig. 5b) permitted the determination of the two glycosidic linkages. The anomeric track of the Gal residue revealed, besides two intraresidual crosspeaks [Gal H-1, H-3, δ 3.71; Gal H-1, H-2 (TOCSY transfer), δ 3.573], an interresidual crosspeak between Gal H-1 and Glc H-4 (δ 3.639). The anomeric track of the Glc residue revealed three intraresidual crosspeaks at δ 3.604 (Glc H-1, H-3), 3.573 (Glc H-1, H-5), and 3.338 [Glc H-1, H-2 (TOCSY transfer)]. Finally, the anomeric track of the GlcNAc residue showed besides three intraresidual crosspeaks [GlcNAc H-1, H-3, δ 3.557; GlcNAc H-1, H-5, δ 3.446; GlcNAc H-1, H-2 (TOCSY transfer), δ 3.74], an interresidual crosspeak between GlcNAc H-1 and Gal H-3 (δ 3.71). In summary, the combined results indicated the formed product to be GlcNAc(β1–3)Gal(β1–4)Glc(β1-OBn).
Figure 5.
600-MHz one-dimensional (a) and two-dimensional (b) ROESY (200 ms) 1H NMR spectra of the isolated trisaccharide GlcNAc(β1–3)Gal(β1–4)Glc(β1-OBn) (C-B-A), recorded at 300 K.
Table 2.
1H NMR data [δ, ppm; J, Hz (in parentheses)] of GlcNAc(β1–3)Gal(β1–4)Glc(β1–OBn), at 300 K, prepared with the human β3GnT enzyme
| Proton* | Residue
|
|||
|---|---|---|---|---|
| Benzyl | β–Glc | β–Gal | β–GlcNAc | |
| C6H5 | 7.431 | — | — | — |
| CH2 | 4.930 (11.6) | — | — | — |
| 4.755 (11.6) | — | — | — | |
| H–1 | — | 4.546 (8.0) | 4.423 (8.0) | 4.673 (8.6) |
| H–2 | — | 3.338 (8.3) | 3.573 (8.9) | 3.74 |
| H–3 | — | 3.604 | 3.71 | 3.557 |
| H–4 | — | 3.639 | 4.135 (2.8) | 3.461 |
| H–5 | — | 3.573 | 3.73 | 3.446 |
| H–6a | — | 3.974 | nd | 3.885 |
| H–6b | — | 3.79 | nd | 3.75 |
| NAc | — | — | — | 2.028 |
Based on data here, the assignment of H–6a and H–6b may have to be interchanged. nd, not determined.
In ppm relative to the signal of internal acetate (δ 1.908; acetone, δ 2.225).
DISCUSSION
We report the isolation of human and mouse β3GnT cDNAs encoding a poly-N-acetyllactosamine synthase activity. The β3GnT protein described here resembles β3GalT enzymes, as both kinds of glycosyltransferases share several conserved motifs. These motifs are spread across the polypeptide chain and are not clustered in distinct regions as observed for example in sialyltransferases (25). This suggests that the conserved motifs do not directly represent a linear binding site for donor or acceptor molecules but rather shape a larger catalytic domain that directs the formation of a (β1–3) glycosidic linkage. Preliminary data obtained from a mutagenesis study confirm the essential role of these conserved motifs in catalyzing the β-1,3-glycosyltransferase activity (A.D., unpublished results). The conservation between the β3GnT and β3GalT enzymes suggests a common evolutionary origin; structural requirements for the catalysis of a (β1–3) glycosidic linkage probably maintained the motifs in the evolving polypeptides.
The establishment of a pattern of (β1–3)-specific motifs should facilitate the recognition of further glycosyltransferases catalyzing a (β1–3) linkage such as the O-glycan core-1 β3GalT and the core-3 β3GnT enzymes. In this respect, it is remarkable that the iGnT described recently (11) does not present any homology with the β3GnT enzyme described here. A functional comparison between both enzymes is difficult to establish based on the data published (11). In fact, different expression systems were applied, which resulted in a great difference at the level of the β3GnT activity measured. It appears that the β3GnT reported here and the poly-N-acetyllactosamine extension enzyme purified by Kawashima et al. (22) likely represent the same enzyme. Closely related enzymatic properties and identical acceptor substrate specificities strengthen this hypothesis in spite of the difference in size reported for both enzymes. The poly-N-acetyllactosamine extension enzyme purified from calf serum has an apparent Mr of 70,000, whereas the relative mass of β3GnT averages 37,000. The cause of this discrepancy is unclear. Extensive posttranslational modifications such as glycosylation of the calf serum enzyme are unlikely to account for such an increase in mass. Alternatively, the secreted serum enzyme may be a disulfide-bridged homodimer. This possibility cannot be ruled out because the mass of the calf serum enzyme was determined under nonreducing conditions (22).
The existence of multiple enzymes capable of synthesizing poly-N-acetyllactosamine chains may indicate a selectivity at the level of the kind of glycoconjugates processed. Poly-N-acetyllactosamine chains occur on glycolipids and glycoproteins as well as on proteoglycans in the form of keratan sulfate (26). It is worth noting that keratan sulfate is the only glycosaminoglycan that is linked to its protein core via O- or N-linkage as found in glycoproteins. It shall be of interest to investigate whether β3GnT and the iGnT enzymes can both synthesize poly-N-acetyllactosamine chains on proteoglycans with the same efficiency as on glycoproteins.
The extent of poly-N-acetyllactosamine chains on tumor cells has directly been related to their metastatic potential (27). It has been previously implied that the branching enzymes β-1,6-N-acetylglucosaminyltransferase-V (28) and core-2 β-1,6-N-acetylglucosaminyltransferase (29) control the generation of poly-N-acetyllactosamine chains by providing the (β1–6) branch favored for initiation of poly-N-acetyllactosamine formation (30, 31). The present Northern blot analysis revealed a variability at the level of β3GnT transcripts detected in human and mouse tissues. This observation suggests that the expression of the β3GnT gene may be regulated to some extent by tissue-specific promoters. This raises the possibility that the formation of poly-N-acetyllactosaminoglycans may in part be modulated by the availability of the β3GnT enzyme itself. Along this line, it will be interesting to investigate the relation between the expression of the β3GnT gene in tumor cells and the emergence of a metastatic phenotype.
Acknowledgments
We thank Minoru Fukuda for generously providing us with the anti i-antigen antiserum. This work was funded by a Roche Research Foundation grant to T.H., by the Swiss National Science Foundation Grant 3100-046836.96 to E.G.B. and T.H., and by the European Union Grant BIO4-CT95-0138 to J.P.K., J.F.G.V., and E.G.B.
ABBREVIATIONS
- β3GalT
β-1,3-galactosyltransferase
- β3GnT
β-1,3-N-acetylglucosaminyltransferase
- iGnT
i-antigen β-1,3-N-acetylglucosaminyltransferase
- EST
expressed sequence tag
- Mco
O(CH2)8CO2Me
- Bn
benzyl
- Lac-N-tetraose
Gal(β1–3)GlcNAc(β1–3)Gal(β1–4)Glc
- Lac-N-neo-tetraose
Gal(β1–4)GlcNAc(β1–3)Gal(β1–4)Glc
- TOCSY
total correlation spectroscopy
- ROESY
rotating frame nuclear Overhauser effect spectroscopy
Footnotes
References
- 1.Kleene R, Berger E G. Biochim Biophys Acta. 1993;1154:283–325. doi: 10.1016/0304-4157(93)90003-7. [DOI] [PubMed] [Google Scholar]
- 2.Harduin-Lepers A, Recchi M-A, Delannoy P. Glycobiology. 1995;5:741–758. doi: 10.1093/glycob/5.8.741. [DOI] [PubMed] [Google Scholar]
- 3.Reguigne-Arnould I, Couillin P, Mollicone R, Faure S, Fletcher A, Kelly R J, Lowe J B, Oriol R. Cytogenet Cell Genet. 1995;71:158–162. doi: 10.1159/000134098. [DOI] [PubMed] [Google Scholar]
- 4.Hagen F K, Nehrke K. J Biol Chem. 1998;273:8268–8277. doi: 10.1074/jbc.273.14.8268. [DOI] [PubMed] [Google Scholar]
- 5.Van den Eijnden D H, Joziasse D H. Curr Opin Struct Biol. 1993;3:711–721. [Google Scholar]
- 6.Yamamoto F, Clausen H, White T, Marken J, Hakomori S. Nature (London) 1990;345:229–233. doi: 10.1038/345229a0. [DOI] [PubMed] [Google Scholar]
- 7.Bakker H, Agterberg M, Van Tetering A, Koeleman C A, Van den Eijnden D H, Van Die I. J Biol Chem. 1994;269:30326–30333. [PubMed] [Google Scholar]
- 8.Miyazaki H, Fukumoto S, Okada M, Hasegawa T, Furukawa K. J Biol Chem. 1997;272:24794–24799. doi: 10.1074/jbc.272.40.24794. [DOI] [PubMed] [Google Scholar]
- 9.Hennet T, Dinter A, Kuhnert P, Mattu T S, Rudd P M, Berger E G. J Biol Chem. 1998;273:58–65. doi: 10.1074/jbc.273.1.58. [DOI] [PubMed] [Google Scholar]
- 10.Kolbinger F, Streiff M B, Katopodis A G. J Biol Chem. 1998;273:433–440. doi: 10.1074/jbc.273.1.433. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki K, Kurata-Miura K, Ujita M, Angata K, Nakagawa S, Sekine S, Nishi T, Fukuda M. Proc Natl Acad Sci USA. 1997;94:14294–14299. doi: 10.1073/pnas.94.26.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul S F, Boguski M S, Gish W, Wootton J C. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- 13.Luckow V A, Lee S C, Barry G F, Olins P O. J Virol. 1993;67:4566–4579. doi: 10.1128/jvi.67.8.4566-4579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malissard M, Borsig L, Di Marco S, Grütter M G, Kragl U, Wandrey C, Berger E G. Eur J Biochem. 1996;239:340–348. doi: 10.1111/j.1432-1033.1996.0340u.x. [DOI] [PubMed] [Google Scholar]
- 15.Ciucanu I, Kerek F. Carbohydr Res. 1984;131:209–217. [Google Scholar]
- 16.Kamerling J P, Vliegenthart J F G. In: Clinical Biochemistry–Principles, Methods, Applications. Lawson A M, editor. Vol. 1. Berlin: de Gruyter; 1989. pp. 176–263. [Google Scholar]
- 17.Jansson P-E, Kenne L, Liedgren H, Lindberg B, Lönngren J. Chem Commun (Stockholm Univ) 1976;8:1–74. [Google Scholar]
- 18.Hård K, Van Zadelhoff G, Moonen P, Kamerling J P, Vliegenthart J F G. Eur J Biochem. 1992;209:895–915. doi: 10.1111/j.1432-1033.1992.tb17362.x. [DOI] [PubMed] [Google Scholar]
- 19.Schuler G D. J Mol Med. 1997;75:694–698. doi: 10.1007/s001090050155. [DOI] [PubMed] [Google Scholar]
- 20.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 21.Paulson J C, Colley K J. J Biol Chem. 1989;264:17615–17618. [PubMed] [Google Scholar]
- 22.Kawashima H, Yamamoto K, Osawa T, Irimura T. J Biol Chem. 1993;268:27118–27126. [PubMed] [Google Scholar]
- 23.Feizi T, Childs R A, Watanabe K, Hakomori S-I. J Exp Med. 1979;149:975–980. doi: 10.1084/jem.149.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merkle R K, Cummings R D. J Biol Chem. 1987;262:8179–8189. [PubMed] [Google Scholar]
- 25.Livingston B D, Paulson J C. J Biol Chem. 1993;268:11504–11507. [PubMed] [Google Scholar]
- 26.Bhavanandan V P, Davidson E A. In: Proteoglycans: Structure, Synthesis, Function. Allen H J, Kisailus E C, editors. New York: Dekker; 1992. pp. 167–202. [Google Scholar]
- 27.Dennis J W, Laferte S, Waghorne C, Breitman M L, Kerbel R S. Science. 1987;236:582–585. doi: 10.1126/science.2953071. [DOI] [PubMed] [Google Scholar]
- 28.Shoreibah M, Perng G S, Adler B, Weinstein J, Basu R, Cupples R, Wen D, Browne J K, Buckhaults P, Fregien N, Pierce M. J Biol Chem. 1993;268:15381–15385. [PubMed] [Google Scholar]
- 29.Bierhuizen M F, Fukuda M. Proc Natl Acad Sci USA. 1992;89:9326–9330. doi: 10.1073/pnas.89.19.9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heffernan M, Lotan R, Amos B, Palcic M, Takano R, Dennis J W. J Biol Chem. 1993;268:1242–1251. [PubMed] [Google Scholar]
- 31.Higgins E A, Siminovitch K A, Zhuang D L, Brockhausen I, Dennis J W. J Biol Chem. 1991;266:6280–6290. [PubMed] [Google Scholar]