Abstract
The enzyme ornithine carbamoyltransferase (OTCase) of Moritella abyssi (OTCaseMab), a new, strictly psychrophilic and piezophilic bacterial species, was purified. OTCaseMab displays maximal activity at rather low temperatures (23 to 25°C) compared to other cold-active enzymes and is much less thermoresistant than its homologues from Escherichia coli or thermophilic procaryotes. In vitro the enzyme is in equilibrium between a trimeric state and a dodecameric, more stable state. The melting point and denaturation enthalpy changes for the two forms are considerably lower than the corresponding values for the dodecameric Pyrococcus furiosus OTCase and for a thermolabile trimeric mutant thereof. OTCaseMab displays higher Km values for ornithine and carbamoyl phosphate than mesophilic and thermophilic OTCases and is only weakly inhibited by the bisubstrate analogue δ-N-phosphonoacetyl-l-ornithine (PALO). OTCaseMab differs from other, nonpsychrophilic OTCases by substitutions in the most conserved motifs, which probably contribute to the comparatively high Km values and the lower sensitivity to PALO. The Km for ornithine, however, is substantially lower at low temperatures. A survey of the catalytic efficiencies (kcat/Km) of OTCases adapted to different temperatures showed that OTCaseMab activity remains suboptimal at low temperature despite the 4.5-fold decrease in the Km value for ornithine observed when the temperature is brought from 20 to 5°C. OTCaseMab adaptation to cold indicates a trade-off between affinity and catalytic velocity, suggesting that optimization of key metabolic enzymes at low temperatures may be constrained by natural limits.
Intracellular biosynthetic enzymes are usually exposed to low substrate concentrations, in contrast to extracellular enzymes. Optimizing their catalytic efficiency (kcat/Km) in psychrophilic (cold-adapted) organisms may thus be challenging, since improving kcat at low temperatures by decreasing the activation enthalpy may have a cost in terms of affinity for the substrate(s) of the reaction (18, 35). The study of cold-active enzymes is thus an important topic in terms of physiology and metabolic evolution (for recent reviews, see references 6, 21, 25, 44, and 60).
No cold-active ornithine carbamoyltransferase (OTCase; EC 2.1.3.3) had been characterized until now. However, the presence of this enzyme in microorganisms adapted to the full range of environments compatible with life makes it an excellent candidate for investigations of protein evolution and of molecular adaptations to extreme conditions. OTCase catalyzes the conversion of ornithine and carbamoyl phosphate (CP) into citrulline and inorganic phosphate in the de novo pathway for arginine biosynthesis and in the detoxifying urea cycle. Biosynthetic and urea cycle OTCases are usually homotrimers of 33- to 40-kDa subunits (7, 9), except in the hyperthermophilic archaeon Pyrococcus furiosus, where OTCase is a dodecamer (51). In Pseudomonas aeruginosa, a dodecameric catabolic OTCase catalyzes the reverse reaction (conversion of citrulline into ornithine and CP in the presence of inorganic phosphate) in the deiminase pathway for arginine degradation; the allosteric properties of this enzyme, which are intimately linked to the dodecameric state, account for this ability despite a very unfavorable equilibrium constant (50). Crystal structures have been established for the trimeric OTCases of Escherichia coli and humans (26, 29, 47) and for the two known dodecameric transferases (50, 51).
This report presents the biochemical characterization of a biosynthetic OTCase from a new species of γ-Proteobacteria, Moritella abyssi (OTCaseMab), isolated from Atlantic Ocean sediments at a depth of 2,815 m and a temperature of 2°C (57). With a maximal growth rate at 4°C and a maximal growth temperature of 13°C, the organism is one of the strictest psychrophilic bacteria isolated until now (25) and was chosen for this reason. OTCaseMab was found to be distinctly psychrophilic and to differ from its homologues by a number of properties, illustrating the natural constraints which may limit the functional adaptation of metabolic enzymes at low temperatures.
MATERIALS AND METHODS
Strains and culture conditions.
E. coli strains were grown at 30 or 15°C in medium 853 (24) as a liquid medium and with 1.5% agar (Difco) as a solid medium. For bacteria containing recombinant plasmids, the media contained kanamycin and chloramphenicol at 50 μg ml−1.
Construction of an overexpressing strain.
Restriction enzymes and T4 ligase were purchased from Boehringer Mannheim and used according to the manufacturer's instructions. The argF gene of M. abyssi strain 2693 (57, 59) was amplified by PCR with oligonucleotides 5′-GGAATTCCATATGGAAAATTTATTATCAGTTAAAGATTTA-3′ (start codon underlined) and 5′-CGGGATCCCTACTTTCTTAACTGTTTGTGTC-3′ to produce NdeI and BamHI restriction sites at the ends of the fragment. The NdeI restriction site was designed to overlap the ATG start codon and the BamHI site after the TAA stop codon. The amplified fragment was inserted into the PCR-2 vector (Invitrogen) to verify the argF sequence (46) and subsequently was cloned into the pET24a vector (Novagen), which was then used to transform E. coli BL21-codonPlus(DE3)-RIL (Stratagene) competent cells; this rare-codon-usage-improved strain was found to be necessary to obtain satisfactory expression.
Expression and purification of recombinant OTCaseMab.
Exponentially growing cells of E. coli BL21-codonPlus(DE3)-RIL harboring plasmid pXY144 were induced with 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG), harvested after 20 h of induction at 15°C, and washed in 0.9% NaCl. Cells were disintegrated with a high-pressure cell disrupter (Basic Z model; Constant Systems Ltd.) at 28 MPa in 40 mM phosphate buffer (pH 7.0). The extracts were freed of cell debris by centrifugation two times, first for 30 min at 12,000 × g and then for 1 h at 100,000 × g. The supernatant was applied to a 15Q XK16/20 column (Pharmacia) preequilibrated with 10 mM phosphate buffer (pH 6.5) and washed with the same buffer. A salt step gradient (0 to 220 and 220 to 450 mM KCl) was then applied to the column, and the activities of eluted fractions were tested. The enzyme eluted as the major peak at about 300 to 400 mM KCl. Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and native PAGE of eluted fractions revealed that the M. abyssi OTCase peak contained more than 95% pure protein. Typically, 100 mg of pure protein was obtained from 1 liter of culture.
Purified E. coli ArgF and P. aeruginosa OTCase, which were used to calibrate gel filtration chromatography, were obtained from Catherine Tricot (J. M. Wiame Institute); δ-N-phosphonoacetyl-l-ornithine (PALO), synthesized by D. Gigot (Brussels University), was a gift from C. Bompain.
N-terminal amino acid analysis.
The N-terminal sequence (six amino acids) of the recombinant enzyme was determined by automated Edman degradation with a Procise 494 protein sequencer (Applied Biosystems).
SDS-PAGE.
SDS-PAGE was performed by using a Pharmacia PhastSystem with a discontinuous buffer system and homogeneous 12.5% gels. Gels were stained with Coomassie brilliant blue. The Benchmark protein ladder (Life Technology) was used as a standard to estimate subunit molecular masses.
Native molecular mass determination.
The molecular mass of native OTCaseMab was determined by gel filtration with a Pharmacia AKTA fast protein liquid chromatography system fitted with a Hiload 16/60 Superdex 200 prep-grade column and the following buffers: 10 mM phosphate buffer (pH 7.0) and this buffer plus 0.1, 0.15, 0.2, 0.3, or 0.5 M NaCl. About 0.174 mg of pure OTCase (in 0.1 ml) was applied to a column that had been equilibrated and calibrated with each buffer by using Pharmacia standards (about 100 μg of each protein), OTCase from E. coli (trimeric, 110 kDa), and P. aeruginosa (dodecameric, 420 kDa).
DSC.
Differential scanning calorimetry (DSC) was performed by using a MicroCal MCS-DSC instrument at a scan rate of 1°C/min and under a nitrogen pressure of 2 atm. Samples were desalted by gel filtration with a PD10 column (Pharmacia) equilibrated with 10 mM phosphate buffer (pH 7.0) supplemented with 0.0 M salt, 0.3 M KCl, 0.3 M NaCl, 0.5 M NaCl, or 0.1 M Na2SO4. These buffers were used in the reference cell and for buffer baseline determination. Thermograms of OTCase were analyzed according to a single non-two-state transition model in which the melting point, calorimetric enthalpy, and effective or van’t Hoff enthalpy of unfolding were fitted independently by using MicroCal Origin software (version 2.9).
Enzyme assays.
Activity was measured by citrulline colorimetry as described previously (3). The reaction mixture contained, in a final volume of 2.0 ml, 30 mM Tris HCl buffer (pH 9.0), 100 mM l-ornithine, and 20 mM CP for the standard assay unless otherwise specified. The reaction was started by OTCase addition. Incubation was carried out for 10 min at 20°C unless otherwise specified. Under the conditions applied, OTCase activity increased linearly with the protein concentration, and the reaction rate remained linear. Specific activity was expressed as micromoles of citrulline produced minute−1 milligram of protein−1. The protein concentration was determined by the method of Lowry et al. (35a).
In order to measure residual OTCase activity after thermal denaturation, the enzyme (final concentration, 0.0035 mg/ml) was incubated in the absence of substrates for 15 min at different temperatures in 10 mM Tris HCl (pH 7.5) or potassium phosphate (pH 7.0); the pH was adjusted at each temperature. Samples were withdrawn and kept at 0°C, and their activity in the same buffer was measured under standard conditions.
Half-lives of activity (t1/2) were determined by incubating the enzyme in 10 mM Tris HCl (pH 7.5), potassium phosphate (pH 7.0), or piperazine-N,N′-bis(2-ethanesulfonic acid (PIPES) buffer (pH 7.0) at different temperatures for different times; the pH was adjusted at each temperature. Samples were withdrawn and kept at 0°C, and their activity in the same buffer was measured under standard conditions.
RESULTS
Sequence analysis of M. abyssi argF.
In M. abyssi, the OTCase-encoding gene argF was found to be part of a large divergent operon of which the genes identified so far are clustered in the order ECBFGH(A), with argE constituting the left wing of the operon (59). Alignment of OTCase sequences by the Clustal program revealed that M. abyssi argF is clearly homologous to other OTCase genes. Remarkably, the amino acid sequence of OTCaseMab seems to be more closely related to those of the trimeric OTCase of Thermus thermophilus (42.6% identity) and the dodecameric OTCase of the hyperthermophilic archaeon P. furiosus (43% identity) than to many others. This paradox is due to the complex phylogeny of OTCases, which appears to reflect the parallel development of subfamilies resulting from the differential loss of paralogous copies already present in the last common ancestor of the three domains (30). That psychrophilic and thermophilic enzymes would appear in the same subgroup is not surprising in itself, since relatively few structural features may account for large differences in temperature profiles (28).
Another striking feature is that the CP and ornithine binding motifs are only partly conserved; T56 (E. coli residue numbering) in the CP site (ST56RTR) is replaced by a leucine in OTCaseMab, and L275 in the ornithine site (HCL275P) is replaced by a glutamine. The other known instances of deviations from these canonical sequences are the substitutions T56G in the phaseolotoxin-resistant OTCase from Pseudomonas syringae (27), T56L in the OTCase from T. thermophilus (40), and T56M in the OTCase from Pisum sativum (53). In no known OTCases other than that of Moritella does the ornithine binding site appear to be modified, except in cold-adapted mutants of P. furiosus OTCase (43). The OTCase from the closely related psychrophilic organism Moritella profunda (57) displays the same characteristics (Y. Xu, unpublished results). As for other genes of M. abyssi (59; Xu, unpublished), the codons AGA, AUA, and CUA, which are rare in E. coli, were found to be used several times in the sequence. This finding explains why an E. coli host improved for rare codon usage was required for M. abyssi argF expression.
Quaternary structure of recombinant OTCase.
As shown by N-terminal sequencing and a nucleotide sequence comparison, the recombinant monomer contains 301 amino acid residues (32,873 Da) starting at Met1. The molecular mass of the recombinant OTCase was estimated by native PAGE and gel filtration. In contrast with all other OTCases examined so far, we found that the pure enzyme was present as a mixture of trimers and dodecamers, with the relative proportions of the two forms depending on the salt concentration (Table 1). In addition, the trimeric fraction could reassociate into dodecamers when the salt concentration was increased. The salt requirement of M. abyssi, which is a marine bacterium, can be met with half-strength seawater (57), but the intracellular salt concentration is not known. Assuming intracellular concentrations of free Na+ and K+ of between 0.15 and 0.3 M in moderate halophiles (23, 49), it appears that in vivo, the main fraction of OCTaseMab is in the dodecameric form.
TABLE 1.
Quaternary structure determination by gel filtration on Superdex 200
| NaCl (mM) | %
|
|
|---|---|---|
| Trimers | Dodecamers | |
| 0 | 100 | 0 |
| 150 | 15 | 85 |
| 300 | 10 | 90 |
| 500 | 2 | 98 |
| 1,000 | 0 | 100 |
Temperature dependence of activity and kinetic parameters.
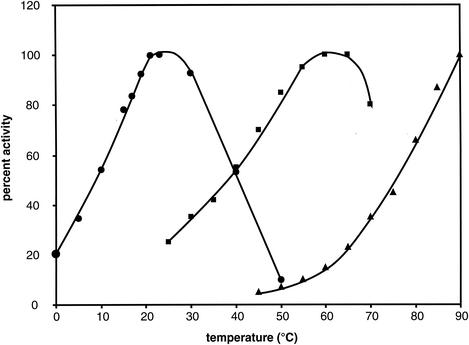
The apparent optimal temperatures for the activity of OTCaseMab were found to be about 23 to 25°C, in sharp contrast to those of its mesophilic and thermophilic homologues (Fig. 1). However, about 37% of this maximal activity could still be recorded at 5°C. Almost identical results were obtained with the native enzyme in crude extracts of Moritella and with the recombinant enzyme in E. coli cell lysates (data not shown).
FIG. 1.
Temperature dependence of OTCaseMab activity (circles) compared to that of E. coli (squares) (56) and P. furiosus (triangles) (32) homologues. The P. furiosus curve represents the reverse reaction. Activity assays were performed under the conditions described in Materials and Methods. The data presented are the means of triplicate measurements; the standard errors of the means were less than 10%.
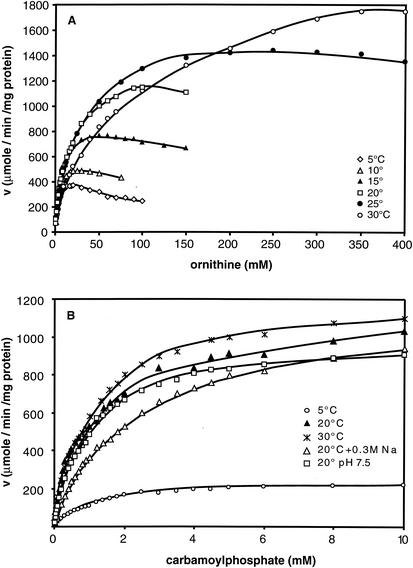
The optimal pHs for the reaction at 20°C were found to be between 9.0 and 10.0 (data not shown), whereas E. coli and T. thermophilus OTCases display broad optimal activity at about pH 8.0 (31, 45). As expected for an anabolic OTCase (9), the reaction catalyzed by OTCaseMab displayed no allosteric kinetics. The enzyme was inhibited by excess ornithine, a common feature of OTCases (9). In E. coli, this effect has been attributed to noncompetitive inhibition of CP (31). Interestingly, a similar substrate inhibition of OTCaseMab by excess ornithine (above 25 mM) was noticeable only at the lowest temperature investigated, 5°C (Fig. 2A), whereas at 20°C, a very high ornithine concentration (above 150 mM) was necessary to induce inhibition. The kinetic parameters for ornithine and CP (Fig. 2) are reported in Table 2, and the results are compared to the cognate values obtained for other OTCases. The Km values for both substrates were, in general, much higher for OTCaseMab, especially for ornithine. The latter parameter displayed a strong temperature dependence, whereas the Km value for CP remained practically unaffected by changes in temperature. When compared at environmental temperatures, the high Km and low kcat values of OTCaseMab at 5°C were responsible for the weak kcat/Km ratios. The bisubstrate analogue PALO, a competitive inhibitor of both CP and ornithine (42), inhibits OTCaseMab only weakly at 20°C, with an apparent Ki of 11.5 ± 0.4 μM (mean and standard error of the mean); the Ki for E. coli OTCase is 0.8 μM. As shown in Table 1, increasing the NaCl concentration favored oligomerization of the trimeric form of the enzyme into a dodecamer. Increasing the NaCl concentration up to 0.5 M in Tris buffer at 20°C decreased the kcat parameter of the reaction by 31%; the corresponding value for phosphate buffer was 24%. This high NaCl concentration had only a limited effect on Km values; a 1.5-fold increase for ornithine and a 2-fold increase for CP were recorded.
FIG. 2.
Effects of the concentrations of l-ornithine (A) and CP (B) on the velocity (v) of the reaction catalyzed by OTCaseMab under standard conditions (unless otherwise indicated) in the presence of 20 mM CP and 100 mM ornithine, respectively. Na, NaCl.
TABLE 2.
Kinetic parameters for different OTCases
| Organism | Max growth rate temp (°C) | Assay conditions (pH) | Assay temp (°C) |
Km Apparent (mM) for:
|
kcat (s−1) |
kcat/Km for:
|
Source or reference | ||
|---|---|---|---|---|---|---|---|---|---|
| Ornithine | CP | Ornithine | CP | ||||||
| M. abyssi | 4 | Tris (9.0)a | 30 | 45.0 | 0.9 | 690 | 15 | 766 | This work |
| 25 | 22.49 | 750 | 33 | This work | |||||
| 20 | 8.0b | 1.0b | 694 | 87 | 694 | This work | |||
| 15 | 5.67 | 546 | 97 | This work | |||||
| 10 | 3.34 | 382 | 114 | This work | |||||
| 5 | 1.78b | 1.1b | 235 | 132 | 214 | This work | |||
| Saccharomyces cerevisiae | 30 | Tris (8.5) | 30 | 0.9 | 0.2 | 354 | 393 | 1,770 | 13 |
| E. coli W | 37 | Tris (8.0) | 37 | 2.4b | 0.2b | 1,732 | 722 | 8,660 | 31 |
| T. thermophilus | 75 | Tris (7.0) | 55 | 0.1 | 0.1 | 74 | 740 | 740 | 45 |
| 70 | 146c | ||||||||
| P. furiosus | 102 | Tris (7.3) or PIPES (7.0) | 55 | 0.13 | 0.13 | 35 | 269 | 269 | 33 |
| 95 | 175c | ||||||||
There were no significant differences in Km values at pH 7.5 and 20°C.
Real Km.
Extrapolated due to CP thermolability.
Thermal inactivation and structural stability.
The inactivation curves, especially in Tris buffer, did not follow first-order kinetics but were distinctly biphasic, suggesting the presence of two populations of enzyme molecules with different stabilities. Very similar results were obtained with the native enzyme in crude extracts of Moritella and with the recombinant enzyme in E. coli cell lysates. These findings can be tentatively related to the equilibrium between the trimeric and the dodecameric forms, which have different stabilities (see below). Table 3 compares the half-lives of activity of OTCaseMab obtained from the first part of the inactivation curves with data obtained from mesophilic and thermophilic homologues. Like other OTCases, OTCaseMab is more stable in potassium phosphate buffer than in Tris HCl buffer; stability in PIPES buffer was 1.5 times higher than that in Tris HCl buffer. Indeed, phosphate is a product of the reaction known to stabilize OTCases against thermal inactivation (31, 33). These half-lives also illustrate the heat-labile character of OTCaseMab.
TABLE 3.
Half-lives of activities of psychrophilic, mesophilic, and thermophilic OTCases
| Organism | Temp (°C) | Half-lifea in the following buffer (pH):
|
||
|---|---|---|---|---|
| Potassium phosphate (7.0) | PIPES-NaOH (7.0) | Tris HCl (7.5) | ||
| M. abyssi | 50 | 60 | 14 | 9 |
| 55 | 40 | 4.5 | 3 | |
| 60 | 2 | |||
| E. coli | 68 | 43b | 5 | |
| P. furiosus | 100 | 65 | 60 | 40 |
Reported in minutes.
Value was determined with 10 mM potassium phosphate in 300 mM Tris HCl (pH 8.0).
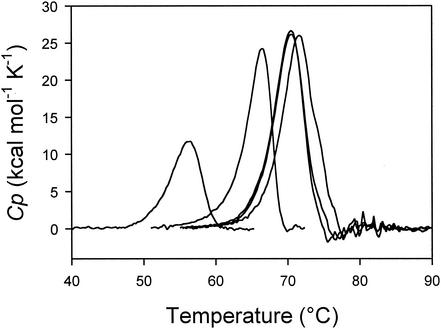
Heat-induced unfolding of OTCaseMab recorded by DSC was found to be irreversible, with a large deviation from the two-state model. Kinetically driven protein unfolding is scan rate and protein concentration dependent (52). Accordingly, OTCase thermograms were recorded at a constant scan rate (1 K/min) and a constant enzyme concentration (Table 4). Heat-induced unfolding was recorded at salt concentrations at which either the trimeric form (no salt added) or the dodecameric form (0.5 M NaCl) of OTCase prevails. As shown in Fig. 3, the latter condition induced strong enzyme stabilization, with marked increases in both the melting point (the midpoint of the transition) and the heat absorbed during unfolding (area under the peak). At a physiological salt concentration (0.3 M), both NaCl and KCl displayed the same stabilization efficiencies. However, the anion concentration played a crucial role in this effect, because at the same ionic strength (0.1 M Na2SO4), the sulfate ion had a weaker stabilization capacity. Table 4 also illustrates the very weak structural stability of OTCaseMab compared to that of its thermophilic homologue as far as the melting point and enthalpy of unfolding are concerned.
TABLE 4.
DSC parameters for thermal unfolding of OTCaseMab and P. furiosus OTCase
| Organism | Buffer supplement | OTCase (μM) | Tm (°C) | Calorimetric enthalpy (kcal mol−1) |
|---|---|---|---|---|
| M. abyssi | None | 48.7 | 56.6 | 66 |
| 0.1 M Na2SO4 | 46.6 | 66.4 | 123 | |
| 0.3 M NaCl | 47.3 | 70.5 | 145 | |
| 0.3 M KCl | 48.7 | 70.5 | 144 | |
| 0.5 M NaCl | 49.0 | 71.7 | 155 | |
| P. furiosusa | 43.2 | 104.0 | 842 |
Data are from B. Clantin (personal communication).
FIG. 3.
Heat-induced unfolding of OTCaseMab recorded by DSC. Curves show the following, from left to right: without added salt, with 0.1 M Na2SO4, with 0.3 M NaCl or KCl (superimposed curves), and with 0.5 M NaCl. The melting point corresponds to the top of the denaturation peak, and the calorimetric enthalpy corresponds to the area under the peak. Thermograms have been baseline subtracted and normalized for protein concentration. Cp, heat capacity change for unfolding; K, degrees Kelvin.
DISCUSSION
OTCaseMab is the first enzyme of its family to have been studied in an obligate psychrophilic organism. Its anabolic function may be inferred from the repressibility of its synthesis and the inclusion of the cognate gene in a biosynthetic operon (59). The apparent optimal temperature of OTCaseMab, 23 to 25°C, is one of the lowest reported for enzymes isolated from psychrophilic organisms (6, 11, 15, 37, 39). At 5°C, a temperature close to that allowing maximal growth of the organism (4°C), the activity of the enzyme is still about 37% maximum. Cold-active enzymes are frequently thermolabile, a fact which is understandable, since they have to be flexible enough to be active at a low energy cost (14). OTCaseMab is indeed considerably less resistant to thermal inactivation (Table 3) and has a weaker conformational stability (Table 4) than its E. coli or thermophilic counterparts (32, 33, 45). Psychrophilic enzymes counteract the inhibitory effect of low temperatures on activity by reducing the temperature dependence of the reaction rate (low activation enthalpy). Because this low activation enthalpy reflects the smaller number of enthalpy-driven interactions that have to be broken to reach the activated transition state, it has been proposed that the activity of a psychrophilic enzyme is heat labile, as these interactions also contribute to the active-site architecture (18, 35). However, the salt-dependent oligomerization state (trimeric or dodecameric) of OTCaseMab also seems to be involved in its stability.
This involvement of quaternary structure in the stability of OTCase activity is particularly clear for P. furiosus OTCase, which is a very stable dodecamer whose architecture and stability are dependent mainly on a network of hydrophobic interactions between trimers (7, 51). A mutant of P. furiosus OTCase (E25Q/M29A/W33A) in which the intertrimeric interactions are lost becomes much less thermostable and exclusively trimeric while retaining the same global kinetic properties of the wild-type enzyme (7). The half-life of the residual activity at 85°C dropped from 150 min (300 min in 0.2 M KCl) for the wild-type dodecameric P. furiosus OTCase to 2.5 min (13 min in 200 mM KCl) for the mutant trimeric enzyme, illustrating the involvement of quaternary structure in the stability of activity. Accordingly, the unusual equilibrium between trimers and dodecamers in OTCaseMab, which arises from weak interactions between trimers, should also contribute to the heat lability of its activity. As shown by DSC (Table 4 and Fig. 3), the salt-dependent oligomerization state of OTCaseMab strongly affects the stability of the molecule, favoring a more stable dodecamer. The difference in stability between the trimeric and the dodecameric forms of OTCaseMab has two possible origins: the preferential exclusion of the cosolvent arising from the high salt concentration (34) and the oligomeric state of the enzyme. As both parameters are interdependent, their relative contributions cannot be firmly established. It should be noted, however, that the increase in stability recorded for OTCase is in the upper range of values reported for the effects of salts on proteins at a neutral pH (2). Accordingly, it can be assumed that the oligomeric state also contributes to the greater stability of the dodecameric OTCase.
What could be the significance of a dodecameric state for OTCaseMab, an enzyme which is neither allosteric, as in P. aeruginosa, nor thermophilic, as in P. furiosus? The fact that it has to function in its natural environment under relatively high pressures (28 MPa at the site of capture) must be considered. The positive effect of salt on the oligomeric state of OTCaseMab suggests that a substantial fraction of the enzyme may be dodecameric in vivo, at least under atmospheric pressures. However, it is not yet known whether increasing pressures will tend to dissociate this enzyme into trimers or, on the contrary, stabilize the dodecameric state. Very little is known about genuinely psychropiezophilic enzymes (25).
Comparing OTCases from organisms whose G+C contents are not divergent enough to obscure biologically significant differences (Moritella, 42%; E. coli, 48 to 52%, Thermotoga maritima, 46%) makes it obvious that as a percentage of the total, the sum of the charged residues (KRED) available to provide stabilizing interactions is slightly lower in Moritella (21.3%) than in E. coli (23.1%) and much lower in Moritella than in the hyperthermophilic bacterium T. maritima (28.8%; M. Van de Casteele, unpublished data, NCBI accession number Y10661), particularly with regard to arginine (3.0, 3.3, and 6.1%, respectively), which can play an important role in protein stabilization through its high level of hydrophilicity and hydrogen-bonding capacity (40). Moreover, the thermolabile residues asparagine and glutamine are more represented in Moritella and E. coli (8.0 and 8.1%, respectively) than in T. maritima (5.7%). These observations are probably related to the psychrophilic nature of OTCaseMab, but direct three-dimensional studies are required to examine their significance.
The comparative analysis of kinetic parameters among various OTCases proved highly significant in terms of functional adaptation to cold. Ideally, adapting an enzyme to cold would mean optimizing both Km and kcat. Such a trend was noted for enzymes from Antarctic or Arctic fishes and some bacterial enzymes (4, 14, 21). However, a survey of cold-adapted enzymes showed that optimization of the kcat/Km ratio is far from universal (25, 60); for instance, markedly high Km values have been reported for psychrophilic glutamate dehydrogenase (12), citrate synthase (22), aspartate carbamoyltransferase (48, 58), aspartate aminotransferase (5), triosephosphate isomerase (1), DNA ligase (20), elongation factor Tu (36), subtilisin (41), xylanase (8), and alpha-amylase (10). OCTaseMab obviously belongs to this group, as shown by the high Km values for both ornithine and CP as well as by the high Ki for the bisubstrate analogue PALO, indicating a low substrate binding affinity. Therefore, it appears that numerous psychrophilic enzymes improve kcat values at low temperatures at the expense of Km values. Several aspects seem to be involved in this adaptive strategy. (i) From the kinetic and thermodynamic theories, it is well known that weak substrate binding is catalytically advantageous (17); indeed, the ground-state enzyme-substrate complex falls in a less deep energy pit, therefore reducing the energy barrier (and increasing kcat) for the reaction (14). (ii) The large activation entropy variation in psychrophilic enzymes suggests large conformational movements between a loose active site in the free state and the tightly bound transition state (18, 35); such loosely structured active sites should bind a substrate weakly. (iii) According to the folding-funnel hypothesis and to achieve the flexibility required for an enzyme to function efficiently at a lower temperature, a larger number of conformational states must be available to the enzyme; therefore, the enzyme may exhibit a higher Km and, possibly, a lower affinity for the substrate(s) of the reaction (18, 35). (iv) Finally, the specific amino acid substitutions noted for the ornithine and CP binding sites of OCTaseMab are possible structural determinants of such weak substrate binding. It is worth mentioning that mutants of P. furiosus OTCase selected in vivo to complement yeast mutants lacking OTCase at 30 and even 15°C displayed a much higher Km value for ornithine and a much higher Ki value for the bisubstrate analogue PALO than the wild type, a lower apparent optimal temperature, a higher kcat value (up to fivefold), and a markedly increased thermolability (43); this example is one of the rare cases (see also references 38 and 54) where it could be observed directly that thermolability was not a property acquired during genetic drift at a low temperature independent of the changes responsible for kinetic adaptation.
In conclusion, OTCaseMab appears to be suboptimal in its physiological temperature range as far as the kcat/Km ratio is concerned. The observed trade-off between affinity and catalytic velocity indicates the limits that can be encountered in optimizing the kinetic parameters of metabolic enzymes at low temperatures. From the point of view of evolution, this means that in many cases, a psychrophilic organism may respond to part of the pressure to adapt to low temperatures by making more of an imperfect enzyme; regulatory effects would therefore be expected to be important elements in evolutionary strategy. Furthermore, by comparing the thermodynamic and kinetic features of different series of homologous enzymes, it may be possible to determine whether such natural shortcomings of optimization result mainly from the barrier set by the enthalpic-entropy balance or are significantly influenced by structural constraints of evolutionary origin, for example, by the fact that the ancestor of a psychrophilic enzyme was adapted to higher temperatures. In this respect, it is interesting that in the polyphyletic pattern of OTCases, OTCaseMab falls in a broad subgroup containing several thermophilic enzymes (30). Although it has become doubtful that the last common ancestor of the three domains of life was a hyperthermophile (19; reviewed in reference 55), it could still have been a moderate thermophile.
Acknowledgments
We thank N. Gérardin for expert technical assistance and S. D'Amico and D. Georlette for kind help. We also thank C. Tricot for providing pure E. coli and P. aeruginosa OTCases and C. Bompain for the gift of PALO.
This work was supported by the Belgian Foundation for Joint and Fundamental Research (FKFO), the EEC programmes Coldzymes and Eurocold, and a Concerted Action between the Belgian State and the Free University of Brussels.
REFERENCES
- 1.Alvarez, M., J. P. Zeelen, V. Mainfroid, F. Rentier-Delrue, J. A. Martial, L. Wyns, R. K. Wierenga, and D. Maes. 1998. Triose-phosphate isomerase (TIM) of the psychrophilic bacterium Vibrio marinus. Kinetic and structural properties. J. Biol. Chem. 273:2199-2206. [DOI] [PubMed] [Google Scholar]
- 2.Arakawa, T., R. Bhat, and S. N. Timasheff. 1990. Why preferential hydration does not always stabilize the native structure of globular proteins? Biochemistry 29:1924-1931. [DOI] [PubMed] [Google Scholar]
- 3.Archibald, R. M. 1944. Determination of citrulline and allantoin and demostration of citrulline in blood plasma. J. Biol. Chem. 156:121-142. [Google Scholar]
- 4.Bentahir, M., G. Feller, M. Aittaleb, J. Lamotte-Brasseur, T. Himri, J. P. Chessa, and C. Gerday. 2000. Structural, kinetic and calorimetric charcterization of the cold-active phosphoglycerate kinase from the antarctic Pseudomonas sp. TACII18. J. Biol. Chem. 275:11147-11153. [DOI] [PubMed] [Google Scholar]
- 5.Birolo, L., M. L. Turino, B. Fontanella, C. Gerday, K. Mainolfi, S. Pascarella, G. Sannia, F. G. Vinci, and G. Marino. 2000. Aspartate aminotransferase from the Antarctic bacterium Pseudoalteromonas haloplanctis TAC 125. Cloning, expression, properties and molecular modelling. Eur. J. Biochem. 267:2790-2802. [DOI] [PubMed] [Google Scholar]
- 6.Cavicchioli, R., K. S. Siddiqui, D. Andrews, and R. K. Sowers. 2002. Low-temperature extremophiles and their applications. Curr. Opin. Biotechnol. 13:253-261. [DOI] [PubMed] [Google Scholar]
- 7.Clantin, B., C. Tricot, T. Lonhienne, V. Stalon, and V. Villeret. 2001. Probing the role of oligomerization in the high thermal stability of Pyrococcus furiosus ornithine carbamoyltransferase by site-specific mutants. Eur. J. Biochem. 268:3937-3942. [DOI] [PubMed] [Google Scholar]
- 8.Collins, T., M. A. Meeuwis, I. Stals, M. Claeyssens, G. Feller, and C. Gerday. 2002. A novel family 8 xylanase: functional and physicochemical characterization. J. Biol. Chem. 277:35815-35818. [DOI] [PubMed] [Google Scholar]
- 9.Cunin, R., N. Glansdorff, A. Piérard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Amico, S., C. Gerday, and G. Feller. 2001. Structural determinants of cold adaptation and stability in a large protein. J. Biol. Chem. 276:25791-25799. [DOI] [PubMed] [Google Scholar]
- 11.Davail, S., G. Feller, E. Narinx, and C. Gerday. 1994. Cold adaptation of proteins. Purification,characterisation and sequence of the heat-labile subtilisin from the antartic psychrophile Bacillus TA41. J. Biol. Chem. 269:17448-17453. [PubMed] [Google Scholar]
- 12.Di Fraia, R., V. Wilquet, M. A. Ciardello, V. Carratore, A. Antignani, L. Camardella, N. Glansdorff, and G. di Prisco. 2000. NADP+-dependent glutamate dehydrogenase in the Antarctic psychrotolerant bacterium Psychrobacter sp. TAD1. Characterization, protein and DNA sequence, and relationship to other glutamate dehydrogenases. Eur. J. Biochem. 267:121-131. [DOI] [PubMed] [Google Scholar]
- 13.Eisenstein, E., J. C. Osborne, Jr., I. M. Chaiken, and P. Hensley. 1984. Purification and characterization of ornithine transcarbamylase from Saccharomyces cerevisiae. J. Biol. Chem. 259:5139-5145. [PubMed] [Google Scholar]
- 14.Feller, G., and C. Gerday. 1997. Psychrophilic enzymes: molecular basis of cold adaptation. Cell. Mol. Life Sci. 53:830-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feller, G., F. Payan, F. Theys, M. Qian, R. Haser, and C. Gerday. 1994. Stability and structural analysis of alpha-amylase from the antarctic psychrophile Alteromonas haloplanctis A23. Eur. J. Biochem. 222:441-447. [DOI] [PubMed] [Google Scholar]
- 16.Feller, G., D. d'Amico, and C. Gerday. 1999. Thermodynamic stability of an cold-active amylase from the Antarctic bacterium Alteromonas haloplanctis. Biochemistry 38:4613-4619. [DOI] [PubMed] [Google Scholar]
- 17.Fersht, A. 1985. Enzyme structure and mechanism. W. H. Freeman & Company, New York, N.Y.
- 18.Fields, P. A., and G. N. Somero. 1998. Hot spots in cold adaptation: localized increases in conformational flexibility in lactate dehydrogenase A (4) orthologs of Antarctic notothenioid fishes. Proc. Natl. Acad. Sci. USA 95:11476-11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galtier, N., N. Tourasse, and M. Gouy. 1999. A non-hyperthermophilic common ancestor to extant life forms. Science 283:220-221. [DOI] [PubMed] [Google Scholar]
- 20.Georlette, D., Z. O. Jonsson, F. Van Petegem, J. Chessa, J. Van Beeumen, U. Hubscher, and C. Gerday. 2000. A DNA ligase from the psychrophile Pseudoalteromonas haloplanctis gives insights into the adaptation of proteins to low temperatures. Eur. J. Biochem. 267:3502-3512. [DOI] [PubMed] [Google Scholar]
- 21.Gerday, C., M. Aittaleb, J. L. Arpigny, E. Baise, J. P. Chessa, J. M. François, I. Petrescu, and G. Feller. 1999. Cold enzymes: a hot topic, p. 257-275. In R. Margesin and F. Schinner (ed.), Cold-adapted organisms. Ecology, physiology, enzymology, and molecular biology. Springer-Verlag, Heidelberg, Germany.
- 22.Gerike, U., M. J. Danson, N. J. Russell., and D. Hough. 1997. Sequencing and expression of the gene encoding a cold-active citrate synthase from an Antarctic bacterium, strain DS2-3R. Eur. J. Biochem. 248:49-57. [DOI] [PubMed] [Google Scholar]
- 23.Gilboa, H., M. Kogut, S. Chalamish, R. Regev, Y. Avi-Dor, and N. J. Russell. 1991. Use of 23Na nuclear magnetic resonance spectroscopy to determine the true intracellular concentration of free sodium in a halophilic eubacterium. J. Bacteriol. 173:7021-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glansdorff, N. 1965. Topography of cotransductible arginine mutations in Escherichiacoli K12. Genetics 51:167-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glansdorff, N., and Y. Xu. 2002. Microbial life at low temperatures: mechanisms of adaptation and extreme biotopes. Implications for exobiology and the origin of life. Recent Res. Dev. Microbiol. 6:1-21. [Google Scholar]
- 26.Ha, Y., M. T. McCann, M. Tuchman, and N. M. Allewell. 1997. Substrate-induced conformational change in a trimeric ornithine transcarbamoylase. Proc. Natl. Acad. Sci. USA 94:9550-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatziloukas, E., and N. J. Panopoulos. 1992. Origin, structure, and regulation of argK, encoding the phaseolotoxin-resistant ornithine carbamoyltransferase in Pseudomonas syringae pv. phaseolicola, and functional expression of argK in transgenic tobacco. J. Bacteriol. 174:5895-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaenicke, R. 1991. Protein stability and molecular adaptation to extreme conditions. Eur. J. Biochem. 202:715-728. [DOI] [PubMed] [Google Scholar]
- 29.Jin, L., B. A. Seaton, and J. F. Head. 1997. Crystal structure at 2.8 Å resolution of anabolic ornithine transcarbamylase from Escherichia coli. Nat. Struct. Biol. 4:622-625. [DOI] [PubMed] [Google Scholar]
- 30.Labedan, B., A. Boyen, M. Baetens, D. Charlier, P. Chen, R. Cunin, V. Durbecq, N. Glansdorff, G. Hervé, C. Legrain, Z. Liang, C. Purcarea, M. Roovers, R. Sanchez, T. L. Thia-Toong, M. Van de Casteele, F. Van Vliet, Y. Xu, and Y. Zhang. 1999. The evolutionary history of carbamoyltransferases: a complex set of paralogous genes was already present in the last universal common ancestor. J. Mol. Evol. 49:461-473. [DOI] [PubMed] [Google Scholar]
- 31.Legrain, C., and V. Stalon. 1976. Ornithine carbamoyltransferase from Escherichia coli W. Purification, structure and steady-state kinetic analysis. Eur. J. Biochem. 63:289-301. [DOI] [PubMed] [Google Scholar]
- 32.Legrain, C., V. Villeret, M. Roovers, D. Gigot, O. Dideberg, A. Piérard, and N. Glansdorff. 1997. Biochemical characterisation of ornithine carbamoyltransferase from Pyrococcus furiosus. Eur. J. Biochem. 247:1046-1055. [DOI] [PubMed] [Google Scholar]
- 33.Legrain, C., V. Villeret, M. Roovers, C. Tricot, B. Clantin, J. Van Beeumen, V. Stalon, and N. Glansdorff. 2001. The ornithine carbamoyltransferase of Pyrococcus furiosus. Methods Enzymol. 331:227-235. [DOI] [PubMed] [Google Scholar]
- 34.Lin, T. Y., and S. N. Timasheff. 1996. On the role of surface tension in the stabilization of globular proteins. Protein Sci. 5:372-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lonhienne, T., C. Gerday, and G. Feller. 2000. Psychrophilic enzymes: revisiting the thermodynamic parameters of activation may explain local flexibility. Biochim. Biophys. Acta 1543:1-10. [DOI] [PubMed] [Google Scholar]
- 35a.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 36.Masullo, M. P. Arcari, B. de Paola, A. Parmeggiani, and V. Bocchini. 2000. PSychrophilic elongation factor Tu from the Antarctic Moraxella sp. Tac II 25: biochemical characterization and cloning of the encoding gene. Biochemistry 39:15531-15539. [DOI] [PubMed] [Google Scholar]
- 37.McDonald, I. J., C. Quadling, and A. K. Chamers. 1963. Proteolytic activity of soime cold-tolerant bacteria from Arctic sediments. Can. J. Microbiol. 9:303-315. [Google Scholar]
- 38.Merz, A., M. C. Yee, H. Szadowski, G. Pappenberger, A. Crameri, W. P. Stemmer, C. Yanofsky, and K. Kirschner. 2000. Improving the catalytic activity of a thermophilic enzyme at low temperatures. Biochemistry 39:880-889. [DOI] [PubMed] [Google Scholar]
- 39.Morita, R. Y. 1975. Psychrophilic bacteria. Bacteriol. Rev. 39:144-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mrabet, N. T., A. Van den Broeck, I. Van den brande, P. Stanssens, Y. Laroche, A. M. Lambeir, G. Matthijssens, J. Jenkins, M. Chiadmi, H. van Tilbeurgh, J. Janin, W. J. Quax, I. Lasters, M. De Maeyer, and S. J. Wodak. 1992. Arginine residues as stabilizing elements in proteins. Biochemistry 31:2239-2253. [DOI] [PubMed] [Google Scholar]
- 41.Narinx, E., E. Baise, and C. Gerday. 1997. Subtilisin from psychrophilic antarctic bacteria: characterization and site-directed mutagenesis of residues possibly involved in the adaptation to cold. Protein Eng. 10:1271-1279. [DOI] [PubMed] [Google Scholar]
- 42.Penninckx, M., and D. Gigot. 1978. Synthesis and interaction with Escherichia coli l-ornithine carbamoyltransferase of two potential transition-state analogues. FEBS Lett. 88:94-96. [DOI] [PubMed] [Google Scholar]
- 43.Roovers, M., R. Sanchez, C. Legrain, and N. Glansdorff. 2001. Experimental evolution of enzyme temperature activity profile: selection in vivo and characterization of low-temperature-adapted mutants of Pyrococcus furiosus ornithine carbamoyltransferase. J. Bacteriol. 183:1101-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell, N. J. 2000. Toward a molecular understanding of cold-activity of enzymes from psychrophiles. Extremophiles 4:83-90. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez, R., M. Baetens, M. Van de Casteele, M. Roovers, C. Legrain, and N. Glansdorff. 1997. Ornithine carbamoyltransferase from the extreme thermophile Thermus thermophilus. Analysis of the gene and characterization of the protein. Eur. J. Biochem. 248:466-474. [DOI] [PubMed] [Google Scholar]
- 46.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi, D., H. Morizono, Y. Ha, M. Aoyagi, M. Tuchman, and N. M. Allewell. 1998. A resolution crystal structure of human ornithine transcarbamoylase complexed with N-phosphonoacetyl-l-ornithine. Catalytic mechanism and correlation with inherited deficiency. J. Biol. Chem. 273:34247-34254. [DOI] [PubMed] [Google Scholar]
- 48.Sun, K., L. Camardella, G. di Prisco, and G. Hervé. 1998. Properties of aspartate transcarbamylase from TAD1, a psychrophilic bacterium from Antarctica. FEMS Microbiol. Lett. 164:375-382. [DOI] [PubMed] [Google Scholar]
- 49.Ventosa, A., J. J. Nieto, and A. Oren. 1998. Biology of moderately halophilic aerobic bacteria. Microbiol. Mol. Biol. Rev. 62:504-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Villeret, V., C. Tricot, V. Stalon, and O. Dideberg. 1995. Crystal structure of Pseudomonas aeruginosa catabolic ornithine transcarbamoylase at 3.0-Å resolution: a different oligomeric organization in the trancarbamoylase family. Proc. Natl. Acad. Sci. USA 92:10762-10766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villeret, V., B. Clantin, C. Tricot, C. Legrain, M. Roovers, V. Stalon, N. Glansdorff, and J. Van Beeumen. 1998. The crystal structure of Pyrococcus furiosus ornithine carbamoyltransferase reveals a key role for oligomerization in enzyme stability at extremely high temperatures. Proc. Natl. Acad. Sci. USA 95:2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogl, V., C. Jatzke, H. J. Hinz, J. Benz, and R. Huber. 1997. Thermodynamic stability of annexin V E17G: equilibrium parameters from an irreversible unfolding reaction. Biochemistry 36:1657-1668. [DOI] [PubMed] [Google Scholar]
- 53.Williamson, C. L., M. R. Lake, and R. D. Slocum. 1996. Isolation and characterization of a cDNA encoding a pea ornithine transcarbamoylase (argF) and comparison with other transcarbamoylases. Plant Mol. Biol. 31:1087-1092. [DOI] [PubMed] [Google Scholar]
- 54.Wintrode, P. L., K. Miyzaki, and F. H. Arnold. 2000. Cold adaptation of a mesophilic subtilisin-like protease by laboratory evolution. J. Biol. Chem. 275:31635-31640. [DOI] [PubMed] [Google Scholar]
- 55.Xu, Y., and N. Glansdorff. 2002. Was our ancestor a hyperthermophilic procaryote? Comp. Biochem. Physiol. A 133:677-688. [DOI] [PubMed]
- 56.Xu, Y., Z. Liang, C. Legrain, V. Villeret, J. Van Beeumen, and N. Glansdorff. 1999. Genes and enzymes involved in arginine and pyrimidine biosynthesis in psychrophilic Vibrio strains from the deep sea, p. 319-333. In R. Margesin and F. Schinner (ed.), Cold-adapted organisms. Ecology, physiology, enzymology, and molecular biology. Springer-Verlag, Heidelberg, Germany.
- 57.Xu, Y., Y. Nogi, Y., C. Kato, Z. Liang, H. J. Rüger, D. De Kegel, and N. Glansdorff. Moritella profunda sp. nov. and Moritella abyssi sp. nov., two psychropiezophilic organisms isolated from deep Atlantic sediments. Int. J. Syst. Evol. Microbiol., in press. [DOI] [PubMed]
- 58.Xu, Y., Y. F. Zhang, Z. Liang, M. Van de Casteele, C. Legrain, and N. Glansdorff. 1998. Aspartate carbamoyltransferase from a psychrophilic deep-sea bacterium, Vibrio strain 2693. Properties of the enzyme, genetic organisation and expression in Escherichia coli. Microbiology 144:1435-1441. [DOI] [PubMed] [Google Scholar]
- 59.Xu, Y., Z. Liang, C. Legrain, H. J. Rüger, and N. Glansdorff. 2000. Evolution of arginine biosynthesis in the bacterial domain: novel gene-enzyme relationships from psychrophilic Moritella strains (Vibrionaceae) and the evolutionary significance of N-α-acetyl ornithinase. J. Bacteriol. 182:1609-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zecchinon, L., P. Claverie, T. Collins, S. d'Amico, D. Delille, G. Feller, D. Georlette, E. Gratia, A. Hoyoux, M. A. Meuwis, G. Sonan, and C. Gerday. 2001. Did psychrophilic enzymes really meet the challenge? Extremophiles 5:313-321. [DOI] [PubMed] [Google Scholar]