Abstract
We present the 4.8-Mb complete genome sequence of Salmonella enterica serovar Typhi strain Ty2, a human-specific pathogen causing typhoid fever. A comparison with the genome sequence of recently isolated S. enterica serovar Typhi strain CT18 showed that 29 of the 4,646 predicted genes in Ty2 are unique to this strain, while 84 genes are unique to CT18. Both genomes contain more than 200 pseudogenes; 9 of these genes in CT18 are intact in Ty2, while 11 intact CT18 genes are pseudogenes in Ty2. A half-genome interreplichore inversion in Ty2 relative to CT18 was confirmed. The two strains exhibit differences in prophages, insertion sequences, and island structures. While CT18 carries two plasmids, one conferring multiple drug resistance, Ty2 has no plasmids and is sensitive to antibiotics.
Salmonella enterica serovar Typhi is a human-specific pathogen causing enteric typhoid fever, a severe infection of the reticuloendothelial system (8, 10, 14). Although difficult to estimate, it is thought that at least 16 million cases and 500,000 deaths occur each year around the world (8). The early administration of antibiotic treatment has proven to be highly effective in eliminating infections, but indiscriminate use of antibiotics has led to the emergence of multidrug-resistant strains of S. enterica serovar Typhi (13, 31). Since typhoid is becoming difficult to treat with conventional drugs, information about the whole genome sequence and genes of S. enterica serovar Typhi will help to reveal more specific targets for drugs aimed at disease treatment and vaccines for prevention. Although S. enterica serovar Typhi can grow in laboratory media and survive in other hosts, such as experimental mice, humans are the only known natural hosts. The mouse model is a poor representive of the human disease. Experimental infections of chimpanzees or human volunteers have been the only way to relate bacterial genetic characteristics with pathogenic effects. Consequently, what is known about S. enterica serovar Typhi pathogenicity has been largely extrapolated from studies of S. enterica serovar Typhimurium infections in mice.
In this work, we present the genome sequence of the well-studied pathogenic strain Ty2. This strain was the foundation for vaccine development and was the parent of mutant strains Ty21a and CVD908 and their derivatives, used in trials of live attenuated vaccines (16). Isolated before the emergence of drug resisitance in the 1970s, it contains no plasmids. In contrast, recently isolated S. enterica serovar Typhi strain CT18 (25) carries multiple drug resistance cassettes on a large plasmid and contains a second large plasmid closely related to pMT1 of Yersinia pestis. The genome sequence of CT18 was recently determined at the Sanger Centre, Hinxton, United Kingdom (GenBank accession number AL513382), and we have used this sequence to perform a detailed comparison of the two genomes.
MATERIALS AND METHODS
Bacterial strain.
S. enterica serovar Typhi strain Ty2 was obtained from K. Sanderson (Salmonella Genetic Stock Center isolate 2666) and was redeposited at the American Type Culture Collection under accession number 700931. The strain was cultured and DNA was kindly prepared by R. A. Welch, University of Wisconsin.
Sequencing.
A whole-genome shotgun library was prepared using nebulization to produce random fragments by mechanically shearing genomic DNA (21). Fragments in the size range of 1 to 2.5 kb were prepared by agarose gel electrophoresis, ends were repaired, and the fragments were cloned into the M13Janus vector (4). Random phage clones were isolated, and purified DNAs were prepared as templates for sequencing reactions by using dye terminator chemistry. Some templates were prepared by PCR directly from M13Janus clones. Sequence data were collected with ABI377 and 3700 automated sequencers, and data were assembled by using Seqman II, Genome Edition (DNASTAR, Madison, Wis.). For the finishing steps, clonal and genomic PCR techniques provided templates for primer walking to link contigs, add complementary reads, and increase coverage. The final coverage was 9.8-fold. The contig order and assembly structure were independently confirmed by using a whole-genome optical map of recognition sites for restriction enzyme NheI (18).
Annotation and analysis.
When the genome of S. enterica serovar Typhi strain CT18 was published, our sequence was completely enclosed in a single contig, but many ambiguities remained and the sequence was not annotated. We made use of the CT18 sequence to accelerate our analysis. Open reading frames (ORFs) in Ty2 were defined by using GeneMark.hmm (1) and compared with CT18 ORFs by using BLAST. Most of the differences were identified from the ORF comparisons and then were checked in detail by using Lasergene DNA analysis tools (DNASTAR). We compared the Ty2 genome with the CT18 genome for all ambiguities and conflicts. When our primary trace data were adequate and consistent with the CT18 sequence at a confidence level of over 70%, we corrected the ambiguities and conflicts to match the CT18 consensus sequence. In total, 2,739 of 4,742 total ambiguities and conflicts were corrected by this process, leaving 2,003 in the Ty2 sequence due to insufficient evidence tosupport such changes or sound data confirming about 450 real differences between the two genomes. Every sequence difference was carefully inspected, and any ambiguity that could indicate a Ty2 feature different from CT18 was resolved. All Ty2 annotations that were different from CT18 were based on sound data. Ty2 ORFs were assigned identifiers in the form t0001.
Nucleotide sequence accession number.
The Ty2 sequence contains cross-references to the equivalent genes in the CT18 sequence and is available at GenBank under accession number AE014613.
RESULTS AND DISCUSSION
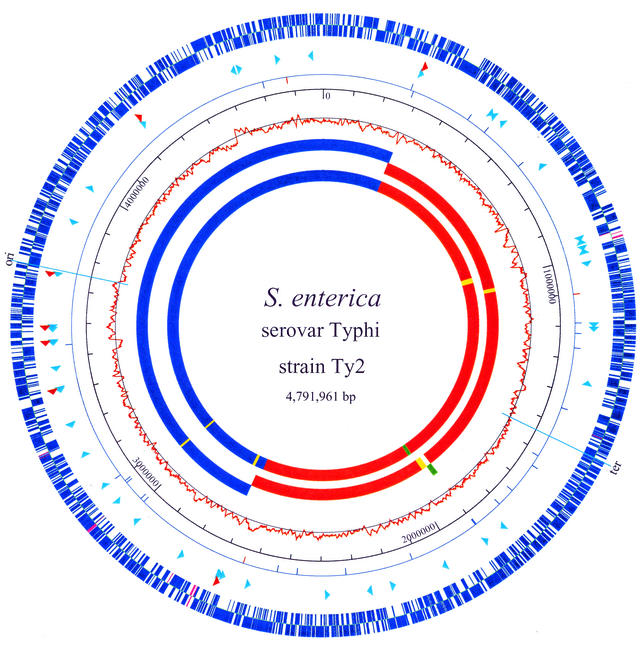
The genome of Ty2 consists of a single, circular chromosome of 4,791,961 bp with an average G+C content of 52.05%. Figure 1 shows the circular genome map of Ty2. Base pair 1 was chosen to correspond to Escherichia coli K-12 minute zero (3). The replication origin and terminus were determined according to their homologies to those in K-12. They are near kb 3750 and 1544, respectively. The C/G distribution switches polarity at both the origin and the terminus. Ty2 has seven rRNA operons, with five copies in one replichore and two in the other (Fig. 1). However, the rRNA operons are located in opposite replichores in Ty2 and CT18 versus K-12 and S. enterica serovar Typhimurium (20, 23, 32). The Ty2 genome is distinguished from that of CT18 by a major interreplichore inversion that spans the terminus of replication and almost half of the Ty2 genome (Fig. 1 and 2). This rearrangement leaves the two replichores slightly uneven in size. The boundaries of the half-genome inversion lie within rRNA operons at the locations of rrnG and rrnH, by comparison with the arrangement in E. coli K-12; both operons were rrnG-rrnH hybrids in Ty2 and were highly likely to have mediated the inversion through homologous recombination (20). Although this recombination did not change the rRNA organization, such genome rearrangements by homologous recombination at rRNA genes often result in distinguishable ribotypes. Variations in ribotypes seem to be linked to host adaptation (32). Besides this major inversion, there is a small inverted region that is also translocated (Fig. 1, green segment in sixth circle).
FIG. 1.
Circular genome map of Ty2. The Ty2 genome has 4,545 ORFs and pseudogenes, 4,516 of which are shared with CT18 (outer circle, blue) and 29 of which are unique (pink). Arrowheads within the second circle show the locations and orientations of rRNA operons (red) and tRNAs (turquoise) (not drawn to scale). The third circle shows insertion element distributions: blue, IS200; red, other IS elements. The fourth circle shows the scale in base pairs. The fifth circle shows the C/G skew, calculated for each sliding window of 10 kb along the genome. The sixth and seventh (innermost) circles show the CT18 and Ty2 genome comparison: blue (and above the axis in CT18) indicates colinear regions, red (and below the axis in CT18) indicates inverted regions, green indicates a region that is translocated and inverted again within the half-genome inversion region, and yellow indicates unique regions. The map was created with GenVision (DNASTAR).
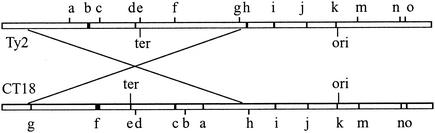
FIG. 2.
Comparative linear maps of Ty2 and CT18 genomes. The region inverted in Ty2 relative to CT18 is indicated by crossed lines. Black segments are unique regions (islands), lettered for cross-reference with Table 1. Tick marks outside the genome bars indicate the corresponding locations where islands are positioned in the other genome.
A comparison of Ty2 with CT18 reveals that more than 98% of the genome sequence is shared in these two S. enterica serovar Typhi strains. Apart from the inversion, the organization of the two genomes is quite similar, unlike the extensive rearrangements seen in a comparison of two Y. pestis strains, KIM and CO92 (7). For Ty2, we predicted 4,339 ORFs, 206 pseudogenes, and 101 RNA genes by comparison with the CT18 annotations (GenBank accession number AL513382). They average 910 bp in length and cover 88% of the genome. While 4,195 ORFs and pseudogenes are identical to those in CT18, 282 differ only in single point mutations from CT18. Together, they account for 97% of the total, a finding consistent with the results of a whole-genome nucleotide comparison. Seven Ty2 ORFs have insertions or deletions relative to CT18 ORFs that are multiples of 3 bp, resulting in longer or shorter proteins without disruption of the reading frames. Ty2 has relatively few insertion sequences (IS), having 26 copies of IS200F and only 3 copies (1 each) of other IS elements: IS1230B, IS285, and IS1351. In addition to these elements, CT18 contains three copies of IS1 (Table 1).
TABLE 1.
Regions of differences between Ty2 and CT18
| Fig. 2 location | tRNA | Ty2
|
CT18
|
|||
|---|---|---|---|---|---|---|
| Coordinates | Features | Length (bp) | Features | Length (bp) | ||
| a | 754495-754496 | 0 | IS1 (STY2419-STY2420) | 769 | ||
| b | Asn | 956415-978952 | Restriction-modification module (t0870-t0872) | 22,536 | 0 | |
| c | 1092785-1092786 | 0 | Part of phage (STY2040-STY2077) | 23,992 | ||
| d | 1489236-1490494 | yneH; glutaminase (t1446) | 1,258 | 0 | ||
| e | 1542863-1542864 | narV and narW fused (t1490) | 0 | narV and narW intact | 746 | |
| f | 1933051-1933052 | 0 | Part of phage (STY1048-STY1073) | 19,969 | ||
| g | 2654902-2654903 | 0 | ORFs (STY311 and STY312) | 972 | ||
| h | ssrA | 2734849-2745476 | Part of phage (t2647-t2656) | 10,626 | 0 | |
| i | Gly | 3039580-3045437 | Hypervariable region; phage? (t2947-t2954) | 5,858 | Hypervariable region; phage? (STY3188-STY3193) | 6,270 |
| j | 3407317-3407836 | Spacer tRNAIle-tRNAAla | 519 | Spacer tRNAGlu | 353 | |
| k | 3732787-3734141 | Spacer tRNAGlu | 1,354 | Spacer tRNAIle-tRNAAla | 585 | |
| m | 3973595-3973596 | 0 | IS1 (STY4124-STY4125) | 770 | ||
| n | 4442808-4442809 | 0 | STY4580-STY4582 | 1,059 | ||
| o | 4501860-4501861 | 0 | IS1 (STY4657-STY4658) | 770 | ||
Ty2 prophages.
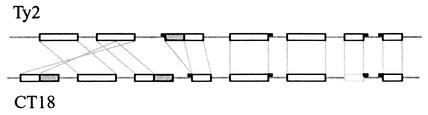
There are 29 ORFs unique to Ty2, whereas 84 are unique to CT18 (Table 2), many of them associated with putative prophages. Like the CT18 genome, the Ty2 genome contains seven regions that are prophage-like. However, they are not all identical. The relationships between the prophage regions of the two genomes are shown by the diagram in Fig. 3. Four prophages are present in identical locations in both genomes relative to the adjacent nonphage genes (Table 3). The prophage integrated near tRNAPhe in Ty2 is probably a remnant prophage; the homologous region in CT18 is not annotated as a prophage in that genome. As shown in Fig. 3, two Ty2 prophages are composed of recombined parts of CT18 prophages (or vice versa), and both genomes also have parts of prophages that are unique to those strains (Table 1). Similar observations for the prophages of O157:H7 suggested that the modular nature of prophage genomes makes a significant contribution to strain variation (24, 27).
TABLE 2.
ORFs unique to each strain
| Strain | ORF | Product |
|---|---|---|
| Ty2 | t0869 | Hypothetical protein |
| t0870 | Hypothetical protein | |
| t0871 | Type III restriction-modification system restriction subunit | |
| t0872 | Type III restriction-modification system methylation subunit | |
| t1446 | Putative glutaminase | |
| t2647 | Putative prophage integrase | |
| t2648 | Phage P4 psu protein | |
| t2649 | Phage P4 delta protein | |
| t2650 | Phage P4 sid protein | |
| t2651 | Phage P4 DNA-binding protein | |
| t2652 | Phage P4 cI protein | |
| t2653 | Phage P4 epsilon protein | |
| t2654 | Phage P4 ORF151 | |
| t2655 | Phage P4 ORF106 | |
| t2656 | Phage P4 alpha protein | |
| t2718 | Hypothetical protein | |
| t2720 | Hypothetical protein | |
| t2722 | Hypothetical protein | |
| t2724 | Hypothetical protein | |
| t2947 | Hypothetical protein | |
| t2948 | Hypothetical protein | |
| t2949 | Hypothetical protein | |
| t2950 | Hypothetical protein | |
| t2951 | Hypothetical protein | |
| t2952 | Hypothetical protein | |
| t2953 | Possible integrase | |
| t2954 | Hypothetical protein | |
| t3166 | Hypothetical protein | |
| t3172 | Putative transcriptional repressor (DeoR family) | |
| CT18a | STY0311 | Hypothetical protein |
| STY0312 | Probable secreted protein | |
| STY1048 | Putative bacteriophage protein | |
| STY1049 | Putative bacteriophage protein | |
| STY1050 | Putative bacteriophage protein | |
| STY1051 | Putative bacteriophage protein | |
| STY1052 | Putative bacteriophage protein | |
| STY1053 | Putative bacteriophage protein | |
| STY1054 | Putative bacteriophage protein | |
| STY1055 | Putative bacteriophage protein | |
| STY1056 | Putative bacteriophage protein | |
| STY1057 | Putative bacteriophage protein | |
| STY1058 | Hypothetical prophage protein | |
| STY1059 | Putative bacteriophage protein | |
| STY1060 | Putative bacteriophage protein | |
| STY1061 | Putative bacteriophage protein | |
| STY1062 | Putative bacteriophage protein | |
| STY1063 | Putative bacteriophage protein | |
| STY1064 | Putative bacteriophage protein | |
| STY1065 | Putative secreted protein | |
| STY1066 | Putative bacteriophage protein | |
| STY1067 | Putative bacteriophage protein | |
| STY1068 | Putative bacteriophage protein | |
| STY1069 | Putative bacteriophage protein | |
| STY1070 | Putative bacteriophage protein | |
| STY1071 | Putative bacteriophage protein | |
| STY1072 | Hypothetical prophage protein | |
| STY1073 | Hypothetical prophage protein | |
| STY1485 | Respiratory nitrate reductase 2 gamma chain; NarV | |
| STY1641 | Alternative bacteriophage tail fiber C terminus | |
| STY20074A | Restriction alleviation and modification enhancement protein | |
| STY2012 | Phage recombinase (pseudogene) | |
| STY2040 | Putative bacteriophage protein | |
| STY2041 | Putative bacteriophage protein | |
| STY2042 | Hypothetical protein | |
| STY2043 | Putative bacteriophage protein | |
| STY2044 | Putative endolysin | |
| STY2045 | Putative bacteriophage protein | |
| STY2046 | Hypothetical protein | |
| STY2047 | Putative membrane protein | |
| STY2048 | Putative bacteriophage protein | |
| STY2049 | Hypothetical protein | |
| STY2050 | Hypothetical protein | |
| STY2051 | Putative bacteriophage protein | |
| STY2052 | Hypothetical protein | |
| STY2053 | Putative bacteriophage cohesive ends | |
| STY2054 | Cell-killing toxin-antitoxin system | |
| STY2054A | Host cell-killing modulation protein | |
| STY2055 | Hypothetical protein | |
| STY2056 | Putative transposase | |
| STY2057 | Hypothetical protein | |
| STY2058 | Putative membrane protein | |
| STY2059 | Putative bacteriophage protein | |
| STY2060 | Hypothetical protein | |
| STY2061 | Conserved hypothetical protein | |
| STY2062 | Putative DNA replication protein | |
| STY2063 | Conserved hypothetical protein | |
| STY2064 | Conserved hypothetical protein | |
| STY2065 | Putative cro repressor | |
| STY2066 | Putative regulator | |
| STY2067 | Conserved hypothetical protein | |
| STY2068 | Conserved hypothetical protein | |
| STY2069 | Hypothetical protein | |
| STY2070 | Putative cell division inhibitor protein | |
| STY2071 | Putative bacteriophage protein | |
| STY2072 | Putative bacteriophage protein | |
| STY2073 | Exodeoxyribonuclease VIII | |
| STY2074 | Conserved hypothetical protein | |
| STY2075 | Conserved hypothetical protein | |
| STY2076 | Putative excisionase | |
| STY2077 | Putative integrase | |
| STY2419 | Insertion element IS1 protein InsB | |
| STY2420 | Insertion element IS1 protein InsA | |
| STY3188 | Hypothetical protein | |
| STY3189 | Hypothetical protein | |
| STY3191 | Hypothetical protein | |
| STY3192 | Hypothetical protein | |
| STY3193 | Possible integrase | |
| STY4124 | Insertion element IS1 protein InsA | |
| STY4125 | Insertion element IS1 protein InsB | |
| STY4580 | Putative membrane protein | |
| STY4582 | Possible exported protein | |
| STY4657 | Insertion element IS1 protein InsB | |
| STY4658 | Insertion element IS1 protein |
Data for CT18 are from GenBank accession number AL513382.
FIG. 3.
Diagram of prophage regions in Ty2 and CT18. Open rectangles outlined in black, cognate prophage and prophage segments; gray rectangles, unique prophage segments; open rectangle outlined in gray, segment not annotated as phage in CT18; small black rectangles, tRNAs near phage integration sites. The diagram is not drawn to scale.
TABLE 3.
Phage regions similar in Ty2 and CT18
| ORFs | tRNA | Comments | |
|---|---|---|---|
| Ty2 | CT18 | ||
| t1346-t1397 | STY1591-STY1642 | ||
| t1867-t1996 | STY1011-STY1047 | ||
| t1897-t1929 | STY2013-STY2039 | ||
| t2657-t2674 | STY2879-STY2897 | ssrA | tmRNA insertion site; contains iro operon |
| t3400-t3435 | STY3657-STY3703 | Trp | |
| t4294-t4338 | STY4600-STY4645 | Contains sopE; virulence factor | |
| t4358-t4371 | STY4667-STY4680 | Phe | Possible phage remnant |
| t4517-t4529 | STY4821-STY4832 | Leu | Part of large island; contains serine/threonine kinases |
Neither the CT18 genome nor the Ty2 genome contains the Gifsy-1 phage identified in S. enterica serovar Typhimurium LT2 (9), and only parts of Gifsy-2 are represented (the leftmost CT18 region in Fig. 3; genes t1904 to t1919 in Ty2). In both genomes, these regions are incorporated into putative prophages different from Gifsy-2 and different from each other. Gifsy-2 in LT2 encodes two proteins important for survival in macrophages, SodC (superoxide dismutase) and MsgA (function unknown). In Ty2 and CT18, the Gifsy-like regions do not contain genes for either of these, but both genes are present elsewhere in nonphage regions that are homologous in the two genomes. The previously identified LT2 phages Fels-1 and Fels-2 are not present in Ty2, although there are some regions of homology where Ty2 prophage genes are similar, as is the case for CT18 (22). Virulence-related genes iroBCDEN (Ty2 ORFs t2668 to t2672, encoding an iron uptake and storage system) and sopE (t4303, encoding an invasion-associated, type III-secreted effector protein) are located in cognate prophages in the two genomes.
Other differences.
Like the phage and IS differences, several lineage-specific islands were found by strain comparison (Table 1). It was not possible to ascertain whether these differences resulted from insertions into one genome or deletions from the other. CT18 ORFs STY0311 and STY0312 are not present in Ty2. The products of these ORFs are not similar to any protein in the database, but the product of STY0312 has characteristics of a secreted protein, as do the products of STY0314 and STY0316 nearby. Since secreted products often interact with extrabacterial targets, these ORFs may be related to pathogenicity. t1446 in Ty2 encodes a homolog of glutaminase (EC.3.5.1.2) that is encoded by yneH in LT2. This enzyme has roles in glutamate and nitrogen metabolism. CT18 has no equivalent gene. A 22.5-kb island at tRNAAsn in Ty2 carries t0871 and t0872, whose products are similar to a restriction enzyme and the C-terminal portion of a modification methylase enzyme of a type III system that probably is not functional, since only 173 out of 525 amino acids (aa) remain. An intact hsdRMS locus encodes the conserved Salmonella and E. coli type I restriction-modification system elsewhere in the genome.
The rRNA loci rrnG and rrnH mediated the large genomic inversion noted above. Recombination between these loci leaves hybrid rrn regions with spacer tRNAs different from those in the analogous K-12 loci, as identified by the DNA sequences flanking the rRNA genes. We found tRNAGlu at Ty2 rrnH (versus tRNAIle-tRNAAla in K-12) and the converse at rrnG. Although CT18 is colinear with K-12 in this part of the genome, it has the same spacers as Ty2 in rrnH and rrnG, suggesting that two events have occurred: an inversion followed by a reinversion restoring colinearity with K-12. Near the origin of replication, intrareplichore inversions in both the CT18 and the Ty2 genomes have resulted in hybrid rrn loci with tRNAIle-tRNAAla in CT18 and tRNAGlu in Ty2 at one hybrid site and the converse at the other site. In this region, the two genomes are colinear, so the difference in spacers again can be explained by a second recombination event. In fact, only rrnB is intact in both genomes. It is interesting that unusual rrn loci with apparently recombined spacer tRNA segments also appear among the annotated genes in Shigella flexneri strain 301 (GenBank accession number AF386526). While these events are admittedly difficult to reconstruct, it is known that rrn-mediated rearrangements are common in S. enterica serovar Typhi (19) and have been used to distinguish Typhi isolates by ribotyping (23).
Differences among pseudogenes in CT18 and Ty2.
Perhaps the most significant difference between Ty2 and CT18 is found in the selective silencing of gene functions in the form of pseudogenes. In CT18, there are 204 verified pseudogenes; 9 of these are intact genes in Ty2 (Table 4). On the other hand, Ty2 has its own unique set of 11 pseudogenes as well as 195 in common with CT18 (Table 5). Seven Ty2-specific pseudogenes result from frameshifts, while four result from disruptions by point mutations that create internal stop codons. One of the pseudogenes in CT18 (STY2012), which encodes a partial phage recombinase and which is located at a phage insertion point, is entirely lost in Ty2. A Ty2 gene (t0235) that encodes a putative chitinase has an internal stop codon and is annotated as a pseudogene in our Ty2 GenBank entry. Chitinase, related to lysozyme, is an enzyme that disrupts cell membranes by digestion of peptidoglycan linkages. In the corresponding CT18 annotation (ORF STY0257), although the DNA sequence for this gene is identical to that in the Ty2 annotation, it is stated that the stop codon is translationally suppressed by insertion of a tryptophan residue. While there has been a report of this mode of suppression at a low efficiency, we know of no evidence to support it in this particular case.
TABLE 4.
Genes that were pseudogenes in CT18a but intact in Ty2
| ORF | Gene | Product | |
|---|---|---|---|
| Ty2 | CT18 | ||
| t2448 | STY0453 | ybaD | Conserved hypothetical protein |
| t2319 | STY0590 | fimI | Fimbrin-like protein FimI |
| t1254 | STY1735 | ttrS | Sensor kinase TtrS protein |
| t1023 | STY1987 | sopE2 | Putative invasion-associated secreted protein |
| t0757 | STY2328 | wcaA | Putative glycosyltransferase |
| t0589 | STY2504 | Putative transcriptional regulator | |
| t0425 | STY2668 | Phosphoenolpyruvate-protein phosphotransferase | |
| t3035 | STY3280 | Bacteriocin | |
| t3695 | STY3955 | torC | Cytochrome c-type protein |
For CT18 mutation details, see Parkhill et al. (25).
TABLE 5.
Genes that were pseudogenes in Ty2 but intact in CT18
| ORF | Gene | Product | Mutation | |
|---|---|---|---|---|
| Ty2 | CT18 | |||
| t2523 | STY0371 | stbC | Outer membrane fimbrial usher protein | Internal stop codon |
| t2474 | STY0423 | aroM | AroM protein | Frameshift |
| t1913 | STY1027 | Hypothetical bacteriophage protein | Internal stop codon | |
| t1549 | STY1423 | Putative exported protein | Frameshift | |
| t1490 | STY1485 | narV | Respiratory nitrate reductase 2 gamma chain | Deletion fuses two ORFs into one |
| STY1486 | narW | Respiratory nitrate reductase 2 delta chain | Deletion fuses two ORFs into one | |
| t1429 | STY1553 | Putative d-mannonate oxidoreductase | Internal stop codon | |
| t1183 | STY1810 | astA | Arginine N-succinyltransferase | Frameshift |
| t1166 | STY1829 | Putative regulatory protein | Frameshift | |
| t0706 | STY2379 | stcC or yehB | Putative outer membrane usher protein | Frameshift |
| t2688 | STY2913 | gabP | GabA permease (4-aminobutyrate transport carrier) | Internal stop codon |
| t4529 | STY4832 | Bacteriophage P4 DNA primase | Frameshift | |
Table 6 shows genes in Ty2 that have changes causing their coding frames to diverge from those in CT18. For example, gltX, encoding glutamyl-tRNA synthetase, is intact in Ty2, but in CT18, a frameshifting deletion results in a predicted protein three residues longer at the C terminus. Since gltX is an essential gene, the extra residues presumably do not affect the synthetic activity appreciably. Some of the pseudogenes listed in Table 4 to 6 also may have a role in pathogenesis. In addition to the loss of 7 out of 12 fimbrial operons in both strains, Ty2 has lost 2 more (stcC and stbC) but has gained fimI. Three other genes that are intact in Ty2 may be associated with pathogenicity. TtrS is a sensor for tetrathionate, an alternative electron acceptor in vitamin B12-dependent anaerobic growth, and may be important for intracellular survival. SopE2 is secreted by a type III mechanism into host cells, where it is involved in actin rearrangements (12), a common first step in bacterial attack or invasion of host cells. WcaA is thought to be a glycosyltransferase involved in the synthesis of colanic acid, which is secreted to form the exopolysaccharide capsule providing protection from, for example, dehydration, acid stress, and osmotic stress, conditions encountered both inside and outside the human host. This capsule is also implicated in another pathogenic mechnism, biofilm formation (6, 28).
TABLE 6.
ORFs whose products have divergent C-terminal sequences
| ORF | Gene | Product | Comments | |
|---|---|---|---|---|
| Ty2 | CT18 | |||
| t0080 | STY0089 | yaaU | Putative major facilitator superfamily transport protein | 59 aa longer; stop codon further downstream in Ty2 |
| t1445 | STY1536 | Putative aldehyde dehydrogenase | 87 aa longer; insertion of 1,260 bp in Ty2 | |
| t1379 | STY1609 | Hypothetical protein | 103 aa longer; stop codon further downstream in Ty2 | |
| t0442 | STY2654 | gltX | Glutamyl-tRNA synthetase | 3 aa longer; frameshift in Ty2 |
| t2825 | STY3049 | rpoS | RNA polymerase sigma subunit transposase | 54 aa longer; frameshift in Ty2 |
| t3447 | STY3706 | tnpA | 18 aa longer; stop codon further downstream in Ty2 | |
| t3828 | STY4105 | sapB | Putative autotransporter | 9 aa longer; frameshift in Ty2 |
| t4435 | STY4740 | sgaB | Putative phosphotransferase system IIB protein | 77 aa longer; frameshift in Ty2 |
NR-Z and RpoS.
Differences between the loci for nitrate reductase Z (NR-Z) and RpoS in Ty2 and CT18 may seem to indicate significant differences in their abilities to thrive in anaerobic conditions, but a detailed investigation yielded more questions than answers. NR-A and NR-Z complexes provide electron transport during anaerobic respiration. CT18 has intact genes in both loci. In Ty2, the NR-A locus is intact, but the Z components encoded by narW and narV are fused by an in-frame deletion. Since both gene products are essential for a functional Z complex, this pathway is probably inactivated in Ty2, although the consequences are unclear. However, S. enterica serovar Typhi mutants deficient in anaerobic respiration are also less capable of intracellular replication (5), a factor which would compromise virulence.
The structure and function of the Z complex are understood, but its role is obscure. The narZYWV genes (product, NR-Z) are homologs of the genes of the NR-A locus, narGHIJ, which are active in anaerobic respiration. The four genes encode the alpha, beta, delta, and gamma components, respectively. By analogy with NR-A, the alpha component contains the catalytic site and requires a molybdenum cofactor for activity. The beta component contains an iron-sulfur center and transfers electrons from the gamma component to the alpha component. Both alpha and beta are cytoplasmic proteins. Paired gamma chains form a membrane anchor, and heme-iron centers are embedded in the membrane. Electrons are accepted by the gamma chains from quinone and then transferred to the beta protein, which in turn transfers them to the alpha subunits, where the reduction reaction is completed. The delta protein is not actually part of the complex but is essential for activity; it is thought to be important in assembling the complex. In Ty2 NarW, the C-terminal 54 aa out of 236 are deleted, and in NarV, aa 1 to 194 out of 225 are lost, including all of the transmembrane segments that form the membrane anchor.
NR-Z was thought to be expressed constitutively at a low level and was known not to be induced during anaerobic growth (11). Recently, the locus in S. enterica serovar Typhimurium was shown to be induced by carbon starvation (stationary phase) and to be RpoS dependent (34). NR-Z is also essential for starvation-induced heat and acid tolerance. NR-Z expression is not induced during anaerobic growth but is actually repressed by Fnr, the nitrogen-sensing regulator. Active hybrid enzymes mixing NR-A and NR-Z subunits have been obtained (2); therefore, it is possible that the NR-A membrane anchor tethers the NR-Z enzymes and that NarJ replaces the mutated NarW, if they are expressed under the same conditions as NR-Z.
However, rpoS is also abnormal in Ty2 (although it is intact in CT18). The alternative sigma factor encoded by this gene regulates more than 30 genes in the stress response. In Ty2, a frameshift at aa 312 replaces the last 12 aa of the wild-type product with 74 different aa. This change was discovered when mutations were introduced into Ty2 in efforts to attenuate its virulence for the creation of a safe vaccine strain, called Ty21a (29) or CVD908 (17). Subsequently, attenuation proved to be due partly to a mutant RpoS which had not been deliberately introduced. The mutation was later shown to be present in the Ty2 parent strain as well (30). Ty21a survived starvation and other stresses poorly, an advantage in a vaccine strain, and these deficiencies were complemented by an introduced wild-type RpoS. Unfortunately, no equivalent data exist for Ty2; therefore, we cannot be sure that other mutations in Ty21a, either engineered or accidental, have influenced the stress response of Ty21a or determine whether there is any direct effect on the virulence of Ty2.
We obtained Ty2 for sequencing from a Salmonella archive to minimize possible effects of repeated passaging. However, variant rpoS genes have been identified in archived stocks of Salmonella and have also been observed in E. coli K-12 W3110 (35). Some spontaneous mutants that arise during prolonged starvation are associated with a growth advantage (38). These observations suggest that mutation of rpoS under extreme stress itself may be a stress response, permitting the selection of a more efficient transcription factor that can improve fitness to respond to adverse conditions.
Interestingly, the genomic region between rpoS and mutS is highly plastic in members of the family Enterobacteriaceae (15), with many rearrangements in this interval. In S. enterica serovar Typhimurium, the expression of the Spv proteins, necessary for bacterial survival inside invaded host cells, is under RpoS control. The Spv genes, carried on a plasmid in serovar Typhimurium, are not present in serovar Typhi, and little is known about how different host tissue invasion mechanisms may be in serovar Typhi or how dependent on RpoS they may be. In fact, the extended RpoS protein in Ty2 retains intact both the RNA polymerase core binding site and the putative DNA binding site that together bring about enhanced binding of the polymerase to the DNA template. Stationary-phase (starvation) responses are under the control of RpoE (a different alternative sigma factor) (36), as are responses to other stimuli important in virulence, such as oxidative stress imposed by macrophage defenses. It is possible that these two transcription factors have some overlapping effects, making it very difficult to predict the effect of this particular mutant RpoS. It is interesting that S. flexneri 2a strain 2457T also has mutations in the narZYWV locus as well as a mutant rpoS (unpublished data). A frameshift changes the C-terminal 30 aa and truncates the transcript, whereas another 2a strain, 301, has also lost narZ but retains rpoS intact. There is much to be learned yet.
Clearly, while genome sequencing has revealed many genes with potentially important contributions to pathogenicity, discovering the details and deciphering the message, if any, in individual sets of pseudogenes will require extensive reseach in many laboratories, each with a specific expertise. A more appropriate model system is also needed. Newer techniques, such as microarray analysis of gene expression, have begun to provide the next level of information about genes involved in pathogenesis. Of the many knockout mutations already constructed in K-12 strain MG1655 in our laboratory, rpoS mutations should be very informative. A set of characterized mutations in an isogenic background should help to unravel the complex aspects of the stress response and provide clues as to how NR-Z may be involved.
Differences in pseudogene content between CT18 and Ty2 fall into no discernible pattern or functional relationship. These differences may have arisen due to variations in stresses applied by human host defense systems and may contribute subtle effects to the complex mechanisms of pathogenesis used by these two strains. They may also reflect a need to adjust the balance of metabolic capabilities to optimize virulence, perhaps achievable by more than one possible combination of genes.When analyzing pseudogenes, as emphasized by McClelland et al. (22), investigators should be aware that pseudogenes may be identified with confidence only when an intact homolog is found in a closely related (and sequenced) genome, and even then, annotation criteria vary. Some, but not all, annotations designate genes disrupted by IS elements as pseudogenes. It is often unclear whether small differences in protein structure, such as the three extra residues in GltX mentioned above, will eliminate function. Criteria based strictly on gene structure are not appropriate for secreted or surface proteins, such as those of fimbriae, whose genes are normally more variable than the conserved genes nearby (27), with the obvious advantage of evading the host immune system or potentially increasing the ability of the bacterium to attach to and affect host tissues.
How can we account for the accumulation of pseudogenes? Although mutagenic processes such as transposase induction are triggered by the kinds of stresses that bacteria undergo in repeated passages in the laboratory, strains that have been used in laboratories for the longest periods, E. coli K-12 and S. enterica serovar Typhimurium strain LT2, have almost 1 order of magnitude fewer pseudogenes than strains of intracellular pathogens that have also been used in laboratories for long periods, Y. pestis KIM, S. flexneri 2457T, and Ty2. The largest numbers of pseudogenes have been observed primarily in host-adapted pathogens that grow intracellularly and are thought to result from an adaptive process (33). In S. enterica serovar Typhi, adaptive changes have limited the host range to humans and (presumably) inactivated metabolic functions that are not needed for intracellular growth (26, 37) or survival in the intestine. These differences, as well as any of the far more numerous differences between S. enterica serovar Typhi and serovar Typhimurium strains, may underlie disease characteristics that are overtly or subtly distinctive of the pathogenic potential of the strains. It is important that each difference is examined with expert knowledge to identify the genetic variables that may yield valuable information through experimental evaluation. This goal may still be costly and difficult to carry out without an animal model that reproduces the human disease accurately and has reasonable costs. Inspection of every pseudogene for the possibility of residual or altered activity is not a trivial task; in many cases, even this initial test is not immediately possible, since researchers may have no idea of the function of encoded proteins or of which parts of encoded proteins may be essential for function.
Acknowledgments
We thank the University of Wisconsin Genome Sequencing Team members for excellent technical work.
This work was supported by NIH/NIAID grant AI44387 to F.R.B.
Footnotes
Paper 3604 from the Laboratory of Genetics, University of Wisconsin.
REFERENCES
- 1.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27:3911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blasco, F., F. Nunzi, J. Pommier, R. Brasseur, M. Chippaux, and G. Giordano. 1992. Formation of active heterologous nitrate reductases between nitrate reductases A and Z of Escherichia coli. Mol. Microbiol. 6:209-219. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 4.Burland, V., D. L. Daniels, G. Plunkett III, and F. R. Blattner. 1993. Genome sequencing on both strands: the Janus strategy. Nucleic Acids Res. 21:3385-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Contreras, I., C. S. Toro, G. Troncoso, and G. C. Mora. 1997. Salmonella typhi mutants defective in anaerobic respiration are impaired in their ability to replicate within epithelial cells. Microbiology 143:2665-2672. [DOI] [PubMed] [Google Scholar]
- 6.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, W., V. Burland, G. Plunkett III, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, S. Zhou, D. C. Schwartz, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. S. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Everest, P., J. Wain, M. Roberts, G. Rook, and G. Dougan. 2001. The molecular mechanisms of severe typhoid fever. Trends Microbiol. 9:316-320. [DOI] [PubMed] [Google Scholar]
- 9.Figueroa-Bossi, N., S. Uzzau, D. Maloriol, and L. Bossi. 2001. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Mol. Microbiol. 39:260-271. [DOI] [PubMed] [Google Scholar]
- 10.Galan, J. E. 1996. Molecular genetic bases of Salmonella entry into host cells. Mol. Microbiol. 20:263-271. [DOI] [PubMed] [Google Scholar]
- 11.Gennis, R. B., and V. Stewart. 1996. Respiration, p. 236-237. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. American Society for Microbiology, Washington, D.C.
- 12.Higashide, W., S. Dai, V. P. Hombs, and D. Zhou. 2002. Involvement of SipA in modulating actin dynamics during Salmonella invasion into cultured epithelial cells. Cell. Microbiol. 4:357-365. [DOI] [PubMed] [Google Scholar]
- 13.Ivanoff, B., and M. M. Levine. 1997. Typhoid fever: continuing challenges from a resilient bacterial foe. Bull. Inst. Pasteur 95:129-142. [Google Scholar]
- 14.Jones, B. D., and S. Falkow. 1996. Salmonellosis: host immune responses and bacterial virulence determinants. Annu. Rev. Immunol. 14:533-561. [DOI] [PubMed] [Google Scholar]
- 15.Kotewicz, M. L., B. Li, D. D. Levy, J. E. LeClerc, A. W. Shifflet, and T. A. Cebula. 2002. Evolution of multi-gene segments in the mutS-rpoS intergenic region of Salmonella enterica serovar Typhimurium LT2. Microbiology 148:2531-2540. [DOI] [PubMed] [Google Scholar]
- 16.Levine, M. M. 1999. Typhoid fever vaccines, p. 781-814. In S. A. Plotkin and W. A. Orenstein (ed.), Vaccines. The W. B. Saunders Co., Philadelphia, Pa.
- 17.Levine, M. M., J. Galen, E. Barry, F. Noriega, S. Chatfield, M. Sztein, G. Dougan, and C. Tacket. 1996. Attenuated Salmonella as live oral vaccines against typhoid fever and as live vectors. J. Biotechnol. 44:193-196. [DOI] [PubMed] [Google Scholar]
- 18.Lim, A., E. T. Dimalanta, K. D. Potamousis, G. Yen, J. Apodoca, C. Tao, J. Lin, R. Qi, J. Skiadas, A. Ramanathan, N. T. Perna, G. Plunkett III, V. Burland, B. Mau, J. Hackett, F. R. Blattner, T. S. Anantharaman, B. Mishra, and D. C. Schwartz. 2001. Shotgun optical maps of the whole Escherichia coli O157:H7 genome. Genome Res. 11:1584-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, S. L., and K. E. Sanderson. 1996. Highly plastic chromosomal organization in Salmonella typhi. Proc. Natl. Acad. Sci. USA 93:10303-10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, S. L., and K. E. Sanderson. 1995. Rearrangements in the genome of the bacterium Salmonella typhi. Proc. Natl. Acad. Sci. USA 92:1018-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahillon, J., H. A. Kirkpatrick, H. L. Kijenski, C. A. Bloch, C. K. Rode, G. F. Mayhew, D. J. Rose, G. Plunkett III, V. Burland, and F. R. Blattner. 1998. Subdivision of the Escherichia coli K-12 genome for sequencing: manipulation and DNA sequence of transposable elements introducing unique restriction sites. Gene 223:47-54. [DOI] [PubMed] [Google Scholar]
- 22.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 23.Ng, I., S. L. Liu, and K. E. Sanderson. 1999. Role of genomic rearrangements in producing new ribotypes of Salmonella typhi. J. Bacteriol. 181:3536-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohnishi, M., C. Tanaka, S. Kuhara, K. Ishii, M. Hattori, K. Kurokawa, T. Yasunaga, K. Makino, H. Shinagawa, T. Murata, K. Nakayama, Y. Terawaki, and T. Hayashi. 1999. Chromosome of the enterohemorrhagic Escherichia coli O157:H7: comparative analysis with K-12 MG1655 revealed the acquisition of a large amount of foreign DNAs. DNA Res. 6:361-368. [DOI] [PubMed] [Google Scholar]
- 25.Parkhill, J., G. Dougan, K. D. James, N. R. Thomson, D. Pickard, J. Wain, C. Churcher, K. L. Mungall, S. D. Bentley, M. T. Holden, M. Sebaihia, S. Baker, D. Basham, K. Brooks, T. Chillingworth, P. Connerton, A. Cronin, P. Davis, R. M. Davies, L. Dowd, N. White, J. Farrar, T. Feltwell, N. Hamlin, A. Haque, T. T. Hien, S. Holroyd, K. Jagels, A. Krogh, T. S. Larsen, S. Leather, S. Moule, P. O'Gaora, C. Parry, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 413:848-852. [DOI] [PubMed] [Google Scholar]
- 26.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 27.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 28.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robbe-Saule, V., C. Coynault, and F. Norel. 1995. The live oral typhoid vaccine Ty21a is a rpoS mutant and is susceptible to various environmental stresses. FEMS Microbiol. Lett. 126:171-176. [DOI] [PubMed] [Google Scholar]
- 30.Robbe-Saule, V., and F. Norel. 1999. The rpoS mutant allele of Salmonella typhi Ty2 is identical to that of the live typhoid vaccine Ty21a. FEMS Microbiol. Lett. 170:141-143. [DOI] [PubMed] [Google Scholar]
- 31.Rowe, B., L. R. Ward, and E. J. Threlfall. 1997. Multidrug-resistant Salmonella typhi: a worldwide epidemic. Clin. Infect. Dis. 24(Suppl. 1):S106-S109. [DOI] [PubMed]
- 32.Sanderson, K. E., and S. L. Liu. 1998. Chromosomal rearrangements in enteric bacteria. Electrophoresis 19:569-572. [DOI] [PubMed] [Google Scholar]
- 33.Sokurenko, E. V., D. L. Hasty, and D. E. Dykhuizen. 1999. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7:191-195. [DOI] [PubMed] [Google Scholar]
- 34.Spector, M. P., F. Garcia del Portillo, S. M. Bearson, A. Mahmud, M. Magut, B. B. Finlay, G. Dougan, J. W. Foster, and M. J. Pallen. 1999. The rpoS-dependent starvation-stress response locus stiA encodes a nitrate reductase (narZYWV) required for carbon-starvation-inducible thermotolerance and acid tolerance in Salmonella typhimurium. Microbiology 145:3035-3045. [DOI] [PubMed] [Google Scholar]
- 35.Sutton, A., R. Buencamino, and A. Eisenstark. 2000. rpoS mutants in archival cultures of Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:4375-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Testerman, T. L., A. Vazquez-Torres, Y. Xu, J. Jones-Carson, S. J. Libby, and F. C. Fang. 2002. The alternative sigma factor sigmaE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol. Microbiol. 43:771-782. [DOI] [PubMed] [Google Scholar]
- 37.Wain, J., D. House, J. Parkhill, C. Parry, and G. Dougan. 2002. Unlocking the genome of the human typhoid bacillus. Lancet Infect. Dis. 2:163-170. [DOI] [PubMed] [Google Scholar]
- 38.Zambrano, M. M., and R. Kolter. 1996. GASPing for life in stationary phase. Cell 86:181-184. [DOI] [PubMed] [Google Scholar]