Abstract
An O-acetylserine sulfhydrylase (OASS) from the hyperthermophilic archaeon Aeropyrum pernix K1, which shares the pyridoxal 5′-phosphate binding motif with both OASS and cystathionine β-synthase (CBS), was cloned and expressed by using Escherichia coli Rosetta(DE3). The purified protein was a dimer and contained pyridoxal 5′-phosphate. It was shown to be an enzyme with CBS activity as well as OASS activity in vitro. The enzyme retained 90% of its activity after a 6-h incubation at 100°C. In the O-acetyl-l-serine sulfhydrylation reaction, it had a pH optimum of 6.7, apparent Km values for O-acetyl-l-serine and sulfide of 28 and below 0.2 mM, respectively, and a rate constant of 202 s−1. In the l-cystathionine synthetic reaction, it showed a broad pH optimum in the range of 8.1 to 8.8, apparent Km values for l-serine and l-homocysteine of 8 and 0.51 mM, respectively, and a rate constant of 0.7 s−1. A. pernix OASS has a high activity in the l-cysteine desulfurization reaction, which produces sulfide and S-(2,3-hydroxy-4-thiobutyl)-l-cysteine from l-cysteine and dithiothreitol.
In bacteria such as Escherichia coli and Salmonella enterica serovar Typhimurium and in plants, l-cysteine is synthesized from l-serine in two steps (16, 27). Serine acetyltransferase (SAT) (EC 2.3.1.30) converts l-serine and acetyl coenzyme A into O-acetyl-l-serine (OAS) and coenzyme A (equation 1):
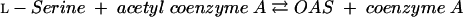 |
(1) |
O-Acetylserine sulfhydrylase (OASS) (EC 4.2.99.8) then catalyzes the formation of l-cysteine and acetic acid from OAS and sulfide (equation 2):
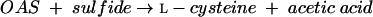 |
(2) |
E. coli and S. enterica serovar Typhimurium produce two OASS isoenzymes (OASS-A and -B). OASS-B is preferentially used for the biosynthesis of l-cysteine during anaerobic growth conditions (11) and is able to utilize thiosulfate instead of sulfide to produce S-sulfo-l-cysteine (33) (equation 3):
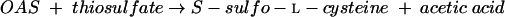 |
(3) |
SATs and OASSs have been purified from several bacterial sources, such as E. coli and S. enterica serovar Typhimurium, as well as from higher plants, and their characteristics have been investigated (16, 27). OASS exists as a dimer and contains pyridoxal 5′-phosphate (PLP).
In mammals, on the other hand, l-cysteine is synthesized from l-methionine (15). l-Methionine is converted to l-homocysteine by the actions of methionine adenosyltransferase (EC 2.5.1.6) (equation 4), various S-adenosylmethionine methyltransferases (EC 2.1.1.?) (equation 5), and adenosylhomocysteinase (EC 3.3.1.1) (equation 6):
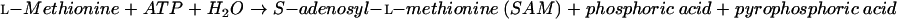 |
(4) |
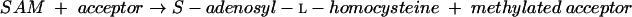 |
(5) |
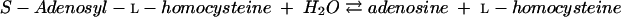 |
(6) |
Cystathionine β-synthase (CBS) (EC 4.2.1.22) is a PLP-dependent enzyme that catalyzes the condensation of l-homocysteine and l-serine to form l-cystathionine (equation 7):
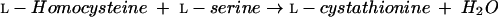 |
(7) |
l-Cystathionine is then converted to l-cysteine by cystathionine γ-lyase (EC 4.4.1.1) (equation 8):
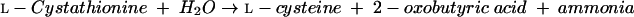 |
(8) |
CBSs from humans and Saccharomyces cerevisiae exist as tetramers and multimers (17, 24). Human CBS contains heme in addition to PLP (22). SAM activates CBSs from human and S. cerevisiae (5, 35). CBSs from S. cerevisiae, mammals, and Trypanosoma cruzi retain activity with respect to l-serine sulfhydrylation reaction (26, 34, 35, 37) (equation 9):
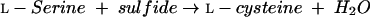 |
(9) |
The three-dimensional structures of S. enterica serovar Typhimurium OASS-A and human CBS without the C-terminal amino acid sequence from position 414 to 551 indicate that the amino acid residues surrounding the PLP binding site are conserved between OASS and CBS (6, 29).
In S. cerevisiae, sulfur is first incorporated into l-homocysteine by means of the enzyme coded for by MET17/MET25, which acts as both O-acetylhomoserine sulfhydrylase (EC 4.2.99.10) and OASS in vitro. l-Cysteine is then synthesized from l-homocysteine as well as from l-methionine via reverse a transsulfuration pathway (equations 4 to 8) (7, 36).
In the case of archaea, little is known concerning the pathway of l-cysteine biosynthesis despite the efforts of several investigators (3, 4, 25, 43). By a complementation analysis using a cysteine auxotrophic E. coli strain, the gene encoding OASS has been identified in the genome of Methanosarcina barkeri (25). The genes encoding SAT and OASS are adjacent to each other in the genomes of both M. barkeri (25) and Methanosarcina thermophila (3). OASS has been purified from cell extracts of M. thermophila and characterized (4). On the other hand, the results of genome sequencing analyses of archaea have shown that the genomes of Aeropyrum pernix, Pyrobaculum aerophilum (12), Pyrococcus abyssi, Pyrococcus furiosus, Sulfolobus solfataricus, Sulfolobus tokodaii (19), Thermoplasma acidophilum, and Thermoplasma volcanium (21) contain genes that have sequence similarity with OASS (25). Both SAT and OASS are required for the synthesis of l-cysteine, according to equations 1 and 2. However, no genes with a sequence similarity to SAT have yet been identified in these archaea.
A. pernix K1 is a hyperthermophilic archaeon that grows optimally in a temperature range of between 90 and 95°C (38). The putative product of the gene (open reading frame [ORF] APE1586) encoding OASS from A. pernix consists of 389 amino acids with a subunit molecular mass of 42 kDa (20). The physiological function of OASS in these archaea, including A. pernix, is presently unclear. We have recently attempted to clarify the physiological role of A. pernix OASS with respect to l-cysteine biosynthesis in A. pernix cells, and the results of this investigation are reported here.
MATERIALS AND METHODS
Materials.
The sources of materials used in this study were as follows: l-cysteine, sodium sulfide, and sodium thiosulfate were from Wako Pure Chemical Industries (Osaka, Japan); HiTrap Q HP and Superdex 200 pg were from Amersham Biosciences (Freiburg, Germany); OAS, PLP, l-homocysteine thiolactone, and l-cystathionine were from Sigma-Aldrich (St. Louis, Mo.); and S-sulfo-l-cysteine was from Tocris Cookson (Bristol, United Kingdom). All other reagents were of reagent grade.
Cloning of the gene.
The gene (ORF APE1586) encoding OASS was amplified from A. pernix genomic DNA by PCR with KOD DNA polymerase (Toyobo, Osaka, Japan) with 5′-GCCCTCGCCGACATAAGTGGTTACCTGGAC-3′ as the upper primer and 5′-TATGGATCCTAGACCGAGTCTCCGGCTCCT-3′ as the lower primer. The BamHI restriction enzyme site in the lower primer is indicated by an underline. To prepare the vector plasmid, pET-3d (Novagen, Madison, Wis.) was digested with NcoI and blunt ended with T4 DNA polymerase, followed by digestion with BamHI. The PCR-amplified gene was digested with BamHI and ligated into the vector plasmid. The resultant plasmid was designated pAPE1586. By using this method, the initial codon of the gene APE1586, GTG, was changed to ATG to permit efficient expression of the enzyme in E. coli.
Cultivation of the cells.
The E. coli Rosetta(DE3) strain (Novagen) was transformed with plasmid pAPE1586. Cultivation of the cells was done by a modification of the method of Kery et al. (23). The transformant cells were grown in 1 liter of NZCYM medium (39) containing 100 μg of ampicillin per ml, 34 μg of chloramphenicol per ml, 0.001% pyridoxine, and 0.001% thiamine, with or without 0.3 mM 5-aminolevulinic acid (ALA), with reciprocal shaking at 100 rpm and 37°C. When the absorbance at 600 nm of the culture medium reached around 0.5, isopropyl-β-d-thiogalactopyranoside was added to the medium at a final concentration of 0.01 mM. The cells were allowed to grow for an additional 19 h and were then washed with 0.9% NaCl and frozen at −80°C.
Enzyme purification.
The cells (5.9 g [wet weight]) were suspended in 150 ml of 50 mM potassium phosphate buffer (pH 7.5) containing 2 mM EDTA, 1 mM dithiothreitol, 0.2 mM PLP, and 0.15 M NaCl (buffer A) and then sonically disrupted with a homogenizer (model 450D; Branson Ultrasonics Corp., Danbury, Conn.) at 90 W for 3 min on ice. Cell debris was removed by centrifugation (18,000 × g for 30 min at 4°C). PLP was added to the supernatant at a concentration of 1 mM, and the mixture was stirred for 30 min at 4°C. This solution was regarded as the crude extract. The crude extract was heated at 85°C for 20 min. The supernatant was recovered by centrifugation (18,000 × g for 20 min). The supernatant was fractionated with ammonium sulfate. The precipitate that formed at 50% ammonium sulfate saturation was recovered by centrifugation and dissolved in 7 ml of 50 mM potassium phosphate buffer (pH 7.5) containing 2 mM EDTA, 1 mM dithiothreitol, and 0.2 mM PLP (buffer B). This solution was dialyzed against buffer B at 4°C. The dialysate was placed on a HiTrap Q HP column (1.6 by 2.5 cm) equilibrated with buffer B and eluted by using a linear increase of NaCl concentration in buffer B from 0 to 1 M at a flow rate of 2 ml/min. The enzyme fractions that eluted between 0.3 and 0.4 M NaCl were collected and concentrated with a Centricon 10 filter (Amicon, Beverly, Mass.). The enzyme solution was placed on a Superdex 200 pg column (2.6 by 61 cm) and eluted with buffer A at a flow rate of 1.5 ml/min. The enzyme fractions were concentrated and dialyzed against buffer A at 4°C. The enzyme solution at a final concentration of 20 mg/ml was stored at −80°C.
Enzyme assays.
PLP was added to all assay solutions at a concentration of 0.2 mM. For the OAS sulfhydrylation reaction, the assay mixture (0.3 ml) contained 100 mM potassium phosphate buffer (pH 6.7), 20 mM OAS, 1 mM sodium sulfide, 1 mM EDTA, and an enzyme concentration of 0.0027 mg/ml. For the S-sulfo-l-cysteine synthetic reaction, the assay mixture was the same as that used for the OAS sulfhydrylation reaction except that the reaction pH and enzyme concentration were pH 6.1 and 0.01 mg/ml, respectively, and sodium thiosulfate was used instead of sodium sulfide at a concentration of 20 mM. Reactions were carried out for 4 min and then stopped by the addition of 0.15 ml of 20% trichloroacetic acid. The solutions were centrifuged at 18,000 × g for 3 min. A 0.3-ml portion of the supernatant was used to determine the amount of l-cysteine or S-sulfo-l-cysteine by the method of Gaitonde (14). For the l-cystathionine synthetic reaction, the assay mixture (0.3 ml) consisted of 100 mM sodium carbonate buffer (pH 8.3), 30 mM l-serine, 0.7 mM l-homocysteine, and an enzyme concentration of 0.067 mg/ml. l-Homocysteine was prepared from l-homocysteine thiolactone as described by Drummond et al. (9). Reactions were carried out for 20 min. Thirty-microliter aliquots of solutions were withdrawn and mixed with 6 μl of 6 N HCl. The solutions were centrifuged at 18,000 × g for 3 min. A 20-μl portion of the supernatant was dried and treated with phenylisothiocyanate. The resulting phenylthiocarbamoyl-cystathionine was measured by using a Wakosil-PTC column (4 by 150 mm) (Wako) connected to a high-pressure liquid chromatography system (DP-8020; Tosoh, Tokyo, Japan) monitored at 254 nm and 40°C. Elution was done by using a linear increase of acetonitrile concentration from 6 to 46% in 60 mM sodium acetate buffer (pH 6.0) at a flow rate of 1 ml/min. Measurement of the activity of the l-serine sulfhydrylation reaction was done under conditions identical to that used for the OAS sulfhydrylation reaction described above except that OAS was replaced with 20 mM l-serine and that 100 mM sodium carbonate buffer (pH 8.5) and 0.1 mg of the enzyme per ml were used. The l-cysteine desulfurization reaction was done by a modification of the method of Flint et al. (13). The assay mixture (0.6 ml) consisted of 100 mM N-tris(hydroxymethyl)methyl-3-aminopropanesulfonic acid (TAPS)-NaOH buffer (pH 8.5), 2.5 mM l-cysteine, 5 mM dithiothreitol, and an enzyme concentration of 0.25 μg/ml. Reactions were carried out for 15 min in a screw-cap vial to avoid leakage of the sulfide vapor produced. The amount of sulfide was measured by the method of Esaki et al. (10).
Reaction mixtures without enzyme were used as blanks. One unit of enzyme activity was defined as the amount of enzyme required to catalyze the formation of 1 μmol of product per min.
Effect of temperature on enzyme activity and stability.
The OASS and CBS activities for A. pernix OASS were measured in the assay solutions described above in a temperature range of 20 to 90°C. A 0.8-ml portion of the enzyme solution (0.3 mg/ml) was incubated at 4 or 100°C for 6 h in 50 mM potassium phosphate buffer (pH 6.1, 6.7, 7.5, or 8.5) containing 2 mM EDTA and 0.2 mM PLP to examine the thermal stability of the enzyme. The relative remaining S-sulfo-l-cysteine synthetic activities were measured at 85°C as described above. The activity prior to incubation was set to 100%.
Kinetic analyses.
The S-sulfo-l-cysteine synthetic reaction, OAS sulfhydrylation reaction, and l-serine sulfhydrylation reaction were carried out at 85, 60, and 70°C, respectively, in 3 ml of the assay mixtures as described above. For the S-sulfo-l-cysteine synthetic reaction, the concentrations of OAS and sodium thiosulfate were varied from 5 to 30 mM. For the OAS and l-serine sulfhydrylation reactions, the concentrations of OAS and l-serine were changed from 5 to 100 mM and from 5 to 120 mM, respectively, at a fixed sodium sulfide concentration of 2 mM. The concentration of sodium sulfide was varied between 0.2 and 2 mM with 20 mM OAS or with 100 mM l-serine. After 40, 80, 120, 160, and 200 s of incubation, 0.22-ml portions of the solutions were withdrawn and mixed with 0.11 ml of 20% trichloroacetic acid. The solutions were centrifuged, and 0.3-ml portions of the supernatants were then used to determine the amount of S-sulfo-l-cysteine and l-cysteine by the method of Gaitonde (14). A kinetic analysis of the l-cystathionine synthetic reaction was done at 85°C with the assay mixture described above. The concentrations of l-serine and l-homocysteine were changed from 2.5 to 30 mM at a fixed l-homocysteine concentration of 1.5 mM and from 0.15 to 1.5 mM at a 30 mM concentration of l-serine, respectively. A 30-μl portion of the reaction mixture was withdrawn after 5, 10, and 15 min of incubation. The l-cystathionine produced was determined as described above.
Kinetic constants for the S-sulfo-l-cysteine synthetic reaction were determined by using an equation that represents a ping-pong bi-bi mechanism (40). Kinetic constants for the l-cystathionine synthetic reaction and the OAS and l-serine sulfhydrylation reactions were determined by using the Michaelis-Menten equation (40).
Purification and identification of a coproduct in the l-cysteine desulfurization reaction.
l-Cysteine (100 mM), dithiothreitol (500 mM), and A. pernix OASS (0.08 mg/ml) were allowed to react for 1.5 h at 85°C in 3 ml of 100 mM TAPS-NaOH buffer (pH 8.5). The reaction was stopped by the addition of 70 μl of 6 N HCl. A 35-μl portion of the solution was injected onto a YMC-Pack ODS-A column (20 by 150 mm) (YMC, Kyoto, Japan) connected to a high-pressure liquid chromatography system (Tosoh CCPS). Elution was accomplished with 0.5% acetonitrile containing 0.1% HCl at a flow rate of 7.9 ml/min and monitored by absorbance at 210 nm. This chromatography was repeated for a total of 10 times. Fractions (80 ml) containing a coproduct were evaporated and then lyophilized. The concentration of the product was determined by means of an amino acid analyzer (L-8500; Hitachi, Tokyo, Japan) with the averaged peak area of an amino acid standard solution (type H; Wako). Approximately 2 mg of the coproduct was subjected to one- and two-dimensional 1H nuclear magnetic resonance (NMR) analyses. One-dimensional 1H NMR spectra of l-cysteine and dithiothreitol were also measured as controls. For the matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOFMS) experiment, approximately 1 μg of the purified product was used.
Spectroscopic measurements.
Absorption spectra of A. pernix OASS were obtained at room temperature with a UV-1600PC spectrophotometer (Shimadzu, Kyoto, Japan). The PLP content of the enzyme was determined by the procedure described by Kaplan and Flavin (18). NMR experiments were done with a JEOL-JNM A-500 spectrometer (500 MHz) (Oxford Instruments, Oxford, United Kingdom). All NMR spectra were measured in D2O solution containing 3-(trimethylsilyl)-1-propanesulfonic acid (Merck, Darmstadt, Germany). MALDI-TOFMS experiments were done with a Voyager DE-STR mass spectrometer (PerSeptive Biosystems, Framingham, Mass.). α-Cyano-4-hydroxy cinnamic acid (Sigma-Aldrich) was used for calibrating the instrument, and 2,5-dihidroxybenzoic acid (Sigma-Aldrich) was used as the matrix.
Sedimentation equilibrium experiment.
The molecular mass of the native enzyme was determined by using an Optima XL-A analytical ultracentrifuge (Beckman Instruments, Fullerton, Calif.) at 20°C.
Protein assay.
Protein concentrations were determined with a DC protein assay kit (Bio-Rad, Hercules, Calif.) with bovine serum albumin as a standard.
SDS-PAGE.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out according to the method of Laemmli (28) with a 12% gel. Phosphorylase b (94 kDa), bovine serum albumin (67 kDa), ovalbumin (43 kDa), carbonic anhydrase (30 kDa), and soybean trypsin inhibitor (20.1 kDa) (Amersham Biosciences LMW electrophoresis calibration kit) were used as markers. The protein bands were stained with Coomassie brilliant blue R-250.
N-terminal amino acid sequence analysis.
Approximately 2 μg of the purified enzyme was developed by SDS-PAGE and then blotted electrically onto a Sequi-Blot polyvinylidene difluoride membrane (Bio-Rad). The N-terminal amino acid sequence was determined by the Takara customer service center.
Multiple-sequence alignment.
Amino acid sequences of the A. pernix proteins were obtained from the database of genomes analyzed at the National Institute of Technology and Evaluation (http://www.bio.nite.go.jp:8080/dogan/MicroTop). The amino acid sequences of known OASSs and CBSs were obtained from the EMBL database. The amino acid sequences were aligned by using the CLUSTAL W software program (42).
RESULTS
Multiple-sequence alignment.
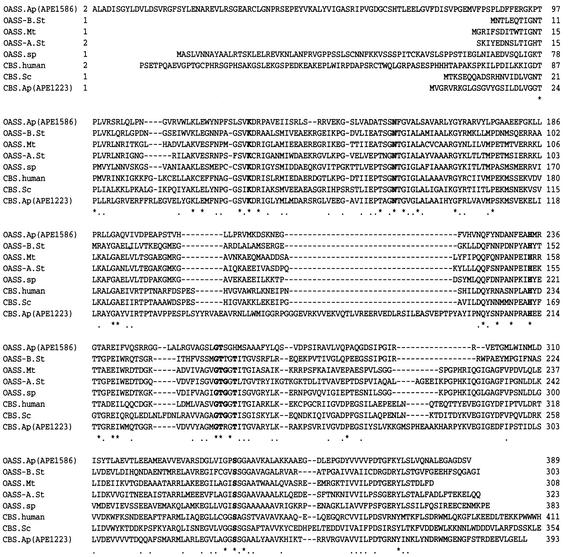
The deduced amino acid sequence of A. pernix OASS (ORF APE1586) had the following sequence identities: 31% with S. enterica serovar Typhimurium OASS-B, 26% with M. thermophila OASS, 24% with S. enterica serovar Typhimurium OASS-A, 21% with spinach chloroplast OASS, 20% with human CBS, 15% with S. cerevisiae CBS, and 21% with an A. pernix protein (ORF APE1223) predicted to be CBS. Alignment of the amino acid sequences of these enzymes showed that the A. pernix OASS shares the PLP binding motif (6, 29) with both OASS and CBS (Fig. 1). The A. pernix OASS has an additional amino acid sequence of around 75 amino acid residues at the N terminus compared to S. enterica serovar Typhimurium OASS-A and -B, M. thermophila OASS, and S. cerevisiae CBS. The amino acid residues, Cys-52 and His-65, responsible for the binding of heme for human CBS (29) were not conserved in A. pernix OASS. The C-terminal region, corresponding to residues 414 to 551 of human CBS and residues 354 to 507 of S. cerevisiae CBS, which are required for both the binding of SAM and the dimer-dimer interaction (17, 24), was absent from the A. pernix OASS.
FIG. 1.
Multiple-sequence alignment of A. pernix OASS with various OASSs and CBSs. Abbreviations: OASS.Ap(APE1586), an A. pernix protein (ORF APE1586) predicted to be OASS; OASS-B.St, S. enterica serovar Typhimurium OASS-B (EMBL accession no. P29848); OASS.Mt, M. thermophila OASS (AAG01804); OASS-A.St, S. enterica serovar Typhimurium OASS-A (P12674); OASS.sp, spinach chloroplast OASS (P32260); CBS.human, N-terminal region of human CBS (P35520); CBS.Sc, N-terminal region of S. cerevisiae CBS (P32582); CBS.Ap(APE1223), an A. pernix protein (ORF APE1223) predicted to be CBS. Hyphens indicate gaps introduced for optimal alignment. Asterisks and dots indicate identical and similar amino acids, respectively. Boldface letters depict the PLP binding motif common to both OASS and CBS.
Purification and physical properties of the enzyme.
E. coli Rosetta(DE3) carrying plasmid pAPE1586 was cultivated in medium without ALA, a precursor of heme. A. pernix OASS was purified by using the recombinant E. coli cells. Table 1 summarizes the purification of A. pernix OASS. Approximately 64 mg of A. pernix OASS was obtained in a yield of 29% on the basis of the activity of the OAS sulfhydrylation reaction. Purification yields calculated from activities of the S-sulfo-l-cysteine and l-cystathionine synthetic reactions and the l-cysteine desulfurization reaction were 30, 29, and 28%, respectively. The purified enzyme was homogeneous on an SDS-polyacrylamide gel, with an apparent subunit molecular mass of 42 kDa (Fig. 2). An N-terminal sequence analysis of the first seven amino acid residues of the purified protein showed that the residues were identical to those obtained from the A. pernix genome sequence database except that an initial Met residue was missing, probably due to the action of methionine aminopeptidase from E. coli (2). A sedimentation equilibrium experiment with A. pernix OASS indicated a molecular mass of 70,583 Da. Considering the subunit molecular mass of A. pernix OASS, it is likely that the A. pernix OASS exists as a dimer. The absorption spectrum of A. pernix OASS exhibited two peaks at 280 and 412 nm. The appearance of the peak at 412 nm indicates the formation of a Schiff base with PLP (30). The peak at 428 nm, typical for the heme observed for human CBS (22), was not present in the absorption spectrum of A. pernix OASS. A. pernix OASS was prepared from recombinant E. coli cells that had been cultivated in medium supplemented with ALA at a purification yield of 24%. The enzyme showed the same spectrum as that of the A. pernix OASS purified previously. The ratio of the absorbance at 280 nm to that at 412 nm for A. pernix OASS was 6.2. This is high compared with those of OASS-As from E. coli (4.0) (13) and S. enterica serovar Typhimurium (3.5) (1). The PLP content of A. pernix OASS was found to be 1.2 per enzyme subunit. Incubation of the purified A. pernix OASS in the presence of PLP at a concentration of 2 mM for 4 h at 4°C failed to increase the activity of the OAS sulfhydrylation reaction. These findings indicate that the purified A. pernix OASS is a holoenzyme.
TABLE 1.
Purification of A. pernix OASS
| Purification step | Vol (ml) | Total activitya (U) | Total protein (mg) | Sp acta (U/mg) | Yield (%) |
|---|---|---|---|---|---|
| Crude extract | 150 | 11,070 | 930 | 11.9 | 100 |
| Heat treatment | 142 | 7,210 | 554 | 13.0 | 65 |
| Ammonium sulfate | 11 | 6,220 | 189 | 32.9 | 56 |
| HiTrap Q HP | 3.9 | 5,760 | 164 | 35.1 | 52 |
| Superdex 200 pg | 3.2 | 3,250 | 63.8 | 50.9 | 29 |
The activity of the OAS sulfhydrylation reaction was measured at 60°C as described in the text.
FIG. 2.
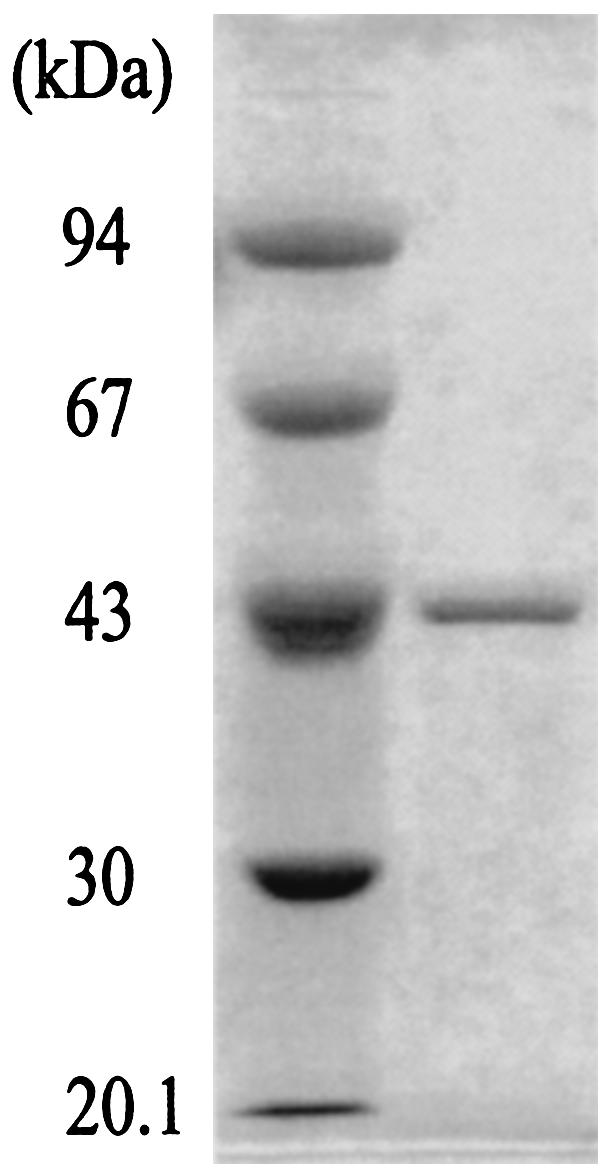
SDS-PAGE of the purified A. pernix OASS. Marker proteins with the indicated molecular masses are shown in the left lane, and the purified A. pernix OASS (1 μg) is in the right lane.
Effect of pH and temperature on enzyme activity and stability.
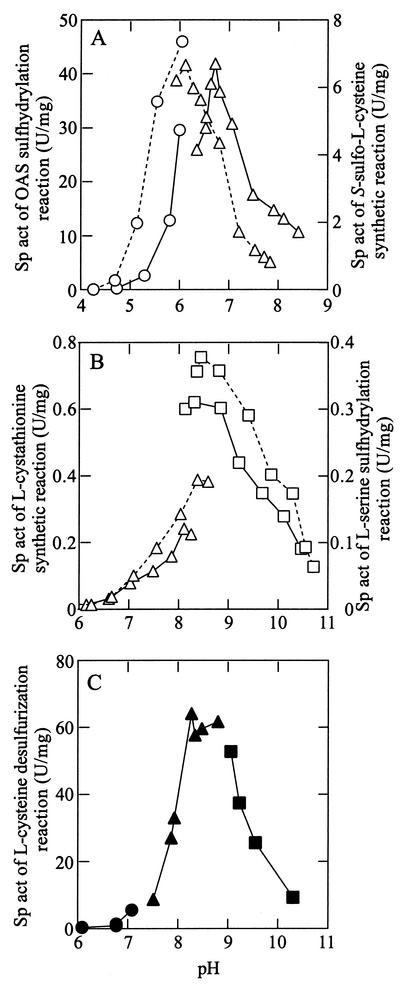
The pH dependencies of the enzyme activities observed for A. pernix OASS are shown in Fig. 3. The optimal pHs for the OAS sulfhydrylation and S-sulfo-l-cysteine synthesis reactions were in a slightly acidic region (Fig. 3A), whereas those for the l-cystathionine synthesis, l-serine sulfhydrylation, and l-cysteine desulfurization reactions were in the alkaline region (Fig. 3B and C). The activity of the l-cystathionine synthetic reaction measured at pH 8.3 was unaffected by the addition of SAM at a concentration of 0.5 mM to the assay solution.
FIG. 3.
pH Dependencies of the enzyme activities for A. pernix OASS at 85°C. (A) Specific activities for the OAS sulfhydrylation reaction (equation 2) and S-sulfo-l-cysteine synthetic reaction (equation 3) are shown by solid and dashed lines, respectively. (B) Specific activities for the l-cystathionine synthetic reaction (equation 7) and l-serine sulfhydrylation reaction (equation 9) are depicted by solid and dashed lines, respectively. (C) Specific activity for the l-cysteine desulfurization reaction. In panels A and B, citrate-NaOH buffer (○), potassium phosphate buffer (▵), and sodium carbonate buffer (□) were used. In panel C, 2-morpholinoethanesulfonic acid-NaOH buffer (•), TAPS-NaOH buffer (▴), and cyclohexylaminopropanesulfonic acid-NaOH buffer (▪) were used.
A plot of the observed activity for the S-sulfo-l-cysteine synthetic reaction against temperature showed that the A. pernix OASS exhibited the highest activity at or above 90°C. On the other hand, the activity of the OAS sulfhydrylation reaction showed maxima at both 70 and 80°C. In the case of the l-cystathionine synthetic reaction and l-serine sulfhydrylation reaction, both activities had a maximum at 80°C. The A. pernix OASS retained around 90% of its activity after the enzyme solutions had been incubated for 6 h at 100°C at pH 6.1 and 6.7. The relative remaining activities of the enzyme solutions incubated at pH 7.5 and 8.5 were 56 and 11%, respectively. No loss of enzyme activities was observed for enzyme solutions stored at 4°C.
Kinetic analyses.
Double-reciprocal plots of the initial velocity for the S-sulfo-l-cysteine synthetic reaction were represented as a series of parallel lines. This indicates that the S-sulfo-l-cysteine synthetic reaction followed a typical ping-pong bi-bi mechanism, which was identical to the kinetic mechanism of the OAS sulfhydrylation reaction for OASS-As from E. coli and S. enterica serovar Typhimurium (31, 41). In this mechanism, OAS first binds to the enzyme to form a Schiff base with PLP, releasing acetic acid. Second, thiosulfate attacks the α-aminoacrylate intermediate of the enzyme to form S-sulfo-l-cysteine. At low concentrations of thiosulfate, a slight inhibition by OAS was observed. The kinetic parameters for the reactions of S-sulfo-l-cysteine synthesis and OAS sulfhydrylation for A. pernix OASS are summarized in Table 2. The kinetic parameters for the reactions of l-cystathionine synthesis and l-serine sulfhydrylation are shown in Table 3. The kinetic constants obtained for known OASSs and CBSs are also shown in Tables 2 and 3 for comparison. It can be seen that the kcat value for the OAS sulfhydrylation reaction for A. pernix OASS differed by two orders of magnitude from those for the l-cystathionine synthetic reaction and l-serine sulfhydrylation reaction for A. pernix OASS. The apparent Km values for sulfide for A. pernix OASS were the lowest among those for the substrates in the reactions catalyzed by A. pernix OASS.
TABLE 2.
Comparison of kinetic parameters of the OAS sulfhydrylation reaction and S-sulfo-l-cysteine synthetic reaction for A. pernix OASS and for known OASSs and CBSs
| Enzyme (reference) | OAS sulfhydrylation
|
S-Sulfo-l-cysteine synthesis
|
||||
|---|---|---|---|---|---|---|
| Km for OAS (mM) | Km for sulfide (mM) | kcat (s−1) | Km for OAS (mM) | Km for thiosulfate (mM) | kcat (s−1) | |
| A. pernix OASS | 28 | <0.2 | 202 | 13 | 21 | 24 |
| S. enterica serovar Typhimurium OASS-B (33) | 9 | 2.7 | 341 | |||
| E. coli OASS-A (31) | 4.8 | 0.006 | 2,030 | |||
| Spinach chloroplast OASS (8) | 1.3 | 0.25 | ||||
| T. thermophilus OASS (32) | 4.8 | 0.05 | 493 | |||
| S. cerevisiae CBS (35) | 1.27 | 16.6 | 19.2 | |||
| T. cruzi CBS (34) | 4.9 | 4.1 | 12.5 | |||
TABLE 3.
Comparison of kinetic parameters of the l-cystathionine synthetic reaction and l-serine sulfhydrylation reaction for A. pernix OASS and for known CBSs
| Enzyme (reference) |
l-Cystathionine synthesis
|
l-Serine sulfhydrylation
|
||||
|---|---|---|---|---|---|---|
| Km for l-serine (mM) | Km for l-homocysteine (mM) | kcat (s−1) | Km for l-serine (mM) | Km for sulfide (mM) | kcat (s−1) | |
| A. pernix OASS | 8.0 | 0.51 | 0.7 | 31 | <0.2 | 0.56 |
| Human CBS (24) | 3.0 | 1.5 | 6.0 | |||
| S. cerevisiae CBS (35) | 2.19 | 2.25 | 7.97 | 4.42 | 16.8 | 20.7 |
| T. cruzi CBS (34) | 1.0 | 0.9 | 21 | 1.1 | 3.1 | 12.8 |
Identification of a coproduct of the l-cysteine desulfurization reaction.
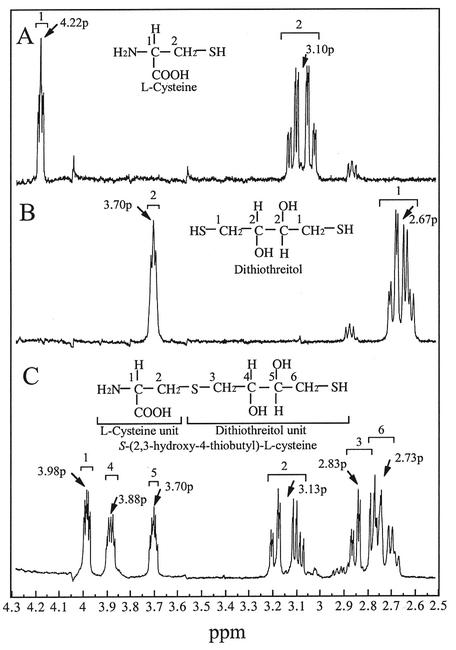
The l-cysteine desulfurization reaction for A. pernix OASS was dependent on the concentration of dithiothreitol in the reaction mixture. The activity reached a maximum at a dithiothreitol concentration of 40 mM when l-cysteine was present at a concentration of 2.5 mM in the assay solution. It has been reported that E. coli OASS-A and -B catalyze the formation of sulfide from l-cysteine in the presence of dithiothreitol (13). Flint et al. proposed a reaction mechanism for l-cysteine desulfurization for E. coli OASS-A and -B (13). In this mechanism l-cysteine first binds to the enzyme to form a Schiff base with PLP. Sulfide is then released from the enzyme-bound l-cysteine. Dithiothreitol then attacks the α-aminoacrylate intermediate to form S-(2,3-hydroxy-4-thiobutyl)-l-cysteine. The second product has never been identified. The coproduct of the l-cysteine desulfurization reaction for A. pernix OASS was purified and subjected to NMR and MALDI-TOFMS analyses. Figure 4A and B show one-dimensional 1H NMR spectra of the substrates l-cysteine and dithiothreitol as controls, respectively. The resonances centered around 4.22 and 3.10 ppm arise from the C-1 and C-2 protons of l-cysteine, respectively (Fig. 4A). The resonances centered around 3.70 and 2.67 ppm are assigned to the C-2 and C-1 protons of dithiothreitol (Fig. 4B). The relative ratios for the areas under each envelope were 1.0 and 2.2, respectively, for l-cysteine and 1.0 and 2.0, respectively, for dithiothreitol. Figure 4C shows a one-dimensional 1H NMR spectrum of the coproduct purified from the reaction mixture of the l-cysteine desulfurization for A. pernix OASS. Considering the NMR spectra of l-cysteine and dithiothreitol (Fig. 4A and B) and the two-dimensional NMR spectrum of the purified coproduct (data not shown), six envelopes of resonances centered around 3.98, 3.88, 3.70, 3.13, 2.83, and 2.73 ppm were identified as the protons belonging to C-1, C-4, C-5, C-2, C-3, and C-6, respectively, of S-(2,3-hydroxy-4-thiobutyl)-l-cysteine (Fig. 4C). The relative ratios for the areas under each envelope were 1.0, 0.88, 0.81, 0.80, 1.5, and 3.6 (sum of the envelopes of the resonances centered around 2.83 and 2.73 ppm), respectively. MALDI-TOFMS data for the purified coproduct showed a single peak corresponding to a mass of 241.98. This is in good agreement with the mass of the protonated molecular ion for S-(2,3-hydroxy-4-thiobutyl)-l-cysteine, 242.052. These results indicate that the coproduct formed with dithiothreitol in the l-cysteine desulfurization reaction for A. pernix OASS was, in fact, S-(2,3-hydroxy-4-thiobutyl)-l-cysteine.
FIG. 4.
1H NMR spectra. (A) l-Cysteine. (B) Dithiothreitol. (C) The coproduct purified from the reaction mixture of l-cysteine desulfurization for A. pernix OASS.
Substrate specificities for the l-cysteine desulfurization reaction.
Substrate specificities for the l-cysteine desulfurization reaction were examined at 85°C in the assay mixture described in Materials and Methods, except that various compounds were added in the place of l-cysteine or dithiothreitol. The l-cysteine desulfurization reaction did not proceed when l-cysteine was replaced with d-cysteine or l-homocysteine. On the other hand, 2-mercaptoethanol, ethanethiol, l-homocysteine, d-cysteine, dl-homocysteine, pyrazole, and glutathione were utilized as nucleophiles instead of dithiothreitol. Their respective relative activities against dithiothreitol were 121, 114, 35, 30, 26, 23, and 8.2%.
DISCUSSION
Little is known concerning the pathway of l-cysteine biosynthesis in archaea, despite several investigations (3, 4, 25, 43). In this study, A. pernix OASS was purified from recombinant E. coli cells and characterized with respect to its physical and catalytic properties.
The purified A. pernix OASS existed as a dimer with one molecule of PLP per subunit of the enzyme, consistent with other OASSs (1, 8, 13, 32). Unlike human CBS, the A. pernix OASS lacked heme and existed as dimmer, and the CBS activity for A. pernix OASS was unaffected by the addition of SAM. The pH optimum for the OAS sulfhydrylation reaction for A. pernix OASS, 6.7 (Fig. 3A), was compatible with that of Thermus thermophilus OASS (pH 7) (32) but was in the acidic region compared to those of E. coli OASS-A (pH 8 to 9.5) (31) and spinach chloroplast OASS (pH 7.5 to 8.5) (8). The pH optimum for the l-cystathionine synthetic reaction for A. pernix OASS, 8.1 to 8.8 (Fig. 3B), was consistent with those for CBSs from humans (pH 8.4 to 9.0) (26) and S. cerevisiae (pH ∼8.5) (17). The difference between the pH optimum for the OAS sulfhydrylation reaction and that for the l-cystathionine synthetic reaction for A. pernix OASS suggests that the catalytic mechanisms of the reactions are different. As is typical of enzymes of hyperthermophiles, the A. pernix OASS was most active at or above 90°C for the S-sulfo-l-cysteine synthetic reaction and was highly stable during incubation at 100°C.
The kinetic parameters for the reactions catalyzed by A. pernix OASS were determined and compared with those for known OASSs and CBSs. The kcat values for the OAS sulfhydrylation and S-sulfo-l-cysteine synthetic reactions for A. pernix OASS were lower than those for known OASSs but higher than those for the OAS sulfhydrylation reaction for known CBSs (Table 2). The kcat values for the l-cystathionine synthetic reaction and l-serine sulfhydrylation reaction for A. pernix OASS were lower than those for known CBSs (Table 3). For the OAS sulfhydrylation reaction, the apparent Km value for sulfide for A. pernix OASS seems to be compatible with those for known OASSs, but that for OAS for A. pernix OASS was high (Table 2). For the S-sulfo-l-cysteine synthetic reaction, the A. pernix OASS showed a higher Km value for thiosulfate than that for S. enterica serovar Typhimurium OASS-B, although the deduced amino acid sequence of A. pernix OASS had the highest identity with that of S. enterica serovar Typhimurium OASS-B. For both the l-cystathionine synthetic reaction and the l-serine sulfhydrylation reaction, the apparent Km values for l-serine for A. pernix OASS were higher than those for known CBSs, whereas that for sulfide for A. pernix OASS was lower than those for the CBSs (Table 3). The fact that A. pernix OASS had a high affinity for sulfide suggests that A. pernix OASS plays a role in the sulfur assimilation pathway. In the genome sequence of A. pernix, hypothetical gene products with sequence similarities to methionine adenosyltransferase (ORF APE1596) (equation 4), adenosylhomocysteinase (APE0624) (equation 6), CBS (APE1223) (equation 7), and cystathionine γ-lyase (APE1226) (equation 8), which are involved in l-cysteine biosynthesis in mammals, have been identified (20). The issues of whether these ORFs have the expected enzyme activities and whether SAT exists in A. pernix cells have not been investigated. Further analyses of these points will be necessary to clarify the pathway of l-cysteine biosynthesis in archaea, including A. pernix, as well as the physiological meaning of A. pernix OASS.
The A. pernix OASS used various thiol compounds and pyrazole in lieu of dithiothreitol to release sulfide from l-cysteine. It has been reported that E. coli OASS-A utilizes several different nucleophiles instead of dithiothreitol in the l-cysteine desulfurization reaction (13). Flint et al. identified the nonproteineous amino acids formed with the nucleophiles homocysteine, cysteine, azide, and 2-mercaptoethanol for E. coli OASS-A by amino acid analysis and mass spectral analysis (13). They also predicted that a nonproteineous amino acid formed with dithiothreitol is S-(2,3-thiohydroxybutyl)-l-cysteine according to the proposed reaction mechanism, but they did not identify it. We first identified the coproduct formed with dithiothreitol as S-(2,3-thiohydroxybutyl)-l-cysteine in the l-cysteine desulfurization reaction for A. pernix OASS by NMR and MALDI-TOFMS analyses. These results suggest that the l-cysteine desulfurization reaction for A. pernix OASS proceeds in the same manner as that for E. coli OASS-A. The rate constant for the l-cysteine desulfurization reaction for A. pernix OASS was 84 s−1 when the assay was carried out at 2.5 mM l-cysteine and 5 mM dithiothreitol at pH 8.5. This is higher than the rate constants for E. coli OASS-A (0.005 s−1) and OASS-B (0.015 s−1) (13).
It has been speculated that OASS and CBS would have diverged from an ancestral enzyme (35). However, the issue of how an ancestral enzyme might have evolved or diverged into the individual enzymes involved in the different pathways of l-cysteine biosynthesis remains unclear. This study revealed that the A. pernix OASS had CBS activity as well as OASS activity in vitro. There are, to our knowledge, only two enzymes that exhibit the activities of both OASS and CBS; these are S. cerevisiae CBS (35) and T. cruzi CBS (34). Based on the findings obtained in this study, it appears that the A. pernix OASS retains the properties of an ancestral enzyme that would have diverged later into individual enzymes with narrower substrate specificities and higher rate constants and by reducing the catalytic efficiencies of other reactions. Information concerning the three-dimensional structure of A. pernix OASS might be helpful in understanding how an ancestral enzyme might have diverged into the individual enzymes involved in the different pathways of l-cysteine biosynthesis.
Acknowledgments
We thank Miyo Sakai (Osaka University) for ultracentrifuge analyses, Atsuyoshi Nakayama for NMR analyses, Tsutomu Nakamura for mass spectroscopy analyses, Ha-Young Kim for technical assistance, and Kazuhiro Nakanishi (Okayama University) for amino acid analyses and for helpful advice.
K.M. was supported by the New Energy Industrial Technology Development Organization (NEDO).
REFERENCES
- 1.Becker, M. A., N. M. Kredich, and G. M. Tomkins. 1969. The purification and characterization of O-acetylserine sulfhydrylase-A from Salmonella typhimurium. J. Biol. Chem. 244:2418-2427. [PubMed] [Google Scholar]
- 2.Ben-Bassat, A., K. Bauer, S.-Y. Chang, K. Myambo, A. Boosman, and S. Chang. 1987. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J. Bacteriol. 169:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borup, B., and J. G. Ferry. 2000. Cysteine biosynthesis in the Archaea: Methanosarcina thermophila utilizes O-acetylserine sulfhydrylase. FEMS Microbiol. Lett. 189:205-210. [DOI] [PubMed] [Google Scholar]
- 4.Borup, B., and J. G. Ferry. 2000. O-Acetylserine sulfhydrylase from Methanosarcina thermophila. J. Bacteriol. 182:45-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukovska, G., V. Kery, and J. P. Kraus. 1994. Expression of human cystathionine β-synthase in Escherichia coli: purification and characterization. Protein Express. Purif. 5:442-448. [DOI] [PubMed] [Google Scholar]
- 6.Burkhard, P., G. S. Jagannatha Rao, E. Hohenester, K. D. Schnackerz, P. F. Cook, and J. N. Jansonius. 1998. Three-dimensional structure of O-acetylserine sulfhydrylase from Salmonella typhimurium. J. Mol. Biol. 283:121-133. [DOI] [PubMed] [Google Scholar]
- 7.Cherest, H., and Y. Surdin-Kerjan. 1992. Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating of the sulfur metabolism pathway. Genetics 130:51-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Droux, M., J. Martin, P. Sajus, and R. Douce. 1992. Purification and characterization of O-acetylserine (thiol) lyase from spinach chloroplasts. Arch. Biochem. Biophys. 295:379-390. [DOI] [PubMed] [Google Scholar]
- 9.Drummond, J. T., J. Jarrett, J. C. González, S. Huang, and R. G. Matthews. 1995. Characterization of nonradioactive assays for cobalamin-dependent and cobalamin-independent methionine synthase enzymes. Anal. Biochem. 228:323-329. [DOI] [PubMed] [Google Scholar]
- 10.Esaki, N., T. Nakamura, H. Tanaka, and K. Soda. 1982. Selenocysteine lyase, a novel enzyme that specifically acts on selenocysteine: mammalian distribution and purification and properties of pig liver enzyme. J. Biol. Chem. 257:4386-4391. [PubMed] [Google Scholar]
- 11.Filutowicz, M., A. Wiater, and D. Hulanicka. 1982. Delayed inducibility of sulphite reductase in cysM mutants of Salmonella typhimurium under anaerobic conditions. J. Gen. Microbiol. 128:1791-1794. [DOI] [PubMed] [Google Scholar]
- 12.Fitz-Gibbon, S. T., H. Ladner, U.-J. Kim, K. O. Stetter, M. I. Simon, and J. H. Miller. 2002. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. USA 99:984-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flint, D. H., J. F. Tuminello, and T. J. Miller. 1996. Studies on the synthesis of the Fe-S cluster of dihydroxy-acid dehydratase in Escherichia coli crude extract: isolation of O-acetylserine sulfhydrylases A and B and β-cystathionase based on their ability to mobilize sulfur from cysteine and to participate in Fe-S cluster synthesis. J. Biol. Chem. 271:16053-16067. [DOI] [PubMed] [Google Scholar]
- 14.Gaitonde, M. K. 1967. A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem. J. 104:627-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffith, O. W. 1987. Mammalian sulfur amino acid metabolism: an overview. Methods Enzymol. 143:366-376. [DOI] [PubMed] [Google Scholar]
- 16.Hell, R. 1997. Molecular physiology of plant sulfur metabolism. Planta 202:138-148. [DOI] [PubMed] [Google Scholar]
- 17.Jhee, K.-H., P. McPhie, and E. W. Miles. 2000. Domain architecture of the heme-independent yeast cystathionine β-synthase provides insights into mechanisms of catalysis and regulation. Biochemistry 39:10548-10556. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan, M. M., and M. Flavin. 1966. Cystathionine γ-synthetase of Salmonella: structural properties of a new enzyme in bacterial methionine biosynthesis. J. Biol. Chem. 241:5781-5789. [PubMed] [Google Scholar]
- 19.Kawarabayasi, Y., Y. Hino, H. Horikawa, K. Jin-no, M. Takahashi, M. Sekine, S.-I. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Kato, T. Yoshizawa, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K.-I. Aoki, S. Masuda, M. Yanagii, M. Nishimura, A. Yamagishi, T. Oshima, and H. Kikuchi. 2001. Complete genome sequence of an aerobic thermoaciodophilic crenarchaeon, Sulfolobus tokodaii strain7. DNA Res. 8:123-140. [DOI] [PubMed] [Google Scholar]
- 20.Kawarabayasi, Y., Y. Hino, H. Horikawa, S. Yamazaki, Y. Haikawa, K. Jin-no, M. Takahashi, M. Sekine, S.-I. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, H. Nakazawa, M. Takamiya, S. Masuda, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K.-I. Aoki, K. Kubota, Y. Nakamura, N. Nomura, Y. Sako, and H. Kikuchi. 1999. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6:83-101. [DOI] [PubMed] [Google Scholar]
- 21.Kawashima, T., N. Amano, H. Koike, S.-I. Makino, S. Higuchi, Y. Kawashima-Ohya, K. Watanabe, M. Yamazaki, K. Kanehori, T. Kawamoto, T. Nunoshiba, Y. Yamamoto, H. Aramaki, K. Makino, and M. Suzuki. 2000. Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium. Proc. Natl. Acad. Sci. USA 97:14257-14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kery, V., G. Bukovska, and J. P. Kraus. 1994. Transsulfuration depends on heme in addition to pyridoxal 5′-phosphate: cystathionine β-synthase is a heme protein. J. Biol. Chem. 269:25283-25288. [PubMed] [Google Scholar]
- 23.Kery, V., D. Elleder, and J. P. Kraus. 1995. δ-Aminolevulinate increases heme saturation and yield of human cystathionine β-synthase expressed in Escherichia coli. Arch. Biochem. Biophys. 316:24-29. [DOI] [PubMed] [Google Scholar]
- 24.Kery, V., L. Poneleit, and J. P. Kraus. 1998. Trypsin cleavage of human cystathionine β-synthase into an evolutionarily conserved active core: structural and functional consequences. Arch. Biochem. Biophys. 355:222-232. [DOI] [PubMed] [Google Scholar]
- 25.Kitabatake, M., M. W. So, D. L. Tumbula, and D. Söll. 2000. Cysteine biosynthesis pathway in the archaeon Methanosarcina barkeri encoded by acquired bacterial genes? J. Bacteriol. 182:143-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraus, J., S. Packman, B. Fowler, and L. E. Rosenberg. 1978. Purification and properties of cystathionine β-synthase from human liver: evidence for identical subunits. J. Biol. Chem. 253:6523-6528. [PubMed] [Google Scholar]
- 27.Kredich, N. M. 1997. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 28.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 29.Meier, M., M. Janosik, V. Kery, J. P. Kraus, and P. Burkhard. 2001. Structure of human cystathionine β-synthase: a unique pyridoxal 5′-phosphate-dependent heme protein. EMBO J. 20:3910-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miles, E. W. 1986. Pyridoxal phosphate enzymes catalyzing β-elimination and β-replacement reactions, p. 253-310. In D. Dolphin, R. Poulson, and O. Avramovic (ed.), Vitamin B6, pyridoxal phosphate: chemical, biochemical, and medical aspects, Part B. Wiley InterScience, New York, N.Y.
- 31.Mino, K., T. Yamanoue, T. Sakiyama, N. Eisaki, A. Matsuyama, and K. Nakanishi. 2000. Effects of bienzyme complex formation of cysteine synthetase from Escherichia coli on some properties and kinetics. Biosci. Biotechnol. Biochem. 64:1628-1640. [DOI] [PubMed] [Google Scholar]
- 32.Mizuno, Y., Y. Miyashita, S. Yamagata, T. Iwama, and T. Akamatsu. 2002. Cysteine synthase of an extremely thermophilic bacterium, Thermus thermophilus HB8. Biosci. Biotechnol. Biochem. 66:549-557. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura, T., H. Iwahashi, and Y. Eguchi. 1984. Enzymatic proof for the identity of the S-sulfocysteine synthase and cysteine synthase B of Salmonella typhimurium. J. Bacteriol. 158:1122-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nozaki, T., Y. Shigeta, Y. Saito-Nakano, M. Imada, and W. D. Kruger. 2001. Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, Trypanosoma cruzi: isolation and molecular characterization of cystathionine β-synthase and serine acetyltransferase from Trypanosoma. J. Biol. Chem. 276:6516-6523. [DOI] [PubMed] [Google Scholar]
- 35.Ono, B.-I., K. Kijima, T. Inoue, S.-I. Miyoshi, A. Matsuda, and S. Shinoda. 1994. Purification and properties of Saccharomyces cerevisiae cystathionine β-synthase. Yeast 10:333-339. [DOI] [PubMed] [Google Scholar]
- 36.Ono, B.-I., T. Hazu, S. Yoshida, T. Kawato, S. Shinoda, J. Brzvwczy, and A. Paszewski. 1999. Cysteine biosynthesis in Saccharomyces cerevisiae: a new outlook on pathway and regulation. Yeast 15:1365-1375. [DOI] [PubMed] [Google Scholar]
- 37.Porter, P. N., M. S. Grishaver, and O. W. Jones. 1974. Characterization of human cystathionine β-synthase. Evidence for the identity of human l-serine dehydratase and cystathionine β-synthase. Biochim. Biophys. Acta 364:128-139. [DOI] [PubMed] [Google Scholar]
- 38.Sako, Y., N. Nomura, A. Uchida, Y. Ishida, H. Morii, Y. Koga, T. Hoaki, and T. Maruyama. 1996. Aeropyrum pernix gen. nov., sp. nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100°C. Int. J. Syst. Bacteriol. 46:1070-1077. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 40.Segel, I. H. 1993. Enzyme kinetics: behavior and analysis of rapid equilibrium and steady-state enzyme systems. John Wiley & Sons, Inc., New York, N.Y.
- 41.Tai, C.-H., S. R. Nalabolu, T. M. Jacobson, D. E. Minter, and P. F. Cook. 1993. Kinetic mechanisms of the A and B isozymes of O-acetylserine sulfhydrylase from Salmonella typhimurium LT-2 using the natural and alternative reactants. Biochemistry 32:6433-6442. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, D., and R. H. White. 1991. Transsulfuration in archaebacteria. J. Bacteriol. 173:3250-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]