Abstract
Bacillus subtilis spores can germinate with a 1:1 chelate of Ca2+ and dipicolinic acid (DPA), a compound present at high levels in the spore core. Using a genetic screen to identify genes encoding proteins that are specifically involved in spore germination by Ca2+-DPA, three mutations were identified. One was in the gene encoding the cortex lytic enzyme, CwlJ, that was previously shown to be essential for spore germination by Ca2+-DPA. The other two were mapped to an open reading frame, ywdL, encoding a protein of unknown function. Analysis of ywdL expression showed that the gene is expressed during sporulation in the mother cell compartment of the sporulating cell and that its transcription is σE dependent. Functional characterization of YwdL demonstrated that it is a new spore coat protein that is essential for the presence of CwlJ in the spore coat. Assembly of YwdL itself into the spore coat is dependent on the coat morphogenetic proteins CotE and SpoIVA. However, other than lacking CwlJ, ywdL spores have no obvious defect in their spore coat. Because of the role for YwdL in a part of the spore germination process, we propose renaming ywdL as a spore germination gene, gerQ.
Bacillus subtilis is a well-studied gram-positive soil organism that when starved for one or more nutrients can initiate the developmental program of sporulation that eventually leads to the production of metabolically dormant spores (16). These spores can survive for long periods of time in the environment (57). However, in response to particular nutrients, termed germinants, the spores can come back to life through the process of germination, followed by outgrowth and conversion to vegetative cells (39). Spore germination has been separated into two distinct and sequential stages (47, 56). The first is germinant recognition by specific receptors. A number of studies have identified and localized the major germinant receptors that sense the nutrients that trigger germination (26, 45). Germinant-receptor interaction then leads to the release of the large depot (∼10% of the spore's dry weight) of pyridine-2,6-dicarboxylic acid (dipicolinic acid [DPA]) that is present in the core region of the dormant spores. This DPA is likely to be in a 1:1 chelate with divalent cations, predominantly Ca2+ (22). After their DPA release, spores can be arrested at this stage if they lack the two major cortex lytic enzymes, CwlJ and SleB, that are needed to progress through the second stage of germination, which requires hydrolysis of the spore's peptidoglycan cortex (47, 56). However, CwlJ and SleB have redundant functions, since in the absence of either one, cortex hydrolysis is complete and spores can eventually become vegetative cells (27). The spore cortex peptidoglycan is formed between the spore's inner and outer membranes, and cortex hydrolysis is necessary for outgrowth to occur. This cortex has one major spore-specific modification that allows its recognition by CwlJ and SleB (2, 50).
It was reported many years ago that in addition to nutrients, a 1:1 chelate of Ca2+-DPA can also trigger spore germination (53). Studies on the mechanism of Ca2+-DPA-induced spore germination have shown that the germinant receptors are not involved (46), and it appears likely that exogenous Ca2+-DPA as well as endogenous Ca2+-DPA released early in spore germination can activate the cortex lytic enzyme, CwlJ, and hence allow progression through the second stage of the germination process (42). While CwlJ has been shown to be essential for spore germination with Ca2+-DPA (42), it is not clear if other proteins are involved in this process. In this work we report the results of a genetic screen to identify mutations in other genes essential for spore germination with Ca2+-DPA. This screen has identified a mutation in cwlJ itself, as expected, and in a new gene, ywdL. Functional analysis of ywdL showed that YwdL is present in the spore coats, a proteinaceous layer surrounding the spore's outer membrane. YwdL was found to be essential for the localization of CwlJ in the spore coats and thus for Ca2+-DPA-induced spore germination.
MATERIALS AND METHODS
Strains and plasmids used in this study.
The B. subtilis strains and plasmids used in this study are listed in Table 1. However, many of the strains used for mapping (obtained from the Bacillus Genetic Stock Center, Ohio State University) are not listed. The B. subtilis strains are all derivatives of strain 168, and strains constructed in this work were prepared by transformation with either chromosomal DNA or plasmid DNA (1). The genotypes of strains arising from transformation with plasmid DNA were confirmed by Southern blot analysis (34). Escherichia coli strains TG1 and DH5α were used for the production of plasmids (34).
TABLE 1.
B. subtilis strains and plasmids used
| Strain or plasmida | Genotype | Location in genome (°) or description | Source or referenceb |
|---|---|---|---|
| Strains | |||
| PS832 | Wild type | Laboratory stock | |
| PY79* | Wild type | 62 | |
| CVO1724* | ywdL-gfp | pCVO289→PY79 | |
| CVO1736* | ΔspoIVA::spc ywdL-gfp | CVO1724→RL2037 | |
| FB72 | ger-3a (ΔgerA::spc ΔgerB::cat ΔgerK::ermC) | 46 | |
| FB104 | ger-3b (ΔgerA::spc ΔgerB::cat ΔgerK::tet) | 46 | |
| FB110 | ΔsleB::tet | 42 | |
| FB111 | ΔcwlJ::tet | 42 | |
| FB112 | ΔsleB::spc | 42 | |
| FB113 | ΔcwlJ::tet ΔsleB::spc | 42 | |
| KB1 | mut-5 (ywdL5) | KB5 + pFE50→PS832 | |
| KB2 | mut-16 ΔyndDEF::tet (ywdL16) | KB16 + pFE170→PS832 | |
| KB3 | ΔsleB::spc ywdL16 | FB112→KB2 | |
| KB4 | ΔsleB::spc ywdL5 | FB112→KB1 | |
| KB5 | ger-3b mut-5 | This work | |
| KB16 | ger-3a mut-16 | This work | |
| KB29 | ΔywdL::spc | pKE28→PS832 | |
| KB30 | ΔsleB::tet ΔywdL::spc | FB110→KB29 | |
| KB33 | cwlJ-His tag ΔywdL::spc | PS3449→KB29 | |
| KB34 | cwlJ-His tag ywdL16 | PS3449→KB2 | |
| KB42 | ywdL-lacZ | pKE41→PS832 | |
| KB43* | ywdL-lacZ | KB42→PY79 | |
| KB44* | spoIVCB::Tn917ΩHU215 ywdL-lacZ | KB43→SC64 | |
| KB45* | spoIIGB::Tn917ΩHU325 ywdL-lacZ | KB43→SC137 | |
| KB46* | spoIIAC1 ywdL-lacZ | KB43→SC1159 | |
| KB47* | ΔspoIIIG::neo ywdL-lacZ | KB43→RL831 | |
| KB48 | ywdL-gfp | CVO1724→PS832 | |
| KB49 | ΔcwlJ::tet ywdL-gfp | CVO1724→FB111 | |
| KB50 | ΔsleB::spc ywdL-gfp | CVO1724→FB112 | |
| KB56 | amyE::cwlJ76(Ts) | pKE55→PS832 | |
| KB57 | amyE::cwlJ76(Ts) ΔcwlJ::tet | pKE55→FB111 | |
| KB62 | ΔcotE ywdL-gfp | CVO1724→PS3328 | |
| KB65 | cwlJ-lacZ | pKE54→PS832 | |
| KB66 | cwlJ-lacZ ΔywdL::spc | KB60→KB29 | |
| KB72 | ywdL3-gfp | pKE70→PS832 | |
| KB73 | ywdL122-gfp | pKE71→PS832 | |
| KB76 | ger-3b mut76(Ts) [cwlJ76(Ts)] | This work | |
| PE239* | spoIIID::tet | 14 | |
| PE436* | ΔspoIIIG::cat spoIIID::tet | RL560→PE239 | |
| PE437* | sigE::ermC spoIIID::tet | RL1061→PE239 | |
| PS3328 | ΔcotE::tet | 42 | |
| PS3449 | cwlJ-His tag | 5 | |
| RL560* | ΔspoIIIG::cat | 28 | |
| RL831* | ΔspoIIIG::neo | 3 | |
| RL1061* | sigE::ermC | 29 | |
| RL2037* | ΔspoIVA::spc | 52 | |
| SC1159* | spoIIAC1 | 3 | |
| SC137* | spoIIGB::Tn917ΩHU325 | 3 | |
| SC64* | spoIVCB::Tn917ΩHU215 | 4 | |
| Additional strains used for mapping | |||
| IB427 | katX-lacZ catc | 338.60 | 3 |
| KB8 | ywdL-cat | 332.50 | pKE7→PS832 |
| KB9 | ywhE-lacZ catc | 328.80 | pPS2994→PS832 |
| KB10 | amyE::cat | 27.90 | pDG364→PS832 |
| KB11 | amyE::ermC | 27.90 | pPS2738→KB10 |
| KB12 | katX-lacZ catc | 338.60 | pPS2738→IB427 |
| KB18 | spsA::ermC | 332.50 | pKE15→PS832 |
| KB25 | spsL::spc | 331.60 | pKE21→PS832 |
| PS2355 | ΔsspF::spc | 4.50 | 33 |
| Plasmids | |||
| pCVO289 | ywdL-gfp | Fusion of ywdL to the 5′ end of gfp | This work |
| pDG364 | amyE::cat | Integrates at the amyE locus in the genome | 9 |
| pFE50 | pUB110-UC19 | Fusion of pUB110 to pUC19 | M. Paidhungat |
| pFE170 | ΔyndDEF::tet | Replacement of the yndDEF operon with tet | 46 |
| pKE7 | ywdL-cat | Fusion of the 3′ end of ywdL to cat | This work |
| pKE15 | spsA::ermC | Insertion of the ermC cassette in spsA | This work |
| pKE21 | spsL::spc | Insertion of the spc cassette in spsL | This work |
| pKE28 | ΔywdL::spc | Replacement of ywdL with spc | This work |
| pKE39 | amyE::ywdL | Integrates the ywdL transcription unit at amyE | This work |
| pKE41 | ywdL-lacZ | Translational fusion of ywdL to lacZ | This work |
| pKE53 | amyE::cwlJ | Integrates the cwlJ transcription unit at amyE | This work |
| pKE55 | amyE::cwlJ76(Ts) | Integrates the cwlJ76ts transcription unit at amyE | This work |
| pKE64 | cwlJ-lacZ | Translational fusion of cwlJ to lacZ | This work |
| pKE70 | ywdL3-gfp | Fusion of the first 3 codons of ywdL to gfp | This work |
| pKE71 | ywdL122-gfp | Fusion of the first 122 codons of ywdL to gfp | This work |
| pPS2738 | cat::ermC | Replacement of the cat cassette with ermC | 58 |
| pPS2994 | ywhE-lacZ cat | Translational fusion of ywhE to lacZ | 48 |
Strains with asterisks are in the PY79 genetic background.
DNA from strains or plasmids to the left of the arrows was used to transform the strains to the right of the arrows.
This strain carries the antibiotic resistance cassette as part of the plasmid integrated into the indicated locus.
Two of the isolated ger mutations, mut-5 and mut-16, were transferred by congression to the wild-type strain PS832 (9). In these congressions, chromosomal DNA from the mutant strain was transformed along with either a nonreplicating plasmid that integrates into the recipient chromosome at a specific locus by a double-crossover event or a derivative of the replicative plasmid pUB110 that does not integrate into the B. subtilis chromosome (7). Plasmid pFE170 was used as the integrative plasmid because it introduces a selectable tetracycline resistance (Tetr) cassette in the middle of an operon (yndDEF) that is not essential for spore germination (46), while plasmid pFE50, which provides kanamycin resistance (Kmr), was used as the replicative plasmid. Transformants in congression experiments were selected by their antibiotic resistance, and these colonies were screened for the acquisition of the ger phenotype by a plate assay (see “Spore germination” below). With congression being a rare event, only 1 to 2% of the selected colonies carried the ger mutation. The resultant strains KB1 and KB2, carrying mutations mut-5 and mut-16, respectively, were used for the genetic mapping experiments.
Plasmid pKE7, containing the 3′ end of the ywdL gene, was constructed as follows. The 3′ region of ywdL (45 bp upstream and 156 bp downstream of the ywdL translation stop codon) was amplified by PCR (primer sequences are available on request) from chromosomal DNA of strain PS832 and cloned into plasmid pCR2.1 (Invitrogen) to give plasmid pCywdL-TA. The insert was sequenced and recovered as a PstI-XbaI fragment (sites introduced into the PCR primers), and the fragment was inserted between the PstI and XbaI sites in plasmid pSGMU2 (20), giving plasmid pKE7. This plasmid was used to transform B. subtilis PS832 to chloramphenicol resistance (Cmr) by a single-crossover event such that the ywdL open reading frame (ORF) is not disrupted by the insertion of the chloramphenicol cassette.
Plasmid pKE15, which carries the internal region of the spsA coding region, was constructed as follows. A 277-bp region within the 768-bp spsA ORF (from bp 127 to 404 downstream of the translation start codon) was amplified by PCR from chromosomal DNA of strain PS832 and cloned into plasmid pCR2.1. The insert was sequenced and recovered as an EagI-XbaI fragment (sites introduced into the PCR primers), and the fragment was inserted between the EagI and XbaI sites in plasmid pFE140 (46), giving plasmid pKE15. This plasmid was used to transform B. subtilis PS832 to macrolide-lincosamide-streptogramin B resistance (MLSr) by a single-crossover event such that the spsA ORF is disrupted by the MLSr cassette. The spsA insertional mutants had no defect in their sporulation or in their spore germination with Ca2+-DPA.
To construct plasmid pKE21, which carries an internal fragment of the spsL coding region, a 261-bp fragment from within the 453-bp spsL ORF (from bp 121 to 382 downstream of the translation start codon) was amplified by PCR from chromosomal DNA of strain PS832 and cloned into plasmid pCR2.1, and the insert was sequenced and recovered as a ClaI-EcoRI fragment (ClaI site introduced into the 5′ PCR primer and EcoRI site present in plasmid pCR2.1). The fragment was inserted between the ClaI and EcoRI sites of plasmid pJL74 (31), giving plasmid pKE21, and this plasmid was used to transform B. subtilis PS832 to spectinomycin resistance (Spr) by a single-crossover event such that the spsL ORF is disrupted by the insertion of the Spr cassette. The spsL insertional mutants had no defect in their sporulation or in their spore germination with Ca2+-DPA.
Plasmid pKE28, used to generate a deletion of ywdL, was constructed in two steps. The 5′ region of ywdL (137 bp upstream and 64 bp downstream of the translation start codon) was amplified by PCR from chromosomal DNA of strain PS832 and cloned into plasmid pCR2.1, and the insert was sequenced and then recovered as a SpeI-BamHI fragment (sites introduced into the PCR primers). The fragment was inserted between the SpeI and BamHI sites of plasmid pJL74 (31) upstream of the Spr cassette, giving plasmid pNywdL-spc. The 3′ region of ywdL was recovered as a PstI-EcoRI fragment from plasmid pCywdL-TA (described above) and was inserted between the PstI and EcoRI sites of plasmid pNywdL-spc, downstream of the Spr cassette, giving plasmid pKE28. Plasmid pKE28 was used to transform B. subtilis strains to Spr by a double-crossover event such that the internal part of the ywdL ORF is deleted and replaced by the Spr cassette.
A translational fusion of ywdL to lacZ was constructed as follows. The region encompassing 194 bp upstream and 17 bp downstream of the translation start codon of ywdL was PCR amplified from chromosomal DNA of strain PS832 and cloned into plasmid pCR2.1, and the insert was sequenced and removed as an EcoRI-BamHI fragment (EcoRI site present in the pCR2.1 vector and BamHI site present in the 3′ PCR primer). This fragment was inserted between the EcoRI and BamHI sites of plasmid pJF751 (19) to create plasmid pKE41, such that the 5′ region including the first seven codons of ywdL is fused in frame to the amino-terminal end of the lacZ gene. Plasmid pKE41 was used to transform B. subtilis strains to Cmr by integration at the ywdL locus through a single-crossover event. Similarly, plasmid pKE64 was constructed so that it carries the 5′ region including the first four codons of cwlJ in frame with the amino-terminal end of the lacZ gene.
The fusion of ywdL to the amino-terminal end of gfp was also in frame and was constructed as follows. The complete coding region of ywdL (minus the first two codons and the stop codon) was amplified by PCR with chromosomal DNA from strain PY79 as the template. The resulting fragment was digested with EcoRI and XhoI (sites introduced into the PCR primers) and was cloned into similarly digested pKL168 (32), giving plasmid pCVO289. This plasmid was used to transform strain PY79 to Kmr by a single-crossover event, producing strain CVO1724. In this strain gfp is fused in frame to the 3′ end of ywdL at the ywdL locus. Therefore, the ywdL-gfp fusion in strain CVO1724 is the only complete ywdL gene.
To fuse either the first 3 or first 122 codons of ywdL to the amino-terminal end of the gfp gene, plasmid pKE69 was constructed as follows. Plasmid pCVO119 (15) was digested with SphI, and the sites were made blunt with T4 DNA polymerase (New England Biolabs) and then digested with KpnI. The fragment released from pCVO119 that carries the gfp gene downstream of a multiple cloning site was ligated to HincII-KpnI-digested pFE140, which carries an MLSr cassette, producing plasmid pKE69. The region encompassing 194 bp upstream and 6 bp downstream of the ywdL translation start codon was amplified by PCR from chromosomal DNA of strain PS832 and cloned into plasmid pCR2.1, and the insert was sequenced and removed as an EcoRI-XhoI fragment (EcoRI site present in pCR2.1 and XhoI site present in the 3′ PCR primer). This fragment was then ligated to EcoRI-XhoI-digested pKE69, giving plasmid pKE70. This plasmid was used to transform strain PS832 to MLSr by a single-crossover event such that the gfp gene is in frame with the first three codons of ywdL. Similarly, the region starting from 99 bp upstream to 363 bp downstream of the ywdL translation start codon was amplified by PCR from chromosomal DNA of strain PS832 and cloned into plasmid pCR2.1, and the insert was sequenced and removed as an EcoRI-XhoI fragment (sites present in the PCR primers). This fragment was ligated to EcoRI-XhoI-digested pKE69, giving plasmid pKE71. This plasmid was used to transform strain PS832 as described above such that the gfp gene is in frame with the first 122 codons of ywdL.
Media, growth, and sporulation under normal conditions.
E. coli and B. subtilis strains were normally grown at 37°C in rich (Luria-Bertani [LB] or 2× YT) medium (34), supplemented when necessary with the following antibiotics (in milligrams per liter): ampicillin (100), spectinomycin (100), erythromycin (1) and lincomycin (25) (MLS), chloramphenicol (5), kanamycin (10), or tetracycline (7).
B. subtilis strains were routinely sporulated at 37°C by nutrient exhaustion on 2× SG medium agar plates for 6 to 8 days, and the spores were harvested by scraping them off the agar surface as previously described (43). The spores were purified by sonication and repeated washing with cold distilled water. All spore preparations were free (>99%) of vegetative and sporulating cells and were stored in distilled water at 12°C in the dark (41). B. subtilis strains used for transduction by phage PBS1 were grown at 37°C in Penassay broth (PAB) medium (24). For studies of gene expression during sporulation at 37°C, B. subtilis strains were grown and induced to sporulate by the resuspension method (59).
Spore germination.
B. subtilis spores were germinated by nutrients or Ca2+-DPA. Spores in water (optical density at 600 nm [OD600] of 50) were heat activated (70°C, 30 min), cooled on ice, diluted to an OD600 of 1 in 2× YT medium plus 10 mM l-alanine, and incubated at 37°C as described previously (42). For Ca2+-DPA germination, spores at an OD600 of 1 were incubated in 60 mM Ca2+-DPA (60 mM CaCl2, 60 mM DPA [pH 8.0]) at either 25 or 42°C (42). Heat activation of spores prior to germination with Ca2+-DPA was not essential and was omitted.
The germination of spores in nutrients was monitored by measurement of the changes in OD600 as described previously (38, 44). This assay was not used for monitoring germination by Ca2+-DPA, since there was a slow precipitation of the mixture of 60 mM Ca2+and DPA at a ratio of 1:1 during incubation (53). Instead, spore germination with Ca2+-DPA was monitored by checking the spores with phase-contrast microscopy or by a plate assay. The plate assay described previously (38, 44) was modified slightly to test well-sporulated colonies for spore germination by Ca2+-DPA. Individual B. subtilis colonies were patched onto 2× SG agar plates and were sporulated by incubation at either 25°C (for 20 days) or 30 or 37°C (for 10 days); the plates were wrapped in plastic bags to reduce drying. The sporulated colonies were lifted onto Whatman 3MM filter paper disks that were then baked at 70°C for 3 h to kill any remaining vegetative cells. After cooling to room temperature, the filters were soaked in the germination medium (60 mM Ca2+-DPA [pH 8.0] and 1 mg of 2,3,5-triphenyltetrazolium chloride per ml, plus 5 mM glucose for color enhancement [44]) and left in a covered petri dish at either 25 or 42°C for 6 to 10 h. The colonies that contain germinated spores turn red in this assay because they reduce the tetrazolium dye, while dormant spores do not (38).
Mutagenesis and enrichment.
Four independent batches of exponentially growing cells of B. subtilis FB72 (ΔgerA::spc Δgerb::cat ΔgerK::ermC) or FB104 (ΔgerA::spc Δgerb::cat Δgerk::tet) were mutagenized with ethylmethanesulfonate (EMS) as follows. Cells were grown at 37°C in PAB medium to mid-exponential phase (OD600 of 0.5 to 0.7), 20 ml of cells was centrifuged (2,600 × g, 10 min, 25°C), and the pellet was resuspended in 20 ml of TSS minimal medium (9). EMS was added, the sample was split into four 5-ml aliquots, and the samples were incubated at 37°C for 1 h. In separate mutageneses the concentrations of EMS used were 1.5, 1.8, and 2.7%. The mutagenized cells were washed twice by centrifugation with 5% sodium thiosulfate, and the percentage of cells killed by the mutagen was determined by spotting appropriate cell dilutions on LB plates before and after treatment with EMS. The level of mutagenesis was estimated from the percentage of the surviving cells that could either not grow on TSS minimal medium plates and/or not sporulate on 2× SG medium plates (17).
Mutagenized cells were grown in PAB medium at 25 or 30°C for 24 h before they were transferred to 2× SG liquid medium for sporulation at either 25 or 30°C. B. subtilis cells grow and sporulate equally well at both 25 and 30°C, but we found the sporulation to be significantly faster at 30 than at 25°C. For this reason only one batch of mutagenized cells was grown and sporulated at 25°C; the others were grown and sporulated at 30°C. Spores were purified and germinated in liquid by Ca2+-DPA at 42°C (nonpermissive temperature) for 1 to 2 h as described above, and the mix was heated at 70°C for 1 h to kill any germinated spores and then pelleted by centrifugation (2,600 × g, 10 min, 25°C). In order to separate the spores from the Ca2+-DPA precipitate that is slowly formed during germination, we took advantage of the fact that the precipitate of a 1:1 chelate of Ca2+-DPA is soluble in excess DPA (53). Therefore, we treated the germinated spore pellet with 60 mM DPA and washed it twice by centrifugation with distilled water. The washed spores were then germinated again with 60 mM Ca2+-DPA at 25°C (permissive temperature) for 1 h, and the germinated spores were pelleted by centrifugation, resuspended in PAB medium, and grown at 25 or 30°C for 24 h. Aliquots of these cells were then sporulated in liquid 2× SG medium at 25 or 30°C. The cycle of elimination of spores that germinated at 42°C and recovery of spores that germinated at 25°C was repeated three or four times to enrich for spores that germinate with Ca2+-DPA at the permissive temperature (25°C) but not at the nonpermissive temperature (42°C). After the final enrichment, 200 individual colonies from each independent mutagenesis were screened by the plate assay described above.
Genetic mapping.
The B. subtilis mapping strains 1A627 to 1A645 (from the Bacillus Genetic Stock Center [60]) and phage PBS1 were used to map our ger mutants as described previously (44). Additional B. subtilis strains (Table 1) were used for the refined mapping of the mutations. Manipulation of PBS1 was as described previously (13), and the number of phage particles was amplified by three cycles of transduction of each individual B. subtilis mapping strain.
Complementation studies.
The transcription units of wild-type and mutant cwlJ were cloned into plasmid pDG364, a plasmid that integrates by a double-crossover event at the B. subtilis amyE locus (9). The cwlJ transcription unit was PCR amplified from either PS832 or KB76 [ger-3b mut76(Ts)] chromosomal DNA with primers designed to include the promoter (475 bp upstream of the translation start site), the ORF, and the terminator sequence (85 bp downstream of the translation stop codon) of cwlJ (27). The PCR product was cloned into plasmid pCR2.1, and the insert was sequenced and recovered as an EcoRI-BamHI fragment (sites introduced into the PCR primers) that was inserted between the EcoRI and BamHI sites of plasmid pDG364, giving plasmids pKE53 and pKE55, carrying the wild-type and mutant version of cwlJ, respectively. These plasmids were used to transform B. subtilis strains to Cmr. Transformants in which the cwlJ transcriptional unit was integrated at the amyE locus were identified by their amy phenotype on starch plates (9).
Since the position of the promoter of ywdL was unknown at the time this work began, the region encompassing 194 bp upstream of the translation start codon as well as the complete ORF and the region 156 bp downstream of the translation stop codon of ywdL were PCR amplified and cloned into plasmid pCR2.1. The insert was sequenced, recovered as a HindIII-EcoRI fragment (sites introduced into the PCR primers), and inserted between the HindIII and EcoRI sites of plasmid pDG364, giving plasmid pKE39. B. subtilis transformants with the ywdL transcription unit integrated at the amyE locus were obtained and identified as described above.
Identification of the ywdL transcription start site by 5′ RACE-PCR.
The 5′ end of the ywdL mRNA was determined by rapid amplification of cDNA ends-PCR (5′ RACE-PCR) (21, 51). Total RNA was extracted from strains PE436 (sigE+) and PE437 (sigE). These strains also had a spoIIID mutation, and strain PE436 had a spoIIIG mutation to eliminate σG- and σK-dependent gene expression (49). Strains PE436 and PE437 were induced to sporulate in parallel by the resuspension method (59). Two and a half hours after resuspension, 25 ml of each culture was harvested and the pellets were immediately mixed with an equal volume of methanol at −20°C. After centrifugation for 5 min at 4,000 × g, the RNA was extracted from the cell pellets by a hot acid-phenol protocol (18). Twenty-five micrograms of RNA was reverse transcribed by using the Superscript first-strand synthesis system for reverse transcription-PCR (Invitrogen) in the presence of 70 pmol of a gene-specific primer located 465 bp downstream of the predicted promoter region. The cDNA was purified with a QiaQuick purification spin column (Qiagen), a homopolymeric T-tail was added to the 3′ end of the cDNA with terminal transferase (Roche) according to the instructions of the manufacturer, and the sample was purified with a QiaQuick column. The tailed cDNA served as the template for PCR amplification in the presence of a poly(dA) primer and a second gene-specific primer located 223 bp downstream from the predicted promoter region. The PCR product was gel purified and sequenced by using a primer located 139 bp downstream of the predicted promoter. The transcription start site was defined as the nucleotide immediately preceding the stretch of A residues complementary to the T tail in the sequence of the PCR product. The transcription start site obtained for ywdL from the RNA isolated from strain PE436 is compatible with this mRNA being transcribed by σE as predicted by a promoter search approach (15). In this approach, the pattern search module of the SubtiList website (http://genolist.pasteur.fr/Subtilist) was used to search the region upstream of ywdL for motifs similar to the defined consensus for σE (25). No ywdL mRNA was identified in RNA isolated from strain PE437 (sigE).
Spore titers and DPA assay.
Spore DPA content was assayed as described previously (41, 55). Spore titers were determined as described previously (46) to determine the number of spores that can form colonies on LB medium agar plates. Briefly, spores at an OD600 of 1 in water were heat activated (70°C, 30 min), and 10-μl aliquots of serial dilutions of spores in water were spotted on LB agar plates. The CFU were counted after overnight incubation at 30°C.
Spore resistance to sodium hypochlorite and lysozyme.
B. subtilis spores at an OD600 of 1 were treated at room temperature with 0.25% sodium hypochlorite (pH 11.5) in distilled water. Reactions were stopped at different times by the addition of 1% sodium thiosulfate, and aliquots of serial dilutions in water were spotted on LB agar plates to determine the number of the survivors as described above.
B. subtilis spores at an OD600 of 1 were treated with lysozyme (1 mg/ml) in buffer (20 mM Tris-HCl [pH 8.0], 300 mM NaCl) and incubated at 37°C for 20 min. Aliquots of serial dilutions in water were then spotted on LB agar plates to determine the number of survivors as described above.
Microscopy and photography.
For the YwdL-green fluorescent protein (GFP) localization experiments, sporulating cells and dormant spores were viewed with a BX40 microscope (Olympus, New Hyde Park, N.Y.) equipped with an Endow GFP filter (Chroma Technology Corp.). Images were taken with an Olympus Plan phase-contrast objective (magnification, ×100; numerical aperture, 1.25) and captured with a Magna-Fire digital camera (Optronics International, Chelmsford, Mass.). Images were processed with Photoshop 3.0 (Adobe, Mountain View, Calif.).
Spore decoating and spore protein extraction and detection.
Spores were decoated by treatment for 30 min at 70°C with 0.1 M NaCl-0.1 M NaOH-1% sodium dodecyl sulfate (SDS)-0.1 M dithiothreitol, and the treated spores were washed as described previously (42, 61).
For CwlJ-His tag detection, ∼12 mg (dry weight) of spores was dry ruptured (18 times for 30 s each at 30-s intervals) with 100 mg of glass beads in a dental amalgamator (Wig-L-Bug). Soluble proteins were extracted with 50 mM HEPES (pH 7.5)-5 mM EDTA-1 mM phenylmethylsulfonyl fluoride for 30 min on ice, followed by a 5-min centrifugation (13,000 × g, 12°C) as previously described (5). Both the supernatant (soluble fraction) and the pellet (insoluble fraction) were assayed for the presence of the CwlJ-His tag by Western blot analysis with anti-His tag antibodies (Novagen) as described previously (5). The CwlJ-His tag protein has been shown to be fully functional and is in the insoluble fraction of spore extracts (5).
For detection of the different YwdL-GFP products, ∼50 mg (dry weight) of sporulating cells was collected and lyophilized 4 h after initiation of sporulation by resuspension (59). The cells were dry ruptured, and the soluble proteins were extracted in 400 μl of the extraction buffer described above. Both the soluble and insoluble fractions were suspended in 1× sample buffer (23), boiled for 15 min, and separated by SDS-12.5% polyacrylamide gel electrophoresis, and the proteins were transferred to an Immobilon-P membrane (Millipore) (23). The YwdL-GFP products were detected with a 1:5,000 dilution of anti-GFP polyclonal antibodies (Molecular Probes) and a 1:10,000 dilution of goat anti-rabbit immunoglobulin G-alkaline phosphatase conjugate (Southern Biotechnology Associates) as described previously (23).
Other methods.
Cells were extracted for β-galactosidase assays as described previously (41). β-Galactosidase was assayed with the use of either o-nitrophenyl-β-d-galactoside (4 mg/ml) or 4-methylumbelliferyl-β-d-galactopyranoside (50 μg/ml) as the substrate, as described previously (41); β-galactosidase specific activities were expressed in Miller units for o-nitrophenyl-β-d-galactoside hydrolysis (36) or in picomoles per milliliter · minute · OD600 unit for 4-methylumbelliferyl-β-d-galactopyranoside hydrolysis (41).
Complete sequences of cwlJ and ywdL from the genomes of Bacillus anthracis strain A2012, Bacillus cereus, Bacillus halodurans, and Bacillus stearothermophilus were downloaded from the National Center for Biotechnology Information website (http://www.ncbi.nih.gov) as DNA sequences in contigs and then translated to the predicted protein sequences by DNA Strider. Protein sequence alignments were done with ClustalW (http://www.ebi.ac.uk/clustalw).
RESULTS
Isolation of mutants defective in Ca2+-DPA-induced spore germination.
Although germination of B. subtilis spores is commonly triggered by nutrients, these spores can also be germinated by other agents, in particular Ca2+-DPA. While spores that lack the cortex lytic enzyme CwlJ germinate relatively normally with nutrients, they can no longer germinate with Ca2+-DPA (42). Indeed it appears that Ca2+-DPA released from the dormant spore soon after nutrient addition normally activates CwlJ and hence triggers cortex hydrolysis (42). While CwlJ is clearly needed for Ca2+-DPA-triggered spore germination, it is not clear if other proteins are also required for this process. Consequently, we initiated a genetic screen to identify proteins other than CwlJ that might be involved in spore germination triggered by Ca2+-DPA.
Since our genetic screen aimed to isolate mutant spores that fail to germinate only with Ca2+-DPA, we used B. subtilis strains (termed ger-3 strains) that lack all of the functional nutrient germinant receptors and consequently can germinate only with Ca2+-DPA (46). Vegetative cells of either the ger-3a or ger-3b strains were mutagenized with EMS at a concentration of 1.5, 1.8, or 2.7%; the percentages of cell killing were 32, 87.5, and 95%, respectively, for the three EMS concentrations tested, and 4 to 50% of the survivors had mutations giving rise to either auxotrophy or asporogeny. The mutagenized cells were then enriched for mutant spores that were unable to germinate with Ca2+-DPA at a nonpermissive temperature (42°C) but were able to germinate with Ca2+-DPA at a permissive temperature (25°C), as described in Materials and Methods. Screening of the final enriched mutant spore population with a plate assay identified five mutants whose spores reduced a tetrazolium dye at the permissive temperature in the presence of Ca2+-DPA, an indication of successful spore germination, but failed to do so at the nonpermissive temperature. Analyses by phase-contrast microscopy further indicated that spores that failed to reduce the dye during Ca2+-DPA treatment remained bright while spores that reduced the dye appeared dark and swollen and thus had germinated. The spores of three mutants failed to germinate significantly at both temperatures. Presumably, these spores were recovered through the enrichment cycles by their low level of spontaneous germination, since even spores lacking all nutrient receptors exhibit a low frequency of spontaneous germination in nutrients (46). The sporulated colonies of the putative ger mutants were picked from a replica plate, resuspended in water, and heat treated at 70°C for 30 min to kill any vegetative cells, and the spores were germinated by Ca2+-DPA at the permissive temperature, plated on nutrient agar, and left to grow at the permissive temperature. As expected, the spores of the five temperature-sensitive mutants gave rise to viable cells at the permissive temperature. In contrast, the spores of the three mutants that did not germinate with Ca2+-DPA at either temperature gave rise to viable colonies on nutrient agar at only a very low frequency, consistent with spontaneous germination (≤ 0.1% of input spores) (46). The mutants that were recovered were then grown further and sporulated at both the permissive temperature and 37°C, the temperature that is routinely used for sporulation of B. subtilis in the laboratory. Four of the temperature-sensitive and one of the absolute Ca2+-DPA ger mutants sporulated poorly at 37°C and were not considered further. The remaining three mutants [mut-5, mut-16, and mut-76(Ts) mutants] grew and sporulated normally at 37°C. Mutations mut-5 and mut-16 were further transferred to the wild-type strain PS832 by congression, giving strains KB1 (mut-5) and KB2 (mut-16 ΔyndDEF). The spores of the congressant strains and the temperature-sensitive mutant [mut-76(Ts)] readily exhibited their ger phenotype in the plate assay (Fig. 1).
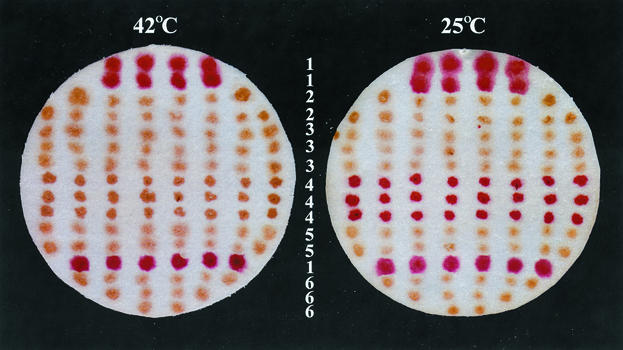
FIG. 1.
Germination of spores with Ca2+-DPA at 42 and 25°C as determined by the plate assay. Well-sporulated colonies were transferred onto a filter paper and incubated with Ca2+-DPA, glucose, and a tetrazolium dye at either 42 or 25°C as described in Materials and Methods. Colonies of germinated spores appear red, due to their ability to carry out metabolism and reduce the tetrazolium dye, while colonies of dormant spores appear brown, because they do not reduce the dye. Horizontal rows of colonies (numbered in the center) are from the following strains: 1, PS832 (wild type); 2, KB1 (mut-5); 3, KB2 (mut-16 ΔyndDEF); 4, KB76 [ger-3b mut-76(Ts)]; 5, KB29 (ΔywdL); and 6, FB111 (ΔcwlJ). Note that ΔcwlJ spores do not germinate with Ca2+-DPA at either temperature.
Mapping of the ger mutations.
We determined the linkage between the ger mutations in strain KB1, strain KB2, and the temperature-sensitive mutant KB76 [ger-3b mut-76(Ts)] and the antibiotic resistance markers in the mapping strains by phage-mediated generalized transduction. Phage PBS1 was initially used to infect the mapping strains, each of which carries a selectable antibiotic marker at a specific locus in the B. subtilis genome. The PBS1 transducing lysates were then used to transduce the various ger strains to antibiotic resistance, and spores from the antibiotic-resistant transductants were tested by the plate assay for the acquisition of the wild-type spore germination phenotype.
We obtained 20% cotransduction of the wild-type alleles of the ger mutations in strains KB1 and KB2 with the MLS resistance marker located at 342° in the chromosome of the mapping strain 1A645. The frequency of cotransduction increased with the use of strains KB12 (katX-lacZ cat) and KB9 (ywhE-lacZ cat), reaching 100% with the use of strain KB8 (ywdL-cat), indicating a close linkage of the ger mutations in strains KB1 and KB2 to a locus around or in the ywdL gene. There was no cotransduction between the ger mutations and the Spr marker in strain PS2355 (ΔsspF::spc). For refined mapping, chromosomal DNAs from strains KB8, KB18 (spsA::ermC), and KB25 (spsL::spc) were used to transform strain KB2 to antibiotic resistance. One hundred of the transformants were sporulated and tested for germination by Ca2+-DPA. The cotransformation frequencies for the ger mutation from strain KB2 were 99% with the Cmr marker in ywdL, 92% with the MLSr marker in spsA, and 32% with the Spr marker in spsL (data not shown), indicating that the ger mutation in strain KB2 lies very close to ywdL. These cotransformation frequencies are consistent with the location of ywdL 300 bp away from spsA and 10 kb away from spsL.
We obtained 72% cotransduction between the temperature-sensitive ger mutation in strain KB76 [ger-3b mut-76(Ts)] and the antibiotic marker at the amyE locus 45 kb from cwlJ in strain KB11 (amyE::ermC). This value is similar to the frequency of cotransduction obtained between the antibiotic marker at the amyE locus and the antibiotic marker in cwlJ when strain FB111 (ΔcwlJ) was infected with a phage lysate from strain KB11 (data not shown).
Identification of Ca2+-DPA ger mutations in cwlJ and ywdL.
Since the mapping experiments had localized the temperature-sensitive ger mutation close to or in cwlJ and the other two ger mutations close to or in ywdL, we PCR amplified the regions encompassing each gene from both the parental (PS832) and mutant (KB1, KB2, and KB76) strains. Two independent PCR amplifications were performed for each mutant and parental DNA, and each PCR product was sequenced in duplicate to minimize the possibility of errors during PCR or DNA sequencing.
The cwlJ allele in strain KB76 differed from the parental gene in a G-to-A transition that changed an aspartate residue (D) to asparagine (N) at position 47 of the cwlJ ORF [D47 (GAT)→N (AAT)] (data not shown). This residue is aspartate in CwlJ homologs from B. anthracis, B. cereus, B. halodurans, and B. stearothermophilus. To confirm that this change alone was responsible for the temperature-sensitive spore germination phenotype of strain KB76, the transcription unit of cwlJ from strain KB76 was inserted at the amyE locus of strain FB111 (ΔcwlJ), giving strain KB57, in which the only copy of cwlJ is that from the mut-76(Ts) isolate. The germination phenotype of spores of strain KB57 [ΔcwlJ cwlJ76(Ts)] with Ca2+-DPA was temperature sensitive as tested by the plate assay (data not shown), indicating that the point mutation giving the D47N change in CwlJ is sufficient to render spores temperature sensitive for germination by Ca2+-DPA. Interestingly, we transferred the cwlJ76(Ts) mutation into the wild-type strain PS832, thus creating a merodiploid (strain KB56) in which both wild-type and mutant forms of CwlJ are present. Spores of the merodiploid strain did germinate with Ca2+-DPA at 42°C as tested by the plate assay, but the overall process was much slower than that of the wild-type spores under the same conditions (data not shown). This result indicates that the temperature-sensitive CwlJ is interfering with the function of the wild-type protein and suggests that functional CwlJ may be oligomeric. Indeed, recent work suggests that CwlJ may be at least dimeric (8). Both the mutant and wild-type versions of CwlJ from our laboratory strain PS832 differed from the published cwlJ sequence (http://genolist.pasteur.fr/SubtiList/) in the presence of an arginine residue at position 86 instead of glycine [R86 (CGG) instead of G86 (GGC)] (data not shown). This residue is also an arginine in the CwlJ homologs from B. cereus, B. anthracis, and B. halodurans and is a glutamine in B. stearothermophilus, and this difference is not a result of our mutagenesis.
Strains KB1 (mut-5) and KB2 (mut-16 ΔyndDEF) were found to carry mutations in the coding sequence of ywdL. In mut-5 a C-to-T change at position 39 of YwdL resulted in a nonsense codon early in the ORF [Gln39 (CAG)→stop codon (TAG)], and in mut-16 a G-to-A change at position 123 of YwdL resulted in a nonsense mutation midway in the ORF [Trp123 (TGG)→stop codon (TAG)] (Fig. 2). The mut-5 and mut-16 mutations will be subsequently referred to as ywdL5 and ywdL16. The ger phenotype of spores of the ywdL5 and ywdL16 mutants was duplicated by deletion of the majority of the coding region of ywdL and its replacement with an Spr cassette in strain KB29. This ΔywdL strain grew and sporulated normally, but the spores were not able to germinate with Ca2+-DPA as tested by both the plate assay and phase-contrast microscopy (Fig. 1 and data not shown). In addition, the Ca2+-DPA-induced germination of spores carrying the ywdL5 and ywdL16 mutations was restored to wild type when the KB1 and KB2 strains were complemented with a DNA fragment containing the putative promoter and ORF of ywdL (data not shown) (Fig. 2). These results confirm that the mutations in isolates KB5 (ger-3b mut-5) and KB16 (ger-3a mut-16) are in ywdL and indicate that spores lacking ywdL do not germinate with Ca2+-DPA.
FIG. 2.
Location of mut-5 and mut-16 mutations within the ywdL coding region. The DNA sequence is shown with the predicted YwdL protein sequence in the one-letter code over the first base in each codon. Upstream of the coding region, the −10 and −35 sequences recognized by the sporulation-specific transcription factor σE are shown in boxes. The ywdL transcription start site was determined by 5′ RACE-PCR as described in Materials and Methods and is in an uppercase boldface letter; the putative ribosome binding site is indicated with the broken underline. A putative transcription terminator sequence downstream of the coding region is doubly underlined. The arrows upstream and downstream of the coding region indicate the beginning and the end of the chromosomal region that complemented the mut-5 (KB1) and mut-16 ΔyndDEF (KB2) mutants, as described in Materials and Methods. The sites of the mut-5 [Gln39 (CAG)→stop codon (TAG)] and the mut-16 [Trp123 (TGG)→stop codon (TAG)] mutations are in boldface. The consensus sequence for σE is ATa (16- to 18-bp gap) cATAcaxT, where x can be any nucleotide, uppercase letters indicate highly conserved positions, and lowercase letters indicate less conserved positions (adapted from reference 15).
Role of YwdL in spore germination.
It is clear from the results presented above that YwdL is essential for spore germination triggered by Ca2+-DPA. The obvious question that arises is whether YwdL also plays a role in spore germination triggered by nutrients. To answer this question, spores of various ywdL mutant strains were incubated with nutrients and their germination was measured by monitoring the change in OD600. With wild-type spores there is normally a fall of ∼50% in the OD600 during the initial minutes of incubation as a result of changes that occur early in spore germination, in particular, DPA release and spore core expansion due to cortex hydrolysis and water uptake (47, 56). The increase in the OD600 at later time points in such incubations is due to spore outgrowth eventually leading to vegetative growth. The ability of the spores to hydrolyze their cortex and ultimately give rise to viable cells in response to nutrients can be further estimated by measurement of their ability to form colonies on nutrient agar plates. These analyses (Fig. 3A; Table 2) indicated that spores with mutations in ywdL were able to germinate relatively normally in nutrients, although slightly slower than wild-type spores.
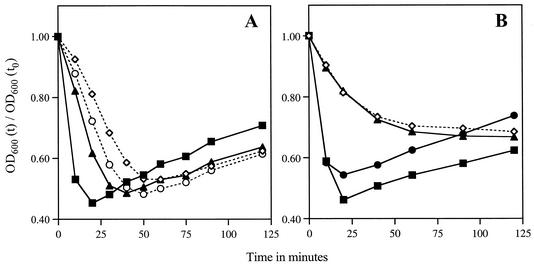
FIG. 3.
Germination of spores in nutrients. (A) Spores of the wild-type strain PS832 (▪) or mutant strain KB1 (ywdL5) (⋄), KB2 (ywdL16) (○), or KB29 (ΔywdL) (▴) were heat activated and incubated in 2× YT medium with 10 mM l-alanine at 37°C. The OD600 of each sample was measured at various time intervals [OD600 (t)] and plotted as the fraction of the initial OD600 at time zero [OD600 (t)/OD600 (t0)] versus time. (B) Spores from wild-type strain PS832 (▪) or mutant strain FB112 (ΔsleB) (•), FB113 (ΔcwlJ ΔsleB) (⋄), or KB30 (ΔsleB ΔywdL) (▴) were germinated in nutrients, and the OD600 of each sample was measured as described above.
TABLE 2.
Germination of spores of various strains with nutrients and Ca2+-DPA
| Genotype (strain) | Spore germination with:
|
|
|---|---|---|
| Nutrients (CFU/ml)a | Ca2+-DPAb | |
| Wild type (PS832) | 3.6 × 107 | + |
| ywdL5 (KB1) | 3.6 × 107 | − |
| ywdL16 (KB2) | 4.6 × 107 | − |
| ΔywdL (KB29) | 5.2 × 107 | − |
| ΔsleB (FB110) | 3.4 × 107 | + |
| ΔsleB ΔywdL (KB30) | 1.8 × 102 | − |
| ΔsleB ywdL5 (KB4) | 1.1 × 103 | − |
| ΔsleB ywdL16 (KB3) | 1.2 × 102 | − |
| ΔcwlJ ΔsleB (FB113) | 1.5 × 102 | − |
| ΔsleB ywdL-gfp (KB50) | 3 × 106 | − |
Heat treated spores were prepared at an OD600 of 1, and then serial dilutions of spores in water were spotted on LB agar plates and incubated at 30°C for 24 h and the CFU per milliliter were determined as described in Materials and Methods. Values shown are the averages from two independent experiments. The individual experimental values were within 45% of the average.
Spores at an OD600 of 1 were incubated in 60 mM Ca2+-DPA (pH 8.0) at 25°C for 1 h and then checked by phase-contrast microscopy for spore germination. + and −, 99% germinated and dormant spores, respectively.
Since it is only the Ca2+-DPA-induced germination that is affected in ywdL mutants, and knowing that one protein essential for Ca2+-DPA germination is CwlJ (42), an obvious possibility is that YwdL is required in some fashion for the function of CwlJ. In an early experiment mutants KB2 and FB111 (ΔcwlJ) were transformed with the cwlJ transcription unit and tested for their germination with Ca2+-DPA. Although spores of FB111 were complemented by cwlJ and did germinate with Ca2+-DPA, KB2 spores were not complemented and did not germinate with Ca2+-DPA as tested by the plate assay (data not shown). This indicated that the extra wild-type copy of cwlJ did not rescue the phenotype of ywdL16 mutant spores and therefore that mutations in ywdL likely affect the function of CwlJ. To determine whether YwdL is acting through CwlJ, we first examined the effect of ywdL mutations on spore germination by nutrients when the only active cortex lytic enzyme present is CwlJ. For this purpose we introduced a sleB mutation into ywdL mutant strains. As found previously, sleB spores germinated normally in nutrients, and the percentage of these spores forming viable colonies on nutrient agar plates was similar to that of wild-type spores, indicating that cortex hydrolysis in sleB spores can be completed (Fig. 3B; Table 2). However, spores from the sleB ΔywdL strain behaved like spores of the cwlJ sleB strain, which previously have been shown to go through the initial stages of germination (27, 56) but are unable to hydrolyze their cortex and hence do not give rise to viable colonies on nutrient agar plates (Fig. 3B; Table 2). Spores of both the sleB ΔywdL and cwlJ sleB strains showed similar changes in OD600 during incubation with nutrients and gave a very low frequency of viable colonies on nutrient agar plates (Fig. 3B; Table 2). The spore DPA content and its release during germination were similar for wild-type, cwlJ sleB, and sleB ΔywdL spores, suggesting that it is at the step of cortex hydrolysis that germination of sleB ΔywdL spores is arrested, as has been shown to be the case for cwlJ sleB spores (27, 56) (data not shown). We obtained the same results for sleB ywdL5 and sleB ywdL16 spores (Table 2), suggesting that the premature stop codon in ywdL5 and ywdL16 results in a ΔywdL phenotype. From these results it is evident that in the absence of YwdL, CwlJ is not functioning. This is why ywdL spores are unable to germinate fully under those conditions when CwlJ is absolutely essential, namely, germination with exogenous Ca2+-DPA or with nutrients when SleB is not present to carry out cortex hydrolysis.
Localization of CwlJ and coat structural integrity in ywdL spores.
The results given above indicated that ywdL mutations act by eliminating CwlJ function. Previous work has shown that cwlJ is expressed in the mother cell compartment of the sporulating cell under the control of the sporulation-specific sigma factor for RNA polymerase, σE (15, 27). It was thus formally possible that YwdL is required in some fashion for expression of cwlJ. To test this possibility, we constructed a translational cwlJ-lacZ fusion and measured the β-galactosidase specific activity in sporulating cells of strains KB65 (cwlJ-lacZ) and KB66 (cwlJ-lacZ ywdL). We found no effect on cwlJ expression when YwdL was absent (data not shown), indicating that YwdL might be required either for CwlJ activity directly or for CwlJ localization or stability in the spore coats. To test whether CwlJ is localized normally in spores in the absence of YwdL, extracts were prepared from spores also carrying a functional His-tagged CwlJ in either wild-type, ΔywdL, or ywdL16 strains, and the extracts were analyzed for the CwlJ-His tag by Western blot analysis. While the CwlJ-His tag protein was readily detected in wild-type spore extracts (Fig. 4, lanes 1), we were unable to detect this protein in spores with the ΔywdL and ywdL16 mutations or in wild-type spores lacking the cwlJ-His tag fusion (Fig. 4, lanes 2, 3, and 4). The CwlJ-His tag protein was also absent from both the soluble and insoluble fraction of the ywdL spore extracts (Fig. 4, lanes 2 and 4, and data not shown). The absence of CwlJ from ywdL spores explains why they are unable to germinate with Ca2+-DPA, since CwlJ is required for this process. The lack of germination of sleB ywdL spores in nutrients is also consistent with the absence of CwlJ from these spores.
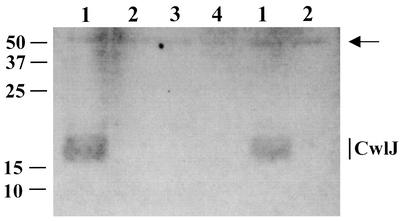
FIG. 4.
Localization of CwlJ-His tag in spore extracts of various strains. The insoluble fraction of the extract from 1 mg of spores of strains PS3449 (cwlJ-His tag) (lanes 1), KB33 (cwlJ-His tag ΔywdL) (lanes 2), KB29 (ΔywdL) (lane 3), and KB34 (cwlJ-His tag ywdL16) (lane 4) was run on an SDS-12.5% polyacrylamide gel. Each lane contains protein from 1 mg (dry weight) of spores. The gel was blotted onto an Immobilon-P membrane (Millipore) that was hybridized with anti-His tag monoclonal antibody (Novagen) as outlined in Materials and Methods. An extract from spores of strain KB29 (lane 4) that lacks the CwlJ-His tag served as a negative control for the anti-His tag antibody. The arrow indicates an immunoreactive product at ∼50 kDa (with no His tag attached) that has been seen previously (5) and that we use here as a rough indication of protein levels. The CwlJ-His tag runs as a wide band of ∼18 kDa as shown previously (5). Only a portion of the Western blot is shown, and the numbers shown at the left indicate the migration positions of protein molecular mass markers in kilodaltons. The same experiment was run for the soluble fraction of the spore extracts, where CwlJ-His tag was not found (data not shown), as reported previously (5).
The finding that ywdL spores lack CwlJ and the knowledge that CwlJ is a coat protein (5, 8) suggest that ywdL spores may have a general spore coat defect. One such general defect is caused by the lack of the coat morphogenetic protein CotE; when CotE is absent, the assembly of the spore's outer coat is perturbed (64). Indeed, levels of CwlJ are greatly reduced in cotE spores (5). To test the integrity of the coats in spores of various strains, spores of wild-type, ywdL5, ywdL16, ΔywdL, and cotE strains were treated with either lysozyme or sodium hypochlorite. Spores that lack the major coat morphogenetic protein CotE are killed rapidly by treatment with either lysozyme (64) or sodium hypochlorite (S. B. Young and P. Setlow, unpublished data). However, the spores of all ywdL mutants exhibited resistance to both lysozyme and sodium hypochlorite that was essentially identical to that of wild-type spores, while cotE spores were much more sensitive, as expected (data not shown). Treatment of spores with sodium hypochlorite was done for both short and longer time periods (2 to 60 min), and both ywdL and wild-type spores showed similar levels of survival (data not shown). These data indicate that the ywdL mutations do not cause a gross defect in spore coat assembly as does a cotE mutation.
Expression of ywdL during sporulation.
Since ywdL mutations affect the level of CwlJ in dormant spores and CwlJ is a coat protein, an obvious suggestion is that YwdL itself is a spore coat protein. To examine this possibility, we first measured ywdL expression during growth and sporulation by using a translational ywdL-lacZ fusion. The expression of β-galactosidase from this fusion was absent in vegetative cells, and it began at the second hour in sporulation, reaching maximum levels at the fourth hour (Fig. 5A). The level of expression was exceptionally high, reaching a peak specific activity of 1,600 Miller units before falling (Fig. 5A). The fall in β-galactosidase specific activity could be due either to the inactivation of the enzyme when it is no longer synthesized or to its synthesis in the forespore and subsequent sequestering in the dormant spore, where β-galactosidase cannot be assayed without removing the spore coats (35). Since there was no β-galactosidase activity in decoated spores of the ywdL-lacZ strain (data not shown), the enzyme is likely not synthesized in the forespore and inactivation of the enzyme is the most probable reason for the fall in its specific activity. Analysis of the RNA polymerase sigma factor dependence of ywdL expression indicated that ywdL expression was abolished when either the σF or σE transcription factor was absent (Fig. 5B). However, when either σG or σK was absent, ywdL expression continued for longer than in the wild-type strain (Fig. 5B), perhaps due to the loss of regulation when σG or σK is absent. Since σE activity depends on the function of σF, but not vice versa (49), these data indicate that the transcription of ywdL depends on σE. This further indicates that ywdL is expressed only in the mother cell compartment of the sporulating cell. In agreement with these studies, a microarray analysis of the transcriptional profile during sporulation of wild-type and sigE strains also indicates that ywdL is a member of the σE regulon (15).
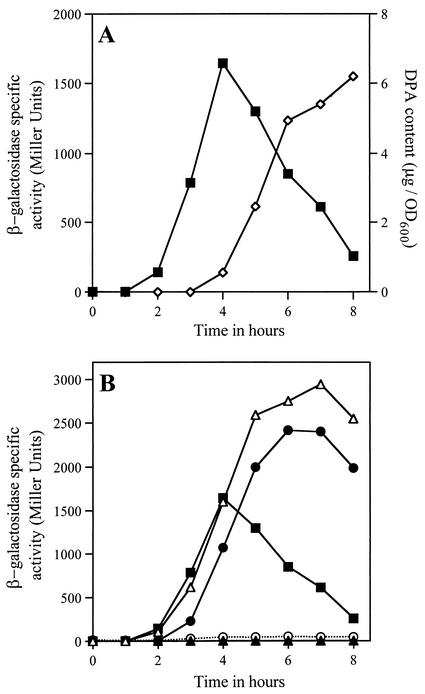
FIG. 5.
(A) Expression of ywdL-lacZ during sporulation. Strain KB43 (ywdL-lacZ) was induced to sporulate by resuspension at time t0, and samples were removed for estimation of β-galactosidase specific activity (▪) and DPA content (⋄) as described in Materials and Methods. The DPA curve is used here as a marker of the progression through sporulation. (B) Expression of ywdL-lacZ in strains that lack one of the sporulation-specific RNA polymerase σ factors. Strains KB46 (sigF ywdL-lacZ) (○), KB45 (sigE ywdL-lacZ) (▴), KB47 (sigG ywdL-lacZ) (•), KB44 (sigK ywdL-lacZ) (▵), and KB43 (ywdL-lacZ) (▪) were induced to sporulate by resuspension, and the β-galactosidase specific activity was determined as described for panel A. The maximum β-galactosidase specific activity of strain PY79, which was induced to sporulate in parallel with the mutant strains, was found to be 4 Miller units.
Localization of YwdL in dormant spores.
Having established that ywdL is expressed during sporulation, we next asked whether YwdL is present in the dormant spores or whether it functions during sporulation without being a spore structural component. To answer this question we studied the localization of the GFP from Aequoria victoria fused to YwdL. The YwdL-GFP construct was inserted in the B. subtilis chromosome such that YwdL-GFP is the only copy of YwdL that is expressed (see Materials and Methods). Midway in sporulation, the YwdL-GFP appeared as a bright dot adjacent to the developing spore (Fig. 6A and B), while at later times the protein appeared to be around the spore's periphery (Fig. 6B and C). No confocal microscopy was done to definitely establish if YwdL-GFP is localized in a complete shell around the spore. The peripheral localization of YwdL-GFP was also seen in the majority of the dormant spores after their release and the mother cell lysis, as 70% of the dormant spores were surrounded by YwdL-GFP, although in a significant number (30%) YwdL-GFP appeared as a bright dot instead (data not shown). The latter spores could have a mislocalized YwdL, as the YwdL-GFP may not be fully functional. To test whether YwdL-GFP functions normally, we moved the ywdL-gfp fusion to a sleB strain and tested the germination of the resultant spores in nutrients. If YwdL-GFP was nonfunctional, we expected a phenotype similar to that of a sleB ywdL strain (Table 2; Fig. 3). However, while sleB ywdL spores exhibited <0.01% of the CFU of wild-type spores in nutrients, sleB ywdL-gfp spores had only a 10-fold decrease in their CFU in nutrients (Table 2). The localization of YwdL-GFP in sleB spores was also essentially identical to that observed in the wild-type spores (data not shown). Presumably the spores from the sleB ywdL-gfp population that did not germinate in nutrients have a mislocalized YwdL-GFP and thus a nonfunctional CwlJ; however, this is only speculation at present. We also observed by phase-contrast microscopy that none (<1%) of the sleB ywdL-gfp spores germinated with exogenous Ca+2-DPA (Table 2). Together, these results suggest that since there is some YwdL function observed in sleB ywdL-gfp spores, at least some YwdL-GFP is functional.
FIG. 6.
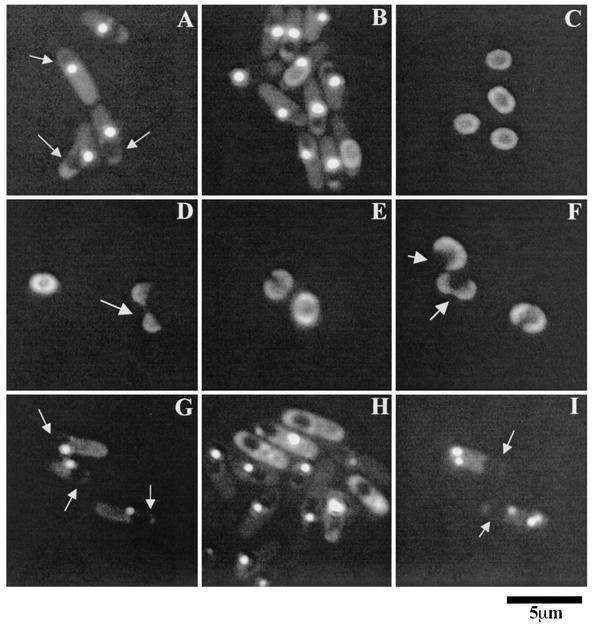
Localization of YwdL-GFP in sporulating cells and dormant spores. (A to C) Strain KB48 (ywdL-gfp), was sporulated by resuspension, and samples were viewed with a fluorescence microscope at 3 h (A), 7 h (B), and 24 h (C) after the onset of sporulation. (D to F) Dormant spores of strain KB48 were germinated in nutrients at 37°C for 1 h, and samples were examined. Arrows indicate germinated spores whose coats have split open. (G to I) Strains KB62 (ΔcotE ywdL-gfp) (G and H) and CVO1736 (ΔspoIVA ywdL-gfp) (I) were sporulated by resuspension, and samples were viewed at 3 h (G and I) and 7 h (H) after the onset of sporulation. Strain KB59 released dormant spores with no visible YwdL-GFP on their periphery, while strain CVO1736 did not release any dormant spores. The arrows pointing to the dark areas enclosed in the sporulating cell in panels A, G, and I indicate the developing forespore.
To further test the localization pattern of YwdL in the ywdL16 mutant, where at most only the first 122 amino acids (aa) of the protein are expressed, we made a construct in which gfp is fused to only that part of YwdL. In contrast to the specific localization we observed when the complete YwdL was fused to GFP, with this truncated YwdL-GFP fusion the GFP fluorescence was uniformly distributed in the cytoplasm of the mother cell compartment during sporulation (data not shown). To check whether the GFP moiety was indeed linked to the truncated YwdL, we analyzed the sizes of these proteins by Western blot analysis with anti-GFP antibodies as described in Materials and Methods, and both the complete and 122-aa YwdL were indeed linked to GFP (data not shown). A control strain that was made with just the first 3 aa of YwdL fused to GFP also gave the same uniform mother cell distribution during sporulation (data not shown), as expected. These results indicate that while the complete YwdL is specifically associated with the developing forespore during sporulation, the truncated protein from the ywdL16 mutant is not.
Results from both the gene expression and YwdL-GFP localization studies indicate that YwdL is specifically expressed during sporulation, is found in the dormant spores, and is likely a coat protein. Examination of YwdL-GFP in germinated spores gave a localization pattern generally similar to that observed in dormant spores, although germinated spores are swollen due to core expansion and cortex hydrolysis (Fig. 6D, E, and F). We also observed a number of outgrowing ywdL-gfp spores in which the YwdL-GFP was localized in the remnants of the spore coats that have cracked open and are in the process of being shed (Fig. 6D and F). These results further support the assignment of YwdL as a spore coat protein.
Dependence of YwdL localization on other spore coat proteins.
The results given above indicate that YwdL is somehow involved in the assembly, localization, or stability of CwlJ and further that YwdL is a spore coat protein itself. An obvious question concerns the dependence of YwdL localization on other proteins important in spore coat morphogenesis. Consequently, we studied the localization pattern of YwdL-GFP in the absence of either of two proteins (CotE and SpoIVA) that are known to have dramatic effects on the assembly of the spore coat (10). SpoIVA is essential for the anchorage of the coats on the forespore (54). Sporulating cells of a spoIVA null mutant fail to synthesize a cortex, and the coat is misassembled as swirls within the mother cell rather than being deposited on the outside surface of the forespore (54). CotE is essential for the assembly of mostly proteins that form the outer spore coat as well as a few proteins of the inner spore coat (10). Midway during sporulation of the spoIVA ywdL-gfp strain, instead of one bright dot next to the forespore we observed multiple dots distributed throughout the mother cell cytoplasm but not adjacent to the developing spore (Fig. 6I). This result is consistent with SpoIVA being needed for YwdL assembly on the spore coat, as in the absence of SpoIVA, any coat protein is expected to be distributed throughout the mother cell compartment (54). Three hours after the onset of cotE ywdL-gfp sporulation, we observed a bright dot next to the forespore as seen in the wild-type strain (Fig. 6G). However, 7 h after the onset of cotE ywdL-gfp sporulation, YwdL-GFP was often diffuse in the mother cell cytoplasm instead of localizing around the periphery of the forespore (Fig. 6H). This result indicates that CotE is also required for YwdL-GFP localization on the periphery of the spore.
Since localization of CwlJ in the dormant spores depends on CotE (5) as well as YwdL, we also checked YwdL-GFP localization in sporulating cells of the cwlJ ywdL-gfp strain. We found no difference in YwdL-GFP localization from that observed in the spores of the wild-type ywdL-gfp strain (data not shown). This finding indicates that CwlJ is not required for localization of YwdL in the spore coat.
DISCUSSION
In our search for proteins that are involved in the germination of B. subtilis spores with Ca2+-DPA, we isolated mutants that no longer respond to this chemical and therefore cannot germinate and generate viable cells. We isolated three mutations that identified two genes that are essential for spore germination with Ca2+-DPA. One mutation that resulted in temperature-sensitive germination with Ca2+-DPA was due to a change of a conserved aspartate residue to asparagine in the cortex lytic enzyme CwlJ. Previous work has shown that endogenous DPA released during spore germination activates CwlJ, providing a link between the germination signal being received by the germinant receptors and the cortex hydrolysis that follows (42). Since spore germination can be triggered by exogenous DPA as well, we reasoned that CwlJ is in a location accessible by exogenous DPA. Indeed, previous studies have shown that CwlJ is localized in the spore coats (5, 8). However, we still do not know if there is some receptor for Ca2+-DPA that might interact with CwlJ, and we also have not established whether the mutation we found in CwlJ affects its catalytic function during cortex hydrolysis or its activation by Ca2+-DPA.
The other two mutations identified an additional component essential for Ca2+-DPA-induced germination, the protein of hitherto-unknown function termed YwdL. The role of YwdL in spore germination appears to be due to its importance in ensuring the presence of CwlJ in the spore coats. However, it is not yet clear how YwdL functions in this process. YwdL might serve as an anchor for CwlJ in the coats, might form some sort of a chaperone ensuring CwlJ assembly in the coats, or might even protect CwlJ from degradation by the large amount of proteases present in the sporulation medium, which potentially have access to CwlJ after mother cell lysis. This last point could be addressed in the future by examining total CwlJ levels in ywdL strains prior to and after mother cell lysis.
YwdL and CwlJ are synthesized in the same compartment of the sporulating cell, at about the same time in sporulation (15, 27; this work). Their coding genes are also transcribed by RNA polymerase with the same sigma factor. While these two genes are well separated on the B. subtilis chromosome, they appear to be in an operon in other Bacillus species whose genomes have been sequenced. This is further evidence, albeit circumstantial, for some functional relationship between the products of these genes. Interestingly, the other B. subtilis spore cortex lytic enzyme, SleB, is also encoded by a gene in a bicistronic operon, with the gene following sleB, termed ypeB, again encoding a protein that is essential for either the localization or stabilization of SleB in the spore (8).
Although YwdL is needed in some way for the presence of CwlJ in the spore coats, YwdL is not a major coat morphogenetic protein like CotE or SpoIVA; ywdL spores are not lysozyme or hypochlorite sensitive, and there was no change in the levels of detectable coat proteins extracted from wild-type and ywdL spores (K. Ragkousi, A. Driks, and P. Setlow, unpublished data). The localization of SleB, which has also been suggested to be in or adjacent to the spore coats (8, 40), is also not affected in ywdL spores, since this enzyme is still functional. However, it is possible that some coat defect may become evident in ywdL spores upon further analysis.
The primary amino acid sequence of YwdL is highly conserved among Bacillus species, especially in the carboxy-terminal region (Fig. 7). Although there is little amino acid homology at the amino terminus, there is a high content of glutamine, glycine, and proline residues present in this part of the protein in all Bacillus species (Fig. 7). However, the amino acid sequence of this part of the protein gives no clear indication of its functional significance.
FIG. 7.
Protein sequence alignment of YwdL from various Bacillus species. Asterisks below the aligned sequences indicate identical residues in that column, double dots indicate well conserved residues, and single dots indicate poorly conserved residues (as determined from the ClustalW website). The residues that were altered in strains KB1 and KB2 (Gln39 and Trp123, respectively) are in boldface in the B. subtilis sequence. No homologs of YwdL were found in Clostridium species (15).
Interestingly, while endogenous Ca2+-DPA can activate the germination of ywdL-gfp spores, indicating normal CwlJ function, exogenous Ca2+-DPA cannot. Unfortunately, we do not know the precise details of CwlJ localization in the spore coats and its activation by Ca2+-DPA. Consequently, it is not clear why the fusion of the carboxy-terminal region of YwdL to GFP interferes with CwlJ activation by exogenous Ca+2-DPA. However, since the ywdL16 mutant does not germinate with Ca2+-DPA and does not contain CwlJ and GFP fused to this truncated YwdL does not localize properly during sporulation, presumably some or all of the 59 aa at the carboxy terminus of YwdL are essential for its proper localization and hence the proper localization of CwlJ as well.
An interesting feature of YwdL is its localization pattern during sporulation. Initially, YwdL-GFP is assembled in a dot adjacent to the forespore. A similar pattern has been observed with the coat morphogenetic proteins CotE and SpoIVA, although the latter proteins appear to localize initially in arc-like structures adjacent to the forespore (11, 52). In the absence of CotE, YwdL-GFP is initially targeted to the forespore but does not assemble around the spore. In the absence of SpoIVA, however, YwdL-GFP is not even targeted adjacent to the forespore. These results fit well with the model in which SpoIVA localizes and attaches the precoat to the forespore before CotE assembles itself and other proteins into the mature coat (10).
Both CwlJ and YwdL seem to be outer coat proteins, since both depend on the presence of CotE for their localization, although some inner coat proteins are CotE dependent (10). If the levels of the translational fusion of ywdL to β-galactosidase are indicative of the protein levels in the cell, we would expect YwdL to be a highly abundant protein. However, until recently YwdL has not been identified in coat protein extracts. YwdL has been recently identified in a proteomic study (30), but it appears to be at most a minor coat protein, which is at odds with the very high level of expression of ywdL-lacZ. One possibility is that the great majority of YwdL in the spore is in that fraction of spore coat protein that cannot be extracted. Approximately 30% of total coat protein is not extracted by procedures using alkali, detergents, or both (63). The reasons for the formation of this insoluble coat fraction are not clear, although both dityrosine cross-links and isopeptide bonds involving glutamine residues via transglutaminases have been suggested to contribute to the formation of a protein complex that is difficult to solubilize (10). It is notable that YwdL has a high content of both glutamine and tyrosine residues (Fig. 7). We have also found that ywdL-gfp spores retain their YwdL-GFP after decoating as seen in the fluorescence microscope (K. Ragkousi and P. Setlow, unpublished data).
Whatever the location of YwdL in the spore coats, it is clear that this protein has an effect on spore germination. Because of the significant role of YwdL in a part of the spore germination process, we propose the renaming of this protein as a Ger protein, GerQ, with the coding gene being renamed gerQ. Other spore coat proteins are also involved in spore germination in some fashion. Thus, gerE spores, which do not make a number of coat proteins and appear to have defective coats are severely impaired in their germination (37). In addition, GerP proteins play a role in germination by enabling the access of germinants to their receptors localized in the spore's inner membrane (6). While coat proteins could be involved in spore germination in distinct ways, it is still unclear how a protein such as CwlJ that is localized in the coat, a layer separated from the cortex by the outer spore membrane, is able to hydrolyze the cortex peptidoglycan. Perhaps the outer membrane is not complete (12) and the cortex is normally accessible to coat proteins such as CwlJ, which are only activated by signals following the release of DPA. Another possibility is that DPA release from the spore core alters the cortex structure in some fashion (47) such that it bulges into the coat area where it becomes exposed to the cortex lytic enzymes. Clearly, understanding of how the spore coats are involved in spore germination has only just begun to take shape.
Acknowledgments
We are grateful to Madan Paidhungat and Irina Bagyan for advice and strains, to Glenn King for the use of the fluorescence microscope, to Susan Young for advice on the sodium hypochlorite assay, to Scott Robson for scanning the tetrazolium assay filters, and to Adam Driks for discussions. We also appreciate the comments of Madan Paidhungat and Richard Losick on the manuscript.
This work was supported by grants from the NIH to P.S. (GM19698), R.L. (GM18568), and C.V.O. (GM20165) and by a postdoctoral fellowship from the Human Frontier Science Program and the Swiss National Science Foundation to P.E. Preliminary genomic sequence data for B. anthracis and B. cereus were made available by the Institute for Genomic Research, where work was supported by the Office of Naval Research, the Department of Energy, the National Institute of Allergy and Infectious Disease, and the National Institutes of Health. B. stearothermophilus sequence data were obtained from the University of Oklahoma Advanced Center for Genome Research, where work was funded by an EPSCoR grant from the National Science Foundation.
REFERENCES
- 1.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:74-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atrih, A. P. Z., G. Allmaier, and S. Foster. 1996. Structural analysis of Bacillus subtilis 168 endospore peptidoglycan and its role during differentiation. J. Bacteriol. 178:6173-6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagyan, I., L. Casillas-Martinez, and P. Setlow. 1998. The katX gene, which codes for the catalase in spores of Bacillus subtilis, is a forespore-specific gene controlled by σF, and KatX is essential for hydrogen peroxide resistance of the germinating spore. J. Bacteriol. 180:2057-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagyan, I., B. Setlow, and P. Setlow. 1998. New small, acid-soluble proteins unique to spores of Bacillus subtilis: identification of the coding genes and regulation and function of two of these genes. J. Bacteriol. 180:6704-6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagyan, I., and P. Setlow. 2002. Localization of the cortex lytic enzyme CwlJ in spores of Bacillus subtilis. J. Bacteriol. 184:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behravan, J., H. Chirakkal, A. Masson, and A. Moir. 2000. Mutations in the gerP locus of Bacillus subtilis and Bacillus cereus affect access of germinants to their targets in spores. J. Bacteriol. 182:1987-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bron, S. 1990. Plasmids, p. 75-174. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 8.Chirakkal, H., M. O'Rourke, A. Atrih, S. J. Foster, and A. Moir. 2002. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology 148:2383-2392. [DOI] [PubMed] [Google Scholar]
- 9.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 10.Driks, A. 1999. The Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Driks, A., S. Roels, B. Beal, C. P. J. Moran, and R. Losick. 1994. Subcellular localization of proteins involved in the assembly of the spore coat of Bacillus subtilis. Genes Dev. 8:234-244. [DOI] [PubMed] [Google Scholar]
- 12.Driks, A., and P. Setlow. 1999. Morphogenesis and properties of the bacterial spore. American Society for Microbiology, Washington, D. C.
- 13.Dubnau, D. 1971. Genetic mapping of Bacillus subtilis. Methods Enzymol. 21:430-438. [Google Scholar]
- 14.Eichenberger, P., P. Fawcett, and R. Losick. 2001. A three-protein inhibitor of polar septation during sporulation in Bacillus subtilis. Mol. Microbiol. 42:1147-1162. [DOI] [PubMed] [Google Scholar]
- 15.Eichenberger, P., S. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J.-E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol., in press. [DOI] [PubMed]
- 16.Errington, J. 1993. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol. Rev. 57:1-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fairhead, H., B. Setlow, and P. Setlow. 1993. Prevention of DNA damage in spores and in vitro by small, acid-soluble proteins from Bacillus species. J. Bacteriol. 175:1367-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fawcett, P., P. Eichenberger, R. Losick, and P. Youngman. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:8063-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrari, E., S. M. Howard, and J. A. Hoch. 1985. Effect of sporulation mutations on subtilisin expression, assayed using a subtilisin-β-galactosidase gene fusion, p. 180-184. In J. A. Hoch and P. Setlow (ed.), Molecular biology of microbial differentiation. American Society for Microbiology, Washington, D.C.
- 20.Fort, P., and J. Errington. 1985. Nucleotide sequence and complementation analysis of a polycistronic sporulation operon, spoVA, in Bacillus subtilis. J. Gen. Microbiol. 131:1091-1105. [DOI] [PubMed] [Google Scholar]
- 21.Frohman, M. A. 1994. On beyond classic RACE (rapid amplification of cDNA ends). PCR Methods Appl. 4:S40-S58. [DOI] [PubMed]
- 22.Gould, G. W. 1969. Germination, p. 397-444. In G. W. Gould and A. Hurst (ed.), The bacterial spore. Academic Press, New York, N.Y.
- 23.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Harwood, C. R., and A. R. Archibald. 1990. Growth, maintenance and general techniques, p. 1-26. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 25.Helmann, J. D., and C. P. J. Moran. 2002. RNA polymerase and sigma factors, p. 289-312. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 26.Hudson, K. D., B. M. Corfe, E. H. Kemp, I. M. Feavers, P. J. Coote, and A. Moir. 2001. Localization of GerAA and GerAC germination proteins in the Bacillus subtilis spore. J. Bacteriol. 183:4317-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa, S., K. Yamane, and J. Sekiguchi. 1998. Regulation and characterization of a newly deduced cell wall hydrolase gene (cwlJ) which affects germination of Bacillus subtilis spores. J. Bacteriol. 180:1375-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karmazyn-Campelli, C., C. Bonamy, B. Savelli, and P. Stragier. 1989. Tandem genes encoding σ-factors for consecutive steps of development in Bacillus subtilis. Genes Dev. 3:150-157. [DOI] [PubMed] [Google Scholar]
- 29.Kenney, T. J., and C. P. Moran, Jr. 1987. Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J. Bacteriol. 169:3329-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuwana, R., Y. Kasahara, M. Fujibayashi, H. Takamatsu, N. Ogasawara, and K. Watabe. 2002. Proteomics characterization of novel spore proteins of Bacillus subtilis. Microbiology 148:3971-3982. [DOI] [PubMed] [Google Scholar]
- 31.LeDeaux, J. R., and A. D. Grossman. 1995. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J. Bacteriol. 177:166-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemon, K. P., and A. D. Grossman. 1998. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 282:1516-1519. [DOI] [PubMed] [Google Scholar]
- 33.Loshon, C. A., P. Kraus, B. Setlow, and P. Setlow. 1997. Effects of inactivation or overexpression of the sspF gene on properties of Bacillus subtilis spores. J. Bacteriol. 179:272-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning, a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 35.Mason, J. M., R. H. Hackett, and P. Setlow. 1988. Regulation of expression of genes coding for small, acid-soluble proteins of Bacillus subtilis spores: studies using lacZ gene fusions. J. Bacteriol. 170:239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Moir, A. 1981. Germination properties of a spore coat-defective mutant of Bacillus subtilis. J. Bacteriol. 146:1106-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moir, A., E. Lafferty, and D. A. Smith. 1979. Genetic analysis of spore germination mutants of Bacillus subtilis 168: the correlation of phenotype and map location. J. Gen. Microbiol. 111:165-180. [DOI] [PubMed] [Google Scholar]
- 39.Moir, A., and D. A. Smith. 1990. The genetics of bacterial spore germination. Annu. Rev. Microbiol. 44:531-553. [DOI] [PubMed] [Google Scholar]
- 40.Moriyama, R., H. Fukuoka, S. Miyata, S. Kudoh, A. Hattori, S. Kozuka, Y. Yasuda, K. Tochikubo, and S. Makino. 1999. Expression of a germination-specific amidase, SleB, of bacilli in the forespore compartment of sporulating cells and its localization on the exterior side of the cortex in dormant spores. J. Bacteriol. 181:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons Ltd., Chichester, England.
- 42.Paidhungat, M., K. Ragkousi, and P. Setlow. 2001. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J. Bacteriol. 183:4886-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paidhungat, M., B. Setlow, A. Driks, and P. Setlow. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J. Bacteriol. 182:5505-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paidhungat, M., and P. Setlow. 1999. Isolation and characterization of mutations in Bacillus subtilis that allow spore germination in the novel germinant d-alanine. J. Bacteriol. 181:3341-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paidhungat, M., and P. Setlow. 2001. Localization of a germinant receptor protein (GerBA) to the inner membrane of Bacillus subtilis spores. J. Bacteriol. 183:3982-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paidhungat, M., and P. Setlow. 2000. Role of Ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J. Bacteriol. 182:2513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paidhungat, M., and P. Setlow. 2002. Spore germination and outgrowth, p. 537-548. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 48.Pedersen, L. B., K. Ragkousi, T. J. Cammett, E. Melly, A. Sekowska, E. Scopick, T. Murray, and P. Setlow. 2000. Characterization of ywhE, which encodes a putative high-molecular-weight class A penicillin-binding protein in Bacillus subtilis. Gene 246:187-196. [DOI] [PubMed] [Google Scholar]
- 49.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-515. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 50.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Muramic lactam in peptidoglycan of Bacillus subtilis spores is required for spore outgrowth but not for spore dehydration or heat resistance. Proc. Natl. Acad. of Sci. 93:15405-15410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 52.Price, K. D., and R. Losick. 1999. A four-dimensional view of assembly of a morphogenetic protein during sporulation in Bacillus subtilis. J. Bacteriol. 181:781-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Riemann, H., and J. Z. Ordal. 1961. Germination of bacterial endospores with calcium and dipicolinic acid. Science 133:1703-1704. [DOI] [PubMed] [Google Scholar]
- 54.Roels, S., A. Driks, and R. Losick. 1992. Characterization of spoIVA, a sporulation gene involved in coat morphogenesis in Bacillus subtilis. J. Bacteriol. 174:575-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rotman, Y., and M. L. Fields. 1967. A modified reagent for dipicolinic acid analysis. Anal. Biochem. 22:168.. [DOI] [PubMed] [Google Scholar]
- 56.Setlow, B., E. Melly, and P. Setlow. 2001. Properties of Bacillus subtilis spores blocked at an intermediate stage in spore germination. J. Bacteriol. 183:4894-4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Setlow, P. 1994. Mechanisms which contribute to the long-term survival of spores of Bacillus species. J. Appl. Bacteriol. Symp. Suppl. 76:49S-60S. [DOI] [PubMed]
- 58.Steinmetz, M., and R. Richter. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142:79-83. [DOI] [PubMed] [Google Scholar]
- 59.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vandeyar, M. A., and S. A. Zahler. 1986. Chromosomal insertions of Tn917 in Bacillus subtilis. J. Bacteriol. 167:530-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vary, J. C. 1973. Germination of Bacillus megaterium spores after various extraction procedures. J. Bacteriol. 95:1327-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1-9. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, J., P. C. Fitz-James, and A. I. Aronson. 1993. Cloning and characterization of a cluster of genes encoding polypeptides present in the insoluble fraction of the spore coat of Bacillus subtilis. J. Bacteriol. 175:3757-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zheng, L., W. P. Donovan, P. C. Fitz-James, and R. Losick. 1988. Gene encoding a morphogenic protein required in the assembly of the outer coat of the Bacillus subtilis endospore. Genes Dev. 2:1047-1054. [DOI] [PubMed] [Google Scholar]