Abstract
Penicillin binding protein (PBP) 5, a dd-carboxypeptidase that removes the terminal d-alanine from peptide side chains of peptidoglycan, plays an important role in creating and maintaining the uniform cell shape of Escherichia coli. PBP 6, a highly similar homologue, cannot substitute for PBP 5 in this respect. Previously, we localized the shape-maintaining characteristics of PBP 5 to the globular domain that contains the active site (domain I), where PBPs 5 and 6 share substantial identity. To identify the specific segment of domain I responsible for shape control, we created a set of hybrids and determined which ones complemented the aberrant morphology of a misshapen PBP mutant, E. coli CS703-1. Fusion proteins were constructed in which 47, 199 and 228 amino-terminal amino acids of one PBP were fused to the corresponding carboxy-terminal amino acids of the other. The morphological phenotype was reversed only by hybrid proteins containing PBP 5 residues 200 to 228, which are located next to the KTG motif of the active site. Because residues 220 to 228 were identical in these proteins, the morphological effect was determined by alterations in amino acids 200 to 219. To confirm the importance of this segment, we constructed mosaic proteins in which these 20 amino acids were grafted from PBP 5 into PBP 6 and vice versa. The PBP 6/5/6 mosaic complemented the aberrant morphology of CS703-1, whereas PBP 5/6/5 did not. Site-directed mutagenesis demonstrated that the Asp218 and Lys219 residues were important for shape maintenance by these mosaic PBPs, but the same mutations in wild-type PBP 5 did not eliminate its shape-promoting activity. Homologous enzymes from five other bacteria also complemented the phenotype of CS703-1. The overall conclusion is that creation of a bacterial cell of regular diameter and uniform contour apparently depends primarily on a slight alteration of the enzymatic activity or substrate accessibility at the active site of E. coli PBP 5.
In Escherichia coli, four dd-carboxypeptidase low-molecular-weight penicillin binding proteins (PBPs), 4, 5, and 6, and DacD modify peptidoglycan by removing the terminal d-alanine from pentapeptide side chains (3). Although all of these proteins are believed to originate from a common ancestor (19), the physiological role of only one of these proteins is known. PBP 5 functions to maintain a regular cell shape and contour in mutants lacking multiple low-molecular-weight PBPs (10, 23), and none of the other dd-carboxypeptidases can take its place in restoring normal morphology to a mutant lacking seven PBPs (22). Previously, we took advantage of the close relationship between PBPs 5 and 6 to create hybrids in which the two major domains of these proteins were fused to one another. Only proteins containing the globular domain I of PBP 5 restored normal shape to the septuple PBP mutant (21). This domain constitutes the core dd-carboxypeptidase and is structurally homologous to the class A β-lactamases (9, 12).
To identify the morphologically important segments of domain I, we constructed hybrid proteins in which portions of domain I from PBPs 5 and 6 were interchanged with one another. A stretch of 20 amino acids around the KTG motif emerged as a major contributor to cellular morphology. PBPs 5 and 6 differ in 8 of these 20 residues, of which 5 were different compared with DacD. Site-directed mutagenesis of mosaic proteins established that amino acids Asp218 and Lys219 were especially important for creating and/or maintaining normal cell shape in E. coli.
MATERIALS AND METHODS
Strains and growth conditions.
E. coli XL-1 Blue (recA endA hsdR supE thi recA gyrA relA lac) (Stratagene, La Jolla, Calif.) and E. coli DH5α (deoR recA endA hsdR supE thi gyrA relA) were used as hosts for constructing recombinant plasmids. Strains used in the morphological experiments were derived from CS109 (W1485 rpoS rph) (C. Schnaitman) via CS701-1 (CS109 Δ[mrcA-yrfE-yrfF] ΔdacB ΔdacA ΔdacC ΔpbpG ΔampC ΔampH) and CS703-1 (CS109 ΔmrcA ΔdacB ΔdacA ΔdacC ΔpbpG ΔampC ΔampH) (10, 20).
PBP genes were expressed under the control of the arabinose promoter of pBAD18-CAM, provided by J. Beckwith (14). Strains were grown on Luria-Bertani (LB) broth or agar plates, with chloramphenicol (20 μg/ml) added as required to maintain selection of pBAD plasmids. Overnight broth cultures of E. coli strains were diluted 1:250 into fresh LB medium and incubated at 37°C until they reached an A600 of 0.2 (ca. five to six doublings) before complementation was assessed by microscopy. Cells were collected, prepared for microscopy, and photographed at 1,000× magnification as described previously (23). When necessary, glucose (0.2 to 0.4%, wt/vol) was added to the medium to inhibit gene expression from the arabinose promoter. To induce protein expression in complementation experiments, E. coli strains were grown in the absence of glucose or in the presence of arabinose (0.0005% or 0.001%) (22). For expression of dd-carboxypeptidases from other species in E. coli, the arabinose concentration for induction was increased (up to 0.1%). Unless otherwise noted, all chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.).
Molecular techniques.
Plasmids were isolated from E. coli by with QIAprep Spin miniprep and midiprep kits (Qiagen Corp., Valencia, Calif.) according to the manufacturer's instructions. Competent cells were prepared and transformed by electroporation with the Gene Pulser apparatus from Bio-Rad (Hercules, Calif.) according to the manufacturer's instructions. CS109 chromosomal DNA for PCR amplifications was prepared by boiling 200 μl of overnight culture with 800 μl of distilled water for 10 min, followed by centrifugation at 14,000 × g for 1 min and collection of the supernatant. DNA agarose gel electrophoresis was performed as described before (24). DNA purification from agarose gels was performed with QIAquick gel extraction kits (Qiagen Corp.) as described by the manufacturer. Restriction digests and ligations were performed with enzymes purchased from New England Biolabs (Beverly, Mass.).
PCR.
PCR was performed in a model 2400 Gene Amp thermal cycler (Perkin Elmer, Boston, Mass.). Oligonucleotide primers for PCR were from MWG Biotech Inc. (High Point, N.C.). Stock solutions of individual deoxynucleoside triphosphates for PCR were from Promega (Madison, Wis.). Deep Vent DNA polymerase was from New England Biolabs (Beverly, Mass.).
Construction of PBP gene fusions.
With a PCR-based strategy, portions of the 5′ terminus of the dacA gene, encoding domain I of PBP 5, were fused to the corresponding 3′ terminus of dacC, encoding PBP 6. The PCR primers are described in Table 1, and their order of use to amplify and assemble each hybrid gene is listed in Table 2. Fragments of PBPs 5 and 6 were fused to one another at three positions where their amino acid sequences were identical or differed by only one of seven residues. Therefore, forward and reverse primers corresponding to amino acid positions 41 to 47, 197 to 203, and 220 to 228 of the mature form of PBP 5 were synthesized to match the nucleotide sequences of each separate gene (Table 1).
TABLE 1.
Primer sequences
| Primera | Sequenceb | Gene or fragmentc |
|---|---|---|
| A | 5′-CTCTCTGCTAGCAGGAGGAATTCACCATGAATACCATTTTTTCCGC-3′ | dacA (F) |
| B | 5′-GCATGCAAGCTTCTAGATTTTTAACCAAACCAGTGATG-3′ | dacA (R) |
| C | 5′-CTCTTTGCTAGCAGGAGGAATTCACATGACGCAATACTCCTCTC-3′ | dacC (F) |
| D | 5′-CTCTCTAAGCTTTTAAGAGAACCAGCTGCC-3′ | dacC (R) |
| E | 5′-GAT CCC GCG AGC CTG ACT AAA-3′ | dacC 41-47 (F) |
| F | 5′-AAC CGT AAC CGT CTG CTG TGG-3′ | dacC 197-203 (F) |
| G | 5′-GCG GGA TAT AAT CTG GTT GCT TCG GCT-3′ | dacC 220-228 (F) |
| H | 5′-TTT GGT CAG GCT GGC AGG ATC-3′ | dacA 41-47 (R) |
| I | 5′-CCA TAA CAG GCC GTT ACG GTT-3′ | dacA 197-203 (R) |
| J | 5′-CGC AGA AGC AAC AAG GTT GTA ACC TGC-3′ | dacA 220-228 (R) |
| K | 5′-GAT CCT GCC AGC CTG ACC AAA-3′ | dacA 41-47 (F) |
| L | 5′-AAC CGT AAC GGC CTG TTA TGG-3′ | dacA 197-203 (F) |
| M | 5′-GCA GGT TAC AAC CTT GTT GCT TCT GCG-3′ | dacA 220-228 (F) |
| N | 5′-TTT AGT CAG GCT CGC GGG ATC-3′ | dacC 41-47 (R) |
| O | 5′-CCA CAG CAG ACG GTT ACG GTT-3′ | dacC 197-203 (R) |
| P | 5′-AGC CGA AGC AAC CAG ATT ATA TCC CGC-3′ | dacC 220-228 (R) |
Primer names are given in Table 2.
Underlined sequences are complementary to the 5′ (forward primer [F]) or 3′ (reverse primer [R]) end of the gene fragment in each fusion construct. Boldfacing and double underlining designate enzyme recognition sites: AAGCTT, HindIII; GCTAGC, NheI.
The gene to which each oligonucleotide anneals in the PCR amplifications is indicated first, followed by the amino acid residues represented by the sequence. Residue numbers refer to positions in the crystal structure of the mature protein (9), from which the amino-terminal 29 amino acids (PBP 5, dacA) or 27 amino acids (PBP 6, dacC) have been removed from the predicted translation products during translocation to the periplasm. Primers A to D anneal to the 5′ or 3′ end of the genes indicated. F, forward primer (P1 or P3); R, reverse primer (P2 or P4).
TABLE 2.
Plasmids and hybrid proteins
| Plasmida | Terminus encodedb
|
Primersc | Junction sequenced | Complementatione | |
|---|---|---|---|---|---|
| Amino | Carboxyl | ||||
| pAG6 | Wild-type PBP 6 | Wild-type PBP 6 | |||
| pPJ5C | Wild-type PBP 5 | Wild-type PBP 5 | |||
| pAGP-7 | 1-47 PBP 5 | 48-378 PBP 6 | A, H, E, D | DPASLTK | − |
| pAGR-10 | 1-199 PBP 5 | 200-378 PBP 6 | A, I, F, D | NRNRLLW | − |
| pAGS-29 | 1-228 PBP 5 | 229-378 PBP 6 | A, J, G, D | AGYNLVASA | + |
| pAGE-1 | 1-47 PBP 6 | 48-374 PBP 5 | C, N, K, B | DPASLTK | + |
| pAGG-1 | 1-199 PBP 6 | 200-374 PBP 5 | C, O, L, B | NRNGLLW | + |
| pAGH-1 | 1-228 PBP 6 | 229-374 PBP 5 | C, P, M, B | AGYNLVASA | − |
Plasmids were constructed by cloning DNA fragments into the NheI and HindIII sites of pBAD18-CAM.
Amino acids and source of the protein encoded by each cloned DNA fragment. Each plasmid expressed a single hybrid PBP consisting of the segments indicated (those derived from PBP 5 are underlined). Numbers refer to the amino acid residues present in the mature protein and correspond to the numbering system used in the crystal structure of PBP 5 (9). Not included in the numbering scheme is the 29-amino-acid signal peptide that is removed from the mature PBP 5 protein. Likewise, the 27-residue signal peptide of PBP 6 is omitted in numbering the mature protein that corresponds to the crystal structure of PBP 5.
Oligonucleotide primers used to construct the two segments of each of the cloned DNA fragments are presented in the order P1, P2, P3, and P4, as described in Materials and Methods. The sequence of each oligonucleotide is given in Table 1.
Partial amino acid sequences (one-letter code) at the junction sites of the hybrid PBPs. Because the fusions were constructed at sequences that were identical or nearly identical between PBPs 5 and 6, no additional or foreign residues are present in the expressed proteins. The pAGR-10 junction is the PBP 6 sequence at that point, and the pAGG-1 junction is the PBP 5 sequence at that point. These two sequences differ only at residue 200 (R or G) (underlined).
Expression of the hybrid PBP did (+) or did not (−) return the aberrant morphology of CS703-1 to normal or near normal.
The strategy for constructing the hybrid genes was described previously (21). Briefly, DNA fragments representing the amino and carboxy termini of each gene were prepared separately with primers P1 and P2 or primers P3 and P4, respectively. The products of these reactions were combined and used to prime one another to create full-length, fused copies of hybrid genes. An NheI site and Shine-Dalgarno sequence were included in the design of the 5′ terminus of the P1 primer, and a HindIII site was included at the 5′ end of the P4 primer, so that these occurred at the 5′ and 3′ ends, respectively, of the final PCR product. The DNA fragments were purified by agarose gel electrophoresis, digested with restriction enzymes NheI and HindIII, and ligated into the vector pBAD18-CAM to place each fusion protein under the control of the arabinose promoter. The nucleotide sequence of all inserts was confirmed by the sequencing services of MWG Biotech Inc. (High Point, N.C.).
Construction of mosaic PBPs.
Two mosaic PBPs were constructed in which amino acids around the active-site KTG motif were moved from one PBP to another. The nucleotides encoding PBP 5 amino acids 200 to 219 were spliced into PBP 6, creating plasmid pAG656-11, in the following manner. Gene fragments were amplified from plasmid pAGG-1 (primers C and J, Table 1) and from pAGS-29 (primers D and L, Table 1). The nucleotide sequence of the first PCR product encoded the first 196 amino acids of PBP 6 fused to residues 197 to 228 of PBP 5, and the nucleotide sequence of the second PCR product encoded residues 197 to 228 of PBP 5 fused to residues 229 to 378 of PBP 6. These two DNA fragments were isolated from agarose gels and used to prime one another in a second PCR, as described previously (21), by hybridization between nucleotides representing amino acid residues 197 to 228. Because the yield in this reaction was poor, the composite full-length PCR product was amplified further with primers C and D (Table 1). This DNA fragment was digested with restriction enzymes NheI and HindIII and cloned between the NheI and HindIII sites of pBAD18-CAM, as described above, creating plasmid pAG656-11. Thus, the final PCR product encoded a mosaic protein composed of PBP 6 residues 1 to 196, PBP 5 residues 197 to 228, and PBP 6 residues 229 to 378. Because amino acid residues 197 to 199 and 220 to 228 are identical in the two proteins, the final mosaic protein is the equivalent of inserting residues 200 to 219 from PBP 5 into full-length PBP 6 (see Table 3).
TABLE 3.
Complementation of morphology by mosaic and mutant PBPsa
| Protein or plasmid | Sequences around KTG motif
|
Complementationb | ||
|---|---|---|---|---|
| 197-203 | 204-219 | 220-228 | ||
| Proteins | ||||
| PBP 5 | NRNGLLW | DNSLNVDGIKTGHTDK | AGYNLVASA | |
| PBP 6 | NRNRLLW | SSNLNVDGMKTGTTAG | AGYNLVASA | |
| DacD | NRNGLLW | DKTMNVDGLKTGHTSG | AGFNLIASA | |
| pAG656-11 derivativesc | ||||
| pAG656-11 | NRNGLLW | DNSLNVDGIKTGHTDK | AGYNLVASA | + |
| pAG656-X2A | NRNRLLW | DNSLNVDGIKTGHTDK | + | |
| pAG656-M1A | DSSLNVDGIKTGHTDK | + | ||
| pAG656-M2A | DNNLNVDGIKTGHTDK | + | ||
| pAG656-M3A | DNSLNVDGMKTGHTDK | + | ||
| pAG656-H | DNSLNVDGIKTGTTDK | + | ||
| pAG656-M4A | DNSLNVDGIKTGHTAK | − | ||
| pAG656-M11A | DNSLNVDGIKTGHTDG | − | ||
| pAG656-M5A | DNSLNVDGIKTGHTAG | − | ||
| pAG565-3 derivativesd | ||||
| pAG565-3 | NRNRLLW | SSNLNVDGMKTGTTAG | AGYNLVASA | − |
| pAG565-X1A | NRNGLLW | SSNLNVDGMKTGTTAG | − | |
| pAG565-M6Ae | SNNLNEDGMKTGTTAG | − | ||
| pAG565-M7Ae | SSSLNEDGIKTGTTAG | − | ||
| pAG565-T | SSNLNVDGMKTGHTAG | − | ||
| pAG565-M9A | SSNLNVDGMKTGTTDG | + | ||
| pAG565-M10A | SSNLNVDGMKTGTTAK | + | ||
| pAG565-X5B | NRNRLLW | SSNLNVDGMKTGTTAG | − | |
| pAG565-X6A | NRNGLLW | SSNLNVDGMKTGTTAG | − | |
| pPJ5 derivativesf (full-length PBP 5 and mutants): | ||||
| pPJ5 | NRNGLLW | DNSLNVDGIKTGHTDK | AGYNLVASA | + |
| pPJ5-DK/AG | DNSLNVDGIKTGHTAG | + | ||
| pAG5-X3A | NRNRLLW | DNSLNVDGIKTGHTDK | + | |
| pAG5-X4A | NRNRLLW | DNSLNVDGIKTGHTAG | + | |
| pPJ5-T217A | DNSLNVDGIKTGHADK | + | ||
| pPJ5-K213E | DNSLNVDGIETGHTDK | − | ||
| pPJ5-K213R | DNSLNVDGIRTGHTDK | − | ||
Residue numbers refer to the mature protein and correspond to the numbering system used in the crystal structure of PBP 5 (9). Amino acids that differ from the sequence of PBP 5 are boldfaced and double underlined. The KTG active site motif is underlined once in the first sequence. Residues that are boldfaced and double underlined indicate differences from the first sequence shown in each section. For clarity, the sequence of residues 197 to 203 and 220 to 228 were omitted from the table when these were identical to the first sequence in each section. The inserted genes in each plasmid were sequenced to confirm that no other alterations were present. The sequences of the primer pairs used to create each mutant can be inferred from the amino acid alterations, but these are also available on request.
Effect of plasmid-encoded proteins on the cellular morphology of E. coli CS701-1 or CS703-1: +, complete or virtually complete restoration of uniform wild-type rod-shaped morphology; −, little or no restoration of uniform wild-type rod-shaped morphology.
These derivatives had PBP 5 residues 200 to 219 inserted into PBP 6, and thus the residues listed in the 204-219 column are from PBP 5.
These derivatives had PBP 6 residues 200 to 219 inserted into PBP 5, and thus the residues listed in the 204-219 column are from PBP 6. The residue at position 141 was D in all these derivatives except the last two, in which it was G. The G141D mutation was present in plasmid pAG565-3 and several of its derivatives. However, this mutation did not affect the complementation properties of the PBP mosaic proteins (compare the results for pAG565-3 with those for pAG565-X5B and pAG565-X6A).
The glutamic acid residue (E) at position 209 in pAG565-M6A and pAG565-M7A is present in the E. coli K-12 genomic sequence (6), but a valine residue (V) is present in E. coli O157:H7 (15) and in the parental E. coli strain CS109 used in this work. Thus, PBP 5 with either glutamic acid or valine in this position has wild-type properties.
These derivatives had full-length PBP 5 with various mutations.
The nucleotides encoding PBP 6 amino acids 200 to 219 were spliced into PBP 5, creating plasmid pAG565-3, in a similar manner. Gene fragments were amplified from plasmid pAGR-10 (primers A and P, Table 1) and from pAGH-1 (primers F and B, Table 1). The nucleotide sequence of the first PCR product encodes the first 196 amino acids of PBP 5 fused to residues 197 to 228 of PBP 6, and the nucleotide sequence of the second PCR product encodes residues 197 to 228 of PBP 6 fused to residues 229 to 374 of PBP 5. The two PCR products were hybridized to one another, amplified, and cloned as described above to create plasmid pAG565-3. The final PCR product encoded a mosaic protein composed of PBP 5 residues 1 to 196, PBP 6 residues 197 to 228, and PBP 5 residues 229 to 374. Because amino acid residues 197 to 199 and 220 to 228 are identical in the two proteins, the final mosaic protein is the equivalent of inserting residues 200 to 219 from PBP 6 into full-length PBP 5 (see Table 3). The nucleotide sequence of each mosaic protein was confirmed by the sequencing services of MWG Biotech Inc. (High Point, N.C.).
Site-directed mutagenesis.
Site-directed mutagenesis of the amino acids in and around the conserved KTG motif was performed with the Quick Change mutagenesis kit from Stratagene (La Jolla, Calif.) as described previously (21). Mutagenesis on supercoiled double-stranded plasmid DNA was carried out exactly according to the manufacturer's instructions, with oligonucleotide primer pairs obtained from MWG Biotech, Inc. The primers ranged from 33 to 47 bases, depending on the number of individual nucleotides to be altered (one to three), and for each primer pair one or two codons at the center were altered to give the amino acid substitutions described in Table 3. The number of PCR cycles was from 12 to 18, increasing according to the number of bases altered. Mutated plasmids were transformed by heat shock into Epicurian XL1-Blue supercompetent cells (Stratagene, La Jolla, Calif.) and plated on LB-chloramphenicol plates. Mutagenesis was confirmed by DNA sequencing (MWG Biotech, Inc.).
PBP labeling, photography, and sequence analysis.
Expression of hybrid PBP proteins from recombinant plasmids was confirmed by labeling equal numbers of cells with [125I]penicillin X, separating total cellular proteins by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and visualizing by autoradiography as described previously (16). Photography was performed and interpreted as described previously (22, 23). Homologous protein sequences were identified and compared by the BlastP 2.1.3 program (1) as supplied on the National Institutes of Health Entrez web site (URL http://www3.ncbi.nlm.nih.gov/Entrez/) and by the ClustalW program (version 1.81) (25) as supplied on the European Bioinformatics Institute web site (URL http://www2.ebi.ac.uk/clustalw/).
RESULTS
A 20-amino-acid sequence is important for morphological activity of PBP 5.
We constructed three hybrid proteins consisting of increasing lengths of the amino terminus of PBP 5 fused to the carboxy terminus of PBP 6, as well as three similar hybrids consisting of the corresponding amino segments of PBP 6 fused to PBP 5 (Table 2). The fusion joints were placed at sequences that were either identical or differed in only one of seven residues in the two proteins to increase the likelihood that the homologous hybrids would represent combinations of naturally occurring internal subdomains. All hybrid proteins bound penicillin (data not shown), indicating that they were functional. In the text and tables, please note that references to amino acid positions correspond to the numbering system adopted to describe the sequence of the crystallized form of mature PBP 5 (9).
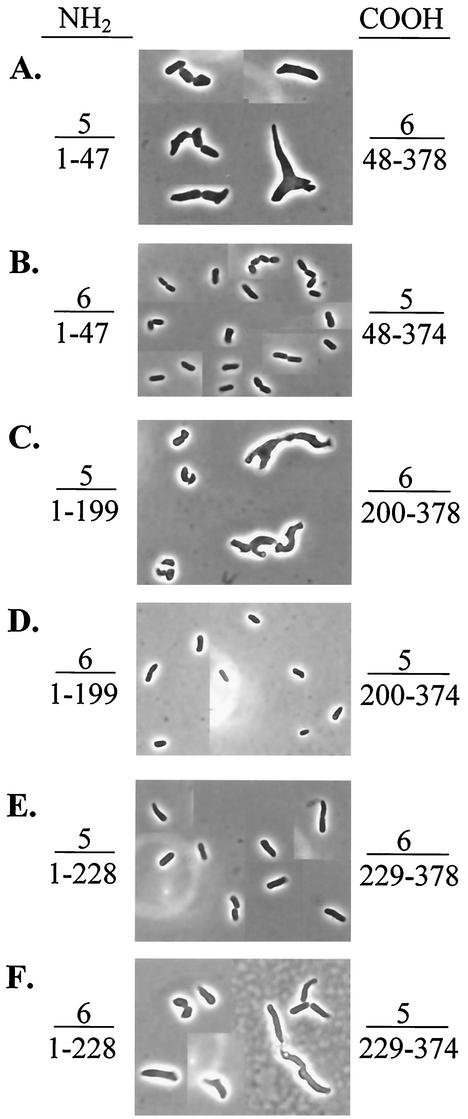
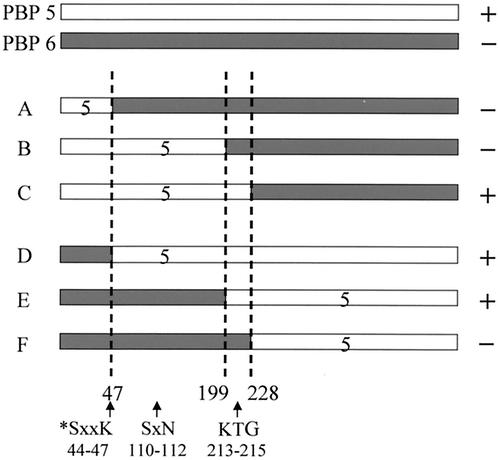
Fusion proteins containing various portions of PBPs 5 and 6 reversed the shape defects of E. coli CS703-1 when residues 200 to 228 of the hybrids were derived from PBP 5 (Table 2 and Fig. 1 and 2). Residues 220 to 228 were identical in PBPs 5 and 6 (AGYNLVASA). Therefore, the ability to return CS703-1 to its normal shape was associated with the presence of a contiguous stretch of 20 amino acids consisting of residues 200 to 219 from PBP 5.
FIG. 1.
Complementation of morphological defects by hybrid dd-carboxypeptidases. Hybrid proteins were expressed in E. coli CS703-1 containing the following plasmids: A, pAGP-7; B, pAGE-1; C, pAGR-10; D, pAGG-1; E, pAGS-29; and F, pAGH-1. Arabinose was not added to any of the cultures. Instead, expression of each protein was induced at a low, nonlethal level by growth in LB medium. For each photograph, the sources (5, PBP 5; 6, PBP 6) and amino acid residues comprising the amino (left side) and carboxy (right side) termini of the hybrid proteins are given. The residue numbers are expressed relative to the mature, crystallized form of PBP 5 (9). All photographs are displayed at equal magnification.
FIG. 2.
Schematic representation of hybrid PBP proteins. Lightly shaded bars represent residues derived from E. coli PBP 5; heavier shaded lines represent residues derived from E. coli PBP 6. Residue numbers refer to positions in the mature protein (9). To the right is noted the ability of each hybrid protein to complement (+) or not complement (−) shape abnormalities of E. coli CS703-1. The plasmids from which each hybrid PBP was produced were (A) pAGP-7, (B) pAGR-10, (C) pAGS-29, (D) pAGE-1, (E) pAGG-1, and (F) pAGH-1. The locations of the three active-site motifs are indicated by arrows. The active-site serine is denoted by an asterisk.
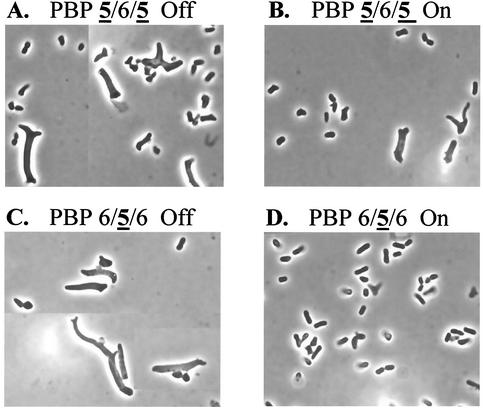
We constructed two additional mosaic proteins to test the importance of this segment in shape determination. Plasmid pAG656-11 expressed PBP 6/5/6, in which residues 200 to 219 from PBP 5 were inserted into PBP 6, and plasmid pAG565-3 expressed PBP 5/6/5, in which residues 200 to 219 from PBP 6 were inserted into PBP 5 (Table 3). Both bound 125I-labeled penicillin X and migrated as expected on SDS-PAGE (data not shown). PBP 6/5/6 (Fig. 3D) complemented the aberrant morphology of CS703-1, whereas PBP 5/6/5 did not (Fig. 3B). The results confirmed that these 20 amino acids from PBP 5 could impart the ability to create or maintain a normal cell shape on wild-type PBP 6.
FIG. 3.
Complementation of morphological defects by mosaic dd-carboxypeptidases. Amino acid residues 200 to 219 of PBP 5 were replaced with the homologous residues from PBP 6 (A and B) or inserted into the homologous positions in PBP 6 (C and D). Mosaic proteins were expressed in E. coli CS703-1 from pAG565-3 (PBP 5/6/5) (A and B) and pAG656-11 (PBP 6/5/6) (C and D). Gene expression was inhibited by growth in LB plus 0.2% glucose (A and C), or the gene products were expressed at a low level by growth in LB alone, without induction by addition of arabinose (B and D). Photographs are displayed at equal magnifications.
Asp218 and Lys219 are important shape-influencing residues in mosaic PBPs.
Eight of the amino acids in positions 200 to 219 differ between PBPs 5 and 6, and of these, five are different between PBP 5 and DacD (Table 3). Thus, one or more of these five amino acids were the most likely candidates for imparting a morphological capability to PBP 5. To determine the relative importance of these sites, we mutated individual residues in the mosaic protein PBP 6/5/6 (Table 3, pAG656-11 derivatives). In each case, one or more of the amino acids originally derived from PBP 5 were replaced with the amino acids normally present in PBP 6. Similarly, we mutated the corresponding sites in PBP 5/6/5, replacing individual residues derived from PBP 6 with amino acids normally present in PBP 5 (Table 3, pAG565-3 derivatives). As before, pAG565-3 and pAG656-11 bound 125I-labeled penicillin X, as did all the mutants derived from them (data not shown).
Five mutants derived from PBP 6/5/6 retained the ability to complement the aberrant shape of CS703-1: G200R (pAG656-X2A), N205S (pAG656-M1A), S206N (pAG656-M2A), I212M (pAG656-M3A), and H216T (pAG656-H) (Table 3). Converting the same positions in PBP 5/6/5 to PBP 5 residues did not restore normal morphology to CS703-1: R200G (pAG565-X1A and pAG565-X6A), S205N (pAG565-M6A), N206S plus M212I (pAG565-M7A), and T216H (pAG656-T) (Table 3). Therefore, the amino acids at positions 200, 205, 206, 212, and 216 did not distinguish the activities of PBPs 5 and 6.
On the other hand, mutation of either of two residues in PBP 6/5/6 destroyed its shape-complementing ability: D218A (pAG656-M4A), K219G (pAG656-M11A), and DK218/219AG (pAG656-M5A) (Table 3). Similarly, converting either of these two positions in PBP 5/6/5 to PBP 5 residues successfully reactivated the shape complementation ability of this hybrid: A218D (pAG565-M9A) and G219K (pAG565-M10A) (Table 3). Thus, at least in these mosaic proteins, the charged amino acids Asp218 and Lys219 were important differential determinants of the ability to create or maintain normal bacterial morphology.
Features other than Asp218 and Lys219 contribute to morphological activity of PBP 5.
It appeared that the ability of PBP 5 to affect cell shape might be mediated by one or two individual residues guarding the entrance to the active site (see Discussion). However, the presence of either of these normally occurring residues (either Asp218 or Lys219) in PBP 5/6/5 created a functional enzyme (Table 3), indicating that one or the other amino acid was sufficient to confer morphological activity on this particular mosaic protein. On the other hand, mutating either or both of these residues in PBP 6/5/6 (e.g., D218A, K219G, or DK218/219AG) (Table 3) inhibited the morphological activity of this hybrid. Thus, while these two residues were obviously vital to the function of the mosaic proteins, their relative importance was still unclear. To understand this further, we mutated residues 218 and 219 simultaneously in wild-type PBP 5, changing them to amino acids normally present in PBP 6 (DK218/219AG, expressed from plasmid pPJ5-DK/AG) as well as in the presence of a third mutation (G200R, expressed from plasmid pPJ5-X4A) (Table 3, pPJ5 derivatives). Unexpectedly, these mutant proteins retained the ability to complement the shape defects of CS703-1 (Table 3). Therefore, although these two amino acids evidently play significant morphological roles in the mosaic proteins, the effects of these residues are moderated in the context of wild-type PBP 5.
Correlation of dd-carboxypeptidase activity with morphological effects of PBP 5.
The lysine at position 213 of PBP 5 is the first residue in the canonical KTG motif of the PBPs (13). A K213E mutation in PBP 5 eliminates both penicillin-binding and dd-carboxypeptidase activity (28), whereas a K213R mutation eliminates the dd-carboxypeptidase activity but leaves intact the penicillin-binding and penicillin-hydrolyzing ability of PBP 5 (18). To see which activity contributed more strongly to the morphological abilities of PBP 5, we constructed mutant proteins containing these alterations and observed their effects on cell shape. Consistent with previous observations (18, 28), the K213E mutant did not bind 125I-labeled penicillin X, but the K213R mutant could still do so (data not shown). Neither mutant complemented the aberrant cell shapes exhibited by E. coli CS703-1 (Table 3, pPJ5 derivatives), indicating that maintenance of cell shape was correlated with dd-carboxypeptidase activity, not with penicillin binding.
As a further test of the supposition that the morphological ability of PBP 5 was related to its dd-carboxypeptidase action, we created a T217A mutant (Table 3, pPJ5 derivatives), which bound penicillin X very well (data not shown). This mutation decreases the dd-carboxypeptidase activity of PBP 5 to 0.5% of normal while leaving intact 45% of the penicillin binding activity (28). Nonetheless, the mutant protein corrected the morphological oddities in E. coli CS703-1 (Table 3, pPJ5 derivatives). Thus, if dd-carboxypeptidase activity is crucial to the function of PBP 5, then either a very small amount of activity suffices when the protein is supplied from a multicopy plasmid or else this particular mutation alters the activity of PBP 5 toward in vitro substrates but not toward the relevant in vivo substrate.
Shape complementation by dd-carboxypeptidases from other bacteria.
Individual gram-negative bacteria express multiple low-molecular-weight PBPs related to PBPs 5 and 6 of E. coli. Until now, sequence comparison has been the only way to predict if one or more of these proteins might perform homologous functions within different species. The morphological assay afforded us the opportunity to test candidate proteins for functional equivalence.
A Blast search was performed to identify proteins most closely related to PBP 5 from E. coli (not shown). The most closely related dd-carboxypeptidase genes from five gram-negative bacteria were amplified by PCR, cloned under control of the arabinose promoter, sequenced, and expressed in E. coli CS703-1 (Table 4 and data not shown). DacA proteins from Salmonella enterica serovar Typhimurium, Vibrio cholerae (Cpase-1), Pasteurella multocida, Haemophilus influenzae, and Yersinia pestis reversed the morphological defects of CS703-1 (Table 4). The DacD protein of S. enterica serovar Typhimurium and the Cpase-2 protein of V. cholerae failed to complement the defects (Table 4). Thus, the shape phenotype of this multiply mutated E. coli provided the first explicit method for measuring homologous function among the dd-carboxypeptidases.
TABLE 4.
Shape complementation by different dd-carboxypeptidases
| Species (protein) | Genea | Sequencesb
|
Complementationc | ||
|---|---|---|---|---|---|
| 197-203 | 204-219 | 220-228 | |||
| E. coli (PBP 5) | dacA | NRNGLLW | DNSLNVDGIKTGHTDK | AGYNLVASA | + |
| S. entericad | dacA | NRNGLLW | DNSLNVDGIKTGHTSK | AGYNLVASA | + |
| V. cholerae | Cpase-1 | NRNGLLW | DKSMNVDGIKTGHTSG | AGYNLVSSA | + |
| P. multocida | dacA | NRNGLLW | DKSIQVDGIKTGHTDK | AGYNLVASA | + |
| H. influenzae | dacA | NRNGLLW | DKTINVDGMKTGHTSQ | AGYNLVASA | + |
| Y. pestis | dacA | NRNGLLW | DTSLNVDGIKTGHTEA | AGYNLVASA | + |
| E. coli (PBP 6) | dacC | NRNRLLW | SSNLNVDGMKTGTTAG | AGYNLVASA | − |
| E. coli (DacD) | dacD | NRNGLLW | DKTMNVDGLKTGHTSG | AGYNLIASA | − |
| S. entericad | dacD | NRNGLLW | DKTMHIDGLKTGHTSG | AGFNLIASA | − |
| V. cholerae | Cpase-2 | NRNGLLR | DRSMNVDGMKTGYTSG | AGYSLVSSA | − |
The Swiss-Prot abbreviations for the two V. cholerae dd-carboxypeptidases are Q9KTF5 (Cpase-1) and Q9KMQ0 (Cpase-2).
Amino acid sequences homologous to positions 197 to 228 in the crystal structure of PBP 5. The KTG active-site motif is underlined once. Residues that are different from those in E. coli PBP 5 are boldfaced and underlined.
Effect of plasmid-encoded proteins on the cellular morphology of E. coli CS703-1: +, complete or virtually complete restoration of uniform wild-type rod-shaped morphology; −, little or no restoration of uniform wild-type rod-shaped morphology.
Serovar Typhimurium.
DISCUSSION
The morphological activity of PBP 5 is a feature of its globular domain I, which is highly similar to β-lactamase and includes the dd-carboxypeptidase active site (21). This activity is essential for complementing the aberrant shape of E. coli CS703-1, because mutating the active-site serine (22) or KTG motif (this work) eliminates the restorative properties of PBP 5. Although PBP 5 must be anchored to the outer surface of the inner membrane, and though it may be part of a multienzyme complex, these factors cannot explain why the physiological function of PBP 5 differs from that of other closely related dd-carboxypeptidases (21-23). Instead, we hypothesize that the singular ability of PBP 5 to influence cell shape derives more from its enzymology than from any other property (21).
In support of this idea, we discovered that splicing a 20-amino-acid segment from PBP 5 into PBP 6 transformed the latter protein into a morphological substitute for PBP 5. The simplest interpretation is that the enzymatic specificity of the mosaic PBP was altered to be more like that of wild-type PBP 5. This explanation is consistent with observations made by Chang et al., who performed an analogous experiment with RTEM β-lactamase (7). When they replaced 28 amino acids around the active-site SxxN motif with homologous sequences from PBP 5, the enzyme lost its ability to hydrolyze penicillin and was transformed into a low-level dd-carboxypeptidase (7). Similarly, as shown here, PBP 6 acquired the ability to complement morphological abnormalities when 20 residues around its KTG motif were replaced with homologous residues from PBP 5.
Further evidence for an enzymatic explanation comes from site-directed mutagenesis of the PBP 5/6/5 and 6/5/6 mosaic proteins. In each case, the functional state of the protein could be toggled between the active and inactive forms by changing either of two amino acids at position 218 or 219, downstream of the conserved KTG motif (residues 213 to 215). There are precedents for considering these positions important determinants of enzymatic specificity. Ubukata et al. found that 9 of 25 β-lactamase-negative, ampicillin-resistant clinical isolates of Haemophilus influenzae accumulated PBP 3 mutations that mapped to just this position, changing from KTGTARK to KTGTAHK (group I mutants) (26). Strains harboring these mutant PBP 3 molecules exhibited very different susceptibilities to a variety of β-lactam antibiotics (26), indicating that the mutations changed the substrate specificity of this PBP. The locations of two charged amino acids at the third and fourth residues downstream of the KTG motif are equivalent to positions 218 and 219 in PBP 5 from E. coli, and the effect of the R-to-H mutation in the third downstream residue parallels the mutation in position 218 that we report for the mosaic proteins created in this work.
In addition, the other 16 isolates of Ubukata et al. (group II mutants) contain an N526K mutation which, compared with the homologous crystal structure of PBP 5, is equivalent to position 227 in PBP 5. This residue is one of nine very highly conserved amino acids that follow the KTG motif (compare positions 220 to 228 for the different species in Table 4). Residues 210 to 228 double back to form a hairpin structure in which amino acids 210 and 216 and 220 and 228 are in contact with one another as parallel β-sheets, with residues 218 and 219 forming the “turn” of the hairpin (9). Thus, both mutations described by Ubekata et al. probably affect the active site in the vicinity of the KTG motif. These results were confirmed and extended by Dabernat et al., who found the same two mutant groups and alterations in β-lactam susceptibility among 108 β-lactamase-negative, ampicillin-resistant clinical isolates of H. influenzae (8). The parallels between these observations for PBP 3 in H. influenzae and the mosaic proteins described here strengthen the conclusion that the physiological difference between PBPs 5 and 6 lies in a specific enzymatic capacity and thus narrows the possible explanations for how E. coli maintains a uniform cell shape.
Given that the mutagenesis data support an enzymatic mechanism, what enzymatic property might explain the divergence in physiological function between PBP 5 and other noncomplementing dd-carboxypeptidases? To begin with, the ability of a PBP to bind β-lactams does not, by itself, create a functional protein. For example, the K213R mutant, which binds penicillin but has no dd-carboxypeptidase activity (18), did not complement the misshapen phenotype. Thus, the dd-carboxypeptidase reaction is necessary, although we cannot say it is sufficient in the absence of β-lactam binding. On the other hand, a T217A mutant, which retains most of its penicillin binding ability but only ≈0.5% of its dd-carboxypeptidase activity (28), remained capable of complementing the odd morphology of CS703-1. So, if dd-carboxypeptidase activity per se is the vital property, then only a small amount is sufficient. This would be consistent with the observation that in the absence of induction, a low-level background expression of cloned PBP 5 complements the morphological phenotype (21, 22). Also, expressing the T217A mutant from a multicopy plasmid may have provided enough activity to restore proper morphology. A final possibility is that the chemical state or environment of the in vivo substrate of PBP 5 may be different than the artificial in vitro substrates used to assay bulk dd-carboxypeptidase activity.
The fact that inserting Asp218 or Lys219 residues changed a nonfunctional PBP 5/6/5 mosaic into a protein that complemented the morphological defects of E. coli CS703-1 suggests that there is a subtle biochemical explanation behind the differences in physiological function. The 20-amino-acid segment surrounding the conserved KTG extends almost linearly across one side of the active site of PBP 5, and the Asp218 and Lys219 residues within this sequence are located in a small loop a short distance away from the active site proper (9). These amino acids make no direct contact with the d-Ala-d-Ala terminus of the peptide side chain, but residues at these positions might modify the structure of the active site or interact with other portions of the peptide side chain or glycan polymer. In the simplest case, substitutions at these positions would change the kinetics of PBPs 5 and 6 toward a common substrate; for example, by moderating substrate access or affinity.
The two PBPs do have different in vitro activities: one group reports that PBP 5 is three to four times more active toward artificial substrates than is PBP 6 (2), though another lab reports that PBP 6 exhibits no activity at all towards the same compounds (27). In vivo evidence of such a difference also exists: mutants lacking PBP 5 accumulate muramyl pentapeptides, while PBP 6 mutants do not (B. Glauner, Ph.D. thesis, quoted and referenced in reference 27). In any case, the physiological difference between PBPs 5 and 6 may reflect nothing more than the enzymatic superiority of PBP 5. Alternatively, the presence of Asp218 and Lys219 might change the substrate specificity entirely. Unfortunately, in vitro assays cannot be trusted to distinguish among these possibilities because no one knows the relevant characteristics of the true in vivo substrates, which may be modified by the three-dimensional structure of peptidoglycan or by the degree of extension of peptide side chains (17). Perhaps these mosaic proteins may be used to discover substrates that mimic more closely what occurs in vivo.
The above considerations beg the following question: if the Asp218 and Lys219 residues are so important, why does mutating these amino acids eliminate the morphological activity of PBP 6/5/6 but not affect the complementation ability of wild-type PBP 5? First of all, it is possible that the mutations in wild-type PBP 5 produced a subtle or quantitative morphological change that was not detected by the gross microscopic assay that we employed. However, we easily detected that mutation of the mosaic proteins dramatically altered their ability to carry out a normal level of cell shape maintenance, indicating clearly that these enzymes were not exactly equivalent to wild-type PBP 5. Instead, the most straightforward explanation for the differences between PBP 5 and the mosaic proteins is that the wild-type PBP 5 active site retains partial activity or substrate specificity because of the nature or strength of interactions elsewhere in the molecule. This is easily understandable if only 0.5% activity is required to correct morphological deficiencies, as exhibited by the T217A mutant of PBP 5.
The active site of the PBPs and β-lactamases contain variations of three major motifs — SxxK, SxN, and KTG in this case — which are brought into close proximity by protein folding. In the hybrids that we created previously (21) and in those reported here, both the SxxK and SxN motifs are derived either from PBP 5 or from PBP 6, and only the KTG motif comes from a different source. So, one possibility is that in PBP 6/5/6, the combination of SxxK and SxN from PBP 6 might make the protein sensitive to alterations in the PBP 5-derived KTG region, whereas mutations near the KTG segment in wild-type PBP 5 are offset by interactions in other parts of the molecule. This interpretation is consistent with the spectrum of β-lactamase-negative, ampicillin-resistant PBP 3 mutants of H. influenzae: in many cases, mutation of residues in the KTG region were accompanied by additional unlinked mutations, which further affected the β-lactam sensitivity of these mutant PBPs (8, 26).
The fact that dd-carboxypeptidase PBPs from other species complement the shape deficiencies of these E. coli mutants enhances the idea that subtle differences in enzymology are responsible. First of all, the degree of homology, as measured along the full length of these enzymes, does not predict which ones will function like PBP 5. For example, two noncomplementing PBPs (S. enterica serovar Typhimurium DacD and V. cholerae Cpase-2) are more closely related to E. coli PBP 5 than is DacA from Y. pestis, which did complement the phenotype (Table 4 and data not shown). The opposite is true for E. coli PBP 6. This protein is more closely related to E. coli PBP 5 than every protein except DacA from S. enterica serovar Typhimurium (data not shown), yet E. coli PBP 6 does not complement the phenotype.
Instead of overall similarity, the sequences equivalent to residues 204 to 219 of mature PBP 5 may be a more predictive measure of function. In this region, complementing dd-carboxypeptidases are generally more similar to PBP 5 than are PBPs that do not complement (Table 4). A second way in which the heterologous enzymes support the idea of a subtle enzymatic mechanism is that the Asp218 and Lys219 residues are present only in DacA from Pasteurella multocida. Nevertheless, the dd-carboxypeptidases from four organisms complemented the shape phenotype even though these enzymes have other amino acids at these positions (Table 4). This indicates, again, that although these two amino acids may play important roles in the PBP 5/6 mosaics, other factors contribute to the function of wild-type enzymes.
An interesting side observation is that the region defined by residues 204 to 219 in PBP 5 is shared by the dd-carboxypeptidases and β-lactamases, and in both families this region is bracketed on either side by sequences having almost complete identity to that of PBP 5 (Table 4 and data not shown). In particular, the neighboring sequences are nearly identical in the dd-carboxypeptidases of E. coli as well as other bacteria (Table 4). Thus, though the KTG region may vary from one enzyme to another and therefore affect enzyme activity, the adjacent structures that hold it in position may be more highly conserved.
Finally, since it seems that the dd-carboxypeptidase activity of PBP 5 moderates bacterial morphology, the following difficult question arises: How does removing the terminal d-alanine from peptide side chains create a uniform cell shape? This minor change evidently regulates the gross structure of peptidoglycan, but the mechanism by which this occurs is unclear. One possibility is that the pentapeptide (which retains the terminal d-alanine) and the tetrapeptide or tripeptide side chains (which have lost one or two d-alanines, respectively) are preferred substrates of different synthetic PBPs. Such an idea was proposed to explain how enzymes might distinguish the synthesis of cylindrical cell wall versus dividing septum (4, 5). In this view, septation mediated by PBP 3 prefers substrates containing tripeptide side chains, and cell elongation mediated by PBP 2 prefers substrates with pentapeptides (4, 5). Thus, deletion of PBP 5, by increasing the amount of pentapeptides, should preferentially increase the relative activity of PBP 2. However, the morphological defects of PBP 5 mutants apparently emerge as a consequence of inappropriately placed patches of “septal” (inert) peptidoglycan (11), which should represent PBP 3 products. On the surface, this suggests that in these mutants PBP 3 activity is enhanced rather than decreased by elevated levels of pentapeptides. If so, this would contradict the previously proposed model and imply that PBP 3 might prefer pentapeptide substrates. On the other hand, if PBP 3 does indeed prefer tripeptides, then the enzyme might react to a relative abundance of pentapeptides with ill-timed or awkward cross-linking of this unfavorable substrate, resulting in the observed morphological oddities.
Acknowledgments
This work was supported by grant GM61019 from the National Institutes of Health.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amanuma, H., and J. L. Strominger. 1980. Purification and properties of penicillin-binding proteins 5 and 6 from Escherichia coli membranes. J. Biol. Chem. 255:11173-11180. [PubMed] [Google Scholar]
- 3.Baquero, M.-R., M. Bouzon, J. C. Quintela, J. A. Ayala, and F. Moreno. 1996. dacD, an Escherichia coli gene encoding a novel penicillin-binding protein (PBP6b) with dd-carboxypeptidase activity. J. Bacteriol. 178:7106-7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begg, K. J., A. Takasuga, D. H. Edwards, S. J. Dewar, B. G. Spratt, H. Adachi, T. Ohta, H. Matsuzawa, and W. D. Donachie. 1990. The balance between different peptidoglycan precursors determines whether Escherichia coli cells will elongate or divide. J. Bacteriol. 172:6697-6703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Botta, G. A., and J. T. Park. 1981. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J. Bacteriol. 145:333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broome-Smith, J. K., I. Ioannidis, A. Edelman, and B. G. Spratt. 1988. Nucleotide sequences of the penicillin-binding protein 5 and 6 genes of Escherichia coli. Nucleic Acids Res. 16:1617.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y.-H., M. R. Labgold, and J. H. Richards. 1990. Altering enzymatic activity: recruitment of carboxypeptidase activity into an RTEM β-lactamase/penicillin-binding protein 5 chimera. Proc. Natl. Acad. Sci. USA 87:2823-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dabernat, H., C. Delmas, M. Seguy, R. Pelissier, G. Faucon, S. Bennamani, and C. Pasquier. 2002. Diversity of β-lactam resistance-conferring amino acid substitutions in penicillin-binding protein 3 of Haemophilus influenzae. Antimicrob. Agents Chemother. 46:2208-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, C., S. W. White, and R. A. Nicholas. 2001. Crystal structure of a deacylation-defective mutant of penicillin-binding protein 5 at 2.3-Å resolution. J. Biol. Chem. 276:616-623. [DOI] [PubMed] [Google Scholar]
- 10.Denome, S. A., P. K. Elf, T. A. Henderson, D. E. Nelson, and K. D. Young. 1999. Escherichia coli mutants lacking all possible combinations of eight penicillin binding proteins: viability, characteristics, and implications for peptidoglycan synthesis. J. Bacteriol. 181:3981-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Pedro, M. A., K. D. Young, J.-V. Höltje, and H. Schwarz. 2003. Branching of Escherichia coli cells arises from multiple sites of inert peptidoglycan. J. Bacteriol. 185:1147-1152. [DOI] [PMC free article] [PubMed]
- 12.Ghuysen, J.-M. 1994. Molecular structures of penicillin-binding proteins and β-lactamases. Trends Microbiol. 2:372-380. [DOI] [PubMed] [Google Scholar]
- 13.Goffin, C., and J.-M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzman, L.-M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yodoyama, C.-G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, T. A., P. M. Dombrosky, and K. D. Young. 1994. Artifactual processing of penicillin-binding proteins 7 and 1b by the OmpT protease of Escherichia coli. J. Bacteriol. 176:256-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch, A. L. 2000. Simulation of the conformation of the murein fabric: the oligoglycan, penta-muropeptide, and cross-linked nona-muropeptide. Arch. Microbiol. 174:429-439. [DOI] [PubMed] [Google Scholar]
- 18.Malhotra, K. T., and R. A. Nicholas. 1992. Substitution of lysine 213 with arginine in penicillin-binding protein 5 of Escherichia coli abolishes d-alanine carboxypeptidase activity without affecting penicillin binding. J. Biol. Chem. 267:11386-11391. [PubMed] [Google Scholar]
- 19.Massova, I., and S. Mobashery. 1998. Kinship and diversification of bacterial penicillin-binding proteins and β-lactamases. Antimicrob. Agents Chemother. 42:1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meberg, B. M., F. C. Sailer, D. E. Nelson, and K. D. Young. 2001. Reconstruction of Escherichia coli mrcA (PBP 1a) mutants lacking multiple combinations of penicillin binding proteins. J. Bacteriol. 183:6148-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson, D. E., A. S. Ghosh, A. L. Paulson, and K. D. Young. 2002. Contribution of membrane-binding and enzymatic domains of penicillin binding protein 5 to maintenance of uniform cellular morphology of Escherichia coli. J. Bacteriol. 184:3630-3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson, D. E., and K. D. Young. 2001. Contributions of PBP 5 and dd-carboxypeptidase penicillin binding proteins to maintenance of cell shape in Escherichia coli. J. Bacteriol. 183:3055-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson, D. E., and K. D. Young. 2000. Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. J. Bacteriol. 182:1714-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ubukata, K., Y. Shibasaki, K. Yamamoto, N. Chiba, K. Hasegawa, Y. Takeuchi, K. Sunakawa, M. Inoue, and M. Konno. 2001. Association of amino acid substitutions in penicillin-binding protein 3 with β-lactam resistance in β-lactamase-negative ampicillin-resistant Haemophilus influenzae. Antimicrob. Agents Chemother. 45:1693-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Linden, M. P., L. de Haan, M. A. Hoyer, and W. Keck. 1992. Possible role of Escherichia coli penicillin-binding protein 6 in stabilization of stationary-phase peptidoglycan. J. Bacteriol. 174:7572-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Linden, M. P. G., L. de Haan, O. Dideberg, and W. Keck. 1994. Site-directed mutagenesis of proposed active-site residues of penicillin-binding protein 5 from Escherichia coli. Biochem. J. 303:357-362. [DOI] [PMC free article] [PubMed] [Google Scholar]