Abstract
There are two interrelated acyl-homoserine lactone quorum-sensing-signaling systems in Pseudomonas aeruginosa. These systems, the LasR-LasI system and the RhlR-RhlI system, are global regulators of gene expression. We performed a transcriptome analysis to identify quorum-sensing-controlled genes and to better understand quorum-sensing control of P. aeruginosa gene expression. We compared gene expression in a LasI-RhlI signal mutant grown with added signals to gene expression without added signals, and we compared a LasR-RhlR signal receptor mutant to its parent. In all, we identified 315 quorum-induced and 38 quorum-repressed genes, representing about 6% of the P. aeruginosa genome. The quorum-repressed genes were activated in the stationary phase in quorum-sensing mutants but were not activated in the parent strain. The analysis of quorum-induced genes suggests that the signal specificities are on a continuum and that the timing of gene expression is on a continuum (some genes are induced early in growth, most genes are induced at the transition from the logarithmic phase to the stationary phase, and some genes are induced during the stationary phase). In general, timing was not related to signal concentration. We suggest that the level of the signal receptor, LasR, is a critical trigger for quorum-activated gene expression. Acyl-homoserine lactone quorum sensing appears to be a system that allows ordered expression of hundreds of genes during P. aeruginosa growth in culture.
Pseudomonas aeruginosa is an opportunistic pathogen of humans, other animals, plants, and lower eukaryotes (16). There are two acyl-homoserine lactone (acyl-HSL) quorum-sensing systems in P. aerugionosa, LasI-LasR and RhlI-RhlR. LasI is responsible for the synthesis of N-3-oxododecanoyl-HSL (3OC12-HSL), and LasR is a 3OC12-HSL-responsive transcription factor. RhlI is responsible for the synthesis of N-butanoyl-HSL (C4-HSL), and RhlR is a C4-HSL-responsive transcription factor. Both of the acyl-HSLs can diffuse through the cell envelope, so a critical cell population density is required to produce signals at levels sufficient for quorum-controlled gene regulation. Acyl-HSL signaling is an important virulence factor in P. aeruginosa (for recent reviews see references 9, 19, and 31).
Previously, Whiteley et al. identified quorum-controlled genes in P. aeruginosa by screening a library of random Tn5-lacZ insertions in the genome of an acyl-HSL synthesis mutant for induction of β-galactosidase by signal addition (33); 35 quorum-controlled genes were identified. Based on the number of mutants screened and the number of insertions in putative operons, it was estimated that there were over 200 additional quorum-controlled genes. The lacZ induction patterns were grouped into four categories depending on the timing of induction and the signal response specificity. Some genes responded to addition of acyl-HSL signals early in culture growth, and others showed a substantial delay, responding to signals only in the stationary phase. Some genes responded specifically to 3OC12-HSL, and others required both signals for the maximal response. The requirement for both signals to obtain a maximal response is thought to be related to the fact that both rhlR and rhlI are induced by LasR-LasI (14, 22). We have studied several quorum-controlled promoters. Some of the promoters show a high level of specificity for 3OC12-HSL and LasR, while others show specificity for C4-HSL and RhlR but also show a substantial response to 3OC12-HSL and LasR (32).
Other investigators have presented evidence showing that additional genes are controlled by quorum sensing in P. aeruginosa (2, 3, 5, 10-13, 15, 18, 21, 26, 27, 29, 35, 36). Furthermore, several reports have shown that a variety of regulatory proteins can influence the timing of quorum-controlled gene expression (1, 4, 6, 24, 26, 34), but mechanistic details for these proteins are scarce.
Here we describe a transcriptome analysis in which we utilized Affymetrix GeneChip genome arrays to test the hypothesis that there are over 200 quorum-controlled genes in P. aeruginosa, to identify as many members of this putative regulon as possible, and to gain insight into the timing and specificity of quorum-controlled gene expression.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The P. aeruginosa strains used were PAO-MW1 (rhlI::Tn501 lasI::tetA) (33) and PAO lasR rhlR (ΔlasR::Tcr ΔrhlR::Gmr), as well as the isogenic PAO1 parent strain (25). Bacteria were grown in buffered Luria-Bertani broth, which contained 10 g of typtone (Difco) per liter, 5 g of yeast extract (Difco) per liter, 5 g of NaCl per liter, and 50 mM 3-(N-morpholino)propanesulfonic acid (pH 7.0). Synthetic acyl-HSLs (Aurora Biosciences) were added to PAO-MW1 cultures at final concentrations of 2 μM for 3OC12-HSL and 10 μM for C4-HSL, as indicated below. To inoculate the cultures used for transcript profiling, cells grown to the mid-logarithmic phase were added to 100 ml of prewarmed medium in 500-ml culture flasks. The initial optical densities at 600 nm (OD600) were 0.05 for PAO-MW1 and 0.01 for PAO1 and PAO lasR rhlR. Cultures were incubated at 37°C in a rotating shaker at 250 rpm. Growth was monitored by determining the OD600.
Expression profiling experiments.
For studies with the signal generation mutant we isolated RNA from cultures at the following optical densities: 0.2, 0.4, 0.8, 1.4, 2.0, 3.0, and 4.0. For studies with the signal receptor mutant and its parent we isolated RNA from cultures at optical densities of 0.05, 0.1, 0.2, 0.4, 0.8, 1.4, 2.0, 3.0, and 4.0. Between 1 × 109 and 2 × 109 cells were mixed with RNA Protect Bacteria reagent (Qiagen) and treated as recommended by the manufacturer's mechanical disruption and lysis protocol. RNA was purified by using RNeasy mini columns (Qiagen), including the on-column DNase I digestion described by the manufacturer. In addition, we treated the eluted RNA for 1 h at 37°C with DNase I (0.1 U per μg of RNA). DNase I was removed by using DNA-Free (Ambion) or by RNeasy column purification. RNA integrity was monitored by agarose gel electrophoresis of glyoxylated samples.
Further sample preparation and processing of the P. aeruginosa GeneChip genome arrays were done as described by the manufacturer (Affymetrix), with minor modifications suggested by M. Wolfgang (Harvard University). For cDNA synthesis we used 12 μg of purified RNA, semirandom hexamer primers with an average G+C content of 75%, and Superscript II reverse transcriptase (Life Technologies). Control RNAs from yeast, Arabidopsis, and Bacillus subtilis genes (provided by S. Lory) were added to the reaction mixtures to monitor assay performance. Probes for these transcripts are tiled on the GeneChip arrays. RNA was removed from the PCR mixtures by alkaline hydrolysis. The cDNA synthesis products were purified and fragmented by brief incubation with DNase I, and the 3′ termini of the fragmentation products were labeled with biotin-ddUTP. Fragmented and labeled cDNA was hybridized to an array by overnight incubation at 50°C. Washing, staining, and scanning of microarrays were performed with an Affymetrix fluidic station.
Analysis of expression profiling experiments.
We used the Affymetrix Microarray Software suite (MAS) (version 5.0) to determine transcript levels and whether there were differences in transcript levels when different samples were compared. Affymetrix scaling was used to normalize data from different arrays. A scale factor is derived from the mean signal of all of the probe sets on an array and a user-defined target signal. The signal from each individual probe set is multiplied by this scale factor. For any given array between 18 and 28% of the mRNAs were considered absent by MAS, indicating that the corresponding genes were not expressed at levels above background levels. Furthermore, the average changes in control transcript intensities were less than twofold for any comparison of array data. This indicates that the efficiency of cDNA synthesis and labeling was similar from sample to sample.
For comparison analyses, the log2 ratio for absolute transcript signals obtained from a given pair of arrays was calculated by using MAS. A statistical algorithm of the software also assigned a change call for each transcript pair, which indicated whether the level of a transcript was significantly increased, decreased, or not changed compared to the level for the baseline sample. The baseline samples were those derived from cultures of PAO-MW1 without added acyl-HSL and from cultures of PAO lasR rhlR. Graphical analysis of the signal log ratios from each experiment (any pair of arrays) revealed a normal distribution with a mean very close to zero (no change). Among the transcripts with significant increases or decreases compared to the baseline in one or more samples, we subjected those that showed a ≥2.5-fold change to further analysis.
For cluster analyses and transcript pattern analyses we used GeneSpring software (Silicon Genetics, Redwood City, Calif.). The fold change values for each gene were normalized independently by defining the half-maximal value for the gene as 1 and representing all other values as a ratio of that value. This scaling procedure allowed direct visual comparison of gene expression patterns within an experiment, as well as between experiments. GeneSpring was also used to sort genes according to the P. aeruginosa genome project (http://www.pseudomonas.com).
Identification of las-rhl box-like sequences.
A 20-bp consensus sequence (ACCTGCCAGATCTGGCAGGT) was derived from the following previously identified las-rhl box-like sequences in quorum-sensing-controlled genes: PA1869 (qsc117), PA1896 (qsc102), hcnA, lasB, lasI, and phzA (32), as well as PA2592 (qsc104), PA3327 (qsc126), PA4217 (qsc132), rhlA, and rhlI (33). To search the entire P. aeruginosa genome for sequences similar to this consensus, we developed a computer program based on the program used previously to search for LexA binding sites (8). The scoring matrix of the program is based on a heterology index (HI), which determines the degree of divergence of any 20-nucleotide sequence from the consensus las-rhl box sequence. Sequences in a region from 400 bp upstream to 50 bp downstream of annotated translational start sites were considered potential las-rhl boxes if they showed an HI of less than 13.
RESULTS
Genes induced by addition of acyl-HSL signals to the P. aeruginosa signal generation mutant: validation of the microarray analysis.
Previously, the effects of acyl-HSLs on chromosomal lacZ insertions in a quorum-sensing signal generation mutant (MW1, a lasI rhlI mutant) were studied (33), and 39 loci that responded to the addition of 3OC12-HSL and C4-HSL were identified. These loci correspond to 35 different genes in the previously published annotation of the P. aeruginosa genome (28). Two insertions originally thought to reside in two adjacent genes (qsc109 and qsc110) are in a single predicted gene (PA2402), and three insertions (qsc114, qsc127, and qsc136) were oriented in a direction opposite that of the predicted open reading frames.
To validate the microarray approach, we grew the signal generation mutant with or without 3OC12-HSL and C4-HSL under conditions identical to those used in the previous study (identical medium, growth temperature, acyl-HSL concentrations, etc.) and examined whether the genes identified previously responded to signal addition in a transcriptome analysis. Most genes in the P. aeruginosa genome showed no significant response. Among 638 genes that showed a maximal response to acyl-HSL addition of at least 2.5-fold, 29 of the 35 previously described qsc genes (33) were identified (Table 1). The six remaining genes all exhibited relatively low induction levels in the previous study (33). Four of these six genes, PA2385 (qsc112), PA2401 (qsc111), PA2402 (qsc109-110), and PA2426 (qsc108), showed a significant response to signal addition in our analysis, but the response was less than 2.5-fold. Two genes, PA0051 (qsc137) and PA4084 (qsc113), showed no response. Taken together, our results showed quite good agreement with the results of the previous study (33).
TABLE 1.
Quorum-activated genes
| Gene no.a | Descriptionb | Maximum changec
|
||
|---|---|---|---|---|
|
lasI rhlI mutant
|
Wild type vs lasR rhlR mutant | |||
| 3OC12-HSL | C4-HSL + 3OC12-HSL | |||
| PA0007 | Hypothetical protein | 4.4 | 5.7 (2.0) | 14 (1.4) |
| PA0026 | Hypothetical proteind | 4.4 | 4.4 (1.4) | 5.9 (0.2) |
| PA0027 | Hypothetical protein | 3.8 | 4.9 (0.8) | 5.7 (0.2) |
| PA0028 | Hypothetical protein | 5.8 | 7.5 (1.4) | 8.2 (1.4) |
| PA0050 | Hypothetical protein | 2.8 | 2.5 (2.0) | 3.0 (1.4) |
| PA0052 | Hypothetical proteind | 4.7 | 8.3 (1.4) | 22 (2.0) |
| PA0059 | osmC, osmotically inducible protein OsmC | 2.5 | 6.7 (2.0) | 8.9 (2.0) |
| PA0105 | coxB, cytochrome c oxidase subunit II | 3.4 | 4.0 (2.0) | 2.6 (2.0) |
| PA0106 | coxA, cytochrome c oxidase subunit I | 4.2 | 4.8 (1.4) | 3.3 (1.4) |
| PA0107 | Conserved hypothetical protein | 4.1 | 4.8 (2.0) | 4.9 (2.0) |
| PA0108 | coIII, cytochrome c oxidase subunit III | 3.0 | 3.6 (2.0) | 2.8 (2.0) |
| PA0109 | qsc115, hypothetical protein | 2.1 | 3.5 (1.4) | 4.1 (1.4) |
| PA0122 | Conserved hypothetical proteine | 13 | 36 (1.4) | 51 (1.4) |
| PA0132 | Beta-alanine-pyruvate transaminase | 1.6 | 3.1 (1.4) | 4.1 (2.0) |
| PA0143 | Probable nucleoside hydrolase | 4.7 | 4.7 (0.4) | 5.4 (0.1) |
| PA0144 | Hypothetical protein | 1.5 | 19 (2.0) | 28 (2.0) |
| PA0158 | Probable RND efflux transporter | 2.6 | 2.6 (2.0) | 2.6 (2.0) |
| PA0175 | Probable chemotaxis protein methyltransferase | 2.0 | 2.6 (3.0) | 4.6 (1.4) |
| PA0176 | Probable chemotaxis transducer | 2.1 | 2.6 (3.0) | 3.9 (1.4) |
| PA0179 | Probable two-component response regulator | 2.7 | 2.8 (1.4) | 3.7 (1.4) |
| PA0198 | exbBI, transport protein ExbB | 7.3 | 10 (4.0) | 3.7 (4.0) |
| PA0263 | hcpC, secreted protein Hcp | 1.7 | 8.9 (1.4) | 9.4 (1.4) |
| PA0355 | pfpI, protease PfpI | 2.3 | 4.8 (2.0) | 8.1 (2.0) |
| PA0364 | Probable oxidoreductase | 2.9 | 3.1 (2.0) | 3.0 (2.0) |
| PA0365 | Hypothetical protein | 2.0 | 2.5 (2.0) | 2.7 (2.0) |
| PA0366 | Probable aldehyde dehydrogenase | 2.4 | 2.8 (2.0) | 2.5 (3.0) |
| PA0534 | Conserved hypothetical protein | 1.5 | 2.9 (4.0) | 9.8 (2.0) |
| PA0567 | Conserved hypothetical protein | 6.9 | 15 (2.0) | 11 (2.0) |
| PA0572 | Hypothetical protein | 19 | 22 (0.4) | 19 (0.05) |
| PA0586 | Conserved hypothetical protein | 2.1 | 2.6 (1.4) | 4.6 (1.4) |
| PA0852 | qsc129, cpbD, chitin-binding protein CbpD precursord | 11 | 43 (0.4) | 94 (0.1) |
| PA0855 | qsc116, hypothetical protein | 2.4 | 2.5 (0.8) | 3.0 (0.8) |
| PA0996 | Probable coenzyme A ligasee | 220 | 90 (0.8) | 42 (0.2) |
| PA0997 | Hypothetical protein | 110 | 96 (0.8) | 200 (0.05) |
| PA0998 | Hypothetical protein | 68 | 40 (0.4) | 200 (0.2) |
| PA0999 | fabHI, 3-oxoacyl-(acyl carrier protein) synthase III | 37 | 25 (0.8) | 45 (0.2) |
| PA1000 | Hypothetical protein | 22 | 12 (0.8) | 44 (0.2) |
| PA1001 | phnA, anthranilate synthase component I | 39 | 23 (0.8) | 290 (0.2) |
| PA1002 | phnB, anthranilate synthase component II | 17 | 13 (1.4) | 28 (0.8) |
| PA1003 | Probable transcriptional regulator | 8.1 | 6.6 (0.2) | 78 (0.05) |
| PA1130 | Hypothetical protein | 2.4 | 9.4 (1.4) | 16 (1.4) |
| PA1131 | Probable MFS transporterd | 1.7 | 5.0 (2.0) | 7.9 (1.4) |
| PA1173 | napB, cytochrome c-type protein NapB precursor | 2.3 | 2.8 (2.0) | 4.1 (1.4) |
| PA1175 | napD, NapD protein of periplasmic nitrate reductase | 2.6 | 2.4 (2.0) | 3.8 (1.4) |
| PA1176 | napF, ferredoxin protein NapF | 2.5 | 2.5 (2.0) | 5.8 (1.4) |
| PA1177 | napE, periplasmic nitrate reductase protein NapE | 2.9 | 3.6 (1.4) | 3.6 (1.4) |
| PA1215 | Hypothetical protein | NC | 18 (1.4) | 55 (1.4) |
| PA1216 | Hypothetical protein | 4.7 | 15 (0.8) | 120 (0.8) |
| PA1217 | Probable 2-isopropylmalate synthase | 2.9 | 41 (1.4) | 380 (1.4) |
| PA1218 | Hypothetical protein | NC | 6.9 (1.4) | 160 (1.4) |
| PA1221 | Hypothetical proteine | NC | 3.1 (3.0) | 11 (1.4) |
| PA1245 | Hypothetical proteine | 8.6 | 10 (0.8) | 11 (0.2) |
| PA1246 | aprD, alkaline protease secretion protein AprD | 8.6 | 9.8 (1.4) | 6.6 (0.8) |
| PA1247 | aprE, alkaline protease secretion protein AprE | 6.2 | 6.4 (1.4) | 9.1 (1.4) |
| PA1248 | aprF, alkaline protease secretion protein AprF | 7.2 | 7.6 (1.4) | 5.2 (1.4) |
| PA1249 | aprA, alkaline metalloproteinase precursor | 25 | 27 (1.4) | 22 (1.4) |
| PA1250 | aprI, alkaline proteinase inhibitor AprId | 20 | 20 (0.2) | 24 (0.05) |
| PA1289 | Hypothetical protein | 2.9 | 5.7 (1.4) | 2.6 (1.4) |
| PA1317 | cyoA, cytochrome o ubiquinol oxidase subunit II | 2.5 | 4.7 (4.0) | 15 (2.0) |
| PA1318 | cyoB, cytochrome o ubiquinol oxidase subunit I | NC | 3.9 (4.0) | 16 (2.0) |
| PA1319 | cyoC, cytochrome o ubiquinol oxidase subunit III | 2.0 | 4.8 (4.0) | 7.9 (3.0) |
| PA1320 | cyoD, cytochrome o ubiquinol oxidase subunit IV | 42 | 71 (4.0) | 9.1 (3.0) |
| PA1323 | Hypothetical protein | 2.8 | 6.1 (2.0) | 9.6 (2.0) |
| PA1324 | Hypothetical protein | 2.4 | 5.3 (2.0) | 8.5 (2.0) |
| PA1404 | Hypothetical protein | 2.0 | 2.7 (2.0) | 3.8 (2.0) |
| PA1431 | rsaL, regulatory protein RsaLd | 350 | 340 (0.2) | 39 (0.8) |
| PA1432 | lasI, autoinducer synthesis protein LasId | NCf | NCf | 7.7 (0.8) |
| PA1656 | Hypothetical proteina | 2.4 | 3.7 (1.4) | 5.7 (0.8) |
| PA1657 | Conserved hypothetical protein | 5.9 | 15 (0.4) | 24 (0.8) |
| PA1658 | Conserved hypothetical protein | 3.9 | 9.3 (0.8) | 17 (0.8) |
| PA1659 | Hypothetical protein | 4.1 | 8.5 (0.8) | 17 (0.8) |
| PA1660 | Hypothetical protein | 2.6 | 7.9 (0.8) | 16 (0.8) |
| PA1661 | Hypothetical protein | 2.3 | 4.4 (1.4) | 4.4 (0.8) |
| PA1662 | Probable ClpA/B-type protease | 2.9 | 6.6 (1.4) | 7.7 (0.8) |
| PA1663 | Probable transcriptional regulator | 2.5 | 4.5 (0.8) | 9.1 (0.8) |
| PA1664 | Hypothetical protein | 5.9 | 16 (0.4) | 22 (0.8) |
| PA1665 | Hypothetical protein | 21 | 55 (1.4) | 28 (0.8) |
| PA1666 | Hypothetical protein | 2.9 | 12 (0.8) | 38 (0.8) |
| PA1667 | Hypothetical protein | 3.1 | 7.6 (0.8) | 12 (0.8) |
| PA1668 | Hypothetical protein | 2.8 | 4.6 (0.8) | 6.3 (0.8) |
| PA1669 | Hypothetical protein | 2.2 | 3.8 (1.4) | 17 (0.8) |
| PA1670 | stp1, serine/threonine phosphoprotein phosphatase Stp1 | NC | 2.8 (1.4) | 3.6 (0.8) |
| PA1745 | Hypothetical protein | 2.1 | 2.6 (2.0) | 2.8 (1.4) |
| PA1784 | Hypothetical proteine | 14 | 15 (1.4) | 18 (1.4) |
| PA1869 | qsc117, probable acyl carrier proteine | 7.8 | 41 (0.2) | 340 (0.2) |
| PA1870 | Hypothetical protein | NC | 3.2 (2.0) | 6.3 (2.0) |
| PA1871 | lasA, LasA protease precursord | 48 | 88 (0.8) | 130 (1.4) |
| PA1881 | Hypothetical protein | 2.4 | 2.6 (2.0) | 2.8 (1.4) |
| PA1888 | Hypothetical protein | 2.7 | 2.3 (2.0) | 4.3 (1.4) |
| PA1891 | Hypothetical protein | 3.3 | 4.3 (2.0) | 6.5 (0.8) |
| PA1893 | Hypothetical protein | 16 | 13 (0.4) | 2.7 (2.0) |
| PA1894 | qsc101, hypothetical protein | 59 | 59 (0.8) | 5.0 (1.4) |
| PA1895 | Hypothetical protein | 36 | 31 (0.8) | 4.2 (1.4) |
| PA1896 | Hypothetical protein | 41 | 49 (1.4) | 3.1 (1.4) |
| PA1897 | qsc102, hypothetical proteine | 130 | 130 (0.4) | 8.5 (0.8) |
| PA1914 | Conserved hypothetical protein | 42 | 190 (2.0) | 700 (2.0) |
| PA1921 | Hypothetical protein | NC | 14 (2.0) | 13 (2.0) |
| PA1930 | Probable chemotaxis transducer | 2.2 | 2.9 (2.0) | 3.8 (1.4) |
| PA1939 | Hypothetical protein | 2.6 | 3.2 (1.4) | 2.9 (2.0) |
| PA2030 | Hypothetical protein | 3.3 | 4.2 (2.0) | 14 (2.0) |
| PA2031 | Hypothetical protein | 4.5 | 6.5 (0.4) | 12 (1.4) |
| PA2066 | Hypothetical protein | 1.9 | 3.7 (2.0) | 12 (1.4) |
| PA2067 | Probable hydrolase | 1.8 | 5.0 (2.0) | 19 (1.4) |
| PA2068 | Probable MFS transporter | NC | 16 (1.4) | 150 (1.4) |
| PA2069 | Probable carbamoyl transferasee | NC | 45 (1.4) | 110 (0.2) |
| PA2076 | Probable transcriptional regulatord | 3.7 | 4.3 (0.2) | 4.3 (0.2) |
| PA2080 | Hypothetical protein | 3.5 | 4.0 (0.2) | 4.0 (0.2) |
| PA2081 | Hypothetical protein | 3.3 | 4.3 (0.2) | 3.6 (0.2) |
| PA2134 | Hypothetical protein | 3.1 | 5.4 (3.0) | 7.9 (2.0) |
| PA2142 | Probable short-chain dehydrogenase | NC | 3.6 (3.0) | 19 (2.0) |
| PA2143 | Hypothetical protein | 21 | 39 (2.0) | 51 (2.0) |
| PA2144 | glgP, glycogen phosphorylase | 2.5 | 4.7 (3.0) | 15 (2.0) |
| PA2146 | Conserved hypothetical protein | 2.7 | 4.8 (3.0) | 11 (2.0) |
| PA2147 | katE, catalase HPII | 3.5 | 7.1 (2.0) | 35 (2.0) |
| PA2148 | Conserved hypothetical protein | NC | 3.1 (2.0) | 3.4 (2.0) |
| PA2151 | Conserved hypothetical protein | 2.6 | 38 (2.0) | 34 (2.0) |
| PA2152 | Probable trehalose synthase | 2.1 | 5.3 (2.0) | 6.1 (2.0) |
| PA2153 | glgB, 1,4-alpha-glucan branching enzyme | 2.1 | 5.6 (2.0) | 16 (2.0) |
| PA2156 | Conserved hypothetical protein | 2.3 | 4.8 (3.0) | 17 (2.0) |
| PA2157 | Hypothetical protein | 2.1 | 2.8 (3.0) | 2.9 (3.0) |
| PA2158 | Probable alcohol dehydrogenase (Zn dependent) | 6.7 | 15 (2.0) | 26 (2.0) |
| PA2159 | Conserved hypothetical protein | 4.1 | 5.9 (2.0) | 10 (2.0) |
| PA2160 | Probable glycosyl hydrolased | 2.3 | 4.4 (3.0) | 5.8 (2.0) |
| PA2161 | Hypothetical proteind | 4.4 | 6.3 (2.0) | 10 (2.0) |
| PA2163 | Hypothetical protein | 2.2 | 6.9 (2.0) | 31 (2.0) |
| PA2164 | Probable glycosyl hydrolase | 2.5 | 4.7 (2.0) | 6.5 (2.0) |
| PA2165 | Probable glycogen synthase | 3.2 | 5.7 (2.0) | 6.3 (2.0) |
| PA2166 | Hypothetical protein | 3.1 | 9.1 (2.0) | 17 (2.0) |
| PA2167 | Hypothetical protein | 2.3 | 2.6 (2.0) | 4.7 (2.0) |
| PA2169 | Hypothetical protein | 2.8 | 5.7 (2.0) | 5.1 (2.0) |
| PA2170 | Hypothetical protein | 3.6 | 6.9 (2.0) | 13 (2.0) |
| PA2171 | Hypothetical protein | 5.2 | 9.1 (2.0) | 22 (2.0) |
| PA2172 | Hypothetical protein | 3.8 | 7.7 (2.0) | 12 (2.0) |
| PA2173 | Hypothetical protein | 3.5 | 6.5 (2.0) | 17 (2.0) |
| PA2176 | Hypothetical protein | 1.4 | 5.3 (2.0) | 27 (2.0) |
| PA2180 | Hypothetical protein | 1.9 | 2.6 (3.0) | 2.7 (3.0) |
| PA2190 | Conserved hypothetical protein | 3.4 | 4.5 (2.0) | 7.5 (2.0) |
| PA2192 | Conserved hypothetical protein | NC | 10 (2.0) | 8.4 (2.0) |
| PA2193 | hcnA, hydrogen cyanide synthase HcnAe | 140 | 190 (0.2) | 88 (0.2) |
| PA2194 | qsc128, hcnB, hydrogen cyanide synthase HcnB | 37 | 51 (0.2) | 59 (0.8) |
| PA2195 | hcnC, hydrogen cyanide synthase HcnC | 16 | 30 (0.4) | 46 (0.8) |
| PA2274 | Hypothetical protein | NC | 3.4 (3.0) | 11 (2.0) |
| PA2300 | chiC, chitinasee | 1.7 | 14 (1.4) | 100 (1.4) |
| PA2302 | qsc100, probable nonribosomal peptide synthetase | 5.2 | 7.9 (0.8) | 130 (1.4) |
| PA2303 | qsc107, hypothetical protein | 25 | 28 (0.4) | 130 (0.2) |
| PA2304 | Hypothetical protein | 8.4 | 12 (0.8) | 29 (0.8) |
| PA2305 | Probable nonribosomal peptide synthetase | 52 | 51 (0.2) | 70 (0.2) |
| PA2327 | Probable permease of ABC transporter | 5.9 | 8.9 (4.0) | 6.9 (4.0) |
| PA2328 | Hypothetical protein | 6.8 | 9.1 (2.0) | 7.5 (3.0) |
| PA2329 | Probable component of ABC transporter | 7.8 | 9.9 (1.4) | 18 (3.0) |
| PA2330 | Hypothetical protein | 7.9 | 11 (0.8) | 15 (2.0) |
| PA2331 | Hypothetical protein | 8.3 | 19 (1.4) | 20 (1.4) |
| PA2345 | Conserved hypothetical proteine | 2.2 | 3.2 (2.0) | 2.6 (2.0) |
| PA2365 | Conserved hypothetical protein | 4.7 | 5.4 (1.4) | 5.9 (1.4) |
| PA2366 | Conserved hypothetical protein | 4.3 | 5.2 (1.4) | 6.9 (1.4) |
| PA2367 | Hypothetical protein | 4.8 | 5.1 (1.4) | 6.4 (1.4) |
| PA2368 | Hypothetical protein | 3.5 | 3.4 (1.4) | 7.5 (1.4) |
| PA2370 | Hypothetical protein | 2.9 | 3.6 (3.0) | 3.5 (1.4) |
| PA2371 | Probable ClpA/B-type protease | 2.4 | 2.6 (3.0) | 5.0 (1.4) |
| PA2372 | Hypothetical protein | 3.2 | 2.7 (2.0) | 3.7 (1.4) |
| PA2414 | l-Sorbosone dehydrogenase | 3.1 | 4.9 (2.0) | 21 (0.2) |
| PA2415 | Hypothetical protein | 3.5 | 5.6 (2.0) | 14 (2.0) |
| PA2423 | Hypothetical protein | 11 | 11 (0.4) | 13 (0.2) |
| PA2433 | Hypothetical protein | 2.8 | 5.9 (2.0) | 11 (2.0) |
| PA2442 | gcvT2, glycine cleavage system protein T2 | 2.0 | 2.6 (3.0) | 3.1 (3.0) |
| PA2444 | glyA2, serine hydroxymethyltransferase | 9.1 | 12 (3.0) | 10 (3.0) |
| PA2445 | gcvP2, glycine cleavage system protein P2 | 6.6 | 7.5 (4.0) | 11 (3.0) |
| PA2446 | gcvH2, glycine cleavage system protein H2 | 12 | 17 (4.0) | 18 (3.0) |
| PA2448 | Hypothetical protein | NC | 4.1 (3.0) | 12 (1.4) |
| PA2512 | antA, anthranilate dioxygenase large subunit | −600 | 43 (2.0) | 27 (3.0) |
| PA2513 | antB, anthranilate dioxygenase small subunit | −96 | 14 (2.0) | 13 (3.0) |
| PA2514 | antC, anthranilate dioxygenase reductase | −67 | 9.3 (2.0) | 3.8 (4.0) |
| PA2564 | Hypothetical protein | 2.9 | 7.8 (2.0) | 21 (1.4) |
| PA2565 | Hypothetical protein | 3.1 | 6.6 (2.0) | 14 (2.0) |
| PA2566 | Conserved hypothetical proteine | 6.5 | 13 (2.0) | 21 (1.4) |
| PA2570 | pa1L, PA-1 galactophilic lectind | NC | 26 (1.4) | 200 (1.4) |
| PA2572 | Probable two-component response regulator | 2.3 | 2.8 (1.4) | 3.3 (1.4) |
| PA2573 | Probable chemotaxis transducer | 2.3 | 4.1 (1.4) | 3.9 (1.4) |
| PA2587 | qsc105, probable FAD-dependent monooxygenase | 12 | 12 (0.2) | 15 (0.1) |
| PA2588 | Probable transcriptional regulator | 15 | 22 (0.2) | 46 (0.8) |
| PA2591 | Probable transcriptional regulatore | 21 | 25 (0.2) | 42 (0.2) |
| PA2592 | qsc104, probable spermidine-putrescine-binding proteine | 5.6 | 8.7 (0.4) | 15 (0.8) |
| PA2593 | Hypothetical protein | NC | 4.6 (2.0) | 29 (0.8) |
| PA2717 | cpo, chloroperoxidase precursor | 2.4 | 2.6 (2.0) | 3.4 (1.4) |
| PA2747 | Hypothetical protein | 3.6 | 7.2 (2.0) | 11 (2.0) |
| PA2927 | Hypothetical protein | 2.6 | 3.4 (2.0) | 14 (1.4) |
| PA2939 | Probable aminopeptidase | 38 | 42 (1.4) | 27 (1.4) |
| PA3022 | Hypothetical protein | 3.5 | 4.7 (2.0) | 4.3 (2.0) |
| PA3032 | qsc135, cytochrome c | 2.3 | 3.3 (2.0) | 9.3 (2.0) |
| PA3104 | xcpP, secretion protein XcpP | 3.1 | 3.2 (2.0) | 4.7 (1.4) |
| PA3181 | 2-Keto-3-deoxy-6-phosphogluconate aldolase | 1.6 | 2.9 (3.0) | 3.2 (3.0) |
| PA3182 | Conserved hypothetical protein | 1.7 | 3.2 (3.0) | 5.0 (3.0) |
| PA3183 | zwf, glucose-6-phosphate 1-dehydrogenase | 2.0 | 3.7 (3.0) | 4.0 (3.0) |
| PA3188 | Probable permease of ABC sugar transporter | 2.9 | 4.2 (2.0) | 6.8 (3.0) |
| PA3189 | Probable permease of ABC sugar transporter | 2.0 | 2.5 (3.0) | 3.0 (3.0) |
| PA3190 | Probable component of ABC sugar transporter | 2.7 | 3.4 (2.0) | 4.1 (3.0) |
| PA3194 | edd, phosphogluconate dehydratase | 2.0 | 3.2 (3.0) | 2.9 (3.0) |
| PA3195 | gapA, glyceraldehyde-3-phosphate dehydrogenase | 3.1 | 5.0 (3.0) | 5.4 (3.0) |
| PA3274 | Hypothetical proteind | 1.9 | 4.3 (2.0) | 10 (2.0) |
| PA3311 | Conserved hypothetical protein | 3.6 | 3.6 (2.0) | 6.0 (1.4) |
| PA3326 | Probable Clp family ATP-dependent proteasee | 6.6 | 20 (0.4) | 19 (0.8) |
| PA3327 | qsc126, probable nonribosomal peptide synthetasee | NC | 6.8 (0.8) | 20 (0.8) |
| PA3328 | qsc125, probable FAD-dependent monooxygenase | NC | 17 (0.4) | 47 (0.8) |
| PA3329 | qsc124, hypothetical protein | NC | 250 (0.4) | 310 (0.8) |
| PA3330 | qsc123, probable short-chain dehydrogenase | NC | 120 (0.4) | 320 (0.8) |
| PA3331 | qsc122, cytochrome P450 | 3.5 | 39 (0.4) | 62 (0.8) |
| PA3332 | Conserved hypothetical protein | 2.3 | 35 (0.8) | 41 (1.4) |
| PA3333 | qsc121, fabH2, 3-oxoacyl-(acyl carrier protein) synthase III | NC | 32 (0.4) | 64 (0.8) |
| PA3334 | Probable acyl carrier protein | 1.8 | 49 (0.4) | 69 (0.8) |
| PA3335 | Hypothetical protein | NC | 9.6 (0.4) | 29 (1.4) |
| PA3336 | qsc120, probable MFS transporter | NC | 22 (1.4) | 24 (0.8) |
| PA3346 | Probable two-component response regulator | 2.7 | 2.8 (2.0) | 4.7 (2.0) |
| PA3347 | Hypothetical proteinc | 2.3 | 2.8 (2.0) | 4.3 (1.4) |
| PA3361 | Hypothetical protein | 10 | 13 (1.4) | 68 (1.4) |
| PA3369 | Hypothetical protein | 1.9 | 3.3 (2.0) | 4.8 (2.0) |
| PA3370 | Hypothetical protein | 1.7 | 3.5 (2.0) | 5.6 (2.0) |
| PA3371 | Hypothetical protein | 1.7 | 3.6 (2.0) | 6.0 (2.0) |
| PA3416 | Probable pyruvate dehydrogenase component | 2.5 | 3.2 (2.0) | 4.1 (1.4) |
| PA3418 | ldh, leucine dehydrogenase | 2.6 | 3.7 (1.4) | 5.0 (1.4) |
| PA3476 | qsc118, rhlI, autoinducer synthesis protein RhlIe | NCf | NCf | 34 (0.05) |
| PA3477 | rhlR, transcriptional regulator RhlR | 8.5 | 9.6 (0.4) | 130 (0.05) |
| PA3478 | qsc119,rhlB, rhamnosyltransferase chain B | 5.3 | 89 (0.8) | 120 (1.4) |
| PA3479 | qsc119,rhlA, rhamnosyltransferase chain Ae | 10 | 120 (0.8) | 200 (0.8) |
| PA3520 | Hypothetical proteind | 2.2 | 13 (1.4) | 32 (1.4) |
| PA3535 | Probable serine protease | 7.5 | 8.1 (0.4) | 5.9 (0.8) |
| PA3676 | Probable RND efflux transporter | 3.9 | 1.9 (2.0) | 5.8 (1.4) |
| PA3677 | Probable RND efflux protein precursor | 3.6 | 3.8 (2.0) | 8.3 (1.4) |
| PA3678 | Probable transcriptional regulator | 2.9 | 1.6 (2.0) | 3.5 (1.4) |
| PA3688 | Hypothetical protein | 3.0 | 5.4 (0.2) | 3.5 (1.4) |
| PA3691 | Hypothetical protein | 2.4 | 4.5 (2.0) | 6.3 (2.0) |
| PA3692 | Probable outer membrane protein | 3.0 | 5.8 (2.0) | 6.9 (2.0) |
| PA3724 | lasB, elastase LasBe | 110 | 180 (0.8) | 240 (0.8) |
| PA3734 | Hypothetical protein | NC | 4.1 (3.0) | 16 (2.0) |
| PA3888 | Probable permease of ABC transporter | NC | 3.2 (2.0) | 3.9 (2.0) |
| PA3890 | Probable permease of ABC transporter | 1.8 | 4.1 (2.0) | 4.7 (2.0) |
| PA3891 | Probable component of ABC transporter | 2.1 | 5.0 (2.0) | 8.2 (2.0) |
| PA3904 | Hypothetical protein | 49 | 42 (0.2) | 46 (0.05) |
| PA3905 | Hypothetical protein | 37 | 59 (0.2) | 87 (0.05) |
| PA3906 | Hypothetical protein | 140 | 130 (0.2) | 71 (0.05) |
| PA3907 | qsc103, hypothetical protein | 20 | 19 (0.2) | 58 (0.05) |
| PA3908 | Hypothetical protein | 10 | 11 (0.2) | 55 (0.05) |
| PA3986 | Hypothetical protein | 2.7 | 3.3 (1.4) | 2.8 (2.0) |
| PA4078 | qsc134, probable nonribosomal peptide synthetased | 3.2 | 4.6 (2.0) | 20 (2.0) |
| PA4117 | Probable bacteriophytochrome | 5.3 | 5.6 (1.4) | 4.3 (1.4) |
| PA4129 | Hypothetical protein | 25 | 31 (0.8) | 15 (0.8) |
| PA4130 | Probable sulfite or nitrite reductase | 23 | 27 (0.8) | 1 (0.8) |
| PA4131 | Probable iron-sulfur protein | 24 | 30 (0.8) | 21 (0.8) |
| PA4132 | Conserved hypothetical protein | 14 | 15 (0.8) | 6.4 (0.8) |
| PA4133 | Cytochrome c oxidase subunit (cbb3 type) | 100 | 100 (0.8) | 37 (0.8) |
| PA4134 | Hypothetical protein | 43 | 47 (0.8) | 21 (0.8) |
| PA4139 | Hypothetical protein | 3.1 | 2.9 (2.0) | 3.9 (2.0) |
| PA4141 | Hypothetical protein | 2.6 | 26 (0.4) | 73 (1.4) |
| PA4142 | Probable secretion protein | NC | 5.2 (2.0) | 16 (1.4) |
| PA4171 | Probable protease | 3.5 | 4.6 (2.0) | 5.1 (2.0) |
| PA4172 | Probable nuclease | 2.0 | 3.4 (2.0) | 14 (2.0) |
| PA4175 | Probable endoproteinase Arg-C precursor | 11 | 15 (2.0) | 23 (1.4) |
| PA4190 | Probable FAD-dependent monooxygenase | 3.0 | 2.5 (1.4) | 4.0 (0.2) |
| PA4205 | Hypothetical protein | 1.9 | 8.7 (3.0) | 56 (2.0) |
| PA4206 | Probable RND efflux protein precursor | 1.7 | 6.3 (3.0) | 30 (2.0) |
| PA4207 | qsc133, probable RND efflux transporter | NC | 2.5 (3.0) | 17 (2.0) |
| PA4208 | Probable outer membrane efflux protein | 1.6 | 3.1 (3.0) | 19 (2.0) |
| PA4209 | Probable O-methyltransferasee | 4.6 | 11 (1.4) | 27 (1.4) |
| PA4210 | Probable phenazine biosynthesis proteine,g | NC | 59 (1.4) | 71 (1.4) |
| PA4211 | Probable phenazine biosynthesis proteind | 10 | 69 (0.8) | 220 (0.8) |
| PA4212 | qsc131, phenazine biosynthesis protein PhzCd | 2.2 | 15 (1.4) | 77 (1.4) |
| PA4213 | Phenazine biosynthesis protein PhzD | 3.7 | 36 (1.4) | 210 (1.4) |
| PA4214 | Phenazine biosynthesis protein PhzE | 2.5 | 18 (1.4) | 59 (1.4) |
| PA4215 | Probable phenazine biosynthesis protein | 3.1 | 24 (1.4) | 110 (1.4) |
| PA4216 | Probable pyridoxamine 5-phosphate oxidase | 3.0 | 21 (1.4) | 56 (1.4) |
| PA4217 | qsc132, probable FAD-dependent monooxygenase | 4.4 | 28 (1.4) | 41 (1.4) |
| PA4296 | Probable two-component response regulator | 2.4 | 3.6 (1.4) | 5.6 (1.4) |
| PA4297 | Hypothetical protein | 2.4 | 3.3 (2.0) | 12 (2.0) |
| PA4298 | Hypothetical protein | 2.3 | 4.7 (2.0) | 8.7 (2.0) |
| PA4299 | Hypothetical protein | 2.1 | 3.6 (2.0) | 7.0 (2.0) |
| PA4300 | Hypothetical protein | 2.0 | 3.5 (2.0) | 7.8 (2.0) |
| PA4302 | Probable type II secretion system protein | 3.2 | 6.1 (2.0) | 7.4 (2.0) |
| PA4304 | Probable type II secretion system protein | 2.2 | 3.1 (2.0) | 6.1 (2.0) |
| PA4305 | Hypothetical protein | 2.1 | 2.7 (2.0) | 5.9 (2.0) |
| PA4306 | Hypothetical protein | 10 | 16 (1.4) | 38 (1.4) |
| PA4311 | Conserved hypothetical protein | 2.5 | 3.2 (2.0) | 2.6 (2.0) |
| PA4384 | Hypothetical protein | NC | 2.7 (3.0) | 4.0 (3.0) |
| PA4498 | Probable metallopeptidase | 1.6 | 4.5 (3.0) | 9.1 (3.0) |
| PA4590 | pra, protein activator | 9.3 | 14 (1.4) | 13 (0.8) |
| PA4648 | Hypothetical protein | 3.4 | 7.7 (2.0) | 17 (1.4) |
| PA4649 | Hypothetical protein | NC | 3.2 (2.0) | 7.4 (1.4) |
| PA4650 | Hypothetical protein | NC | 3.3 (2.0) | 8.8 (2.0) |
| PA4651 | Probable pilus assembly chaperoned | NC | 4.6 (2.0) | 15 (2.0) |
| PA4652 | Hypothetical protein | 6.0 | 13 (2.0) | 9.6 (2.0) |
| PA4677 | Hypothetical protein | 16 | 13 (0.2) | 36 (0.1) |
| PA4703 | Hypothetical protein | 3.1 | 4.3 (2.0) | 3.5 (1.4) |
| PA4738 | Conserved hypothetical protein | 3.8 | 9.1 (2.0) | 11 (2.0) |
| PA4739 | Conserved hypothetical protein | 4.2 | 9.4 (2.0) | 14 (2.0) |
| PA4778 | Probable transcriptional regulator | 5.4 | 4.9 (0.4) | 8.6 (0.1) |
| PA4869 | qsc106, hypothetical proteind | 5.0 | 5.7 (0.4) | 3.8 (0.1) |
| PA4876 | osmE, osmotically inducible lipoprotein OsmE | 2.3 | 3.6 (2.0) | 4.9 (2.0) |
| PA4880 | Probable bacterioferritin | 2.2 | 4.6 (2.0) | 5.8 (2.0) |
| PA4916 | Hypothetical protein | 1.5 | 4.3 (4.0) | 6.1 (2.0) |
| PA4917 | Hypothetical proteind | 1.4 | 5.8 (2.0) | 7.7 (2.0) |
| PA4925 | Conserved hypothetical protein | 3.8 | 3.7 (2.0) | 5.7 (1.4) |
| PA5027 | Hypothetical proteine | 1.5 | 2.8 (2.0) | 3.2 (3.0) |
| PA5058 | phaC2, poly(3-hydroxyalkanoic acid) synthase 2e | 4.5 | 4.7 (1.4) | 9.2 (1.4) |
| PA5059 | Probable transcriptional regulator | 4.4 | 5.9 (2.0) | 9.3 (1.4) |
| PA5061 | Conserved hypothetical protein | 1.7 | 2.5 (4.0) | 2.6 (4.0) |
| PA5161 | rmlB, dTDP-d-glucose 4,6-dehydratase | NC | 2.9 (4.0) | 5.9 (3.0) |
| PA5162 | rmlD, dTDP-4-dehydrorhamnose reductase | NC | 2.5 (4.0) | 4.9 (3.0) |
| PA5164 | rmlC, dTDP-4-dehydrorhamnose 3,5-epimerase | NC | 2.6 (3.0) | 5.6 (2.0) |
| PA5220 | qsc138, hypothetical protein | 2.8 | 18 (0.8) | 26 (1.4) |
| PA5352 | Conserved hypothetical protein | 2.0 | 2.8 (1.4) | 2.9 (1.4) |
| PA5353 | glcF, glycolate oxidase subunit GlcF | 1.9 | 3.4 (1.4) | 3.5 (1.4) |
| PA5354 | glcE, glycolate oxidase subunit GlcE | 2.0 | 2.6 (1.4) | 3.2 (1.4) |
| PA5355 | glcD, glycolate oxidase subunit GlcD | 2.1 | 3.6 (1.4) | 3.8 (1.4) |
| PA5356 | qsc130, glcC, transcriptional regulator GlcC | 2.4 | 4.1 (1.4) | 2.8 (1.4) |
| PA5415 | glyA1, serine hydroxymethyltransferase | 2.6 | 2.8 (3.0) | 5.0 (3.0) |
| PA5481 | Hypothetical protein | 4.1 | 11 (2.0) | 15 (1.4) |
| PA5482 | Hypothetical protein | 5.4 | 15 (2.0) | 18 (1.4) |
Gene number from the Pseudomonas genome project (http://www.pseudomonas.com).
Boldface type indicates genes or gene products previously reported to be controlled by quorum sensing. RND, resistance-nodulation-cell division; FAD, flavin adenine dinucleotide.
Maximum changes (fold) in gene expression (rounded to two significant figures) in the signal generation mutant in the presence of the signal(s) indicated compared with the absence of signal and in the wild-type P. aeruginosa strain compared with the receptor mutant. The values in parentheses are the OD600 at which the earliest change of ≥2.5-fold was observed (for the signal generation mutant both time courses were considered). NC, no change.
There is a las-rhl box-like sequence with an HI of ≥10 and <13.
There is a las-rhl box-like sequence with an HI of <10.
The transcript levels for lasI and rhlI were close to the background level in the signal generation mutant due to the disruption of both loci by insertional mutagenesis.
The GeneChip probes for PA4210 to PA4216 are identical to those for PA1899 to PA1905. Although the sequences for the genes in these two clusters are almost identical, the region upstream of PA4210 contains a las-rhl box-like sequence, but the region upstream of PA1899 does not.
Some of the genes identified in the previous study have also been determined by other workers to be quorum controlled (for example, hcnABC) (23). There are genes other than those described in the previous study that have been reported to be quorum controlled. Most of these genes (for example, lasA [20, 29] and rsaL [5]) were confirmed by our transcriptome study. A few were not confirmed, including toxA (11) and sodA (13). It is not possible to draw conclusions about the genes that were not confirmed by the transcriptome analysis. The experimental conditions and strains that were used previously were different from those that we used.
Quorum-activated regulon.
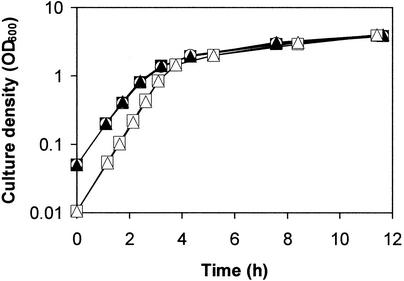
To identify a larger group of genes in the quorum-sensing regulon of P. aeruginosa, we used the results of the experiment described above, and we performed an additional independent experiment in which we compared transcripts in a quorum-sensing signal receptor double mutant to transcripts in the parent strain. This was an independent procedure to assess whether genes are controlled by quorum sensing. We reasoned that genes showing differential regulation with both approaches (addition of signals to a signal generation mutant and a parent compared to a signal receptor mutant) were likely influenced by quorum sensing. The wild-type P. aeruginosa strain, the signal receptor mutant, and the signal generation mutant grown with and without added acyl-HSL signals showed similar growth patterns under the conditions of our experiments (Fig. 1).
FIG. 1.
Growth of wild-type P. aeruginosa strain PAO1 (□), growth of the receptor mutant PAO lasR rhlR (▵), and growth of the signal generation mutant PAO-MW1 without added acyl-HSL (▴), with 3OC12-HSL (○), and with C4-HSL and 3OC12-HSL (▪).
As mentioned above, we identified 638 genes that were induced or repressed by addition of the acyl-HSL signals to the signal generation mutant. We identified 810 genes that were differentially expressed in the parent compared to the signal receptor mutant. In all, there was an overlapping set of 411 genes. Visual inspection of the expression patterns of individual genes led to exclusion of 58 genes. These genes either showed expression levels close to the background level or showed inconsistent regulatory patterns when the two experimental approaches were compared. An interesting example of an inconsistent regulatory pattern was observed with a few genes identified and classified as late 3OC12-HSL-dependent genes in the previous study. These genes, PA2401, PA2402, and PA2385 (qsc109-110, qsc111, and qsc112) showed the predicted regulatory pattern in our transcriptome analysis of the signal generation mutant (although they showed low response levels of 2.0, 2.1, and 2.3, respectively), but they showed quorum-controlled repression when the parent was compared to the signal receptor mutant. In all then, we identified 315 genes which we believe to be quorum activated. These genes and some information regarding their expression are shown in Table 1.
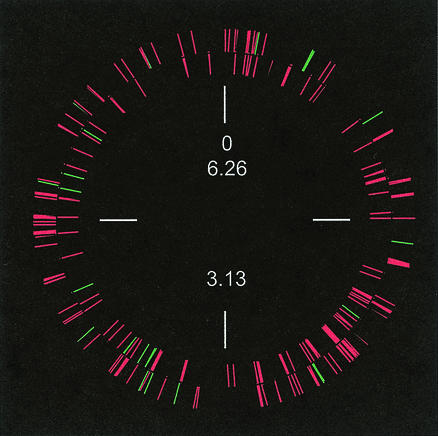
There is no obvious chromosomal clustering of the genes identified (Fig. 2). The final set of quorum-induced genes represents about 6% of the genome. This is somewhat higher than but not too different from the previous prediction that around 2 to 4% of the genome is quorum induced. However, the genes that we have identified are likely a subset of the quorum regulon. For example, we used one standard set of growth conditions for all of our experiments; it is not unreasonable to believe that other genes in the regulon might be revealed by altering the growth medium or culture conditions. In our experiments about 20 to 30% of the transcripts were undetectable; some of these transcripts might be quorum controlled or expressed at higher levels under different conditions. As discussed above, we also filtered the data set, and we do not believe that the genes that survived the filtering procedure represent an exhaustive compilation of quorum-controlled genes. Rather, the data provide a conservative estimate of quorum-controlled genes. Among the genes listed in Table 1, the most overrepresented categories consist of genes known or predicted to be involved in the production of secreted products. Genes in the adaptation and protection categories and in the central intermediary metabolism categories are also overrepresented.
FIG. 2.
Map of quorum-regulated genes on the P. aeruginosa chromosome. Red, activated genes; green, repressed genes. Genes in the inner circle are transcribed in a counterclockwise direction. The numbers indicate map positions (in megabases).
Quorum-repressed genes.
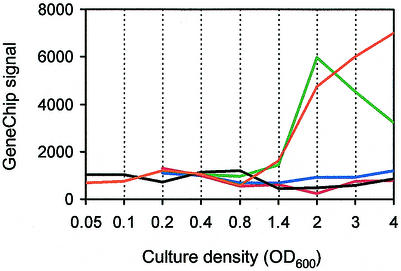
We identified 38 quorum-repressed genes (Table 2). These genes showed lower transcript levels in the late logarithmic and stationary phases in the wild type than in the receptor mutant, as well as in the signal generation mutant in the presence of signals than in the mutant grown without added signal. All of the repressed genes responded as well or nearly as well to 3OC12-HSL alone as they did to 3OC12-HSL and C4-HSL together. The expression patterns of a representative quorum-repressed gene, PA0433, are shown in Fig. 3. A curiousity is that these genes are expressed at low levels throughout growth of the parent strain. They are derepressed only in the mutants and only during the late logarithmic and stationary phases. Among the quorum-repressed genes with known or predicted functions, those involved in carbohydrate utilization or nutrient transport appeared to be the most abundant (Table 2).
TABLE 2.
Quorum-repressed genes
| Gene no.a | Description | Maximum changeb
|
||
|---|---|---|---|---|
|
lasI rhlI mutant
|
Wild type vs lasR rhlR mutant | |||
| 3OC12-HSL | C4-HSL + 3OC12-HSL | |||
| PA0165 | Hypothetical protein | −2.7 | −2.9 (2.0) | −4.8 (2.0) |
| PA0433 | Hypothetical protein | −6.8 | −20 (2.0) | −8.9 (1.4) |
| PA0434 | Hypothetical protein | −7.7 | −8.5 (2.0) | −5.6 (2.0) |
| PA0435 | Hypothetical protein | −9.4 | −26 (2.0) | −34 (2.0) |
| PA0485 | Conserved hypothetical proteinc | −1.7 | −3.4 (1.4) | −3.0 (3.0) |
| PA0887 | acsA, acetyl-coenzyme A synthetase | −3.3 | −4.2 (2.0) | −3.6 (3.0) |
| PA1559 | Hypothetical protein | −2.4 | −3.5 (2.0) | −3.2 (1.4) |
| PA2007 | maiA, maleylacetoacetate isomerase | −3.2 | −1.4 (4.0) | −3.2 (3.0) |
| PA2008 | fahA, fumarylacetoacetase | −3.7 | −1.5 (4.0) | −2.6 (3.0) |
| PA2009 | hmgA, homogentisate 1,2-dioxygenase | −4.0 | −1.5 (4.0) | −2.7 (3.0) |
| PA2250 | lpdV, lipoamide dehydrogenase-Val | −3.1 | −1.8 (4.0) | −2.6 (3.0) |
| PA2338 | Probable component of ABC maltose transporter | −5.0 | −3.2 (3.0) | −4.2 (3.0) |
| PA2339 | Probable maltose-mannitol transport protein | −1.9 | −6.8 (3.0) | −4.1 (3.0) |
| PA2340 | Probable maltose-mannitol transport protein | −3.4 | −2.0 (3.0) | −3.7 (3.0) |
| PA2341 | Probable component of ABC maltose transporter | −3.1 | −2.0 (3.0) | −4.2 (3.0) |
| PA2343 | mtlY, xylulose kinase | −1.7 | −4.0 (3.0) | −3.2 (4.0) |
| PA3038 | Probable porin | −2.3 | −3.5 (2.0) | −4.4 (3.0) |
| PA3174 | Probable transcriptional regulator | −2.1 | −3.5 (4.0) | −6.5 (3.0) |
| PA3205 | Hypothetical protein | −1.3 | −3.1 (4.0) | −3.1 (4.0) |
| PA3233 | Hypothetical protein | −2.2 | −2.7 (3.0) | −5.1 (3.0) |
| PA3234 | Probable sodium-solute symporter | −4.5 | −3.4 (2.0) | −7.0 (3.0) |
| PA3235 | Conserved hypothetical protein | −3.9 | −4.2 (3.0) | −6.6 (3.0) |
| PA3281 | Hypothetical protein | −5.7 | −6.4 (1.4) | −25 (1.4) |
| PA3282 | Hypothetical protein | −8.5 | −8.8 (1.4) | −21 (1.4) |
| PA3283 | Conserved hypothetical protein | −9.0 | −8.8 (1.4) | −28 (1.4) |
| PA3284 | Hypothetical protein | −7.1 | −10 (2.0) | −24 (1.4) |
| PA3364 | amiC, aliphatic amidase expression-regulating protein | −2.7 | −1.8 (4.0) | −2.7 (1.4) |
| PA3365 | Probable chaperone | −3.0 | −1.7 (4.0) | −4.0 (1.4) |
| PA3575 | Hypothetical protein | −1.6 | −2.7 (1.4) | −3.3 (2.0) |
| PA3790 | oprC, outer membrane protein OprC | −2.7 | −3.7 (2.0) | −4.6 (2.0) |
| PA4359 | Conserved hypothetical protein | −1.4 | −2.7 (2.0) | −2.8 (1.4) |
| PA4371 | Hypothetical protein | −1.9 | −4.1 (2.0) | −2.8 (1.4) |
| PA4442 | cysN, ATP sulfurylase GTP-binding subunit | −2.8 | −3.4 (3.0) | −7.6 (2.0) |
| PA4443 | cysD, ATP sulfurylase small subunit | −3.1 | −3.4 (3.0) | −6.5 (2.0) |
| PA4691 | Hypothetical protein | −2.5 | −2.8 (2.0) | −2.9 (2.0) |
| PA4692 | Conserved hypothetical protein | −3.8 | −3.4 (2.0) | −5.0 (1.4) |
| PA4770 | IldP, l-lactate permease | −1.8 | −3.7 (2.0) | −5.0 (2.0) |
| PA5168 | Probable dicarboxylate transporter | −2.7 | −1.9 (4.0) | −5.8 (2.0) |
Gene number from the Pseudomonas genome project (http://www.pseudomonas.com).
Maximum changes in gene expression (rounded to two significant figures) in the signal generation mutant in the presence of the signal(s) indicated compared with the absence of signal and in the wild-type P. aeruginosa strain compared with the receptor mutant. The values in parentheses are the OD600 at which the earliest change of ≥2.5-fold was observed (for the signal generation mutant both time courses were considered).
There is a las-rhl box-like sequence with an HI of ≥10 and <13.
FIG. 3.
Transcript abundance of the quorum-repressed gene PA0433 in wild-type P. aeruginosa (black line), in the receptor mutant (orange line), and in the signal generation mutant without added acyl-HSL (green line), with 3OC12-HSL (blue line), and with C4-HSL and 3OC12-HSL (red line). The values on the y axis represent transcript abundance as determined by the array software.
Operons and las-rhl box-like sequences.
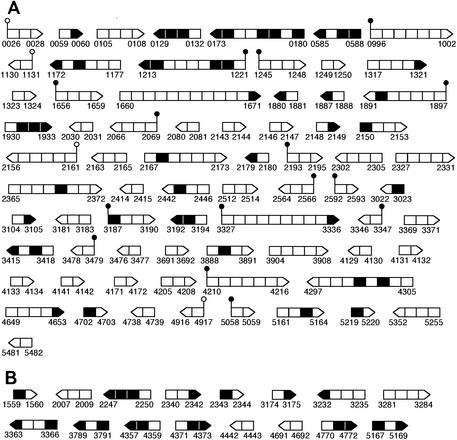
One would expect that all of the genes in an operon should show similar quorum control. In fact, we observed that strings of genes appeared (Tables 1 and 2). These strings often represent known or suspected operons, and the genes in a given string show similar quorum responses (signal responses and timing of induction). For example, the hcn genes (PA2193 to PA2195) are known to exist in an operon (23). Consistent with this, our transcriptome analysis indicated these genes were coinduced by quorum sensing. PA2365 to PA2372 represent a string of quorum-controlled genes with unknown functions. We suggest that these genes represent an operon. On the other hand, many of the quorum-controlled genes are not adjacent to other quorum-controlled genes listed in Tables 1 and 2. To assess whether these genes may also be organized in operons (neighboring genes would have been eliminated if they showed induction just under the 2.5-fold threshold) and to confirm the hypothesis that strings of adjacent quorum-controlled genes are in operons, we performed a more systematic analysis. Operon organization was allowed only if every gene within a gene cluster was in the same orientation, if every gene was activated or every gene was repressed, if there was less than 250 bp between two adjacent open reading frames, and if the absolute transcript profiles of the candidate genes in the parent P. aeruginosa strain showed patterns similar to each other (correlation coefficient of ≥0.95 with the GeneSpring standard correlation algorithm). By using these criteria, we identified 87 possible operons, 71 of which were activated and 16 of which were repressed (Fig. 4). More than 60 additional genes showing coregulation with the genes listed in Tables 1 and 2 were identified by this analysis.
FIG. 4.
Predicted quorum-regulated operons. Genes not listed in Tables 1 and 2 are depicted as black boxes. The arrows indicate the directions of transcription. The solid and open circles indicate putative las-rhl boxes with an HI of less than 10 and less than 13, respectively. (A) Quorum-activated operons; (B) quorum-repressed operons. Numbers are PA gene numbers from the Pseudomonas genome project.
We used a computer algorithm to search for las-rhl boxes in regions upstream of quorum-regulated genes. On the basis of a stringent criterion (an HI of <10), 55 of the P. aeruginosa genes contain a box in the upstream regulatory region. Twenty-five (45%) of these genes are quorum controlled, and 15 represent the first gene in a predicted operon (Table 1 and Fig. 4). At a lower stringency (an HI of <13), we identified 185 genes with las-rhl box-like sequences. Forty-eight (26%) of these genes are quorum controlled, and 19 represent the first gene in a predicted operon. Only one las-rhl box-like sequence was found upstream of a quorum-repressed gene. We did not identify potential boxes for all of our quorum-activated genes. This suggests that some of the genes might be controlled indirectly by quorum sensing or that the search criteria were not sufficiently refined. These possibilities are not mutually exclusive. We also found las-rhl boxes for genes that did not appear to be quorum controlled. Again, this suggests that these genes might be quorum controlled under other conditions or that the search criteria need further refinement. We believe that the search algorithm was biased because it was based on the relatively small subset of quorum-controlled genes with established las-rhl boxes (see Materials and Methods). There was no apparent bias, however, with respect to the timing of induction or signal specificity of the identified genes.
Signal specificity.
In a previous limited analysis of quorum-controlled gene expression, genes were classified into the following categories based on their responses to the signals: genes that responded equally well to 3OC12-HSL and to 3OC12-HSL and C4-HSL together and genes that responded best only when both 3OC12-HSL and C4-HSL were present (33). It appears from the microarray data that the responses are on a continuum, with some genes responding no better to both signals than they do to 3OC12-HSL alone and other genes showing progressively greater responses to both signals compared to the responses to 3OC12-HSL alone. For example, PA2423 responded no better to both signals then it did to 3OC12-HSL alone, PA0122 responded well to 3OC12-HSL alone but showed an approximately threefold-greater response with both signals, and PA2069 did not respond at all to 3OC12-HSL alone but showed a large response in the presence of both signals (Table 1). This suggests that some genes respond to 3OC12-HSL specifically, others respond with various specificities to either signal, and others respond to C4-HSL specifically. The genes encoding anthranilate dioxygenase, antABC (PA2512 to PA2514), are an exceptional case. These genes were strongly repressed in the presence of 3OC12-HSL alone but were activated in the presence of both signals.
Timing of quorum-controlled gene activation.
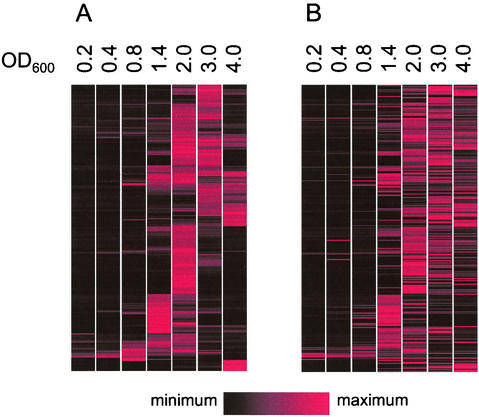
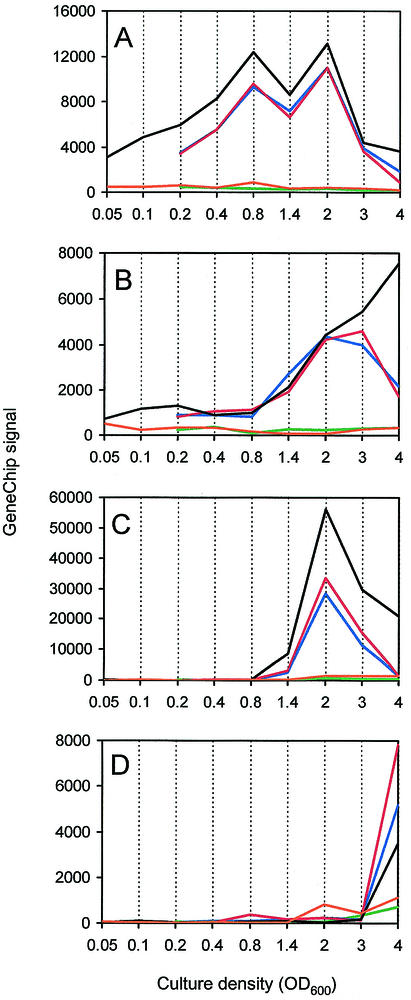
The previous analysis (33) showed that induction of some genes, early genes, could be triggered early in the logarithmic phase by addition of signals. With other genes, late genes, there was a delay in induction even in the presence of excess added signal (33). By examining the microarray data we were able to learn about the timing of quorum-sensing-controlled gene induction in the wild-type strain, and we were able to obtain a broader understanding of the influence of acyl-HSL signal addition on control of the quorum regulon. The patterns of quorum-controlled gene expression were remarkably similar in the parent and in the signal generation mutant grown in the presence of 3OC12-HSL and C4-HSL (Fig. 5). A small number of transcripts showed the greatest induction early in growth. Other genes exhibited low levels of induction early in growth but did not reach maximum levels of induction until later in growth. Most transcripts were induced at culture optical densities between 0.8 and 2.0. Some transcripts showed increased abundance relative to the baseline level only at culture optical densities greater than 2.0 (stationary phase). Thus, the transcriptome analysis suggests that the timing of quorum-controlled gene induction is on a continuum, although most genes in the regulon appeared to be activated during the transition from the logarithmic phase to the stationary phase (optical densities between 0.8 and 2.0). The timing of induction for most genes was not affected by exogenous addition of 3OC12-HSL and C4-HSL. Examples of each of the gene expression patterns described above are shown in Fig. 6.
FIG. 5.
Relative expression profiles for quorum-activated genes during growth of P. aeruginosa. (A) Signal generation mutant with C4-HSL and 3OC12-HSL versus signal generation mutant with no acyl-HSL. (B) Wild-type P. aeruginosa versus the receptor mutant. Depicted are fold changes for each gene normalized to the half-maximal level. The genes in panel A are displayed in the order of the hierarchical clustering of their expression profiles, and the genes in panel B are shown in the same order as those in panel A.
FIG. 6.
Transcript levels for selected quorum-activated genes in the wild-type P. aeruginosa strain (black lines), in the receptor mutant (orange lines), and in the signal generation mutant without added acyl-HSL (green lines), with 3OC12-HSL (blue lines), and with C4-HSL and 3OC12-HSL (red lines). (A) PA3904; (B) PA4677; (C) PA2939; (D) PA0198. For simplicity, the examples shown are all 3OC12-HSL-specific genes. The values on the y axis represent transcript abundance as determined by the array software.
We hypothesize that even in the parent strain at the earliest sampling time (optical density, 0.05) there were sufficient acyl-HSL levels for induction of the early genes and that some other factor was limiting expression of transcripts that were triggered to accumulate later in growth (because of the large volume of culture that would be required, it was impractical to examine cultures at lower optical densities). What other factor might account for the acyl-HSL-independent triggering of quorum gene induction? An obvious possibility is that the acyl-HSL receptors are limiting in the early logarithmic phase and that the abundance of these factors increases during culture growth. We hypothesize that quorum-controlled promoters that are active in the early logarithmic phase bind the transcription factors and effectively titrate them away from other quorum-controlled promoters. As the level of LasR increases, additional quorum-controlled genes should show expression. A prediction of this hypothesis is that lasR, and consequently rhlR, should show increased transcript abundance as a culture grows. Thus, we examined the microarray data with respect to lasR and rhlR (Fig. 7). Starting at an optical density of around 0.8 the levels of the lasR and rhlR transcripts increased markedly, which is consistent with previous results obtained with reporter fusions (1, 22). For lasR, some increase was observed in the wild-type strain and in the signal generation mutant with or without added signal. Thus, the increase was, at least in part, independent of quorum sensing. This result is consistent with but does not constitute proof of the model for timing of quorum-controlled gene expression described above.
FIG. 7.
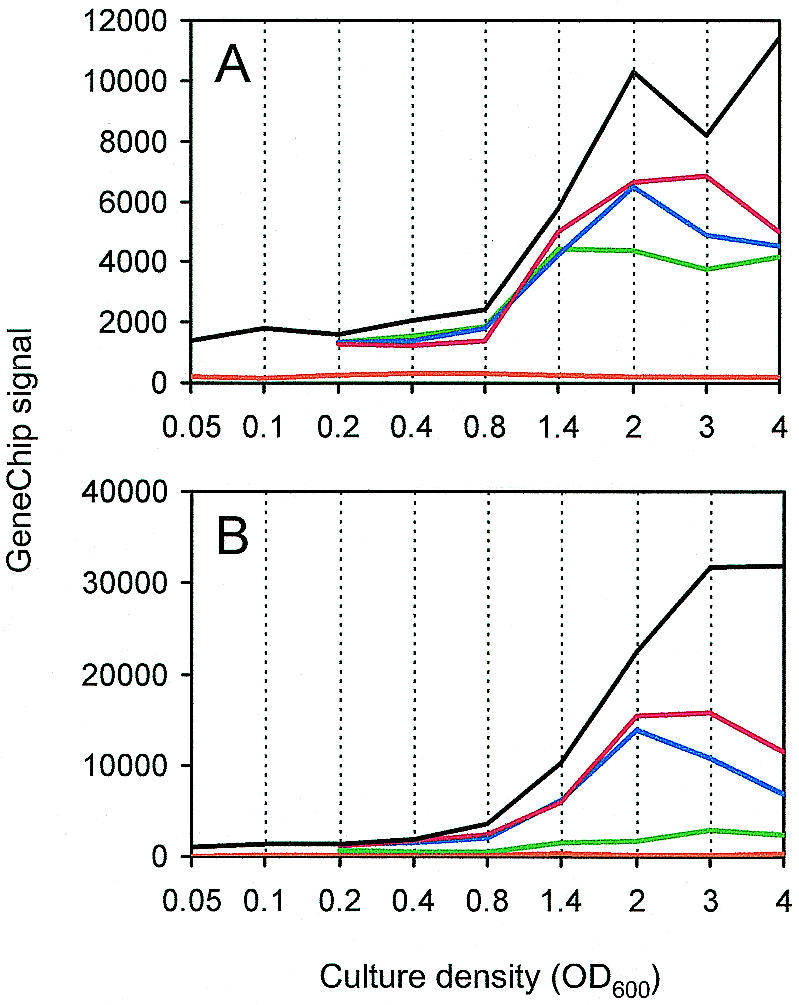
Expression of lasR (A) and rhlR (B) in the wild-type P. aeruginosa strain (black line), the receptor mutant (orange line), and the signal generation mutant without added acyl-HSL (green line), with 3OC12-HSL (blue line), and with C4-HSL and 3OC12-HSL (red line). The values on the y axis represent transcript abundance as determined by the array software.
DISCUSSION
We used Affymetrix GeneChip technology to expand the list of genes reported to be controlled by quorum sensing in P. aeruginosa. In all, we have identified over 300 genes (about 6% of the P. aeruginosa genome) that appear to be part of the quorum regulon (Tables 1 and 2 and Fig. 2 and 4). The fact that these genes were identified in two different types of analyses (one of which involved a comparison of a signal generation mutant with itself in the presence of added signals and the other of which involved a comparison of a signal receptor mutant with its parent) lends confidence to our conclusions. Because timing is important in quorum-controlled gene expression, we examined cultures at several different points during growth. Thus, we minimized problems in identifying genes with relatively unstable transcripts. Nevertheless, the genes that we have identified represent a conservative estimate of the quorum-controlled regulon in P. aeruginosa. We filtered the data in several ways, and of course we examined only cultures grown in one medium at one temperature. Additional experiments in which quorum sensing is examined under different conditions are necessary to more fully understand the breadth of quorum sensing as a global regulator of gene expression in P. aeruginosa. An accompanying paper (30a) provides further insight in this regard.
Of the genes which we identified, most were induced by quorum sensing, although some were repressed (Tables 1 and 2). Most repressed genes showed a curious response. They showed derepression only in mutant backgrounds (Fig. 3). Perhaps these genes are activated in the wild type under specific conditions other than those which we used. Expression of these genes might require inducers not present in our experiments or might require other environmental conditions.
Although the most strongly activated genes reported by Whiteley et al. (33) were among the most strongly activated genes according to the microarray analysis, we do not want to attach great significance to the levels of induction we observed. For some genes the levels of induction varied substantially between the signal addition analysis and the comparison of the receptor mutant and parent. In the most extreme cases, some genes showed activation by addition of a signal to the signal generation mutant and repression in the comparison of the receptor mutant with the parent. This indicates that there may be other factors in P. aeruginosa that contribute to control by acyl-HSLs. One possible factor is the LasR-RhlR signal receptor homolog QscR. We know that this protein somehow influences quorum-controlled gene expression in P. aeruginosa, but we do not know how it functions mechanistically (4).
In general, the representation of functional classes in the quorum-controlled regulon was similar to that in the entire P. aeruginosa genome. For example, 43% of the genes in Tables 1 and 2 have unknown functions, compared with 46% of the genes in the entire P. aeruginosa genome. As expected, many quorum-controlled genes are in the secreted-factor category. There are several genes with adaptation and protection functions, and perhaps more surprising is the finding that there are genes coding for general metabolic functions. Caution should be used when significance is attached to individual genes in a functional group. However, there are genes in the general metabolic function group with well-established functions; for example, PA3183 codes for glucose-6-phosphate dehydrogenase, an enzyme essential for glucose catabolism in P. aeruginosa. Why might this gene show quorum control? This is an example of a gene coding for an enzyme with multiple cellular functions. Glucose-6-phosphate dehydrogenase is an NADP-dependent dehydrogenase. As such, it is an NADPH generator, and in fact through this activity it is known to protect P. aeruginosa from oxidative stress by enhancing glutathione synthesis (17). We imagine that without quorum sensing glucose-6-phosphate dehydrogenase can be produced in a quantity sufficient for glucose catabolism but that quorum sensing can boost the levels for alternate functions of this enzyme.
We found that the specificities of responses to 3OC12-HSL versus the specificities of responses to the two signals together showed great variability (although genes in apparent operons showed responses similar to each other). Some of the genes shown in Table 1 appeared to respond specifically to 3OC12-HSL; other genes seemed to respond to 3OC12-HSL, but activation was boosted by addition of C4-HSL; and still other genes seemed to respond to C4-HSL, showing no response to 3OC12-HSL alone. A previous view was that some genes show specificity for 3OC12-HSL and others show specificity for C4-HSL (9, 31). Some of the C4-HSL-dependent genes show some relaxation of specificity (32). The array data suggest that there is a continuum of specificity responses.
The idea of a continuum of responses extends to the timing of quorum-controlled gene induction. We were more interested in the timing of induction or repression than the maximal transcript levels because the maximal levels result from a complex set of factors that presumably include rates of transcription and transcript stability. When we examined timing we saw that there was great variability (Table 1 and Fig. 5 and 6). Some genes showed increased expression early in growth; for most genes the onset of induction was in the late logarithmic to early stationary phase; and some genes were not induced until the stationary phase. In general, timing was similar in the wild type and in the signal generation mutant grown in the presence of saturating levels of added signals. This indicates that over the range of culture densities in our experiments, the trigger for quorum-controlled gene activation was not signal accumulation. The P. aeruginosa quorum-sensing elements are the two signals synthesized by the signal generators LasI and RhlI and the two signal receptors, LasR and RhlR. If the levels of the signals do not govern the onset of induction, then one might hypothesize that receptor levels govern the onset of induction. In fact, we observed that lasR and rhlR transcript levels increased during the late logarithmic and early stationary phases (Fig. 7), which coincided with the induction of most quorum-activated genes. This finding is consistent with the hypothesis, but more evidence is required to determine its validity. The lasR transcript levels were largely independent of quorum sensing. There are other factors in P. aeruginosa that have been reported to control lasR transcription. These factors include the regulators Vfr (1) and GacA (26), as well as the stringent response (30).
Our results for the timing of gene expression raise an interesting question. Are genes whose expression is not directly triggered by exogenous signals really quorum controlled? We believe that there is no evidence to the contrary. Activation of any of the genes which we identified requires sufficient signal. Signal can accumulate only when a critical population density has been reached. The fact that under at least some conditions additional criteria must be met for transcriptional activation of many genes in the regulon does not alter the fact that a quorum is nevertheless required. In fact, a culture medium-dependent delay in the induction of Vibrio fischeri luminescence, even with ample signal, was observed by Eberhard in 1972 (7).
The finding that signal specificity is on a continuum and the hypothesis that levels of LasR and RhlR might control the precise timing of quorum-controlled gene expression lead to the idea that this regulatory system consisting of two signal generators and two signal receptors can allow for an elaborate pattern of expression of hundreds of genes in P. aeruginosa. Genes can be triggered at different times during culture growth, and they can respond to one or both of the signals to various degrees. The simplistic view that quorum sensing leads to the coordinate expression of genes in the quorum-controlled regulon at a critical population density at which the signals have accumulated to a requisite concentration underestimates the complexity of this regulatory circuitry.
Acknowledgments
We thank Matt Wolfgang and Steve Lory for sharing information and tools for GeneChip use and analysis, we thank Lora Huang from the University of Iowa DNA Facility for array processing, and we thank Niels Bagge for helpful discussions.
This study was supported by grant GM59026 from the National Institute of General Medicine. Support was also provided through a therapeutic development grant from the Cystic Fibrosis Foundation and by a grant from The Procter & Gamble Company, Cincinnati, Ohio. C.P.L. was supported by U.S. Public Health Service training grant T32-AI 07511.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of General Medicine.
Footnotes
For a commentary on this article, see page 2061 in this issue.
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brint, J. M., and D. E. Ohman. 1995. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177:7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapon-Herve, V., M. Akrim, A. Latifi, P. Williams, A. Lazdunski, and M. Bally. 1997. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol. Microbiol. 24:1169-1178. [DOI] [PubMed] [Google Scholar]
- 4.Chugani, S. A., M. Whiteley, K. M. Lee, D. D. Argenio, C. Manoil, and E. P. Greenberg. 2001. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Kievit, T., P. C. Seed, J. Nezezon, L. Passador, and B. H. Iglewski. 1999. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J. Bacteriol. 181:2175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diggle, S. P., K. Winzer, A. Lazdunski, P. Williams, and M. Camara. 2002. Advancing the quorum in Pseudomonas aeruginosa: MvaT and the regulation of N-acylhomoserine lactone production and virulence gene expression. J. Bacteriol. 184:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eberhard, A. 1972. Inhibition and activation of bacterial luciferase synthesis. J. Bacteriol. 109:1101-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernandez De Henestrosa, A. R., T. Ogi, S. Aoyagi, D. Chafin, J. J. Hayes, H. Ohmori, and R. Woodgate. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35:1560-1572. [DOI] [PubMed] [Google Scholar]
- 9.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 10.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gambello, M. J., S. Kaye, and B. H. Iglewski. 1993. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 61:1180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glessner, A., R. S. Smith, B. H. Iglewski, and J. B. Robinson. 1999. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J. Bacteriol. 181:1623-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hassett, D. J., J. F. Ma, J. G. Elkins, T. R. McDermott, U. A. Ochsner, S. E. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, L. Passador, and B. H. Iglewski. 1999. Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34:1082-1093. [DOI] [PubMed] [Google Scholar]
- 14.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 15.Latifi, A., M. K. Winson, M. Foglino, B. W. Bycroft, G. S. Stewart, A. Lazdunski, and P. Williams. 1995. Multiple homologues of LuxR and LuxI control expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol. Microbiol. 17:333-343. [DOI] [PubMed] [Google Scholar]
- 16.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051-1060. [DOI] [PubMed] [Google Scholar]
- 17.Ma, J. F., P. W. Hager, M. L. Howell, P. V. Phibbs, and D. J. Hassett. 1998. Cloning and characterization of the Pseudomonas aeruginosa zwf gene encoding glucose-6-phosphate dehydrogenase, an enzyme important in resistance to methyl viologen (paraquat). J. Bacteriol. 180:1741-1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ochsner, U. A., and J. Reiser. 1995. Autoinducer-mediated regulation of rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:6424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parsek, M. R., and E. P. Greenberg. 2000. Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. Proc. Natl. Acad. Sci. USA 97:8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 21.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pessi, G., and D. Haas. 2000. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182:6940-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. Holden, M. Camara, D. Haas, and P. Williams. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahim, R., U. A. Ochsner, C. Olvera, M. Graninger, P. Messner, J. S. Lam, and G. Soberon-Chavez. 2001. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 40:708-718. [DOI] [PubMed] [Google Scholar]
- 26.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Haas. 1997. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 27.Stintzi, A., K. Evans, J. M. Meyer, and K. Poole. 1998. Quorum-sensing and siderophore biosynthesis in Pseudomonas aeruginosa: lasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol. Lett. 166:341-345. [DOI] [PubMed] [Google Scholar]
- 28.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:947-948. [DOI] [PubMed] [Google Scholar]
- 29.Toder, D. S., M. J. Gambello, and B. H. Iglewski. 1991. Pseudomonas aeruginosa LasA: a second elastase under the transcriptional control of lasR. Mol. Microbiol. 5:2003-2010. [DOI] [PubMed] [Google Scholar]
- 30.van Delden, C., R. Comte, and A. M. Bally. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 183:5376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed]
- 31.Whitehead, N. A., A. M. Barnard, H. Slater, N. J. Simpson, and G. P. Salmond. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 32.Whiteley, M., and E. P. Greenberg. 2001. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J. Bacteriol. 183:5529-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winson, M. K., M. Camara, A. Latifi, M. Foglino, S. R. Chhabra, M. Daykin, M. Bally, V. Chapon, G. P. Salmond, B. W. Bycroft, et al. 1995. Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 92:9427-9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winzer, K., C. Falconer, N. C. Garber, S. P. Diggle, M. Camara, and P. Williams. 2000. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J. Bacteriol. 182:6401-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]