Abstract
Bacterial communication via quorum sensing (QS) has been reported to be important in the production of virulence factors, antibiotic sensitivity, and biofilm development. Two QS systems, known as the las and rhl systems, have been identified previously in the opportunistic pathogen Pseudomonas aeruginosa. High-density oligonucleotide microarrays for the P. aeruginosa PAO1 genome were used to investigate global gene expression patterns modulated by QS regulons. In the initial experiments we focused on identifying las and/or rhl QS-regulated genes using a QS signal generation-deficient mutant (PAO-JP2) that was cultured with and without added exogenous autoinducers [N-(3-oxododecanoyl) homoserine lactone and N-butyryl homoserine lactone]. Conservatively, 616 genes showed statistically significant differential expression (P ≤ 0.05) in response to the exogenous autoinducers and were classified as QS regulated. A total of 244 genes were identified as being QS regulated at the mid-logarithmic phase, and 450 genes were identified as being QS regulated at the early stationary phase. Most of the previously reported QS-promoted genes were confirmed, and a large number of additional QS-promoted genes were identified. Importantly, 222 genes were identified as being QS repressed. Environmental factors, such as medium composition and oxygen availability, eliminated detection of transcripts of many genes that were identified as being QS regulated.
Many bacteria have been shown to communicate by using a process known as quorum sensing (QS) to coordinate population behavior in response to environmental cues. Small molecules, termed autoinducers, are produced by the bacterial cell and accumulate in the environment at a high population density. Once an intracellular threshold level of an autoinducer is reached, the autoinducer binds to its cognate transcriptional regulator protein to activate or repress target genes. This behavior was first identified in Vibrio fischeri as a mechanism regulating the lux genes required for bioluminescence (41). Since this discovery, QS has been described in both gram-negative and gram-positive microorganisms (41) and has been implicated in the regulation of cellular behavior and virulence (10).
Pseudomonas aeruginosa is a highly adaptable bacterium that can colonize various environmental niches, including soil and marine habitats, plants, animals, and humans (10). P. aeruginosa possesses one of the best-studied models of QS, and two complete lux-like QS systems, las and rhl, have been identified (28). The las system consists of the transcriptional regulatory protein LasR and its cognate signaling molecule, N-(3-oxododecanoyl) homoserine lactone (3O-C12-HSL), whose production is directed by the autoinducer synthase encoded by lasI. The rhl system consists of the RhlR protein and an autoinducer synthase (RhlI), which is involved in production of the cognate autoinducer N-butyryl homoserine lactone (C4-HSL). These systems are intertwined in a hierarchical manner, and the las system controls the rhl system at both the transcriptional and posttranslational levels. QS has been shown to regulate the production of P. aeruginosa virulence factors (such as proteases, exotoxin A, rhamnolipids, and pyocyanin) and to be involved in biofilm formation and development, and it has been implicated in antibiotic resistance (7, 10). Given these findings, it has been suggested that QS may contribute to the ability of P. aeruginosa to initiate infection and to persist in a host. Data from many models of both acute infection and chronic infection have supported the hypothesis that QS is important in P. aeruginosa pathogenesis (10, 30, 32, 37).
Recent technological advances have made it possible to study global gene expression in both prokaryotic and eukaryotic organisms by using high-density oligonucleotide microarrays. Completion of the entire wild-type P. aeruginosa PAO1 genome sequence (36), as well as the availability of high-density oligonucleotide arrays (Affymetrix P. aeruginosa GeneChip arrays), permit global gene expression profiling of strain PAO1. The ability of P. aeruginosa to colonize and thrive in diverse environments is reflected by its relatively large genome (6.3 Mbp) and genetic complexity (5,570 open reading frames [ORFs]) (36). Compared to known sequenced bacterial genomes, the genome of P. aeruginosa possesses a greater overall number of genes, as well as a larger number of genes encoding outer membrane proteins, resistance-nodulation-cell division efflux systems, and multiple, complex chemotaxis systems (36), which may contribute to its pathogenesis. More than 9% of assigned ORFs are classified as transcriptional regulators and two-component systems, reflecting the ability of P. aeruginosa to respond and adapt to myriad environments (36). In previous studies workers have identified 50 P. aeruginosa genes that are positively regulated by the las and/or rhl QS systems (8, 18, 19, 28, 29, 38, 39, 44, 46). In this study, P. aeruginosa DNA microarrays were utilized to identify QS-controlled genes by using a mutant deficient in production of the autoinducers 3O-C12-HSL and C4-HSL. Most of the previously described QS-promoted genes were confirmed by this technique, thereby establishing the utility of the DNA microarrays. Furthermore, numerous novel QS-regulated genes were identified, including genes that are repressed by QS in P. aeruginosa. A systems approach to understanding QS regulons may elucidate the mechanism(s) that modulates phenotypic changes in response to different environmental stimuli.
MATERIALS AND METHODS
Bacterial strains, media, and chemicals.
P. aeruginosa wild-type strain PAO1 and the ΔlasI ΔrhlI double-null, autoinducer-deficient strain PAO-JP2 (28) were used for expression analysis studies. The strains were maintained at 37°C on PTSB (26) agar plates. Overnight aerobic cultures (PAO1 or PAO-JP2) were grown with agitation in 125-ml Erlenmeyer flasks at 37°C in 20 ml of modified FAB [0.1 mM CaCl2, 0.01 mM Fe-EDTA, 0.15 mM (NH4)2SO4, 0.33 mM Na2HPO4, 0.2 mM KH2PO4, 0.5 mM NaCl, 1 mM MgCl2, 40 mM KNO3, 10 mM glucose; pH 7.0 (9)]. When appropriate, exogenous 3O-C12-HSL (1 μM) and C4-HSL (2 μM) were added. The autoinducers were synthesized and purified as previously reported at the University of Rochester, Rochester, N.Y. (21), and were used as received. For other studies, PAO1 cultures were grown in NY (nutrient broth-yeast extract [mass ratio, 5:1]; pH 7.0) or NY supplemented with KNO3 at a final concentration of 100 mM (pH 7.0). For anaerobic studies, overnight cultures were grown at 37°C in an anaerobic chamber (85% nitrogen, 5% CO2, 10% H2; Coy Laboratory Products Inc., Grass Lake, Mich.) in 20-ml portions of NY containing KNO3 in 50-ml culture bottles; each bottle contained a magnetic stir bar for agitation. All medium components other than autoinducers were reagent grade, were obtained from Sigma (St. Louis, Mo.), and were used as received.
Aerobic overnight cultures were used to inoculate 200-ml portions of medium in 500-ml Erlenmeyer flasks to obtain a starting optical density at 660 nm (OD660) of 0.05, and the resulting cultures were grown at 37°C with agitation to the mid-logarithmic or early stationary phase. The mid-logarithmic phase was defined as the OD660 at the midpoint of the linear range of exponential growth (OD660, 0.4 to 0.5), and the early stationary phase was defined as the OD660 at which the culture exhibited a growth rate close to zero (OD660, 1.0 to 1.4). Anaerobic overnight cultures were subcultured in 200 ml of NY containing KNO3 in 250-ml culture bottles, agitated with a magnetic stir bar, and grown to the mid-logarithmic phase (OD660, 0.5) or the early stationary phase (OD660, 1.4) in an anaerobic chamber. All experiments were performed in triplicate.
RNA extraction, cDNA probe generation, and microarray processing.
Culture samples were collected at the mid-logarithmic and early stationary growth phases. Bacterial cells were pelleted by centrifugation at 4°C for 5 min at 10,000 × g, and the supernatant was removed. GramCracker reagent (Ambion, Austin, Tex.) was used to degrade the bacterial cell walls, and total RNA was isolated by using RNAwiz (Ambion) according to the manufacturer's instructions. RNA was purified from residual DNA by using a DNA-Free kit (Ambion), and RNA purity was assessed by reverse transcriptase PCR by using RETROscript and SuperTaq kits (Ambion) with primers specific for pilA (sense primer 5′-GAATCGCAGAAGGGCTATTG-3′ and antisense primer 5′-CCTAACGCCTGAACCTCAAC-3′). Synthesis of cDNA, target hybridization, staining, and scanning were performed at the Microarray Core Facilities in the Functional Genomics Center at the University of Rochester. Briefly, cDNA was generated by using random hexamers as primers for reverse transcription (Invitrogen Life Technologies, Carlsbad, Calif.). The primers were annealed (70°C for 10 min, followed by 25°C for 10 min) to total RNA (10 μg) and to exogenous transcripts (described in the Affymetrix technical manual for P. aeruginosa expression products [www.affymetrix.com]) (130 pM) that were added to each sample, which served as a control for monitoring transcriptional efficiency and array performance, and they were extended with SuperScript II reverse transcriptase (Invitrogen) (25°C for 10 min, 37°C for 60 min, 42°C for 60 min, and 70°C for 10 min). Residual RNA was removed by alkaline treatment followed by neutralization, and cDNA was purified with a QIAquick PCR purification kit (QIAGEN, Valencia, Calif.). Purified cDNA was fragmented with DNase I (Amersham Pharmacia Biotech, Piscataway, N.J.) and end labeled with Biotin-ddUTP by using an Enzo BioArray terminal labeling kit (Affymetrix, Santa Clara, Calif.). Target hybridization, staining, and scanning were performed by using an Affymetrix GeneChip system (workstation with Microarray Suite [MAS] software, version 5.0, a hybridization oven, a fluidics workstation, and a Hewlett-Packard GeneArray scanner). Individual microarrays were scanned at two intensity levels to provide a greater dynamic range of gene expression (www.affymetrix.com). Data were stored in an Affymetrix LIMS database for analysis.
Microarray data analysis.
Microarray data were analyzed by using protocols described by Affymetrix (www.affymetrix.com). All data were globally scaled to a target intensity of 500, and absolute expression levels of transcripts were generated for each chip. Briefly, in this procedure three metrics (detection, call change, signal log ratio) were used to identify genes whose transcripts were different in an individual control sample and an experimental sample. Transcript expression for a particular gene (presence or absence) was detected by using default parameters in the MAS software (version 5.0; Affymetrix). We performed pairwise comparisons with Data Mining Tools (DMT) (version 3.0; Affymetrix) to determine whether a transcript was present (positive call) and, if it was present, whether the transcript changed. We defined a positive call as 66% present across the pairwise comparisons (six of nine pairwise comparisons for three chips) since we performed three biological replicate experiments for each condition. Saturating probes which may have incorrectly appeared to be absent when they were scanned at a higher intensity level were identified by a comparison of microarray scans performed at two intensity levels. Transcripts that were absent under both control and experimental conditions were eliminated from further consideration. We then used statistical tools available in DMT, version 3.0, to perform t tests with absolute data to identify genes that showed significant changes in transcript levels (P ≤ 0.05) when the results obtained under control and experimental conditions were compared. Comparative analyses of experimental and control chips were also performed by using MAS, version 5.0, to generate log2 signal ratio values in order to quantify the direction and magnitude of differential transcript expression, as well as the statistical significance of the difference (change call). We performed pairwise comparisons of change call data for each condition and defined a positive call as a 66% increase or decrease among the nine pairwise comparisons for the three chips. Transcripts that exhibited statistically significant change calls (increases or decreases) were combined with transcripts that showed significant changes by using a t test (P ≤ 0.05) to obtain a final list of genes whose transcripts exhibited differential expression under the two conditions. Log2 signal ratio values were converted and expressed as fold changes.
Data were also analyzed by using Significance of Analysis of Microarrays (SAM) software (version 1.13) (44) to determine the possible number of false positives in each data set (the false discovery rate). Tuning parameters were adjusted to minimize the false discovery rate for each data set. Additionally, lists of transcripts obtained by using SAM were compared to lists of transcripts generated by using MAS (version 5.0) and DMT (version 3.0) software tools to obtain an estimate of the difference between the two statistical approaches and to provide greater confidence in the transcripts that were determined to be differentially expressed under the different conditions. Further information about these statistical tests is available elsewhere (44; www.affymetrix.com).
Microarray validation: QRT-PCR and data analysis.
Differential expression of genes was examined by quantitative real-time PCR (QRT-PCR) by using the MGB Eclipse expression system chemistry with probes and primers designed by using sequences from the Pseudomonas database associated with the probe designations in the Affymetrix library files. Probes and primers were designed by using the Epoch Biosciences MGB Eclipse gene expression design software in the custom mode to allow for the development of assays that have efficiencies commensurate with a comparative threshold cycle (Ct) analysis (www.epochpharm.com). The following dye combination for probe generation was used for detection and data normalization: 6-carboxyfluorescein (FAM) (3′ reporter) and MGB-Q (minor grove binder-quencher) for genes of interest and for normalization control genes. The following probe and primer sequences were used: for PA0122, sense primer 5′-TCGTCGTCCTTGTTGGTGTA-3′, antisense primer 5′-ATGGATCGCAGTGAAGGT-3′, and probe (MGB-Q)CAATGGGGCAAGTTCTA(FAM); for PA1897, sense primer 5′-GTAGGTGACGAAGGTGTTA-3′, antisense primer 5′-CCTGGAACAAATGGATGA-3′, and probe (MGB-Q)TGCCCTGCCACCACTG(FAM); for PA1978, sense primer 5′-GATGCCGTGTTCTTCGTG-3′, antisense primer 5′-AAGCCATCGAGCAGATCCT-3′, and probe (MGB-Q)TGCCCTCGGACGTGAT(FAM); for PA3391, sense primer 5′-GTACAGGGTGAAGGCGTT-3′, antisense primer 5′-CCGCTACGTGTTCTGCAA-3′, and probe (MGB-Q)ATCCCCCATCCCAACGA(FAM); for PA3874, sense primer 5′-TGTTGAACCAGGCGTACT-3′, antisense primer 5′-GTGCTGAACCTCGACAAA-3′, and probe (MGB-Q)CCACACCTGCTCGATC(FAM); for PA3877, sense primer 5′-GGTAGGAGAGGAAGAACA-3′, antisense primer 5′-ACCGCATCCTTCATCACCAT-3′, and probe (MGB-Q)TTCTGGGTCTGCCTGG(FAM); for PA3904, sense primer 5′-CCGACGCTACCTACCA-3′, antisense primer 5′-CGTCTTCTTCGACCATCT-3′, and probe (MGB-Q)CCACCCTCAGCGCCAA(FAM); for PA4651, sense primer 5′-ACCGAATAGCGCATCTGGAA-3′, antisense primer 5′-GTGCTGATCGACGAAGTG-3′, and probe (MGB-Q)ACGCAACGAAGCCCCG(FAM); and for 16S RNA, sense primer5′-GCTTGCACCCTTCGTATT-3′, antisense primer 5′-CCTTGCTGTTTTGACGTTACC-3′, and probe (MGB-Q)GCACCGGCTAACTTCGT(FAM). In short, the MGB Eclipse probe system incorporates a minor groove binder, dark quenchers, and modified bases which work in concert to generate probes for difficult templates. In a nonhybridized state MGB Eclipse probes assume a random coil structure in solution, which causes a quenching of the reporter due to the proximity of the quencher molecule to the reporter molecule (for specific thermodynamic and physical characteristics of the quencher and its ability to efficiently quench reporters in the nonhybridized state, see the MGB Eclipse technical manual [www.epochpharm.com]). When hybridized to complementary targets at lower annealing temperatures, the quencher and the fluorophore are spatially separated, which allows the reporter to be accessible to excitation and emission from a light source. During the extension phase of the PCR, the forward primer causes the MGB Eclipse probe to be removed from the target, which returns the probe to a nonfluorescent state. Data are collected only during the annealing phase of the assays, and a melting curve is completed following each reaction to confirm that no template-independent amplification was present. Although the chemistry is slightly different, it is important to note that the principle of this approach is the same as that of Taqman chemistry used for quantitative PCR and that in fact, this approach is more sensitive and allows for the design of probes for thermodynamically undesirable regions (A. I. Brooks, unpublished observations). Prior to comparative analysis each probe and primer pair were tested to determine their relative efficiency compared with that of the probe and primer pair for 16S RNA (referred to as the normalizer gene) to ensure that the Ct data analysis approach could be employed (for a detailed description of the procedure used to measure efficiency see Applied Biosystems technical bulletin 2, Relative Gene Expression Analysis). The absolute value of the slope of log input amount versus ΔCt was less than 0.1 for all comparisons, which allowed us to use the ΔΔCt calculation to determine the relative levels of gene expression in all experimental cultures compared to the levels in controls. De novo cDNAs from RNA used in microarray target preparation were employed for all analyses. Following probe and primer optimization all cDNAs were diluted to a concentration of 2 ng/μl, and 5 μl was used for each 10-μl PCR mixture containing each deoxynucleoside triphosphate at a concentration of 400 μM, 1× MGB Eclipse buffer, 1× primer mixture, 1× MGB Eclipse probe mixture, and 0.4 U of JumpStart Taq (Sigma). All reactions were performed in triplicate, and the experiment was replicated three times; therefore, the data presented below are a reflection of nine individual reactions for each sample tested. All reactions were performed with an ABI 7900 HT with the following cycle parameters: one cycle of 95°C for 2 min, followed by 50 cycles of 95°C for 5 s, annealing at 56°C for 20 s, and extension at 76°C for 20 s. Data were collected only during the annealing phase, and melting curves were obtained following completion of the reaction. Raw data were analyzed by using the Sequence Detection software (ABI, Foster City, Calif.), while relative quantitation with the Ct method was performed by using Microsoft Excel.
Identification of las boxes upstream of QS-regulated genes.
The DNA regions (nucleotides −500 to 50 relative to the translational start site) surrounding differentially expressed genes were searched for the presence of putative activator protein-autoinducer complex binding elements (las boxes [43]) by using RSA tools (http://bioinformatics.bmc.uu.se/∼jvanheld/rsa-tools). The search was conducted by using a consensus sequence based on known QS-regulated promoters, and the total number of matches, sequence identity, and position are reported below.
RESULTS AND DISCUSSION
Identification of QS-regulated genes in PAO-JP2 in the presence and absence of exogenous autoinducers.
To determine the contribution of QS in regulating P. aeruginosa gene expression, samples of total RNA were extracted at the mid-logarithmic and early stationary phases from three independent cultures of a mutant (PAO-JP2) deficient in 3O-C12-HSL and C4-HSL production grown aerobically in modified FAB with or without exogenously added autoinducers (1 μM 3O-C12-HSL and 2 μM C4-HSL). RNA samples were processed and hybridized to P. aeruginosa DNA microarrays. An average of 3,628 (65%) of the 5,570 PAO1 ORFs were expressed in PAO-JP2 at the mid-logarithmic phase, and 4,204 ORFs (75%) were expressed in PAO-JP2 at the early stationary phase. When exogenous autoinducers were added to culture media, 4,168 ORFs (75%) were expressed at the mid-logarithmic phase, and 4,333 ORFs (78%) were expressed at the early stationary phase. The gene expression pattern of PAO-JP2 was compared to profiles generated from PAO-JP2 grown in the presence of both autoinducers, and transcripts which exhibited significant differential expression (P ≤ 0.05) under the two culture conditions were defined as QS regulated. A comparison of the gene expression profiles of the PAO-JP2 mutant cultivated with and without exogenous autoinducers eliminated possible erroneous interpretations of QS regulation that might have been due to secondary mutations in PAO-JP2 that may have arisen to compensate for disruption of QS regulons (2, 3). The effect of secondary mutations would confuse direct comparisons of PAO-JP2 with the PAO1 parent strain. By using this approach, 616 genes (more than 10% of the known ORFs) were identified as being QS regulated. This value is more than double the 200 to 300 genes predicted previously (44). Table 1 shows genes that were identified as being QS regulated (P ≤ 0.05) and that also showed a change of fivefold or more (QS promoted) or threefold or more (QS repressed). A complete list of all genes whose expression was identified as being QS regulated that exhibited a statistically significant change (P ≤ 0.05) is available online (http://www.urmc.rochester.edu/smd/mbi/bhi/). Using SAM, version 1.1.3, we determined that our data sets have a false discovery rate of approximately 9%. Lists of genes whose expression was determined to exhibit statistically significant changes in response to autoinducer signals generated by MAS (version 5.0) and DMT (version 3.0) were compared to similar lists of genes generated by SAM (version 1.13). Overall, only 24 genes (4%) generated by using SAM (version 1.13) did not match the 616 genes generated by using MAS (version 5.0) and DMT (version 3.0), which substantiated our statistical analysis.
TABLE 1.
Genes whose expression was identified as being QS regulated by using microarraysa
| ORF or intergenic region (ig) | Gene name | Change in PAO-JP2 (induced to uninduced) (fold)b | Growth stagec | Protein descriptiond | las boxe |
|---|---|---|---|---|---|
| QS-promoted genes | |||||
| PA0052 | 6.7 | ES | Hypothetical protein | ||
| PA0105 | coxB | 5.7 | ES | Cytochrome c oxidase subunit II | ACCTGGCTGATCTGATAGCC (R) (−456 to −437) |
| PA0107 | 6.1 | ES | Conserved hypothetical protein | ||
| PA0122 | 29 | ES | Conserved hypothetical protein | GCCTACCAGATCTGGCAGGC (D), GCCTGCCAGATCTGGTAGGC (R) (−166 to −147) | |
| PA0144 | 17 | ES | Hypothetical protein | ||
| PA0399 | 5.2 | ML | Cystathionine beta-synthase | ||
| PA0447 | gcdH | 7.8 | ML | Glutaryl-coenzyme A dehydrogenase | |
| PA0572 | 7.8 | B | Hypothetical protein | ||
| PA0588 | 7.4 | ML | Conserved hypothetical protein | ||
| PA0744 | 7.0 | ML | Probable enoyl-coenzyme A hydratase/isomerase | ||
| PA0745 | 8.2 | ML | Probable enoyl-coenzyme A hydratase/isomerase | ||
| PA0852f | cpbD | 18 | ES | Chitin-binding protein CbpD precursor | |
| PA0997 | 5.9 | B | Hypothetical protein | ||
| PA0998 | 7.2 | B | Hypothetical protein | ||
| PA0999 | fabH1 | 5.1 | B | 3-Oxoacyl-(acyl carrier protein) synthase III | |
| PA1130f | 7.4 | B | Hypothetical protein | ||
| PA1131f | 5.4 | B | Probable MFS transporter | ||
| PA1212 | 6.9 | ES | Probable MFS transporter | ||
| PA1214 | 5.0 | ES | Hypothetical protein | TCCTGGGTGTTCAGCCAGCC (D) (−381 to −362) | |
| PA1217 | 30 | ES | Probable 2-isopropylmalate synthase | GCCTACCCGATGTTCAAGTG (D) (−438 to −419) | |
| PA1219 | 5.3 | ES | Hypothetical protein | ||
| PA1246 | aprD | 6.3 | ML | Alkaline protease secretion protein AprD | |
| PA1249f | aprA | 6.9 | B | Alkaline metalloproteinase precursor | |
| PA1250 | aprI | 12 | B | Alkaline proteinase inhibitor AprI | |
| PA1318 | cyoB | 5.7 | ES | Cytochrome o ubiquinol oxidase subunit I | |
| PA1431f,g | rsaL | 7.7 | B | Regulatory protein RsaL | AACTAGCAAATGAGATAGAT (D) (−77 to −58)h |
| PA1657 | 6.6 | ML | Conserved hypothetical protein | ||
| PA1784 | 7.3 | ES | Hypothetical protein | CCCTATCCGTTCTGGGAGCT (R) (−140 to −121) | |
| PA1869f | 79 | B | Probable acyl carrier protein | CACTGCCAGATCTGGCAGTT (D), AACTGCCAGATCTGGCAGTG (R) (−90 to −71) | |
| PA1871f | lasA | 29 | ES | LasA protease precursor | |
| PA1874 | 9.9 | ES | Hypothetical protein | ||
| PA1875 | 9.1 | ES | Hypothetical protein | ||
| PA1894f | 25 | ES | Hypothetical protein | ||
| PA1895 | 11 | ES | Hypothetical protein | ||
| PA1896f | 11 | ES | Hypothetical protein | ||
| PA1897g | 33 | B | Hypothetical protein | ACCTGCCCGGAAGGGCAGGT (D), ACCTGCCCGGAAGGGCAGGT (R) (−288 to −269) | |
| PA1901f | 16 | ES | Phenazine biosynthesis protein PhzC | ||
| PA1902 | 42 | ES | Phenazine biosynthesis protein PhzD | ||
| PA1903 | 26 | B | Phenazine biosynthesis protein PhzE | ||
| PA1904 | 51 | B | Probable phenazine biosynthesis protein | ||
| PA1905 | 22 | B | Probable pyridoxamine 5-phosphate oxidase | ||
| PA1927 | metE | 20 | ES | Methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase | |
| PA1999 | 32 | ML | Probable coenzyme A transferase subunit A | ||
| PA2000 | 46 | ML | Probable coenzyme A transferase subunit B | ||
| PA2001 | atoB | 23 | ML | Acetyl-coenzyme A acetyltransferase | |
| PA2014 | 7.5 | ML | Probable acyl-coenzyme A carboxyltransferase beta chain | ||
| PA2030 | 7.3 | B | Hypothetical protein | ||
| PA2068 | 8.6 | ES | Probable MFS transporter | ||
| PA2069 | 8.8 | ES | Probable carbamoyl transferase | GCCTGCGAAATCTGGCAGGG (D) (−109 to −90) | |
| PA2193f | hcnA | 210 | B | Hydrogen cyanide synthase HcnA | ACCTACCAGAATTGGCAGGG (D), CCCTGCCAATTCTGGTAGGT (R) (−112 to −93)h |
| PA2194f | hcnB | 52 | B | Hydrogen cyanide synthase HcnB | |
| PA2195 | hcnC | 30 | B | Hydrogen cyanide synthase HcnC | |
| PA2196 | 5.3 | ML | Probable transcriptional regulator | ||
| PA2250 | lpdV | 7.8 | ML | Lipoamide dehydroger ase-Val | |
| PA2300 | chiC | 5.1 | ES | Chitinase | ACCTACCACAATTGGCAGAG (D) (−193 to −174) |
| PA2302f | 27 | ES | Probable nonribosomal peptide synthetase | ||
| PA2303 | 10 | B | Hypothetical protein | ||
| PA2304 | 14 | ES | Hypothetical protein | ||
| PA2305 | 28 | B | Probable nonribosomal peptide synthetase | ||
| PA2306 | 13 | ES | Conserved hypothetica protein | ||
| PA2331 | 9.4 | ES | Hypothetical protein | ||
| PA2366 | 7.5 | ES | Conserved hypothetical protein | /PICK> | |
| PA2367 | 6.3 | ES | Hypothetical protein | ||
| PA2423 | 12 | B | Hypothetical protein | ||
| PA2552 | 10 | ML | Probable acyl-coenzyme A dehydrogenase | ||
| PA2553 | 14 | ML | Probable acyl-coenzyme A thiolase | ||
| PA2554 | 11 | ML | Probable short-chain dehydrogenase | ||
| PA2555 | 6.2 | ML | Probable AMP-binding enzyme | ||
| PA2564 | 11 | ES | Hypothetical protein | ||
| PA2565 | 5.5 | ES | Hypothetical protein | ||
| PA2566 | 10 | ES | Conserved hypothetical protein | CCCTGCCAGTTCTGGTAGGC (D), GCCTACCAGAACTGGCAGGG (R) (−157 to −138) | |
| PA2570f | palL | 60 | ES | PA-I galactophilic lectin | |
| PA2587f | 16 | B | Probable FAD-dependent monooxygenase | CCCTATCAGAAGATCGAGTT (D) (−126 to −107) | |
| PA2588 | 6.9 | B | Probable transcriptional regulator | ||
| PA2591f | 5.7 | B | Probable transcriptional regulator | AACTACCAGTTCTGGTAGGT (D) (−115 to −96) | |
| PA2592f,g | 5.6 | B | Probable periplasmic spermidine-putrescine- binding protein | AACTACCAGTTCTGGTAGGT (D) (−59 to −40) | |
| PA2939 | 16 | ES | Probable aminopeptidase | ||
| PA3032f | snr-1 | 10 | B | Cytochrome c | |
| PA3326f | 8.7 | B | Probable Clp family ATP-dependent protease | ACCTAACAGATTTGTAAGTT (D) (−241 to −222) | |
| PA3328f | 15 | ES | Probable FAD-dependent monooxygenase | ||
| PA3329f | 55 | ES | Hypothetical protein | ||
| PA3330f | 38 | ML | Probable short-chain dehydrogenase | ||
| PA3331f | 16 | B | Cytochrome P450 | ||
| PA3332 | 15 | B | Conserved hypothetical protein | ||
| PA3333f | fabH2 | 32 | B | 3-Oxoacyl-(acyl carrier protein) synthase III | |
| PA3334 | 23 | B | Probable acyl carrier protein | ||
| PA3335 | 5.1 | B | Hypothetical protein | ||
| PA3361 | 51 | ES | Hypothetical protein | ||
| PA3477f | rhlR | 5.9 | ES | Transcriptional regulator RhlR | |
| PA3478f | rhlB | 37 | ES | Rhamnosyltransferase chain B | |
| PA3479f | rhlA | 100 | ES | Rhamnosyltransferase chain A | TCCTGTGAAATCTGGCAGTT (D) (−288 to −269)h |
| PA3519 | 16 | ES | Hypothetical protein | ||
| PA3520 | 70 | ES | Hypothetical protein | ||
| PA3709 | 5.2 | ML | Probable MFS transporter | ||
| PA3721 | 5.8 | ES | Probable transcriptional regulator | ||
| PA3724f | lasB | 39 | B | Elastase LasB | ACCTGCCAGTTCTGGCAGGT (D), ACCTGCCAGTTCTGGCAGGT (R) (−194 to −175)h |
| PA3904 | 18 | ML | Hypothetical protein | ||
| PA3906 | 5.8 | B | Hypothetical protein | ||
| PA3908 | 5.9 | B | Hypothetical protein | ||
| PA3923 | 12 | ML | Hypothetical protein | ||
| PA4078f | 7.6 | ES | Probable nonribosomal peptide synthetase | ||
| PA4129 | 14 | B | Hypothetical protein | ||
| PA4130 | 12 | B | Probable sulfite or nitrite reductase | ||
| PA4131 | 6.8 | ES | Probable iron-sulfur protein | ||
| PA4132 | 8.1 | ES | Conserved hypothetical protein | ||
| PA4133 | 46 | ES | Cytochrome c oxidase subunit (cbb3 type) | ||
| PA4134 | 22 | ES | Hypothetical protein | ||
| PA4141 | 25 | B | Hypothetical protein | ||
| PA4142 | 9.6 | B | Probable secretion protein | ||
| PA4175 | 5.5 | ES | Probable endoproteinase Arg-C precursor | ||
| PA4205 | mexG | 8.0 | ES | Hypothetical protein | |
| PA4206 | mexH | 5.5 | ES | Probable RND efflux membrane fusion protein precursor | |
| PA4207f | mexI | 6.2 | ES | Probable RND efflux transporter | |
| PA4208 | opmD | 6.2 | ES | Probable outer membrane efflux protein precursor | |
| PA4209 | 14 | B | Probable O-methyltransferase | ACCTACCAGATCTTGTAGTT (D) (−327 to −308) | |
| PA4211 | 74 | B | Probable phenazine biosynthesis protein | ||
| PA4217f | 19 | ES | Probable FAD-dependent monooxygenase | ||
| PA4293 | 6.6 | ES | Probable two-component sensor | ||
| PA4294 | 8.6 | ES | Hypothetical protein | ||
| PA4297 | 7.5 | ES | Hypothetical protein | GCCTGGCTGGCCATCCAGGC (R) (−190 to −171) | |
| PA4298 | 6.8 | ES | Hypothetical protein | ||
| PA4299 | 8.9 | ES | Hypothetical protein | ||
| PA4300 | 7.7 | ES | Hypothetical protein | ||
| PA4302 | 9.6 | ES | Probable type II secretion system protein | ||
| PA4303 | 5.2 | ES | Hypothetical protein | ||
| PA4304 | 6.7 | ES | Probable type II secretion system protein | ||
| PA4305 | 7.3 | ES | Hypothetical protein | ||
| PA4306 | 28 | ES | Hypothetical protein | ||
| PA4496 | 10 | ML | Probable binding protein component of ABC transporter | ||
| PA4648 | 14 | ES | Hypothetical protein | ||
| PA4651 | 8.8 | ES | Probable pilus assembly chaperone | ||
| PA4677 | 15 | B | Hypothetical protein | ||
| PA4878 | 8.8 | B | Probable transcriptional regulator | ||
| PA5059 | 11 | ES | Probable transcriptional regulator | ||
| PA5220 | 19 | ML | Hypothetical protein | ||
| ig 4713795-4713098 | 47 | ES | Intergenic region between PA4280 and PA4281, 4713098-4713795, (−) strand | ||
| QS-repressed genes | |||||
| PA0040 | 5.6 | ES | Conserved hypothetical protein | ||
| PA0509 | nirN | 4.7 | ES | Probable c-type cytochrome | |
| PA0510 | 4.1 | ES | Probable uroporphyrin-III c-methyltransferase | ||
| PA0512 | 3.2 | ES | Conserved hypothetical protein | ||
| PA0714 | 3.2 | ES | Hypothetical protein | ||
| PA1978 | 6.1 | ES | Probable transcriptional regulator | ||
| PA1983 | exaB | 12 | ES | Cytochrome c550 | |
| PA1984 | 5.5 | ES | Probable aldehyde dehydrogenase | ||
| PA2259 | ptxS | 3.6 | ML | Transcriptional regulator PtxS | |
| PA2260 | 13 | ML | Hypothetical protein | ||
| PA2261 | 18 | ML | Probable 2-ketogluconate kinase | ||
| PA2321 | 3.2 | ML | Gluconokinase | ||
| PA2539 | 4.5 | ML | Conserved hypothetical protein | ||
| PA2540 | 3.0 | ML | Conserved hypothetical protein | ||
| PA2711 | 3.0 | ES | Probable periplasmic spermidine-putrescine- binding protein | ||
| PA2903 | cobJ | 3.3 | ML | Precorrin-3 methylase CobJ | |
| PA3391 | nosR | 3.0 | ES | Regulatory protein NosR | |
| PA3392 | nosZ | 3.8 | ES | Nitrous oxide reductase precursor | TACTGCCTCGACTGCCAGAT (D) (−168 to −149) |
| PA3393 | nosD | 3.5 | ES | NosD protein | |
| PA3394 | nosF | 4.4 | ES | NosF protein | |
| PA3395 | nosY | 4.8 | ES | NosY protein | |
| PA3396 | nosL | 4.3 | ES | NosL protein | |
| PA3662 | 4.6 | ES | Hypothetical protein | ||
| PA3870 | moaA1 | 5.1 | ES | Molybdopterin biosynthetic protein A1 | |
| PA3871 | 6.4 | B | Probable peptidyl-prolyl cis-trans isomerase, PpiC type | ||
| PA3872 | narI | 6.1 | ES | Respiratory nitrate reductase gamma chain | |
| PA3873 | narJ | 6.1 | B | Respiratory nitrate reductase delta chain | |
| PA3874 | narH | 8.5 | ES | Respiratory nitrate reductase beta chain | |
| PA3875 | narG | 8.3 | ES | Respiratory nitrate reductase alpha chain | |
| PA3876 | narK2 | 11 | ES | Nitrite extrusion protein 2 | |
| PA3877 | narK1 | 13 | ES | Nitrite extrusion protein 1 | ATCTGTCCCGCTGGTCAGCG (R) (−217 to −198 in ORF of narX) |
| PA3911 | 3.7 | ES | Conserved hypothetical protein | ||
| PA3912 | 5.1 | ES | Conserved hypothetical protein | ||
| PA3913 | 6.5 | ES | Probable protease | ||
| PA3914 | moeAI | 6.9 | ES | Molybdenum cofactor biosynthetic protein A1 | |
| PA3916 | moaE | 7.5 | ES | Molybdopterin converting factor large subunit | |
| PA3917 | moaD | 8.0 | ES | Molybdopterin converting factor small subunit | |
| PA3918 | moaC | 6.1 | ES | Molybdopterin biosynthetic protein C | |
| PA4587 | ccpR | 3.9 | ES | Cytochrome c551 peroxidase precursor | |
| PA4628 | lysP | 3.4 | ML | Lysine-specific permease | |
| PA4918 | 3.0 | ML | Hypothetical protein | ||
| ig 2923366-2922568 | 6.4 | ML | Intergenic region between PA2587 and PA2588, 2922568-2923366, (−) strand |
Only genes identified as being QS regulated that were differentially expressed with a magnitude of change of ≥5-fold (QS promoted) or of ≥3-fold (QS repressed) are included. A list of all genes identified as being QS regulated exhibiting a statistically significant change (P ≤ 0.05) is available online (http://www.urmc.rochester.edu/smd/mbi/bhi/). Genes are identified by ORF designation, gene name, and protein description (http://www.pseudomonas.com).
The magnitude of transcript induction was calculated for PAO-JP2 grown with exogenous autoinducers (1 μM 3O-C12-HSL and 2 μM C4-HSL) compared to PAO-JP2 cultures grown aerobically in modified FAB. The values are reported for the early stationary phase when differential expression was observed in both gr
Growth stage(s) during which statistically significant differential transcript expression (P ≤ 0.05) was exhibited. ML, mid-logarithmic phase; ES, early stationary phase; B, both mid-logarithmic and early stationary phases.
FAD, flavin adenine dinucleotide; RND, resistance-nodulation-cell division; MFS, major facilitator superfamily; ABC, ATP-binding cassette.
The orientation of the consensus sequence is indicated in parentheses (D, directly upstream of the ORF; R, opposite strand). The numbers in parentheses are sequence positions.
Gene possessing overlapping las boxes upstream of adjacent ORFs of currently identified QS-regulated genes.
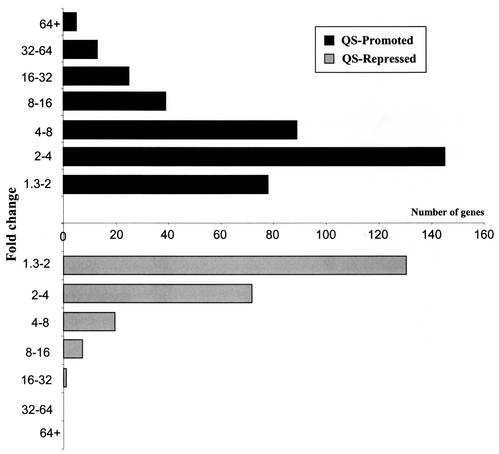
Of the 616 genes whose expression was identified as being QS regulated, 394 were promoted and 222 were repressed. In addition, nine intergenic regions (six promoted regions and three repressed regions) responded to the presence of exogenous autoinducers. The expression of 43 genes was increased more than 16-fold, and five genes exhibited increases of more than 64-fold (Table 1 and Fig. 1). In comparison, only one gene (PA2261) showed a decrease in expression of more than 16-fold, and the expression of 27 genes decreased more than fourfold. Approximately equivalent numbers of genes exhibited changes in expression between 1.3- and 4-fold (the expression of 223 genes increased, while the expression of 197 genes decreased). The temporal patterns of QS-regulated gene expression were also compared at the mid-logarithmic and early stationary phases. The expression of 244 genes was found to be QS regulated at the mid-logarithmic phase, and 450 genes were identified as being QS regulated at the early stationary phase. Seventy-eight genes were identified as being QS regulated at both phases. Interestingly, the transcripts of four genes (PA1760, PA1988, PA4520, and PA4912) exhibited a biphasic response (expression was QS promoted at the mid-logarithmic phase but QS repressed at the early stationary phase). Time dependence of induction of QS-regulated genes in response to exogenous autoinducers has been observed previously in QS mutants (44) and is discussed in an accompanying paper (34a).
FIG. 1.
Changes in the distribution of QS-regulated genes. The numbers of QS-regulated genes displaying specific magnitudes of differential expression (PAO-JP2 induced versus uninduced) are shown. Genes were placed in groups according to the magnitude of differential expression (1.3- to 2.0-, 2.0- to 4.0-, 4.0- to 8.0-, 8.0- to 16-, 16- to 32-, 32- to 64-, and more than 64-fold changes).
In some instances, for genes that formed an operon, such as the hcn cluster (PA2193 to PA2195), the changes for the first gene in the operon were much greater than the changes for the downstream genes when transcript expression in induced PAO-JP2 (grown in the presence of 1 μM 3-O-C12-HSL and 2 μM C4-HSL) was compared with transcript expression in uninduced PAO-JP2 (Table 1) (http://www.urmc.rochester.edu/smd/mbi/bhi/). This phenomenon has also been noted by other workers and has been attributed to RNA degradation from the 3′ end of the transcript (25). Not all individual genes in known operons exhibited statistically significant changes in transcript expression. For example, at least three genes involved in type III secretion systems, pscQ, pscI, and pcrH, exhibited statistically significant decreases in transcript expression in induced PAO-JP2 compared to transcript expression in uninduced PAO-JP2, but other genes in these operons did not change. We observed some fluctuations in transcript levels in replicates which may have been due to biological variation, as well as RNA degradation, and which resulted in large standard deviations for some genes in operons that precluded their identification as being QS regulated in this study. Although standard deviations in transcript levels tended to be lower as the value for the transcript level decreased, the intensities of transcripts present at very low levels (<500 intensity units) were often close to the average background intensities (∼200 intensity units), and thus the transcripts were defined as absent and were excluded from further analyses. These transcripts included some of the genes in type III secretion operons (data not shown), which precluded detection of such genes as genes that exhibit statistically significant changes in response to added autoinducers.
Correlation of microarray data to previously identified QS-regulated genes.
Of 52 previously reported QS-regulated genes, 37 were confirmed by using microarrays. These 37 genes are PA0852 (cpbD), PA1130 (rhlC), PA1131, PA1249 (aprA), PA1431 (rsaL), PA1869, PA1871 (lasA), PA1894, PA1896, PA1901, PA2193 and PA2194 (hcnAB), PA2302, PA2570 (pa1L), PA2587 (designated pqsH [17]), PA2592, PA3032 (snr-1), PA3103 (xcpR), PA3104 (xcpP), PA3327 to PA3331, PA3333 (fabH2), PA3336, PA3477 (rhlR), PA3478 and PA3479 (rhlAB), PA3724 (lasB), PA3907, PA4078, PA4084 (cupB3), PA4207 (designated mexI [1]), PA4217 (phzS), PA4236 (katA), and PA4869 (Table 1) (http://www.urmc.rochester.edu/smd/mbi/bhi/) (8, 18, 19, 28, 29, 38, 39, 44, 46). Interestingly, PA4084 was previously reported to be QS promoted (44) but in this study was found to be QS repressed. In addition, we found that PA3622 (rpoS) is QS promoted (2.7-fold change[P = 0.002] for mid-logarithmic-phase samples and 1.4-fold change [P = 0.026] for early-stationary-phase samples), which agreed with several previous reports (22, 40) but differed from one study in which it was concluded that rpoS was not QS regulated (45). Two genes (PA0179 and PA3904) which were only recently identified as being QS promoted by using a fluorescence-activated cell sorting approach (J. Lamb and B. H. Iglewski, unpublished data) were also determined to be QS promoted by using microarrays.
In addition to the lasI and rhlI genes, which could not be detected in this study because they are mutated in PAO-JP2, 13 genes previously identified as being QS regulated were not identified in this analysis (Table 2). Discrepancies between previously reported QS-regulated genes and the genes identified in the present study could be explained by several observations. Transcripts were not detected for 6 of the 13 genes (Table 2). A high coefficient of variation was observed for an additional 4 of the 13 genes (Table 2). One of these four genes, PA1148, encodes toxin A. This gene is known to be repressed in the presence of elevated iron concentrations, such as the concentration present in modified FAB (16). Although we previously reported that lasR is QS regulated, we did not find that expression of the lasR transcript was QS regulated under the conditions used in the present study, which were different from those used in the previous study (28). Inconsistencies between the results of the present study and the previously reported results for QS-regulated genes that were obtained by using transposon mutagenesis (44) might be explained by the fact that in some cases the orientation of the transposon insertions was found to be opposite that of the ORF, as shown in Table 2 for PA1129, PA2770, and PA3325. We could not detect the transcript for PA1129 under the experimental conditions used in this study; however, we detected transcripts for PA1130 and PA1131, which are immediately downstream of PA1129, which we identified as being QS promoted using microarrays. Interestingly, although we did not determine that PA2770 is QS regulated in the present study, we noted that the intergenic region between PA2770 and PA2771 that corresponded to the reported transposon insertion location (position 3128663 in the intergenic region from position 3122585 to position 3123598 on the opposite strand [44]) showed an increase in expression at the early stationary phase when PAO-JP2 and PAO-JP2 grown in the presence of both autoinducers were compared (P = 0.054). Similarly, even though we did not determine that PA3325 was QS regulated in the present study, we found that PA3326, the gene immediately downstream of PA3325 and on the opposite strand, was QS regulated. Additionally, the temporal expression of QS-regulated genes may influence their behavior under various growth conditions. For example, seven genes (Table 2) not identified as being QS regulated in this study reportedly showed very-late-phase QS regulation in studies performed with lacZ promoter fusions (44). Extraction of RNA from cultures in the early stationary phase, as performed in the present study, may have excluded determination of these genes as being QS regulated. Taking into account these explanations, we found that the results of the present study matched previously reported results for QS-regulated genes. The high degree of correlation between data obtained in this study and data for previously identified QS-regulated genes provided considerable validation for the use of microarrays.
TABLE 2.
Previously reported QS-regulated genes not identified by using microarraysa
| Gene or ORF | Protein description | Transcript detectionb | CVc | Comment | Reference |
|---|---|---|---|---|---|
| PA0051 | Probable glutamine amidotransferase | A | NA | Late induction | 44 |
| PA0109 | Hypothetical protein | P | High | Late induction | 44 |
| PA0855 | Hypothetical protein | PL | High | Late induction | 44 |
| PA1129 | Probable fosfomycin resistance protein | A | NA | Insertion in opposite orientation | 44 |
| PA1148 | Exotoxin A precursor | PL | High | Iron regulated | 16 |
| PA1430 | Transcriptional regulator LasR | P | Low | 28 | |
| PA2385 | Probable acylase | PL | Low | Late induction | 44 |
| PA2401 | Probable nonribosomal peptide synthetase | A | NA | Late induction | 44 |
| PA2402 | Probable nonribosomal peptide synthetase | A | NA | Late induction | 44 |
| PA2424 | Probable nonribosomal peptide synthetase | A | NA | Late induction | 44 |
| PA2770 | Hypothetical protein | P | Low | Insertion in opposite orientation | 44 |
| PA3325 | Conserved hypothetical protein | A | NA | Insertion in opposite orientation | 44 |
| PA5356 | Transcriptional regulator GlcC | P | High | 44 |
Genes are identified by ORF designation and protein description (http://www.pseudomonas.com).
Transcript detection of genes by using microarrays. P, present; A, absent; PL, present at low levels. A low level of transcript abundance was defined as a transcript abundance of less than 500 intensity units.
A high coefficient of variation (CV) was defined as greater than 20%. NA, not applicable.
Independent microarray validation by using QRT-PCR.
In addition to the high correlation between previously identified QS-regulated genes and the results of the present study, we chose eight newly identified QS-regulated genes (four promoted genes and four repressed genes) that represented a range of changes observed in microarray studies (8.0- to 33-fold changes for QS-promoted genes and −3.0- to −13-fold changes for QS-repressed genes) to be validated by QRT-PCR. The direction of regulation for all of these genes matched the direction of regulation (promotion or repression) determined from the microarray data (data not shown). The magnitudes of the changes for the QS-promoted genes (PA0122, PA1897, PA3904, and PA4651) obtained from the QRT-PCR analysis agreed well with the magnitudes of the changes determined in the microarray analysis. For the QS-promoted genes, the greatest difference in changes was obtained for PA0122; the change determined in the QRT-PCR analysis was 34.7-fold ± 8.1-fold at the early stationary phase, compared to the change of 29-fold ± 1.4-fold determined in the microarray analysis. The QRT-PCR analysis resulted in larger changes (approximately three to four times larger) for the QS-repressed genes (PA1978, PA3391, PA3874, and PA3877). For example, PA3877 (narK1) was reported to show a change of −13-fold ± 1.3-fold when the microarrays were used, but the change was −50.3-fold ± 24.3-fold when QRT-PCR was used. This was not unexpected since a bias towards underestimating the magnitude of mRNA change has previously been observed for oligonucleotide microarray data (48). This suggests that our stringent analysis of the microarray data may have underestimated the true number of QS-repressed genes. The excellent agreement (100%) between the results obtained with the QRT-PCR and the results obtained with the microarrays further substantiated both the statistical analysis approach that we utilized in this study and the DNA microarray technology.
Identification of las boxes upstream of QS-regulated genes.
A las box was detected upstream of 7% of the QS-regulated genes identified in this study (Table 1) (http://www.urmc.rochester.edu/smd/mbi/bhi/). Among the genes whose expression was QS regulated, at least 111 gene clusters were discerned that might form possible operons, and 17 of clusters these included a las box upstream of the first gene in the cluster. The las boxes identified exhibit homology with the palindromic lux box DNA elements identified in V. fischeri, which are located upstream of LuxR-regulated genes and have been proposed to act as binding sites for the regulatory protein-autoinducer complex (14). The consensus sequence used in the present study was based on alignment of several known las boxes (Fig. 2). Most boxes were found directly upstream of the ORF on the same strand, but a number of genes possessed las boxes upstream of the ORF but on the antisense strand. The presence of overlapping las boxes on both the sense and antisense strands upstream of some QS-regulated genes is reminiscent of the putatively shared las box of lasI and rsaL (8). Additionally, a las box was located within the first 50 bp internal to the coding region (ORF) of one QS-repressed gene (PA3811). This observation may hint that binding of the regulatory protein-autoinducer complex to this region might interfere with expression of this gene. However, some QS genes that are negatively regulated have las boxes upstream of their ORFs. It would be particularly interesting to determine where these las boxes are relative to the start of transcription. The relative paucity of las boxes upstream of genes and operons that we found to be QS regulated suggests that regulation of a large number of these genes or operons may be indirect. In this regard, it is interesting that we found 37 QS-regulated genes known or thought to be transcriptional regulators or members of two-component systems (Table 1) (http://www.urmc.rochester.edu/smd/mbi/bhi/). Thus, target genes of these regulators would have been identified as being QS regulated in the present study, although their regulation may be via an indirect mechanism. We are currently constructing mutants with mutations in these known and putative transcriptional regulatory genes to determine the effects of such mutations on other genes that we identified as being QS regulated.
FIG. 2.
Alignment of las boxes upstream of QS-regulated genes. Previously identified las boxes were aligned to form a consensus sequence (11, 29, 43). This sequence was used to search all upstream regions in front of the translational start site of all QS-regulated genes. The consensus sequence NHCTRNSNNDHNDKNNAGNB was derived from inspection, where H = C, T, or A; R = A or G; S = C or G; D = G, A, or T; K = G or T; B = G or T; and N = A, C, G, or T. Sequences that matched the consensus sequence were identified, and the position of the las box is shown in Table 1.
Characterization of QS-regulated genes and involvement in physiological processes.
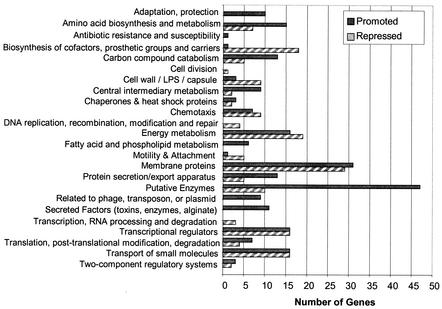
Expression of many genes involved in basic cellular processes, such as DNA replication, RNA transcription and translation, cell division, and amino acid biosynthesis, and genes involved in group behavior, such as chemotaxis and biofilm formation, were found to be QS regulated (Table 1 and Fig. 3) (http://www.urmc.rochester.edu/smd/mbi/bhi/). The large effect of QS on such diverse physiological functions has been observed previously in microarray studies with Escherichia coli, in which approximately 5.6% of the E. coli genome responded to the presence of autoinducer-2 in an autoinducer-2 signaling-deficient mutant (13). In the present study with P. aeruginosa, the largest percentage of QS-regulated transcripts (34%) encoded proteins having hypothetical, unclassified, and unknown functions (Table 1) (http://www.urmc.rochester.edu/smd/mbi/bhi/). Expression of 56 of these genes was QS repressed, and expression of 147 genes was QS promoted. Approximately 31% (62 genes) of the hypothetical proteins exhibited homology with known or similar hypothetical proteins in other microorganisms. Expression of 60 genes (31 QS-promoted genes and 29 QS-repressed genes) (Fig. 3), representing 40% of the 150 known or predicted ORFs encoding membrane proteins (36), was determined to be QS regulated. Membrane proteins are of particular interest due to their involvement in virulence factor secretion (36). Other functional classes with large numbers of QS-regulated genes included the genes encoding putative enzymes (14%), the genes encoding transcriptional regulators and two-component systems (9%), the genes encoding energy metabolism (9%), the genes encoding transport of small molecules (8%), and the genes encoding secreted factors and the protein secretion and/or export apparatus (7%) (Fig. 3). Interestingly, expression of 44 genes involved in nitrogen metabolism was identified as being QS regulated (Table 3) (49). Expression of the transcripts of several well-characterized genes involved in the denitrification system (49) was repressed by QS activation (nar, nos, mod, moa, and moe clusters), while expression of the molybdate-independent shared nitrate reductase (snr-1) gene, the napBC genes, and a probable gene cluster involved in nitrite reduction (PA4129 and PA4130) was promoted by QS (Table 3). This highlights the importance of QS as a switch for preferential usage of cellular pathways and is important when the design of therapeutic agents that disrupt a particular metabolic pathway(s) is considered. It also suggests that QS plays a pivotal role in the anaerobic growth of P. aeruginosa.
FIG. 3.
Functional classes of QS-regulated genes. QS-regulated genes were grouped according to a functional classification. The number of genes either promoted or repressed for each category is shown. LPS, lipopolysaccharide.
TABLE 3.
QS-regulated genes involved in nitrogen metabolism
| Gene(s) or operona | Description | Change (fold)b |
|---|---|---|
| PA0509-PA5018 (nirNJLFCM) | Nitrite respiration and heme D1 synthesis | −4.7 |
| PA0520-PA0522 (nirQ, nirS) | Denitrification | 3.9 |
| PA0523-PA0525 (norBD) | Nitrous oxide (NO) respiration | 2.2 |
| PA1172-PA1173 (napCB) | Periplasmic nitrate reduction | 1.5 |
| PA1861-PA1863 (modCBA) | Molybdenum transporters | 1.8 |
| PA3032 | snr-I (shared nitrate reduction) | 10 |
| PA3391-PA3396 (nosRZDFYL) | Nitrate and N2O respiration | −3.0 |
| PA3872-PA3877 (narK1K2IJHG) | Nitrate respiration | −13 |
| PA3870 (moaA1) | Molybdenum cofactor biosynthesis | −5.1 |
| PA3914-PA3918 (moeA1, moaEDC) | Molybdenum cofactor biosynthesis | −6.9 |
| PA4129-PA4130 | Probable sulfite of nitrite reductase | 14 |
| PA4131-PA4134 | Probable nitrogen fixation proteins | 6.8 |
Genes are identified by ORF designation, gene name, and protein description or known metabolic pathway (http://www.pseudomonas.com).
The magnitude and direction of gene expression change are indicated for PAO-JP2 grown with exogenous autoinducers (1 μM 3O-C12-HSL and 2 μM C4-HSL) compared to PAO-JP2 cultures grown aerobically in modified FAB. The values are the values for the first gene in the known or probable operons.
Microbial pathogenesis has been intimately linked to QS regulatory circuits, and several P. aeruginosa virulence factors are known to be QS regulated (10). More than 50 probable or known virulence genes (10), such as those encoding proteins involved in attachment to and colonization of host tissues and surfaces (type IV pili, PA-I galactophilic lectin) or dissemination (chemotaxis) or those that may be involved in tissue destruction (PA4142 to PA4144, PA1877), were identified as being QS regulated (Table 4). Interestingly, we identified three genes (PA3337, PA4996, and PA5450) important in lipopolysaccharide biosynthesis whose expression was repressed by QS. We also identified genes involved in type II secretion of virulence factors previously reported to be QS regulated (PA3095 to PA3103, PA3104, and PA3105 [4]), as well as a novel type II secretion gene (PA4304). In addition to type II secretion genes, we found that type III secretion genes (PA1694, PA1707, and PA1722) were QS regulated. Expression of type III secretion systems has been previously reported to be QS promoted in E. coli (35). Interestingly, expression of genes associated with the general secretion pathway and the type II pathway (e.g., xcp) was upregulated, and expression of genes involved in type III secretion (e.g., pcrH) was downregulated by QS. Likewise, gene products known to be secreted by the type II pathway (e.g., elastase [LasB] and the staphylolytic protease LasA) were also upregulated (38, 39), while the exoenzyme S synthesis protein B, which is involved in production of exoenzyme S, a known substrate for the type III secretion system, was downregulated (Table 4) (47). We also found that tatC, a gene involved in the twin-arginine translocation system that has only recently been implicated in virulence (23), was QS regulated.
TABLE 4.
Genes involved in virulence and genes differentially expressed in biofilms identified as being QS regulated by using microarraysa
| ORF or operon | Gene name | Protein description or function | Change (fold)b |
|---|---|---|---|
| Motility and attachment | |||
| PA0411 | pilJ | Twitching motility protein PilJ | −1.5 (ES) |
| PA0413 | Still frameshift probable component of chemotactic signal transduction system | −1.7 (B) | |
| PA2570c | palL | PA-I galactophilic lectin | 60 (ES) |
| PA4527 | pilC | Still framshift type 4 fimbrial biogenesis protein PilC | −1.9 (ES) |
| PA4651 | Probable pilus assembly protein | 8.8 (ES) | |
| PA5040-PA5043 | pilNOQ | Type 4 fimbrial biogenesis proteins | −1.4 (ES) |
| Cell wall synthesis, LPS and capsule | |||
| PA3337 | rfaD | ADP-l-glycero-d-mannoheptose 6-epimerase | −1.7 (ES) |
| PA4996 | rfaE | LPS biosynthesis protein RfaE | −1.8 (ML) |
| PA5161 | rmlB | dTDP-d-glucose 4,6-dehydratase | 1.5 (ES) |
| PA5163 | rmlA | Glucose-1-phosphate thymidylyltransferase | 1.6 (ES) |
| PA5450 | wzt | ABC subunit of A-band LPS efflux transporter | −1.6 (ES) |
| PA5451 | wzm | Membrane subunit of A-band LPS efflux transporter | 2.2 (ML) |
| Chemotaxis | |||
| PA0174-PA0179 | Probable chemotaxis cluster | 1.9 (ES) | |
| PA1458 | Probable two-component sensor | −1.3 (ES) | |
| PA1608 | Probable chemotaxis transducer | −2.2 (ES) | |
| PA2573 | Probable chemotaxis transducer | 2.7 (ES) | |
| PA2867 | Probable chemotaxis transducer | −1.6 (ES) | |
| PA4307 | pctC | Probable chemotaxis transducer | −2.4 (ML) |
| PA4309 | pctA | Probable chemotaxis transducer | −1.6 (ES) |
| PA4520 | Probable chemotaxis transducer | 2.1 (ML), −1.5 (ES) | |
| Protein secretion export apparatus, and secreted factors | |||
| PA1901-PA1906 | Probable phenazine biosynthesis cluster | 16 (ES) | |
| PA4209, PA4211, PA4217 | Probable phenazine biosynthesis cluster | 14 (B), 74 (B), 19 (ES) | |
| PA1250 | aprI | Alkaline proteinase inhibitor AprI | 12 (B) |
| PA1249c | aprA | Alkaline metalloproteinase precursor | 6.9 (B) |
| PA1246-PA1248 | aprDEF | Alkaline protease secretion proteins AprDEF | 6.3 (ML) |
| PA1245 | aprX | Membrane protein | 4.1 (B) |
| PA1871c | lasA | LasA protease precursor | 29 (ES) |
| PA2193-PA2195c | hcnABC | HCN biosynthesis genes | 210 (B) |
| PA2300c | chiC | Chitinase | 5.1 (ES) |
| PA3724c | lasB | Elastase LasB | 39 (B) |
| PA3478-PA3479c | rhlAB | Rhamnosyltransferase chain A, chain B | 100 (ES) |
| PA4142-PA4144 | Probable toxin secretion cluster | 9.6 (B) | |
| PA4302 | Probable type II secretion system protein | 9.6 (ES) | |
| PA4304 | Probable type II secretion system protein | 6.7 (ES) | |
| PA1877 | Probable secretion protein | 4.3 (ES) | |
| PA3096-PA3103c | xcpRSTUVWXY | General secretion pathway | 3.5 (ML) |
| PA3104-PA3105c | xcpPQ | General secretion pathway | 2.0 (ES) |
| PA1694 | pscQ | Translocation protein in type III secretion | −1.7 (ML) |
| PA1722 | pscI | Type III secretion protein PscI | −1.8 (ML) |
| PA1712 | exsB | Exoenzyme S synthesis protein B | −2.4 (ES) |
| PA4559 | lspA | Prolipoprotein signal peptidase | −1.4 (ES) |
| PA5070 | tatC | Transport protein TatC | −1.5 (ES) |
| RND efflux transporters or drug efflux transporter | |||
| PA0158 | Probable RND efflux transporter | 2.5 (ML) | |
| PA0425-PA0426 | mexAB | RND multidrug efflux system | 1.9 (B) |
| PA4205-PA4208 | mexGHI-opmD | Probable RND efflux transporter system | 8.0 (ES) |
| PA5160 | Drug efflux transporter | 2.2 (ES) | |
| Transcriptional regulators and sigma factors | |||
| PA0764 | mucB | Negative regulator for alginate biosynthesis MucB | −1.6 (ES) |
| PA0762 | algU | Sigma factor AlgU | −1.5 (ES) |
| PA1003 | mvfR | Regulator of multiple virulence factors | 3.0 (B) |
| PA1707 | pcrH | Regulatory protein PcrH | −2.2 (B) |
| PA1898c | qscR | Quorum-sensing control repressor | 3.0 (ML) |
| PA2259 | ptxS | Transcriptional regulator PtxS | −3.6 (ML) |
| PA3477c | rhlR | Regulatory protein RhlR | 5.9 (ES) |
| Genes expressed in biofilms | |||
| PA0105-PA0108 | Cytochrome c oxidases | 5.7 (ES) | |
| PA0713 | Hypothetical protein | −2.4 (ES) | |
| PA0714 | Hypothetical protein | −3.4 (ES) | |
| PA0722 | Hypothetical protein of bacteriophage Pf1 | −1.2 (ES) | |
| PA0971 | tolA | TolA protein | −1.4 (ES) |
| PA3038 | Probable porin | −2.3 (ES) | |
| PA3234 | Probable sodium:solute symporter | −2.8 (ES) | |
| PA3235 | Conserved hypothetical protein | −2.8 (ES) | |
| PA3418 | ldh | Leucine dehydrogenase | 2.0 (ES) |
| PA3622c | rpoS | Stationary-phase sigma protein RpoS | 2.7 (B) |
| PA4296 | Probable two-component response regulator | 3.4 (ES) | |
| PA4765 | omlA | Outer membrane lipoprotein OmlA precursor | −1.6 (ES) |
Genes are identified by ORF designation, gene name, and protein description (http://www.pseudomonas.com). LPS, lipopolysaccharide; RND, resistance-nodulation-cell division.
The magnitude and direction of gene expression change, as well as the growth phase (ML,mid-logarithmic phase; ES, early stationary phase; B, both mid-logarithmic and early stationary phases), are indicated for PAO-JP2 grown with exogenous autoinducers (1 μM 3O-C12-HSL and 2 μM C4-HSL) compared to PAO-JP2 cultures grown aerobically in modified FAB.
Intrinsic antibiotic and biocide resistance of P. aeruginosa is associated with the ability of this organism to persist as an infectious agent. We found that genes involved in three resistance-nodulation-cell division efflux systems (PA0158, PA0425 and PA0426 [mexAB], and PA4205 to PA4208 [mexGHI-opmD]) were upregulated by QS (Table 4). The MexAB-OprM efflux pump is constitutively expressed in PAO1 and has been correlated with increased antibiotic resistance (12). MexAB-OprM is also required for active efflux of 3O-C12-HSL from the cell (15). P. aeruginosa readily forms biofilms, both in vitro and in vivo (6). Even though in this study planktonic cultures were used exclusively, 15 of 73 genes previously demonstrated to be differentially expressed in PAO1 biofilms (42) were identified as being QS regulated in our analysis (Table 4). This is not surprising since it has been reported that QS plays a role in the formation of PAO1 biofilms (7). Interestingly, previous proteomic analysis of biofilm development suggested that there are distinct phases of biofilm initiation and maturation, and cells that are ready to disperse from a biofilm tend to have proteomic profiles similar to those of a planktonic state (34). In the present study, the expression profiles for genes identified as biofilm-regulated genes and the expression profiles for genes that are QS regulated were generally inversely correlated (11 of 15 genes). For example, genes that were upregulated in biofilms were downregulated by QS (PA0713, PA0714, PA0722, PA0971, and PA4765). The converse was also true (PA0105, PA0107, PA0108, PA3418, PA3622, and PA4296). Interestingly some genes identified as being QS and biofilm regulated are primarily involved in resistance to anionic detergents and/or antibiotics (e.g., low expression of tolA [31] and low expression of omlA [24]) or in the production and secretion of virulence factors (e.g., high expression of exsB [47]). Intriguingly, this suggests that QS is decreased during biofilm maturation, which agrees with the results of previously reported biofilm studies that showed that lasI expression and rhlI expression decrease after 4 to 6 days of biofilm development (9).
Effect of culture conditions on QS-regulated gene expression.
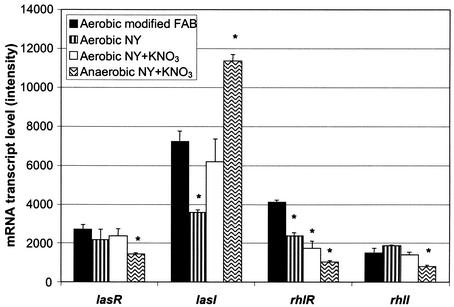
The few discrepancies between the present study and previous studies of QS-regulated genes could be due to the growth media and culture conditions employed in the present study. Therefore, we investigated the influence of media and culture conditions on expression of QS-regulated genes. PAO1 was grown aerobically in modified FAB, NY, or NY containing KNO3 and anaerobically in NY containing KNO3, and the growth curves are available online (http://www.urmc.rochester.edu/smd/mbi/bhi/). The logarithmic growth rates of PAO1 grown aerobically in the three media were equivalent (doubling times, 59 ± 0.1 min) but were ∼40% lower than the growth rates obtained when the strain was grown anaerobically in NY containing KNO3 (doubling time, 92 ± 4.7 min). Mid-logarithmic- and early-stationary-phase samples were obtained by using the criteria described above. Early-stationary-phase growth occurred at an OD660 of 1.0 for PAO1 grown in modified FAB or in NY and at an OD660 of 1.4 for PAO1 cultivated aerobically or anaerobically in NY containing KNO3. Samples from three individual cultures of PAO1 grown aerobically in modified FAB, NY, or NY containing KNO3 expressed averages of 66, 86, and 77% of the ORFs, respectively, at the mid-logarithmic and early stationary phases. PAO1 grown anaerobically in NY containing KNO3 expressed an average of 68% of the total ORFs at the mid-logarithmic and early stationary phases. These values are similar to those obtained in this study with PAO-JP2 cultures, as well as to those previously reported for PAO1 grown in the presence of low or high iron concentrations (25). The presence of QS-regulated gene transcripts was determined at the early stationary phase in PAO1 when it was grown either aerobically in modified FAB, NY, or NY containing KNO3 or anaerobically in NY containing KNO3 (Table 5). As expected, all 616 genes found to be QS regulated by using the PAO-JP2 mutant grown in modified FAB were expressed in PAO1 grown aerobically in modified FAB (data not shown). However, growth in a rich medium (NY) eliminated the expression of 24 QS-regulated genes (4% of the QS-regulated genes). Aerobic growth of PAO1 in NY containing KNO3 reduced the number of detectable QS-regulated gene transcripts by 11% (65 of 616 genes), and anaerobic growth in NY containing KNO3 eliminated detectable transcripts for 18% of the QS-regulated genes identified by using the microarrays (110 of 616 genes). These observations suggest that there may be more than 616 QS-regulated genes that could be identified by using different experimental conditions. Likewise, other studies in which different growth conditions are used may not identify all of the genes that we found to be QS regulated using modified FAB. For example, medium components (KNO3 or glucose) that specifically induce expression of certain genes, such as the nar genes and the glucose-inducible oprB gene, preclude detection of these QS-regulated genes in studies in which these constituents are absent from the medium (33, 49). Interestingly, we observed that three of the previously reported QS-regulated genes for which we did not detect transcripts (PA2401, PA2402, and PA2424 [44]) in modified FAB supplemented with nitrate were detected in medium without nitrate (NY). This most likely explains why we failed to detect these genes as QS-regulated genes in our experiments, since in our study QS-regulated genes were identified exclusively in modified FAB supplemented with nitrate.
TABLE 5.
QS-regulated transcripts detected at the early stationary phase in PAO1 grown under various nutrient and oxygen conditionsa
| ORF or intergenic region | Gene name | Presence inb:
|
Protein descriptionc | ||
|---|---|---|---|---|---|
| NY, aerobic | NY + KNO3 aerobic | NY + KNO3, anaerobic | |||
| PA0007 | + | − | − | Hypothetical protein | |
| PA0040 | + | − | + | Conserved hypothetical protein | |
| PA0103 | + | − | − | Probable sulfate transporter | |
| PA0105 | coxB | + | + | − | Cytochrome c oxidase subunit II |
| PA0107 | − | − | − | Conserved hypothetical protein | |
| PA0108 | coxIII | + | − | − | Cytochrome c oxidase subunit III |
| PA0111 | + | − | − | Hypothetical protein | |
| PA0144 | − | − | − | Hypothetical protein | |
| PA0196 | pntB | + | − | + | Pyridine nucleotide transhydrogenase beta subunit |
| PA0228 | pcaF | − | − | − | Beta-ketoadipyl-coenzyme A thiolase PcaF |
| PA0239 | + | − | − | Hypothetical protein | |
| PA0244 | + | − | − | Hypothetical protein | |
| PA0365 | + | + | − | Hypothetical protein | |
| PA0524 | norB | − | + | + | Nitric oxide reductase subunit B |
| PA0996 | + | + | − | Probable coenzyme A ligase | |
| PA0997 | + | + | − | Hypothetical protein | |
| PA0998 | + | + | − | Hypothetical protein | |
| PA1063 | + | − | + | Hypothetical protein | |
| PA1212 | + | − | − | Probable MFS transporter | |
| PA1214 | + | − | − | Hypothetical protein | |
| PA1217 | + | − | − | Probable 2-isopropylmalate synthase | |
| PA1221 | + | − | + | Hypothetical protein | |
| PA1318 | cyoB | + | − | − | Cytochrome o ubiquinol oxidase subunit 1 |
| PA1327 | + | + | − | Probable protease | |
| PA1356 | − | − | − | Hypothetical protein | |
| PA1608 | + | + | − | Probable chemotaxis transducer | |
| PA1679 | − | + | + | Hypothetical protein | |
| PA1694 | pscQ | + | − | − | Translocation protein in type III secretion |
| PA1771 | + | − | − | Probable esterase/lipase | |
| PA1869 | + | + | − | Probable acyl carrier protein | |
| PA1875 | + | − | − | Hypothetical protein | |
| PA1877 | − | − | − | Probable secretion protein | |
| PA1892 | + | + | − | Hypothetical protein | |
| PA1893 | + | − | − | Hypothetical protein | |
| PA1894 | + | + | − | Hypothetical protein | |
| PA1895 | + | + | − | Hypothetical protein | |
| PA1896 | − | − | − | Hypothetical protein | |
| PA1897 | + | + | − | Hypothetical protein | |
| PA1898 | + | + | − | Probable transcriptional regulator | |
| PA1901 | + | + | − | Phenazine biosynthesis protein PhzC | |
| PA1902 | + | − | − | Phenazine biosynthesis protein PhzD | |
| PA1903 | + | + | − | Phenazine biosynthesis protein PhzE | |
| PA1904 | + | + | − | Probable phenazine biosynthesis protein | |
| PA1927 | metE | − | − | + | 5-Methyltetrahydropteroyltriglutamate-homocysteine S-methyltransferase |
| PA1976 | + | − | − | Probable two-component sensor | |
| PA1978 | + | + | − | Probable transcriptional regulator | |
| PA1983 | exaB | − | − | − | Cytochrome c550 |
| PA1992 | − | − | − | Probable two-component sensor | |
| PA2068 | + | − | − | Probable MFS transporter | |
| PA2081 | + | − | − | Hypothetical protein | |
| PA2190 | + | + | − | Conserved hypothetical protein | |
| PA2194 | hcnB | + | + | − | Hydrogen cyanide synthase HcnB |
| PA2196 | + | + | − | Probable transcriptional regulator | |
| PA2238 | + | + | − | Hypothetical protein | |
| PA2260 | + | − | + | Hypothetical protein | |
| PA2261 | + | − | − | Probable 2-ketogluconate kinase | |
| PA2262 | + | − | − | Probable 2-ketogluconate transporter | |
| PA2266 | + | − | − | Probable cytochrome c precursor | |
| PA2300 | chiC | + | + | − | Chitinase |
| PA2303 | + | + | − | Hypothetical protein | |
| PA2331 | + | + | − | Hypothetical protein | |
| PA2369 | + | − | − | Hypothetical protein | |
| PA2414 | + | + | − | l-Sorbosone dehydrogenase | |
| PA2415 | + | + | − | Hypothetical protein | |
| PA2454 | + | − | + | Hypothetical protein | |
| PA2539 | + | − | − | Conserved hypothetical protein | |
| PA2540 | + | − | + | Conserved hypothetical protein | |
| PA2564 | + | − | − | Hypothetical protein | |
| PA2566 | + | − | − | Conserved hypothetical protein | |
| PA2570 | palL | + | + | − | PA-I galactophilic lectin |
| PA2571 | + | + | − | Probable two-component sensor | |
| PA2572 | + | + | − | Probable two-component response regulator | |
| PA2588 | + | + | − | Probable transcriptional regulator | |
| PA2699 | − | + | − | Hypothetical protein | |
| PA2763 | + | − | − | Hypothetical protein | |
| PA2862 | lipA | + | + | − | Lactonizing lipase precursor |
| PA2903 | cobJ | + | − | + | Precorrin-3 methylase CobJ |
| PA2939 | + | + | − | Probable aminopeptidase | |
| PA2974 | + | + | − | Probable hydrolase | |
| PA3242 | + | + | − | Probable lauroyl acyltransferase | |
| PA3311 | + | − | − | Conserved hypothetical protein | |
| PA3328 | + | + | − | Probable FAD-dependent monooxygenase | |
| PA3329 | + | + | − | Hypothetical protein | |
| PA3330 | + | + | − | Probable short-chain dehydrogenase | |
| PA3332 | + | + | − | Conserved hypothetical protein | |
| PA3333 | fabH2 | + | + | − | 3-Oxoacyl-(acyl carrier protein) synthase III |
| PA3334 | + | + | − | Probable acyl carrier protein | |
| PA3335 | + | + | − | Hypothetical protein | |
| PA3336 | + | + | − | Probable MFS transporter | |
| PA3391 | nosR | − | + | + | Regulatory protein NosR |
| PA3394 | nosF | − | + | + | NosF protein |
| PA3395 | nosY | − | + | + | NosY protein |
| PA3478 | rhlB | + | + | − | Rhamnosyltransferase chain B |
| PA3479 | rhlA | + | + | − | Rhamnosyltransferase chain A |
| PA3519 | + | + | − | Hypothetical protein | |
| PA3709 | − | − | − | Probable MFS transporter | |
| PA3719 | + | + | − | Hypothetical protein | |
| PA3870 | moaAI | − | + | + | Molybdopterin biosynthetic protein A1 |
| PA3877 | narKI | − | + | + | Nitrite extrusion protein 1 |
| PA3914 | moeAI | − | + | + | Molybdenum cofactor biosynthetic protein A1 |
| PA4078 | + | + | − | Probable nonribosomal peptide synthetase | |
| PA4142 | + | − | − | Probable secretion protein | |
| PA4175 | + | − | − | Probable endoproteinase Arg-C precursor | |
| PA4209 | + | + | − | Probable O-methyltransferase | |
| PA4211 | + | + | − | Probable phenazine biosynthesis protein | |
| PA4293 | + | − | − | Probable two-component sensor | |
| PA4297 | − | − | − | Hypothetical protein | |
| PA4298 | + | + | − | Hypothetical protein | |
| PA4300 | + | − | + | Hypothetical protein | |
| PA4302 | − | − | − | Probable type II secretion system protein | |
| PA4303 | + | + | − | Hypothetical protein | |
| PA4304 | + | − | − | Probable type II secretion system protein | |
| PA4305 | + | − | − | Hypothetical protein | |
| PA4307 | pctC | + | − | − | Chemotactic transducer PctC |
| PA4384 | + | − | − | Hypothetical protein | |
| PA4498 | + | − | + | Probable metallopeptidase | |
| PA4648 | + | − | − | Hypothetical protein | |
| PA4649 | + | − | − | Hypothetical protein | |
| PA4651 | + | + | − | Probable pilus assembly chaperone | |
| PA4681 | + | − | + | Hypothetical protein | |
| PA4908 | + | + | − | Hypothetical protein | |
| PA4910 | + | + | − | Probable ATP-binding component of ABC transporter | |
| PA4911 | − | − | − | Probable permease of ABC branched-chain amino acid transporter | |
| PA4912 | + | − | − | Probable permease of ABC branched-chain amino acid transporter | |
| PA4979 | − | − | − | Probable acyl-coenzyme A dehydrogenase | |
| PA5059 | + | + | − | Probable transcriptional regulator | |
| PA5097 | − | − | − | Probable amino acid permease | |
| PA5181 | + | − | + | Probable oxidoreductase | |
| PA5375 | betTI | + | − | − | Choline transporter BetT |
| PA5411 | + | − | + | Probable ferredoxin | |
| PA5543 | + | − | + | Hypothetical protein | |
| ig 1427453-1428080 | + | − | − | Intergenic region between PA1372 and PA1373, 1427453-1428080, (+) strand | |
| ig 4713795-4713098 | + | + | − | Intergenic region between PA4280 and PA4281, 4713098-4713795, (−) strand | |
| ig 5820909-5820113 | + | − | + | Intergenic region between PA5321 and PA5322, 5820113-5820909, (−) strand | |
Genes are identified by ORF designation, gene name, and protein description (http://www.pseudomonas.com).
The presence of each transcript was analyzed by using MAS (version 5.0) and DMT (version 3.0) software for duplicate samples. Transcripts identified as absent on at least two of three chips were considered absent. +, present; −, absent.
FAD, flavin adenine dinucleotide.
Variable responses in the expression of QS-regulated genes to media and environments may be due to alterations in the expression of QS regulators themselves. Thus, we examined the absolute mRNA transcript abundance of lasR, lasI, rhlR, and rhlI in PAO1 under different growth conditions (Fig. 4). The levels of mRNA transcript for lasR at the early stationary phase in modified FAB did not differ significantly from the levels observed during aerobic cultivation in NY or NY containing KNO3, but the level was significantly decreased (46%) when PAO1 was grown anaerobically in NY containing KNO3 (Fig. 4). Of the four regulatory genes, rhlR was the most sensitive to media and oxygen conditions. The level of the rhlR transcript in PAO1 grown to the early stationary phase decreased by 42% when the organism was grown in NY aerobically, by 57% when it was cultivated aerobically in NY containing KNO3, and by 75% when it was grown in anaerobic cultures in NY containing KNO3 compared to the levels in samples obtained at the early stationary phase from cells cultivated aerobically in modified FAB. The large decrease (75%) in rhlR transcript abundance in anaerobic cultures may account for the absence of several transcripts whose genes are known to be primarily rhl regulated, such as rhlAB and pa1L (28, 46). No significant difference in lasI transcript level was observed between PAO1 grown aerobically in NY containing KNO3 and PAO1 grown in modified FAB. However, growth in rich medium (NY) decreased the expression of the lasI transcript by 50%, while anaerobic growth in NY containing KNO3 increased the level of the lasI transcript by 57% compared to the level after growth in modified FAB. Interestingly, a statistically significant decrease in expression of PA1898 (qscR) (P < 0.01, as determined by one-way analysis of variance with the Tukey post-hoc test) (data not shown), a purported negative regulator of lasI (5), was also observed at the early stationary phase for PAO1 cultivated anaerobically in NY containing KNO3 compared with the expression in PAO1 grown aerobically in modified FAB, NY, and NY containing KNO3. QscR is believed to inhibit a premature increase in 3O-C12-HSL production, thereby preventing activation of the QS modulon at low cell densities. The decrease in qscR expression may have caused the concomitant increase in lasI expression observed in PAO1 at the early stationary phase when the organism was grown anaerobically.
FIG. 4.
Effect of media and oxygen conditions on lasR, lasI, rhlR, and rhlI expression. The levels of transcript expression for mRNA encoding the known key elements of the las and rhl regulatory system are shown for PAO1 grown to the early stationary phase. Statistically significant differences between the transcript levels for PAO1 grown in modified FAB and the transcript levels for PAO1 grown under other conditions are indicated by an asterisk (P ≤ 0.05, as determined by one-way analysis of variance with the Tukey post-hoc test).
Conclusions.
The P. aeruginosa DNA microarrays permitted a glimpse into gene regulation mediated by the las and/or rhl QS modulons. The large percentage of the genome (>10%) identified in this study as being QS regulated reflects the global level at which QS influences cellular behavior. Many novel activated genes (394 genes), as well as 222 repressed genes, were discovered. To our knowledge, no QS-repressed genes have been reported previously for P. aeruginosa. The high correlation between the QS-regulated genes identified in this study and those previously identified by alternate approaches provided a measure of confidence for both the P. aeruginosa microarray and data analysis methods and demonstrated the potential of this technology for investigating other regulons or modulons in P. aeruginosa. This conclusion was further supported by independent validation with QRT-PCR. Numerous novel genes identified as being QS regulated are themselves known or putative transcriptional regulators, and we are currently investigating their position in the QS hierarchy. The important effect of the bacterial environment on expression and detection of QS-regulated genes was highlighted by the study of PAO1 cultivated aerobically and anaerobically in various media, and the results underscore the importance of such variables when results from different laboratories are compared. This effect may account for conflicting reports concerning QS regulation of genes, such as rpoS, or concerning group behavior, such as biofilm formation (20). The variation in expression of QS-regulated genes dependent upon culture conditions emphasizes the need to clearly understand and mimic the environment that P. aeruginosa encounters in vivo when therapies are designed. The large number of QS-regulated genes that encode known or probable virulence genes identified in this study further emphasizes the role of QS in P. aeruginosa pathogenesis.
Acknowledgments
We thank K. Miller and K. Wahowski at the Microarray Core Facility in The Functional Genomics Center for superb technical support, H. Kelkar for technical assistance, J. Robinson, D. Frank, and J. Lam for helpful discussions, and J. Lamb and M. Filiatrault for critical evaluation of the manuscript.
We gratefully acknowledge Cystic Fibrosis Foundation Therapeutics, Inc. for subsidizing the P. aeruginosa Affymetrix GeneChip arrays. This work was supported by grant IGLEWS00V0 from Cystic Fibrosis Foundation Therapeutics and by grant NIHR37AI37713 from the National Institutes of Health.
Footnotes
For a commentary on this article, see page 2061 in this issue.
REFERENCES
- 1.Aendekerk, S., B. Ghysels, P. Cornelis, and C. Baysse. 2002. Characterization of a new efflux pump, MexGHI-OpmD, from Pseudomonas aeruginosa that confers resistance to vanadium. Microbiology 148:2371-2381. [DOI] [PubMed] [Google Scholar]
- 2.Beatson, S. A., C. B. Whitchurch, J. L. Sargent, R. C. Levesque, and J. S. Mattick. 2002. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 184:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beatson, S. A., C. B. Whitchurch, A. B. Semmler, and J. S. Mattick. 2002. Quorum sensing is not required for twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 184:3598-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapon-Herve, V., M. Akrim, A. Latifi, P. Williams, A. Lazdunski, and M. Bally. 1997. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol. Microbiol. 24:1169-1178. [DOI] [PubMed] [Google Scholar]
- 5.Chugani, S. A., M. Whiteley, K. M. Lee, D. D'Argenio, C. Manoil, and E. P. Greenberg. 2001. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 98:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton, J., P. Stewart, and E. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 8.de Kievit, T., P. Seed, J. Nezezon, L. Passador, and B. Iglewski. 1999. RsaL, a novel repressor of virulence gene expression in Pseudomonas aeruginosa. J. Bacteriol. 181:2175-2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Kievit, T. R., Y. Kakai, J. K. Register, E. C. Pesci, and B. H. Iglewski. 2002. Role of the Pseudomonas aeruginosa las and rhl quorum-sensing systems in rhlI regulation. FEMS Microbiol. Lett. 212:101-106. [DOI] [PubMed] [Google Scholar]
- 12.de Kievit, T. R., M. D. Parkins, R. J. Gillis, R. Srikumar, H. Ceri, K. Poole, B. H. Iglewski, and D. G. Storey. 2001. Multidrug efflux pumps: expression patterns and contribution to antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 45:1761-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeLisa, M. P., C. F. Wu, L. Wang, J. J. Valdes, and W. E. Bentley. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egland, K. A., and E. P. Greenberg. 1999. Quorum sensing in Vibrio fischeri: elements of the luxl promoter. Mol. Microbiol. 31:1197-1204. [DOI] [PubMed] [Google Scholar]
- 15.Evans, K., L. Passador, R. Srikumar, E. Tsang, J. Nezezon, and K. Poole. 1998. Influence of the MexAB-OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 180:5443-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank, D. W., D. G. Storey, M. S. Hindahl, and B. H. Iglewski. 1989. Differential regulation by iron of regA and toxA transcript accumulation in Pseudomonas aeruginosa. J. Bacteriol. 171:5304-5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallagher, L. A., S. L. McKnight, M. S. Kuznetsova, E. C. Pesci, and C. Manoil. 2002. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J. Bacteriol. 184:6472-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gambello, M., S. Kaye, and B. Iglewski. 1993. LasR of Pseudomonas aeruginosa is a transcriptional activator of the alkaline protease gene (apr) and an enhancer of exotoxin A expression. Infect. Immun. 61:1180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heydorn, A., B. Ersboll, J. Kato, M. Hentzer, M. R. Parsek, T. Tolker-Nielsen, M. Givskov, and S. Molin. 2002. Statistical analysis of Pseudomonas aeruginosa biofilm development: impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl. Environ. Microbiol. 68:2008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kline, T., J. Bowman, B. H. Iglewski, T. de Kievit, Y. Kakai, and L. Passador. 1999. Novel synthetic analogs of the Pseudomonas autoinducer. Bioorg. Med. Chem. Lett. 9:3447-3452. [DOI] [PubMed] [Google Scholar]
- 22.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchal quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 23.Ochsner, U. A., A. Snyder, A. I. Vasil, and M. L. Vasil. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA 99:8312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochsner, U. A., A. I. Vasil, Z. Johnson, and M. L. Vasil. 1999. Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J. Bacteriol. 181:1099-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochsner, U. A., P. J. Wilderman, A. I. Vasil, and M. L. Vasil. 2002. GeneChip® expression analysis of the iron starvation response in Pseudomonas aeruginosa: identification of novel pyoverdine biosynthesis genes. Mol. Microbiol 45:1277-1287. [DOI] [PubMed] [Google Scholar]
- 26.Ohman, D. E., S. J. Cryz, and B. H. Iglewski. 1980. Isolation and characterization of Pseudomonas aeruginosa PAO mutant that produces altered elastase. J. Bacteriol. 142:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson, J., E. Pesci, and B. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pesci, E., J. Pearson, P. Seed, and B. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pessi, G., and D. Haas. 2000. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. J. Bacteriol. 182:6940-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preston, M. J., P. C. Seed, D. S. Toder, B. H. Iglewski, D. E. Ohman, J. K. Gustin, J. B. Goldberg, and G. B. Pier. 1997. Contribution of proteases and LasR to the virulence of Pseudomonas aeruginosa during corneal infections. Infect. Immun. 65:3086-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rivera, M., R. E. Hancock, J. G. Sawyer, A. Haug, and E. J. McGroarty. 1988. Enhanced binding of polycationic antibiotics to lipopolysaccharide from an aminoglycoside-supersusceptible tolA mutant strain of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 32:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rumbaugh, K. P., J. A. Griswold, B. H. Iglewski, and A. N. Hamood. 1999. Contribution of quorum sensing to the virulence of Pseudomonas aeruginosa in burn wound infections. Infect. Immun. 67:5854-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saravolac, E. G., N. F. Taylor, R. Benz, and R. E. Hancock. 1991. Purification of glucose-inducible outer membrane protein OprB of Pseudomonas putida and reconstitution of glucose-specific pores. J. Bacteriol. 173:4970-4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauer, K., A. K. Camper, G. D. Ehrlich, J. W. Costerton, and D. G. Davies. 2002. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184:1140-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed]
- 35.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 37.Tang, H. B., E. DiMango, R. Bryan, M. Gambello, B. H. Iglewski, J. B. Goldberg, and A. Prince. 1996. Contribution of specific Pseudomonas aeruginosa virulence factors to pathogenesis of pneumonia in a neonatal mouse model of infection. Infect. Immun. 64:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Toder, D., S. Ferrell, J. Nezezon, L. Rust, and B. Iglewski. 1994. lasA and lasB genes of Pseudomonas aeruginosa: analysis of transcription and gene product activity. Infect. Immun. 62:1320-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toder, D., M. Gambello, and B. Iglewski. 1991. Pseudomonas aeruginosa lasA, a second elastase gene under transcriptional control of lasR. Mol. Microbiol. 5:2003-2010. [DOI] [PubMed] [Google Scholar]
- 40.van Delden, C., R. Comte, and A. M. Bally. 2001. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J. Bacteriol. 183:5376-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitehead, N. A., A. M. Barnard, H. Slater, N. J. Simpson, and G. P. Salmond. 2001. Quorum-sensing in Gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 42.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 43.Whiteley, M., and E. P. Greenberg. 2001. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J. Bacteriol. 183:5529-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winzer, K., C. Falconer, N. C. Garber, S. P. Diggle, M. Camara, and P. Williams. 2000. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J. Bacteriol. 182:6401-6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yahr, T. L., J. Goranson, and D. W. Frank. 1996. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol. Microbiol. 22:991-1003. [DOI] [PubMed] [Google Scholar]
- 48.Yuen, T., E. Wurmbach, R. L. Pfeffer, B. J. Ebersole, and S. C. Sealfon. 2002. Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res. 30:e48. [DOI] [PMC free article] [PubMed]
- 49.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]