Abstract
A DNA fragment carrying the genes coding for a novel EcoT38I restriction endonuclease (R.EcoT38I) and EcoT38I methyltransferase (M.EcoT38I), which recognize G(A/G)GC(C/T)C, was cloned from the chromosomal DNA of Escherichia coli TH38. The endonuclease and methyltransferase genes were in a head-to-head orientation and were separated by a 330-nucleotide intergenic region. A third gene, the C.EcoT38I gene, was found in the intergenic region, partially overlapping the R.EcoT38I gene. The gene product, C.EcoT38I, acted as both a positive regulator of R.EcoT38I gene expression and a negative regulator of M.EcoT38I gene expression. M.EcoT38I purified from recombinant E. coli cells was shown to be a monomeric protein and to methylate the inner cytosines in the recognition sequence. R.EcoT38I was purified from E. coli HB101 expressing M.EcoT38I and formed a homodimer. The EcoT38I restriction (R)-modification (M) system (R-M system) was found to be inserted between the A and Q genes of defective bacteriophage P2, which was lysogenized in the chromosome at locI, one of the P2 phage attachment sites observed in both E. coli K-12 MG1655 and TH38 chromosomal DNAs. Ten strains of E. coli TH38 were examined for the presence of the EcoT38I R-M gene on the P2 prophage. Conventional PCR analysis and assaying of R activity demonstrated that all strains carried a single copy of the EcoT38I R-M gene and expressed R activity but that diversity of excision in the ogr, D, H, I, and J genes in the defective P2 prophage had arisen.
Restriction (R)-modification (M) systems (R-M systems) serve as bacterial immune systems that destroy foreign DNA entering a cell. Typically, type II R-M systems consist of two enzymes: an endonuclease that recognizes and cleaves a specific DNA sequence and a methyltransferase that modifies the same sequence to protect the host chromosome from cleavage. More than 3,000 restriction endonucleases and methyltransferases have been isolated from many species of bacteria and have been subjected to biochemical and genetic studies (30). E. coli strains produce a variety of restriction endonucleases, including those of type I and III R-M systems. Type I and III systems are located on the chromosome; in contrast, most of the structural genes of type II R-M systems are located on a plasmid (5, 20, 27, 38, 39), and quite a few are located on chromosomal DNA (16, 21). Escherichia coli TH38, isolated from a pig, produces a type II restriction endonuclease, R.EcoT38I, that recognizes and cleaves the nucleotide sequence 5′-G(A/G)GC(C/T) ↓C-3′ (23). The occurrence of Hsd (host specificity for DNA) plasmids in E. coli TH38 has been investigated; one large plasmid was isolated from E. coli TH38, but Hsd+ transformants were not detected by use of a plasmid (38). These findings strongly suggest the possibility of chromosomal localization of the EcoT38I R-M system. Restriction endonucleases that show the same specificity as R.EcoT38I, such as R.BanII (14), have been isolated from a variety of bacteria, but none of the gene structures has been determined yet.
The EcoO109I R-M system has been cloned from chromosomal DNA, and it was discovered that a P4 integrase-like gene is present in the region adjacent to the R-M system (16). Genes encoding proteins involved in DNA mobility, such as transposases, integrases, and invertases, are sometimes found in the vicinity of R-M systems located on chromosomal DNA (1, 7, 12, 15, 21, 33). These genes might facilitate the transfer of R-M genes among different bacterial strains, one of the indices of the chromosomal location of R-M systems. In addition to the integrase gene, the complete P4 phage genome, except for the cII, β, and gop genes, and the leuX gene from E. coli K-12 chromosomal DNA were found in the flanking region; this is the first evidence of the horizontal transfer of a type II R-M system to E. coli chromosomal DNA (16).
In addition to these genes involved in DNA mobility, a small open reading frame (ORF) (C) that is known to regulate the expression of the R and/or M genes has been found next to some R-M genes (31, 35). For the EcoO109I (16) and PvuII (34) R-M systems, it has been shown that the C and R genes form an operon and that each product of the C gene binds to a specific site upstream of its translational start site and triggers the expression of each C-R operon. Both R-M systems have been found to be transferred to cells on a mobile genetic element: the PvuII system on a plasmid (24, 27) and the EcoO109I system on the P4 phage (16). As C proteins are assumed to generate a timing delay, allowing a methyltransferase to appear before a cognate restriction endonuclease in new host cells, the C gene is closely associated with the mobility of the R-M system.
In this article, we report the cloning and characterization of the EcoT38I R-M system and indicate the location of the system. The nucleotide sequence adjacent to the R-M system leads to horizontal transfer of the EcoT38I R-M system through P2 phage integration and indicates the diversity of imprecise P2 excision events.
MATERIALS AND METHODS
Bacterial strains, phages, and plasmids.
The E. coli strains used in this study were TH38 (23), HB101 (6), and JM109 (37). E. coli cultures were incubated at 37°C in Luria-Bertani medium containing 1% Bacto Tryptone (Difco Laboratories, Detroit, Mich.), 0.5% Bacto Yeast Extract (Difco), and 1% NaCl (pH 7.0). When needed, ampicillin (100 μg/ml) and chloramphenicol (25 μg/ml) were added to the cultures.
Enzymes and chemicals.
Restriction enzymes and λ DNA were purchased from Takara Shuzo Co., Ltd. (Kyoto, Japan); Toyobo (Osaka, Japan); and Nippon Gene (Toyama, Japan).
Assays of enzyme activities.
R.EcoT38I activity was assayed by adding 2 μl of enzyme solution to a 15-μl reaction mixture containing 10 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 1 mM dithiothreitol, 50 mM NaCl, and 0.5 μg of λ DNA. Incubation was performed at 37°C for 1 h. Restriction fragments were separated by electrophoresis on a 1% agarose gel. M.EcoT38I activity was assayed by adding 2 μl of enzyme solution to a 20-μl reaction mixture containing 10 mM Tris-HCl (pH 8.0), 10 mM 2-mercaptoethanol, 10 mM EDTA, 80 μM S-adenosyl-l-methionine, and 0.5 μg of λ DNA. After incubation at 37°C for 1 h, the reaction was terminated by phenol-chloroform extraction; the DNA fragments then were precipitated with ethanol. Following the addition of 15 μl of 10 mM Tris-HCl (pH 7.5)-10 mM MgCl2-1 mM dithiothreitol-50 mM NaCl-4 U of R.EcoT38I, the solution was incubated at 37°C for 1 h; aliquots then were analyzed by gel electrophoresis. Modification was judged to have occurred from the lack of digestion. M.EcoT38I activity in vivo was assayed as the susceptibility to R.EcoT38I of plasmid DNA prepared from the cells being tested.
Determination of the N-terminal amino acid sequence of R.EcoT38I.
To determine the N-terminal amino acid sequence of R.EcoT38I, wild-type R.EcoT38I was purified from E. coli TH38 cells by chromatography on DEAE-Sephacel (Pharmacia), heparin-Sepharose (Pharmacia), HiTrap Q (Pharmacia), and HiTrap heparin (Pharmacia) columns. The active fractions were pooled, concentrated with Centricon (Amicon), and stored at −20°C. Enzymes at the final purification step were blotted from the sodium dodecyl sulfate (SDS)-polyacrylamide gel onto a polyvinylidene difluoride membrane (Millipore) (21). Sequential degradation of a protein of interest was performed with an ABI491 protein sequencer.
Selection of EcoT38I R-M clones.
On the basis of the amino acid sequence of R.EcoT38I, oligonucleotide N-ter [5′-GTIAA(C/T)CA(C/T)GA(A/G)CA(A/G)GCITA(C/T)AA(C/T)GTIAT-3′] was synthesized as a probe for Southern hybridization analysis of genomic DNA and screening of a library. Hybridization was performed at 37°C for 16 h with modified Church-Gilbert buffer (9), comprising 0.5 M phosphate buffer (pH 7.2), 7% SDS, 10 mM EDTA, 0.05 mg of denatured herring sperm DNA per ml, and 32P-labeled oligonucleotides. Washes were carried out at 50°C with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS and then with 2× SSC-0.1% SDS. Hybridization was carried out with a PCR product encoding a portion of the M.EcoT38I gene, which was labeled with a PCR digoxigenin probe synthesis kit (Boehringer Mannheim) by using two oligonucleotides (5′-GTCGACGTAAACGACTAGGTAC-3′ and 5′-CGTTCCTCAGAATAGGGAACG-3′) as primers. PCR was carried out according to the manual for the kit.
Amplification of DNA fragments by PCR.
Oligonucleotides were synthesized according to the nucleotide sequences of the target sites and used as PCR primers. PCR was carried out with an LA-PCR kit (Takara Shuzo Co.) as recommended by the manufacturer. Amplification of DNA fragments flanking the EcoT38I R-M system was performed by inverse PCR.
Nucleotide sequencing and analysis.
DNA strands were sequenced with a model 3700 DNA sequencer (Applied Biosystems). Oligonucleotide primers were then synthesized and used to walk along the DNA template. Amino acid sequences were compared with all of the sequences in the GenBank database by using the BLAST program.
Expression and purification of M.EcoT38I and R.EcoT38I in recombinant E. coli cells.
M.EcoT38I was purified from E. coli HB101 cells carrying pUCEV by chromatography on HiTrap Q and HiTrap heparin columns. The active fractions were pooled and stored at −20°C.
To express R.EcoT38I in recombinant E. coli cells, the M.EcoT38I gene was subcloned into pACYC184. The M.EcoT38I gene was digested with SmaI and SphI from pUCEV and then ligated into pACYC184 cleaved with EcoRV and SphI; the resulting plasmid, p184 M38, was transformed into E. coli HB101. The R.EcoT38I gene was amplified by PCR with two primers (5′-GAATTCTTACTAAAGGACACCTATGAAAG-3′ and 5′-CCCGGGAGTATTAATTTTTAAATATGG-3′) and then cloned into the EcoRI and HincII sites of pUC118; the resulting plasmid, pUCR38, was transformed into E. coli HB101 carrying p184 M38. R.EcoT38I was purified from recombinant E. coli HB101 cells carrying pUCR38 and p184 M38 by chromatography on HiTrap Q and HiTrap heparin columns.
Expression of C.EcoT38I and measurement of promoter activity.
pBSCEcoT38I-His6 was constructed as follows. Primers C38N (5′-TCTAGATTTTGGAATAAAACCAATGATAGG-3′) and C38C (5′-GGATCCTCAGTGGTGGTGGTGGTGGTGTGATTTACTAATACTTTCATAGGTGTC-3′), which was flanked by a BamHI site and which added a six-amino-acid (His6) tag to the C-terminal end of C.EcoT38I, were used to subclone the His6-tagged C.EcoT38I gene. The PCR-generated DNA fragment was digested with XbaI and BamHI, ligated into pBluescript II SK cleaved with XbaI and BamHI, and then transformed into E. coli JM109. Ni-nitrilotriacetic acid-alkaline phosphatase conjugates (Qiagen) were used for detection of the His6-tagged protein according to the manufacturer's instructions.
The DNA fragment to be assayed for promoter activity was amplified by PCR with selected primers and then ligated into vector pGEM-T (Promega). The DNA fragment was excised with ApaI and SpeI and then ligated into ApaI/SpeI-digested pMCLTerAR (17). The activity of NADPH-dependent aldehyde reductase (AR1) (EC 1.1.1.2) at 37°C was determined by measuring the rate of decrease in the absorbance at 340 nm as described previously (17).
Nucleotide sequence accession number.
The GenBank accession number for the DNA sequence of the gene encoding the EcoT38I R-M system is AF545861.
RESULTS
Purification of R.EcoT38I from E. coli TH38.
R.EcoT38I was purified to homogeneity from E. coli TH38, giving a major band of 40 kDa (data not shown). The band was blotted onto a polyvinylidene difluoride membrane and then subjected to N-terminal amino acid sequence analysis. The first 26 amino acids of R.EcoT38I obtained by Edman degradation were Met-Lys-Val-Leu-Val-Asn-His-Glu-Gln-Ala-Tyr-Asn-Val-Ile-Ile-Asn-Ala-Xxx-Asn-Asp-Ala-Lys-Lys-Ile-Thr-Asp (Xxx, notidentified).
Isolation of EcoT38I R-M genes.
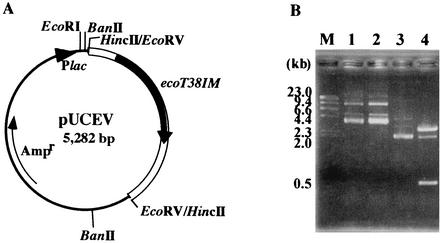
To isolate the R-M genes, oligonucleotide N-ter was synthesized from the N-terminal amino acid sequence of R.EcoT38I and used as a probe for Southern hybridization with E. coli TH38 chromosomal DNA digested with various restriction endonucleases. The 1.4-kb HincII fragment was cloned into the HincII site of the pUC118 vector to obtain pUC14. The nucleotide sequence of the 1.4-kb fragment indicated that of the three ORFs, one was identical to the sequence encoding the N-terminal region of R.EcoT38I and the others were located upstream of the R.EcoT38I gene, one on the same DNA strand and the other on the opposite DNA strand. As the deduced amino acid sequence of the ORF located on the opposite DNA strand exhibits significant identity with those of 5-methylcytosine methyltranferases, we assumed that the ORF encodes M.EcoT38I. In order to find large DNA fragments carrying the complete M.EcoT38I gene within E. coli TH38 chromosomal DNA, the 2.2-kb EcoRV fragment was cloned into the HincII site of pUC118 to obtain pUCEV, in which the M.EcoT38I gene was under the control of the lac promoter. The finding that pUCEV showed resistance to cleavage by R.EcoT38I indicated that the 2.2-kb region is essential for encoding of the methyltransferase.
Sequence analysis of the complete R-M system.
In order to obtain the complete nucleotide sequence of the EcoT38I R-M system, the 1.8-kb DNA fragment downstream of the R.EcoT38I gene was amplified by inverse PCR, and then its nucleotide sequence was analyzed. The DNA sequence of the 2,700-bp region that covers the entire EcoT38I R-M system in the 4,127-bp EcoRV/PstI fragment analyzed is shown in Fig. 1. The R.EcoT38I and M.EcoT38I genes were aligned head to head, and putative palindromic sequences that were seen downstream of each gene could be the transcriptional termination sites for individual genes.
FIG.1.
Nucleotide sequence of the 2,700-bp region. The amino acid sequences assigned to the C.EcoT38I (ecoT38IC) and R.EcoT38I (ecoT38IR) genes are given below the nucleotide sequences, and that assigned to the M.EcoT38I (ecoT38IM) gene is given above the nucleotide sequence. The nucleotide sequence is numbered from the leftmost end, and the deduced amino acid sequences of R.EcoT38I, M.EcoT38I, and C.EcoT38I are numbered from the initiation codon of each gene. The potential ribosome-binding sequence is indicated by dots. Pairs of arrows indicate palindromic sequences characteristic of the termination signal.
In the ORF assigned to the endonuclease gene, an ATG codon at nucleotide position 1,533 and a termination codon at nucleotide position 2,586 were found. In addition, an appropriate ribosome-binding sequence, AGGA, was present 9 bp upstream of the ATG codon. The ORF consisted of 1,053 bp and encoded a 351-residue polypeptide. The predicted mass, 38,898 Da, was close to the value estimated for the enzyme purified from E. coli TH38 by SDS-polyacrylamide gel electrophoresis (PAGE). The deduced amino acid sequence of R.EcoT38I exhibited 28% identity with that of R.SacI (36), which recognizes GAGCT↓C, one of the variants of the recognition sequence of R.EcoT38I.
In the ORF assigned to the methylase gene, an ATG codon at nucleotide position 1,157 and a termination codon at nucleotide position 113 were found. In addition, an appropriate ribosome-binding sequence, AGGA, was present 7 bp upstream of the ATG codon. The ORF consisted of 1,044 bp and encoded a 348-residue polypeptide. The predicted sequence of the M.EcoT38I protein included 10 sequence motifs characteristic of all prokaryotic 5-methylcytosine methyltransferases (29) but did not show significant similarity to that of M.SacI, as expected from the results obtained for the cognate restriction enzyme.
As shown in Fig. 1, a third ORF was discovered upstream of the R.EcoT38I gene. In the ORF, the ATG codon was located at nucleotide position 1,241 and the termination codon was located at nucleotide position 1,553, partially overlapping the R.EcoT38I gene. In addition, an appropriate ribosome-binding sequence, GGA, was present 12 bp upstream of the ATG codon. The ORF consisted of 312 bp and encoded a 104-residue polypeptide. As shown in Fig. 2, the central region (36 to 74 amino acid residues) of the deduced amino acid sequence exhibited significant similarity not only to those of members of the helix-turn-helix 3 and helix-turn-helix XRE (xenobiotic response element) families of DNA-binding proteins, including Cro and cI, but also to that of C.EcoRV, an activator protein for the R.EcoRV gene (25). The ORF therefore was designated the C.EcoT38I gene, which might produce a control protein for the EcoT38I R-M gene. Neither a conserved DNA sequence element termed a “C box,” which is found immediately upstream of some C genes and is at least one target of C.PvuII binding (31), nor a C.EcoO109I-binding sequence (17) was found upstream of the C.EcoT38I gene translational start site. The G+C contents of the R.EcoT38I, M.EcoT38I, and C.EcoT38I genes were 35, 42, and 38%, respectively. There was no significant similarity between the nucleotide and deduced amino acid sequences of R.EcoT38I and M.EcoT38I.
FIG. 2.
Alignment of C.EcoT38I and C.EcoRV. Each line represents one sequence. Shading indicates conserved amino acid residues. Gaps in the aligned sequences are indicated by dashes.
Expression of the R.EcoT38I and M.EcoT38I genes in recombinant E. coli and characterization of the gene products.
We inserted the R.EcoT38I gene under the control of the lac promoter, and the resulting plasmid, pUCR38, was transformed into E. coli HB101 cells carrying p184 M38, which expresses M.EcoT38I. To characterize R.EcoT38I, E. coli HB101 cells carrying p184 M38 and pUCR38 were cultured in the presence of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and then the enzyme was purified from the cells. The specific activity of R.EcoT38I in extracts of recombinant cells was 100 times higher than that in extracts of wild-type cells. The purified sample gave a single protein band corresponding to a molecular mass of 40 kDa. The molecular mass of the native enzyme, which was measured by HiPrep Sephacryl S-100 gel filtration, was estimated to be 73 kDa. Like other type II R enzymes, R.EcoT38I is a homodimeric protein. The sequence of the first five amino acids of the enzyme, Met-Lys-Val-Leu-Val, obtained by Edman degradation exactly corresponded to that of wild-type R.EcoT38I and that predicted from the nucleotide sequence. The restriction pattern obtained on digestion with recombinant R.EcoT38I was the same as that obtained with the wild-type enzyme.
To characterize M.EcoT38I, we purified the enzyme from E. coli HB101 cells carrying pUCEV. The purified sample gave a single protein band corresponding to a molecular mass of 41 kDa. The molecular mass of the native enzyme, which was measured by HiPrep Sephacryl S-100 gel filtration, was estimated to be 39 kDa; this value was consistent with the value determined by SDS-PAGE. Like other type II DNA methyltransferases, M.EcoT38I is a monomeric protein. The start of the methylase gene was confirmed by N-terminal amino acid analysis. The sequence of the first 10 amino acids of the enzyme, Met-Gln-Lys-Ile-Ser-Ala-Val-Ser-Leu-Phe, obtained by Edman degradation exactly corresponded to that predicted from the nucleotide sequence starting at a GTG codon appearing at nucleotide position 1,202 but not that starting from the ATG codon described above. The ORF consisted of 1,092 bp and encoded a 364-residue polypeptide. The predicted mass, 39,803 Da, was close to the value determined by SDS-PAGE. No appropriate ribosome-binding sequence was present upstream of the GTG codon.
In order to characterize the methylation specificity of M.EcoT38I, pUCEV, carrying the M.EcoT38I gene and two EcoT38I sites, GAGCTC and GGGCTC, was incubated with R.BanII, an isoschizomer of R.EcoT38I which can cut G(A/G)GC(C/T)m5C but not G(A/G)Gm5C(C/T)C (14, 26). Digestion of pUC118 with R.BanII gave 2.7- and 0.45-kb fragments (Fig. 3B, lane 4), in contrast, pUCEV was not digested with R.BanII (Fig. 3B, lane 2). The deduced amino acid sequence of M.EcoT38I contained all 10 motifs that are conserved in bacterial 5-methylcytosine methyltransferases. These results suggested that M.EcoT38I methylated the inner cytosines in the recognition sequence to give 5′-G(A/G)Gm5C(C/T)C-3′.
FIG. 3.
Analysis of the methylation specificity of M.EcoT38I (ecoT38IM). (A) Structure of pUCEV, in which the 2.2-kb EcoRV fragment carrying the M.EcoT38I gene was cloned into the HincII site of pUC118. (B) Resistance to R.BanII of M.EcoT38I-modified DNA. pUCEV and pUC118 were incubated with R.BanII. The products were separated by 1% agarose gel electrophoresis. Lane 1, pUCEV; lane 2, pUCEV plus R.BanII; lane 3, pUC118; lane 4, pUC118 plus R.BanII. λ HindΙΙΙ digests were used for molecular size calibration (lane M).
Function of C.EcoT38I.
To determine the C.EcoT38I responsiveness of EcoT38I promoters, we placed the His6-tagged C.EcoT38I gene under the control of the lac promoter of pBluescript II SK to supply C.EcoT38I in trans. The production of C.EcoT38I was examined by Western blot analysis with anti-His tag antibodies; a protein band corresponding to a molecular mass of 16 kDa was detected for E. coli JM109 carrying pBSCEcoT38I-His6 (data not shown). We cloned the putative promoter region upstream of the promoterless AR1 gene in plasmid pMCLTerAR. Fragments to be assayed for promoter activity were PCR amplified with selected oligonucleotide primers, and DNA products were inserted between the SpeI and ApaI sites of the promoter screening vector, pMCLTerAR, in both orientations. Cotransformants of E. coli JM109 carrying plasmids derived from pMCLTerAR and from pBluescript II SK or pBSCEcoT38I-His6, which generates C.EcoT38I, were grown in the presence of IPTG. Reporter gene assays then were carried out for bacterial cell extracts with 4-chloro-3-oxobutanoate ethyl ester as a substrate.
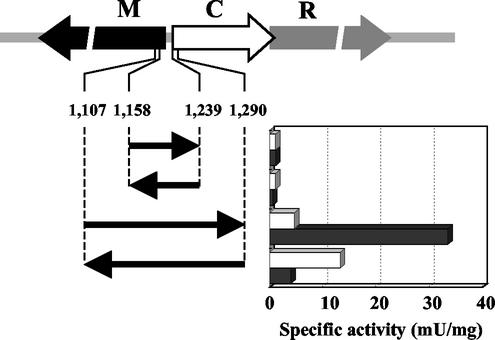
As shown in Fig. 4, the promoter activity of an 82-bp fragment including the intergenic region of the M.EcoT38I and C.EcoT38I genes was below the limit of detection and was not affected by C.EcoT38I in either orientation. The promoter activity of a 184-bp fragment was high in the absence of C.EcoT38I and increased to seven times that in the absence of C.EcoT38I when the fragment from bp 1107 to 1290 was inserted in the same orientation as the reporter gene. In contrast, when the 184-bp fragment was inserted in the opposite orientation, the promoter activity increased to three times that in the absence of C.EcoT38I and decreased in the presence of C.EcoT38I. These results suggested that C.EcoT38I acts as both a positive regulator of R.EcoT38I expression and a negative regulator of M.EcoT38I expression.
FIG. 4.
Effect of C.EcoT38I on the promoter activities of the R.EcoT38I and M.EcoT38I genes. The segments from 1,158 to 1,239 bp and from 1,107 to 1,290 bp shown in Fig. 1 were cloned upstream of a promoterless AR1 gene in vector pMCLTerAR. Promoter activities associated with various DNA fragments were measured for both orientations. Each clone was assayed for AR1 activity in the absence (open bars) and presence (closed bars) of C.EcoT38I (graph). R, M, and C represent R.EcoT38I, M.EcoT38I, and C.EcoT38I, respectively.
Nucleotide sequences flanking the R-M system.
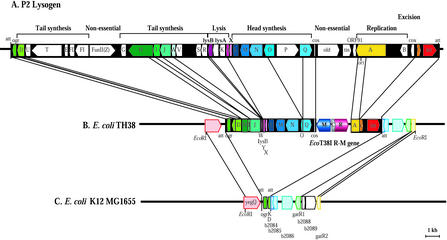
In the analyzed 4,127-bp region, one partial ORF was identified downstream of the R.EcoT38I gene. The product of this ORF showed significant homology to the N-terminal region encoded by the A gene from phage P2 (GenBank accession number AF063097). A P2 cos sequence, followed by a nucleotide sequence similar to that of the Q gene from phage P2, was also found downstream of the M.EcoT38I gene. These findings suggest that the EcoT38I R-M system was inserted between the genes from the P2 prophage integrated in E. coli chromosomal DNA. In order to characterize the gene arrangement of the P2 prophage on E. coli TH38 chromosomal DNA, we amplified the DNA fragments flanking the R-M system and determined the entire nucleotide sequence of the 16-kb EcoRI region. BLAST searches of the GenBank database revealed that DNA downstream of the R.EcoT38I gene exhibited a high level of similarity with the nucleotide sequences of the A, C, and int genes and the attL site from phage P2, followed by the sequence from b2084 through gatR1 of E. coli K-12 MG1655 (4). The DNA downstream of the M.EcoT38I gene exhibited a high level of similarity with the nucleotide sequences of the Q, O, N, M, L, X, Y, lysB, W, J, I, H, D, and ogr genes and the attR site from phage P2, followed by the yegQ sequence of E. coli K-12 MG1655. These results indicated that on E. coli TH38 chromosomal DNA, a P2 attachment site similar to locI found at 46.7 min on E. coli K-12 MG1655 was occupied by a defective P2 prophage carrying the EcoT38I R-M system. The Q, O, Y, lysB, W, J, I, D, and A genes seemed to be pseudogenes, as truncation at the 5′ and/or 3′ ends or a frameshift mutation was found in these genes. In contrast, other genes could code for proteins corresponding to phage P2. Gene duplications, rearrangements, inversions, or insertions have not been found in the region analyzed so far. The gene organization is summarized in Fig. 5.
FIG. 5.
Gene organization of the EcoT38I R-M system and flanking regions on the E. coli TH38 chromosome. (A) Gene order of the P2 phage integrated into the E. coli chromosome. (B) Gene order of the 16-kb EcoRI region flanking the EcoT38I R-M genes on the E. coli TH38 chromosome. The genes and their directions of transcription, cos, and phage attachment sites (att) are shown. R, M, and C represent R.EcoT38I, M.EcoT38I, and C.EcoT38I, respectively. (C) Gene order of P2 att (locI) and flanking regions in E. coli K-12 MG1655. att indicates att (locI).
Variability of the P2 prophage in E. coli TH38.
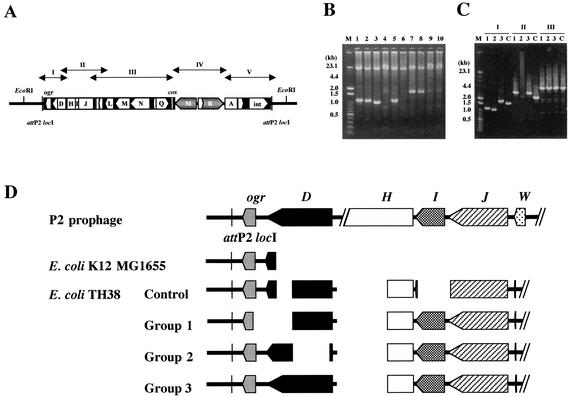
In order to determine whether or not E. coli TH38 has (an)other P2 prophage(s) at (an)other attB site(s), as in E. coli K-12 MG1655, we amplified possible attB sites by using PCR. Four pairs of primers were designed to amplify the locI, locII, locIII, and locH sites based on the nucleotide sequence of E. coli K-12 MG1655 (Table 1). The lengths of the DNA fragments amplified by using the locI and locIII primers from E. coli TH38 were the same as those obtained for E. coli K-12 MG1655. In contrast, a DNA fragment was not amplified from E. coli TH38 with the locH primers (data not shown). These findings indicated the presence of locII and locIII sites and the absence of a locH site or a mutation of the locH flanking region. With the locI primers, 0.7- and 12-kb fragments were amplified from E. coli K-12 MG1655 and E. coli TH38, respectively. These fragments were consistent with the sizes expected from the nucleotide sequences. In addition to the 12-kb fragment, DNA fragments of smaller sizes were amplified from E. coli TH38. A sample from a freezer vial was streaked, and 10 colonies were taken from the same streak. The numbers and sizes of these fragments differed from those of the colony from which the template DNA was prepared. We examined samples for the presence of R.EcoT38I activity and the P2 prophage carrying the EcoT38I R-M system. R.EcoT38I activity was detected in all colonies, and the 12-kb DNA fragment was amplified from all samples, but the patterns of DNA fragments of smaller sizes could be categorized into four groups: 1.7 kb, 1.1 kb, 1.0 kb, and none (Fig. 6B). We determined the partial nucleotide sequences of these fragments and found that each fragment carried the P2 att locI through D genes, but the nucleotide sequence was not identical to that of E. coli TH38 determined so far. These results suggested the variability of defective P2 prophages among E. coli TH38 strains. We synthesized additional PCR primers (Table 1 and Fig. 6A) and amplified DNA segments covering the defective P2 prophage integrated at locI. The lengths of the DNA fragments amplified with the E-F, G-H, and I-J primers were similar (data not shown), but those amplified with the A-B and C-D primers were different and could be categorized into four groups (Fig. 6C). These results suggested a difference in genetic organization from the ogr through L genes in the defective P2 prophage. DNA sequences were determined and compared to those of phage P2, E. coli K-12 MG1655, and the E. coli TH38 control strain analyzed above (Fig. 6D). The gene order and the orientation were identical to those found for the E. coli TH38 control strain. The region from 25 bp upstream of the D gene to 455 bp downstream of the initiation codon of the H gene was deleted at the same positions in all E. coli TH38 strains. In addition, the entire 5′ end of the W gene was truncated at the same position. However, the D gene showed diversity. The N-terminal part of the ogr gene product and the C-terminal part of the D gene product were deleted in group 1, the N-terminal half of the D gene product was deleted in group 2, and the ogr and D gene products were conserved in group 3. The 3′ end of the D gene was truncated at the same positions in the group 1 and control strains, but the 5′ end of the D gene was truncated at different positions in E. coli K-12 MG1655 and the control strain.
TABLE 1.
PCR primers used in this study
| Target DNA | Primer | Sequence (5′-3′) | Positionsa |
|---|---|---|---|
| P2 att locI | I61 | TGTACCAGGAACCACCTCCTTAGCCTGTGT | 1354-1383b |
| I62 | GAAGACGGTGACCAGGGATAGGGCTTATGC | 13398-13369b | |
| P2 att locII | II30 | GAACTGACACAACCGCTATTTATCGCCGCG | 7168-7197c |
| II31 | GTAGAGAATGAGCCACCAAACGCGAATCGC | 7256-7227c | |
| P2 att locIII | III28 | ACTGCCTTTATTCAACAACGGAAATTCCAG | 2814-2843d |
| III29 | TGGAACCAAAAACAAAAAAACAGCGTTCGC | 2902-2873d | |
| P2 att locH | H32 | TTTGTCGCGAAACAGAAACACTGTGTCAGG | 1167-1196e |
| H33 | CATGAGAATCAGACCATTCGCCGTTGCATC | 1261-1232e | |
| If | A | CAGGAACCACCTCCTTAGCC | 1359-1378b |
| B | GGGTATGACGGGGGCGGG | 2713-2696b | |
| IIf | C | GCATCAGCATGTAATCCGGCGTC | 2632-2654b |
| D | ACCGCGTCGCTTTATGAGCG | 4766-4747b | |
| IIIf | E | AATCAGTGTCGCCTTGCGTTC | 3957-3977b |
| F | CGGCCTTTCGACTTCACCATGTTTTCGCG | 7622-7594b | |
| IVf | G | CGCGAAAACATGGTGAAGTCG | 7594-7614b |
| H | GAGTGGTCGGCATTTACGCGC | 11414-11394b | |
| Vf | I | CAATCTCATAATTCATACGCTCTCC | 10865-10889b |
| J | GAAGACGGTGACCAGGGATAG | 13398-13378b |
FIG. 6.
Variability of defective P2 prophage genomes in E. coli TH38 DNA. (A) Locations of target DNAs for PCR amplification. (B) Agarose gel electrophoresis of PCR products obtained from E. coli TH38 strains with primers I61 and I62. The PCR mixture (50 μl) comprised 25 pmol of each primer, 400 μM each deoxynucleotide triphosphate, 2.5 mM MgCl2, LA-PCR buffer (Mg2+ free), template DNA, and 2.5 U of LA Taq DNA polymerase. The reaction mixture was overlaid with mineral oil, and the reaction was carried out with a Perkin-Elmer Cetus thermal cycler. The initial template denaturation step consisted of 1 min at 94°C. The amplification profile (20 s at 98°C and 25 min at 68°C) was repeated for 30 cycles. Lanes 1 to 10 correspond to products from 10 E. coli TH38 colonies picked randomly. A mixture of λ HindIII digests and 100-bp DNA ladder markers (Toyobo) was used for molecular size calibration (lane M). The PCR products were subjected to 1% agarose gel electrophoresis and then stained with ethidium bromide. (C) Agarose gel electrophoresis of E. coli DNA fragments amplified by PCR with primers A and B (I), C and D (II), and E and F (III). The PCR mixture (50 μl) comprised 10 pmol of each primer, 200 μM each deoxynucleotide triphosphate, 2.5 mM MgCl2, LA-PCR buffer (Mg2+ free), template DNA, and 2.5 U of LA Taq DNA polymerase. The reaction mixture was overlaid with mineral oil, and the reaction was carried out with a Perkin-Elmer Cetus thermal cycler. The initial template denaturation step consisted of 1 min at 94°C. The amplification profile (30 s at 94°C, 1 min at 60°C, and 5 min at 72°C) was repeated for 25 cycles. Lanes C correspond to the products from the E. coli TH38 control strain, for which the complete nucleotide sequence of the 16-kb EcoRI region was determined in this study. Lanes 1 to 3 correspond to products with different molecular sizes from the E. coli TH38 strains analyzed in panel B. (D) Schematic diagram of the defective P2 prophage genome in various E. coli TH38 strains. Groups 1 to 3 correspond to lanes 1 to 3 in panel C.
DISCUSSION
We analyzed the gene organization of the novel EcoT38I R-M system on the chromosomal DNA of E. coli TH38. Many R-M systems recognize the same DNA sequence as EcoT38I; however, this is the first report of the gene structure of an R-M system that recognizes [G(A/G)GC(C/T)C]. The 2.7-kb region encompassed all of the R-M genes, which were aligned in a head-to-head orientation. R.EcoT38I and M.EcoT38I could be expressed in E. coli HB101 and purified to homogeneity. The molecular masses of both proteins were consistent with those predicted from the nucleotide sequences. R.EcoT38I was a homodimeric protein; in contrast, M.EcoT38I was a monomeric protein. The results presented in this report suggest that M.EcoT38I methylated the inner cytosines in the recognition sequence to yield 5-methylcytosine.
Between the R and M genes, one small ORF (C.EcoT38I gene) was present upstream of the R.EcoT38I gene and partially overlapped that gene. The C.EcoT38I gene encodes a protein of 104 amino acids with a molecular mass of 12,014 Da, a size which is in good agreement with the predicted sizes of other C proteins that associate with several type II R-M systems and regulate the expression of R-M genes. In this study, we have shown that C.EcoT38I acts both as an activator of R.EcoT38I expression and as a repressor of M.EcoT38I expression. In both the EcoO109I and PvuII systems, it has been shown that the C protein binds to DNA upstream of its structural gene and triggers the transcription of its own gene and the cognate restriction endonuclease gene but has little effect on the transcription of the methyltransferase gene. In contrast, C.BamHI acts both as an activator of endonuclease expression and as a repressor of methyltransferase expression in E. coli (13). No significant homology of the deduced amino acid sequences of C.EcoT38I, C.EcoO109I, C.PvuII, and C.BamHI was found. Furthermore, no sequence similar to the binding sites determined for C.EcoO109I and C.PvuII was found upstream of C.EcoT38I. These results suggest that C.EcoT38I may bind to a novel site and regulate the transcription of the EcoT38I R-M genes. Clarification of the control mechanism awaits the purification of C.EcoT38I as well as in vitro DNA-binding assays. The C protein is assumed to generate a timing delay of R gene expression when the R-M system enters a new host. It is quite reasonable that a gene encoding a putative C protein was found in the EcoT38I R-M system, which was shown to be horizontally transferred by phage P2.
The structure of the DNA adjacent to the EcoT38I R-M system was analyzed in detail, and we found that almost 30% of the phage P2 DNA was arranged sequentially on both sides of the system. Furthermore, a DNA sequence identical to that of locI of the phage P2 attachment site followed by E. coli K-12 MG1655 chromosomal DNA was found in the adjacent region. P2 is a temperate phage that forms stable lysogens in several enterobacteria, including E. coli C and K-12. In the lysogenic stage, P2 has always been found at an integrated prophage. The integration occurs through site-specific recombination between a bacterial attachment site, attB, and the attachment site of P2, attP. The P2 prophage has been found at at least 10 sites on the E. coli genome. The preferred site varies with the strain used, but multiple lysogens are possible, with integration occurring at separate locations. In E. coli C, one site, locI, is preferred, being occupied before any of the others (3). This is also true of E. coli K-12, in which four attB sites, locI, locII, locIII, and locH, were found; however, locI was occupied by a cryptic remnant of P2 (Fig. 5C) (2). The core sequence of locI completely matched the 27-nucleotide core sequence of attP; the sequences of locII, locIII, and locH exhibited 20, 17, and 17 matches, respectively. Both attR and attL found in E. coli TH38 completely matched the core sequence of locI; in addition, nucleotides outside the core region of attR and attL were almost identical to those in E. coli K-12. We showed that E. coli TH38 carries additional P2 attB sites, locII and locIII, but not locH, although it was not determined whether these sites are at the same map positions as in E. coli K-12 MG1655. These results strongly support the proposal that a hybrid phage P2, in which the old, tin, ORF91, and partial A genes were substituted by R-M genes, infected E. coli TH38 cells that carried multiple P2 attB sites and integrated the DNA into chromosomal DNA at the locI site through site-directed recombination catalyzed by P2 integrase, followed by excision of some P2 genes. The G+C content of the genes in the EcoT38I R-M system was low, as in other R-M systems. It is quite interesting that the P2 genes old, tin, and ORF91, which were replaced by the EcoT38I R-M system, and nonessential genes for the lytic cycle, such as FunII(Z), which were deleted from the defective prophage on the E. coli TH38 genome, have low G+C contents. P2 replication is initiated by a strand-specific nick at ori, which is located within the A gene, and replication proceeds unidirectionally (8). The position of the nick site, position 29,892 in the P2 complete sequence (GenBank accession number AF063097), was found in the A gene of the defective P2 prophage and was located about 450 nucleotides downstream of the junction of phage P2 and the R-M system. These results support the possibility that the single-stranded tail (corresponding to the partial A gene, ORF91, tin, old, and other genes) produced by gene A products promotes recombination between phage P2 genes and the EcoT38I R-M system.
In addition to E. coli TH38, it has been shown that E. coli K-12 derivatives, such as MG1655, C600r−m+, K-207, LG102, and the W3110 strain Kohara clone (19), contain a 639-nucleotide cryptic remnant of P2 at a site with a sequence similar to that of locI (2). The P2 remnant consists of the C-terminal part of the D gene, the complete ogr gene, and attR. This finding suggests that an ancestor of E. coli K-12 was lysogenized by phage P2 and that an imprecise excision event that removed most of the phage DNA then occurred, leaving only part of the D gene, the complete ogr gene, and attR on the chromosome. We found four types of variants in this region of the E. coli TH38 chromosome. The region from ogr through the W gene in the P2 prophage appeared to be a hot spot for the excision event in E. coli TH38. With respect to the region from ogr to the J gene, the genomic DNA of the control strain suffered from at least one more genetic recombination event than that of the other E. coli TH38 strains. Further investigation may provide additional insight into the connection between the presence of the R-M system and the stability of the defective prophage in E. coli TH38 strains. The P2 int transcript starts at the Pc promoter, located upstream of the C repressor gene (2). The −35 and −10 sequences of the Pc promoter and the amino acid sequence deduced from the sequence of the int gene on E. coli TH38 chromosomal DNA were completely conserved. These results suggested that P2 integrase was expressed in E. coli TH38.
In this report, we have provided the first evidence of horizontal transfer of the R-M system by phage P2. In an earlier study, Kita et al. provided evidence that the EcoO109I R-M system was inserted between the genes from the P4 prophage on E. coli H709c chromosomal DNA (16). In addition to the P4 phage, other prophages are also known to carry type II R-M systems. For example, the HindIII R-M system was found on a cryptic prophage, φflu, in Haemophilus influenzae Rd (10); the BsuMI R-M system was found in the prophage 3 region in Bacillus subtilis Marburg 168 (28); and the Sau42I R-M system was found on the φ42 prophage in Staphylococcus aureus 42CR3-L (18). EcoprrI type I R-M (32) and EcoP1 type III (11) systems were found on a P1 prophage in E. coli. These systems are assumed to be transferred to the chromosomal DNA by the corresponding phage. It is quite reasonable from the standpoint of the physiological role of the R-M system that there is no target sequence for R.EcoT38I in the phage P2 genome, as shown in the EcoO109I R-M system. Although more than 150 type II R-M genes have been cloned and their nucleotide sequences have been analyzed, DNA sequencing has been limited to structural genes. Sequencing of neighboring regions of the R-M system and a variety of bacterial genomes will provide evidence supporting the proposed phage-mediated mobility of the R-M system.
Acknowledgments
E. coli TH38 was obtained from Nippon Gene. We are greatly indebted to Eiji Namba and Kaori Adachi, Gene Research Center, Tottori University, for the DNA sequence analysis and to Nobuhiro Mori, The United Graduate School of Agricultural Science, Tottori University, for the amino acid sequence analysis. We are also grateful to Dhruba K. Chattoraj for many helpful suggestions.
REFERENCES
- 1.Anton, B. P., D. F. Heiter, J. S. Benner, E. J. Hess, L. Greenough, L. S. Moran, B. E. Slatko, and J. E. Brooks. 1997. Cloning and characterization of the BglII restriction-modification system reveals a possible evolutionary footprint. Gene 187:19-27. [DOI] [PubMed] [Google Scholar]
- 2.Barreiro, V., and E. Haggard-Ljungquist. 1992. Attachment sites for bacteriophage P2 on the Escherichia coli chromosome: DNA sequences, localization on the physical map, and detection of a P2-like remnant in E. coli K-12 derivatives. J. Bacteriol. 174:4086-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bertani, G., and E. Six. 1958. Inheritance of prophage P2 in bacterial crosses. Virology 6:357-381. [DOI] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Man, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1461. [DOI] [PubMed] [Google Scholar]
- 5.Bougueleret, L., M. Schwarzstein, A. Tsugita, and M. Zabeau. 1984. Characterization of the genes coding for the EcoRV restriction and modification system of Escherichia coli. Nucleic Acids Res. 12:3659-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, H. W., and D. Roulland-Dussoix. 1969. Complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 7.Brassard, S., H. Paquet, and P. H. Riym. 1992. A transposon-like sequence adjacent to the AccI restriction-modification operon. Gene 157:69-72. [DOI] [PubMed] [Google Scholar]
- 8.Chattoraj, D. K. 1978. Strand-specific break near the origin of bacteriophage P2 DNA replication. Proc. Natl. Acad. Sci. USA 75:1685-1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendrix, R. W., M. C. M. Smith, R. N. Burns, M. E. Ford, and G. F. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humbelin, M., B. Suri, D. N. Rao, D. P. Hornby, H. Eberle, T. Pripfl, S. Kenel, and T. A. Bickle. 1988. Type III DNA restriction and modification systems EcoP1 and EcoP15. Nucleotide sequences of the EcoP1 gene, the EcoP15 mod gene and some EcoP1 mod mutants. J. Mol. Biol. 200:23-29. [DOI] [PubMed] [Google Scholar]
- 12.Inoue, S., M. Sunshine, E. W. Six, and M. Inoue. 1991. Retronphage φR73: an E. coli phage that contains a retroelement and integrates into a tRNA gene. Science 256:969-971. [DOI] [PubMed] [Google Scholar]
- 13.Ives, C. L., P. D. Nathan, and J. E. Brooks.1992. Regulation of the BamHI restriction-modification system by a small intergenic open reading frame, bamHIC, in both Escherichia coli and Bacillus subtilis. J. Bacteriol. 174:7194-7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwabuchi, M., S. Tajima, T. Inoue, T. Shibata, and T. Ando. 1992. A restriction enzyme, BpuI, is an isoschizomer of BanII. Nucleic Acids Res. 20:5850.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karreman, C., and A. de Waard. 1988. Cloning and complete nucleotide sequences of the type II restriction-modification genes of Salmonella infantis. J. Bacteriol. 170:2527-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kita, K., J. Tsuda, T. Kato, K. Okamoto, H. Yanase, and M. Tanaka. 1999. Evidence of horizontal transfer of the EcoO109I restriction-modification gene to Escherichia coli chromosomal DNA. J. Bacteriol. 181:6822-6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kita, K., J. Tsuda, and S. Nakai. 2002. C.EcoO109I, a regulatory protein for production of EcoO109I restriction endonuclease, specifically binds to and bends DNA upstream of its translational start site. Nucleic Acids Res. 30:3558-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi, I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:3742-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohara, Y., K. Akiyama, and K. Isono. 1987. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell 50:495-508. [DOI] [PubMed] [Google Scholar]
- 20.Kossykh, V., A. Repyk, A. Kaliman, and Y. Buuryanov. 1989. Nucleotide sequence of the EcoRII restriction endonuclease gene. Biochim. Biophys. Acta 1009:290-292. [DOI] [PubMed] [Google Scholar]
- 21.Lee, K. F., P. C. Shaw, S. J. Picone, G. G. Wilson, and K. D. Lunnen. 1998. Sequence comparison of the EcoHK31I and EaeI restriction-modification systems suggests an intergenic transfer of genetic material. Biol. Chem. 379:437-441. [DOI] [PubMed] [Google Scholar]
- 22.Matsudaira, P. 1987. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J. Biol. Chem. 262:10035-10038. [PubMed] [Google Scholar]
- 23.Mise, K., K. Nakajima, N. Terakado, and M. Ishidate. 1986. Production of restriction endonucleases using multicopy Hsd plasmids occurring naturally in pathogenic Escherichia coli and Shigella boydii. Gene 44:165-169. [DOI] [PubMed] [Google Scholar]
- 24.Naderer, M., J. R. Brust, D. Knowle, and R. M. Blumenthal. 2002. Mobility of a restriction-modification system revealed by its genetic context in three hosts. J. Bacteriol. 184:2411-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakayama, Y., and I. Kobayashi. 1998. Restriction-modification gene complexes as selfish entities: role of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc. Natl. Acad. Sci. USA 95:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson, M., C. Christ, and I. Schildkraut. 1984. Alteration of apparent restriction endonuclease recognition specificities by DNA methylases. Nucleic Acids Res. 12:5165-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman, A. K., R. A. Rubin, S.-H. Kim, and P. Modrich. 1981. DNA sequences of the structural genes for EcoRI DNA restriction and modification enzymes. J. Biol. Chem. 256:2131-2139. [PubMed] [Google Scholar]
- 28.Ohshima, H., S. Matsuoka, K. Asai, and Y. Sadaie. 2002. Molecular organization of intrinsic restriction and modification genes BsuM of Bacillus subtilis Marburg. J. Bacteriol. 184:381-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posfai, J., A. S. Bhagwat, G. Posfai, and R. J. Roberts. 1989. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 17:2421-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts, R. J., and D. Macelis. 2001. REBASE—restriction enzymes and methylases. Nucleic Acids Res. 29:268-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao, T., J. C. Bourne, and R. M. Blumenthal. 1991. A family of regulatory genes associated with type II restriction-modification systems. J. Bacteriol. 173:1367-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyndall, C., H. Hehnherr, U. Sandmeier, E. Kulik, and T. A. Bickle. 1997. The type IC hsd loci of the enterobacteria are flanked by DNA with high homology to the phage genome: implications for the evolution and spread of DNA restriction systems. Mol. Microbiol. 23:729-736. [DOI] [PubMed] [Google Scholar]
- 33.Vaisvilla, R., G. Vilkaitis, and A. Janulaitis. 1995. Identification of a gene encoding a DNA invertase-like enzyme adjacent to the PaeR7I restriction-modification system. Gene 157:81-84. [DOI] [PubMed] [Google Scholar]
- 34.Vijesurier, R. M., L. Carlock, R. M. Blumenthal, and J. C. Dunbar. 2000. Role and mechanism of action of C.PvuII, a regulatory protein conserved among restriction-modification systems. J. Bacteriol. 182:477-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson, G., and N. E. Murray. 1991. Restriction and modification systems. Annu. Rev. Genet. 25:585-627. [DOI] [PubMed] [Google Scholar]
- 36.Xu, S.-Y., J.-P. Xiao, L. Ettwiller, M. Holden, J. Aliotta, C. J. Poh, M. Dalton, D. P. Robinson, T. R. Petronzio, L. Moran, M. Ganatra, J. Ware, B. Slatko, J. Benner, B. Slatko, and E. P. Guthrie. 1998. Cloning and expression of the ApaLI, NspI, NspHI, SacI, ScaI and SapI restriction-modification systems in Escherichia coli. Mol. Gen. Genet. 260:226-231. [DOI] [PubMed] [Google Scholar]
- 37.Yanisch-Perron, C., J. Vieiera, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida, Y., and K. Mise. 1986. Occurrence of small Hsd plasmids in Salmonella typhi, Shigella boydii, and Escherichia coli. J. Bacteriol. 165:357-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zakharova, M. Z., I. V. Beletskaya, A. N. Kravetz, A. V. Pertzev, S. G. Mayorov, M. G. Shlyapnikov, and A. S. Solonin. 1998. Cloning and sequence analysis of the plasmid-borne genes encoding the Eco29kI restriction and modification enzymes. Gene 208:177-182. [DOI] [PubMed] [Google Scholar]