Abstract
Fst is a peptide toxin encoded by the par toxin-antitoxin stability determinant of Enterococcus faecalis plasmid pAD1. Intracellular overproduction of Fst resulted in simultaneous inhibition of all cellular macromolecular synthesis concomitant with cell growth inhibition and compromised the integrity of the cell membrane. Cells did not lyse or noticeably leak intracellular contents but had specific defects in chromosome partitioning and cell division. Extracellular addition of synthetic Fst had no effect on cell growth. Spontaneous Fst-resistant mutants had a phenotype consistent with changes in membrane composition. Interestingly, overproduction of Fst sensitized cells to the lantibiotic nisin, and Fst-resistant mutants were cross-resistant to nisin and the pAD1-encoded cytolysin.
Postsegregational killing or addiction systems were originally identified as determinants ensuring proper segregational stability of bacterial plasmids (19). In this context, they function by programming any daughter cell that loses the plasmid for death. Postsegregational killing systems are ubiquitous on low-copy bacterial plasmids and have been identified on plasmids native to both gram-negative and gram-positive species. They uniformly encode at least two components, a stable toxin and an unstable antitoxin. As long as the plasmid is present, the antitoxin prevents postsegregational killing toxicity. If the plasmid is lost, the antitoxin is rapidly degraded and the toxin kills the cell (for comprehensive reviews, see references 17 and 27).
The discovery that restriction-modification systems can perform a postsegregational killing function when present on plasmids (32) and the observation that postsegregational killing homologs are widespread among bacterial chromosomes (15) have forced a reexamination of the roles that such systems play in the bacterial cell. Obviously, chromosomally located postsegregational killing-like systems would not be useful for postsegregational killing. Potential roles in genome shuffling (30), an expanded stringent response (6, 33), and bacterial apoptosis (1) have been proposed for these loci. Since toxin-antitoxin pairs clearly play roles in addition to postsegregational killing, a more general designation as toxin-antitoxin modules (15) is probably more appropriate.
The targets for toxins of toxin-antitoxin modules vary widely, and many remain undefined. The first clearly defined target was that of the CcdB toxin of the F-encoded ccd locus. CcdB targets the GyrA subunit of DNA gyrase and appears to poison a gyrase-DNA complex, trapping it as a DNA-cleaved intermediate. Quinolone antibiotics function in a similar manner, but CcdB binds to a different GyrA site, and mutations resulting in resistance to CcdB and the quinolones map to different locations on GyrA (3). Recently, the ParE toxin of the parDE toxin-antitoxin system of RK2 has also been shown to target DNA gyrase (28). The Kid toxin of the R1-encoded Kis/Kid locus inhibits DnaB-dependent initiation of DNA replication (34). The RelE protein of the chromosomally encoded Escherichia coli RelBE locus targets global translation by an as yet undefined mechanism (7). Finally, the Hok toxin of the R1-encoded hok/sok system leads to the formation of “ghost” cells (19) and collapse of the membrane potential (16).
In most toxin-antitoxin systems, both toxin and antitoxin are proteins, but in a few systems the antitoxin is an unstable antisense RNA that binds to and inhibits the translation of a stable toxin-encoding mRNA. The prototype of the antisense RNA-regulated systems is the plasmid R1-encoded hok/sok system (19). In this system, the message for the Hok toxin is transcribed in a translationally inactive conformation that also does not bind the sok antisense RNA (36). The hok message is slowly degraded from the 3′ end, which results in a conformational change that allows sok binding and rapid RNase III-mediated degradation (18). In the absence of sok, the change in conformation allows translation of Hok and killing of the cell (13). Recently, an antisense-regulated toxin-antitoxin system, designated ldr, has been identified on the long direct repeats on the E. coli chromosome. Ldr expression leads to cell killing and chromosome condensation. The function of ldr is not known, but it does not have a postsegregational killing phenotype when cloned on a plasmid (29).
The third antisense-regulated toxin-antitoxin system is the par system encoded by the pAD1 plasmid of Enterococcus faecalis (42). pAD1 is the prototype of a family of plasmids whose conjugative systems are induced by peptide sex pheromones secreted by potential recipients (8, 12). In addition, it encodes a peptide lantibiotic, cytolysin (4), which has been implicated in enterococcal virulence (20). par is the only known antisense-regulated toxin-antitoxin system in gram-positive bacteria, and with the exception of restriction-modification systems, is one of only two defined toxin-antitoxin systems, the other being the proteic ɛ-ζ system of pSM19035 (11). The par system is regulated by a unique antisense RNA mechanism that involves interaction at multiple, dispersed regions of complementarity rather than a single contiguous complementary region at the 5′ end of its target (21-23). In this way, regulation of par is more analogous to the trans-encoded chromosomal antisense RNA systems than most well-studied plasmid-encoded systems (38). The par toxin, Fst, is a 33-amino-acid peptide encoded on the stable RNAI message. Fst is the smallest known toxin-antitoxin toxin. Both regulated overexpression of Fst (40) and expression due to plasmid loss (44) have been shown to lead to the death of host E. faecalis cells.
In this study, we determined the effects of Fst toxin at the cellular level. We describe the isolation and characterization of mutants resistant to the Fst toxin in this report. Overexpression of Fst was shown to result in the simultaneous inhibition of all macromolecular synthesis coincident with cessation of cell growth. Cytological examination demonstrated that Fst overexpression compromised the integrity of the cell membrane but did not lead to the formation of “ghost” cells. Rather, specific effects on cell division were observed. Synergistic effects with the pore-forming lantibiotic nisin further indicated a cell membrane-localized target for Fst. Fst-resistant mutants had changes in membrane permeability and sensitivity to lantibiotics and glycine indicative of changes in membrane composition.
MATERIALS AND METHODS
Strains and plasmids.
Most experiments were performed with E. faecalis strain OG1X, a streptomycin-resistant, gelatinase-negative derivative of strain OG1 (26). Fst-resistant host mutants m1 to m7 were derived from OG1X by selection of spontaneous mutants as described below. FA2-2 is a rifampin- and fusidic acid-resistant E. faecalis strain (9). Strain FA3333 is a derivative of FA2-2 defective in the production of the cAD1 pheromone due to a mutation in the chromosomal cad gene for cAD1 production, kindly provided by D. Clewell (2).
The key plasmid in this study is pAM2005K (40), an erythromycin resistance-encoding pAD1 miniplasmid in which the fst gene is fused to the pheromone-inducible promoter of the traE1 gene. A terminatorless lacZ gene is located between the pheromone-inducible promoter and fst, providing a useful independent indicator of cAD1 signal detection and transduction. Plasmid pAM2005KR is a derivative of pAM2005K which contains a deletion between direct repeats within the par region, resulting in loss of the fst gene (40, 43). Plasmid pCGC contains the complete cytolysin gene cluster from pAD1 (24) cloned in the Escherichia coli-E. faecalis shuttle vector pAT28, carrying a selectable spectinomycin resistance gene (37). pCGC and pAT28 were kindly provided by M. Gilmore.
Media and growth conditions.
In most experiments, E. faecalis strains were cultured in N2GT, which is nutrient broth no. 2 (Oxoid Ltd.) supplemented with 0.1 M Tris buffer (pH 7.5) and 0.2% glucose. Agar (Difco) was added at 1.8% to form plates. Where indicated, M9 medium (35) supplemented with 0.3% (wt/vol) yeast extract and 1% (wt/vol) Casamino Acids was used for the reasons described. Except where indicated, all cultures were inoculated at 2% from cultures grown overnight in the same medium and grown at 37°C. Cultures were grown in tubes with shaking at 200 rpm in a series 25 shaker incubator (New Brunswick Scientific Co., Edison, N.J.). Optical density was measured on a Milton Roy Spectronic 21D (Fisher Scientific).
The fst gene of OG1X(pAM2005K) was induced by addition of synthetic cAD1 (Sigma-Genosys) at 200 ng/ml, 100 ng/ml, or 20 ng/ml, depending on the purpose of the experiment, from a 200-μg/ml stock in dimethyl sulfoxide. Synthetic Fst (United Biochemical Research) was prepared with the predicted sequence MKDLMSLVIAPIFVGLVLEMISRVLDEEDDSRK derived from the par DNA sequence (42). Synthetic Fst was used at a final concentration of 200 μg/ml from a 20-mg/ml stock in dimethyl formamide. In controls, an equal volume of solvent was added to cultures. Nisin was filter sterilized as a 250-μg/ml stock solution. Nisin concentrations varied depending on the purpose of the experiment. Antibiotics were added at the following concentrations: erythromycin, 10 μg/ml; spectinomycin, 100 μg/ml; and streptomycin, 1 mg/ml. β-Galactosidase assays were performed on toluene-treated cell samples as previously described (41).
Radionuclide incorporation.
OG1X(pAM2005K) was grown overnight in M9 diluted 1:4 with 1× minimal salts, with erythromycin selection. This culture was used to inoculate dilute M9 at 4%. Cultures were grown at 27°C. Although cultures in M9 medium are less sensitive to Fst, reduction of the available nutrients was required to facilitate uptake of the radiolabel. Growth at 27°C makes cells more sensitive to Fst and slows growth and the onset of inhibition enough that better comparisons can be made. This combination of conditions proved optimal for incorporation experiments (unpublished observations). l-[35-S]methionine, [2-14C]uridine, and [methyl-3H]thymidine (ICN) were added to parallel cultures at 10 μCi/ml, 0.1 μCi/ml, and 10 μCi/ml, respectively. Then 200 ng of cAD1 per ml was added after 1 h of incubation, and 400-μl samples were taken at the time points indicated and added to 400 μl of 10% trichloroacetic acid. Precipitates were collected by filtration, and the filters were inserted into scintillation vials containing 10 ml of Ecolume scintillation fluid (ICN). Counting was performed on a Beckman LS6000SC liquid scintillation counter for 1 min on the appropriate channel.
Fluorescence microscopy and flow cytometry.
OG1X(pAM2005K) was grown in N2GT at 37°C for 1 h prior to addition of 100 ng of cAD1 per ml of culture. Incubation was continued for another hour, at which point the cells were harvested by centrifugation and resuspended in 1 ml of staining buffer (0.1 M glucose, 0.05 M Tris [pH 8], 0.01 M EDTA). Then 1.5 μl of a 5 mM solution of Sytox Green nucleic acid stain (Molecular Probes) was added to the cells and incubated for 15 min at room temperature. Cells were then washed once and resuspended in an equal volume of staining buffer.
For fluorescence microscopy, wet mounts were prepared and examined on an Olympus BX61 laser scanning confocal microscope with a 10× ocular and a 100× UPlanApo 1.35 oil immersion objective. Excitation was achieved with an argon laser through a fluorescein isothiocyanate filter. Image capture and analysis were done with Fluoview 3.3 software. All equipment was obtained from Olympus.
Flow cytometry was performed with a FACSVantage SE (Becton Dickinson) equipped with a 488 Interprise II coherent laser. The laser was turned on 20 min before analyzing samples. Emission from the fluorochrome signal was passed through a 530/30 bandpass filter and collected as FL1 versus FSC. The laser was operated at 180 mW and 19.0 A current. Excitation and emission wavelengths were set as suggested for Sytox Green by Molecular Probes. Data were collected from 20,000 events and analyzed with CellQuest software. No attempt was made to break up cell chains by sonication due to concerns about unintentionally disrupting membranes and affecting staining. Therefore, the counted events probably represent staining within various-sized chains rather than individual cells. Microscopic examination indicated that most cells were present in chains of from two to six cells.
Electron microscopy.
OG1X(pAM2005K) was cultured for 1 h in N2GT prior to addition of 200 ng of cAD1 per ml of culture. Cells were grown for 1 additional hour before preparation for microscopic examination. Plasmid-free OG1X and mutant m7 were cultured overnight in N2GT or N2GT supplemented with 2% (wt/vol) glycine (Fisher Scientific).
For transmission electron microscopy, cells were harvested by centrifugation and fixed by addition of 3 ml of 3% paraformaldehyde-0.5% gluteraldehyde to the dry pellet. Cells were allowed to stand at room temperature for 1.5 h, centrifuged as before, and resuspended in 5 ml of phosphate buffered saline. Cells were then washed twice in 3 ml of 0.1 M phosphate buffer and placed in 1 ml of 1% osmium tetroxide-0.1 M phosphate buffer for 1 h at room temperature and then harvested by centrifugation. Cells were then dehydrated in increasing serial concentrations of ethanol in the following series: 1 ml of 50% ethanol for 4 min, 1 ml of 80% ethanol for 4 min, 1 ml of 95% ethanol for 4 min, and 1 ml of absolute ethanol for 4 min. Infiltration with Spurr embedding medium (Electron Microscopies Sciences) was performed by allowing harvested cells to stand in the following mixtures at room temperature: 1:1 Spurr-ethanol for 1 h; 3:1 Spurr-ethanol for 1 h; and 100% Spurr overnight. Following overnight infiltration, cells were placed in fresh 100% Spurr for 1 h and then placed in a 70°C oven overnight. Samples were sectioned with a diamond knife, attached to copper grids, and examined on a Jeol 1210 electron microscope.
For scanning electron microscopy, washed bacteria were placed on glass chips coated with 0.1% poly-l-lysine for 10 min. Nonadherent bacteria were rinsed off with Hanks' balanced salt solution, and glass chips were placed in fixative consisting of 0.1 M cacodylate buffer containing 3% glutaraldehyde and 7% sucrose. Fixed samples were dehydrated in ethanol, critical point dried with CO2, and sputter coated with a discontinuous 1-nm layer of platinum. Specimens were viewed at 3 kV in a Hitachi S-900 field emission scanning electron microscope (Hitachi Scientific Instruments Ltd., Mountain View, Calif.).
Isolation of Fst-resistant mutants.
In previous experiments, selection of mutants by immediate exposure to high levels of pheromone resulted in the isolation of only plasmid-localized mutations (43). Considering the possibility that host mutations resistant to high levels of Fst might be inimical to cell growth, spontaneous Fst-resistant mutants were obtained by selection for the ability to grow in the presence of increasing concentrations of pheromone, i.e., mutants that were resistant to Fst induction by pheromone were isolated. First, cells were grown overnight in N2GT-erythromycin supplemented with cAD1 at concentrations of 1.25, 2.5, 5, or 10 ng/ml. Then 200 μl of each enrichment culture was spread onto N2GT-erythromycin agar plates and allowed to dry. A 10-μl spot of concentrated (200 μg/ml) synthetic cAD1 was placed on one side of the plate and dried. Plates were incubated overnight at 37°C. Plates contained erythromycin to select against any cells that had lost the plasmid and with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to ensure that mutants were not defective in sensing and responding to cAD1.
A zone of inhibition developed around the pheromone spot that was rimmed with blue cell growth, presumably because cells at the edge of the zone of inhibition were exposed to sufficient pheromone to produce detectable levels of β-galactosidase but not sufficient Fst to inhibit growth. On the lawns derived from enrichment in 2.5, 5, and 10 ng of cAD1, isolated blue colonies grew inside the zone of inhibition. Seven of these colonies were selected for further study. To ensure that the mutations were located on the host chromosome, plasmid-cured derivatives were obtained by overnight growth in liquid medium without erythromycin selection. Wild-type pAM2005K was then reintroduced into all mutants by electroporation (10), and transformants were retested for pheromone resistance. All mutants remained resistant to cAD1 induction. As further confirmation, pAM2005K plasmid DNA was purified by modified alkaline lysis (41) from each mutant and electroporated into wild-type, plasmid-free OG1X. Transformants were sensitive to cAD1 induction, indicating that the mutation was not plasmid encoded (data not shown).
Determination of cytolysin sensitivity.
Unlike nisin, purified cytolysin is not available. Therefore, it was necessary to determine cytolysin sensitivity by coculture of the indicator strain with a cytolysin-producing strain. For this purpose, each strain was grown overnight with appropriate selection where necessary. Strains were washed twice with an equal volume of phosphate-buffered saline to remove any remaining antibiotic and resuspended in the same volume of phosphate-buffered saline. Fresh N2GT was inoculated at 1% with the indicator strain [OG1X(pAM2005K), OG1X, m1, or m7] and at 1% with the producing strain [FA2-2(pCGC), FA3333(pCGC), or the same strains carrying pAT28 as a control]. Immediately after inoculation and at 1-h intervals, serial 10-fold dilutions were made, and 10-μl aliquots from each dilution were spotted on N2GT agar plates selective for the indicator strain and allowed to dry. Colony counts were made after overnight incubation at 37°C. At the last time point, 10-μl aliquots were also spotted on plates selective for the producing strain to make sure they were growing equally in each experiment. In general, the producers had approximately half as many CFU per milliliter when grown with resistant cells, probably due to increased competition for nutrients.
RESULTS
Fst effects on macromolecular synthesis and membrane integrity.
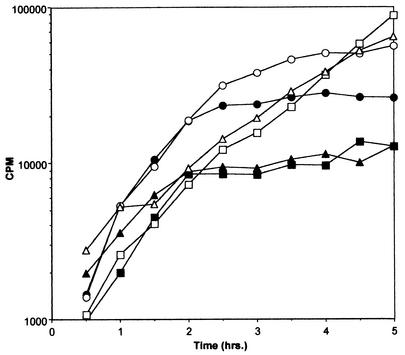
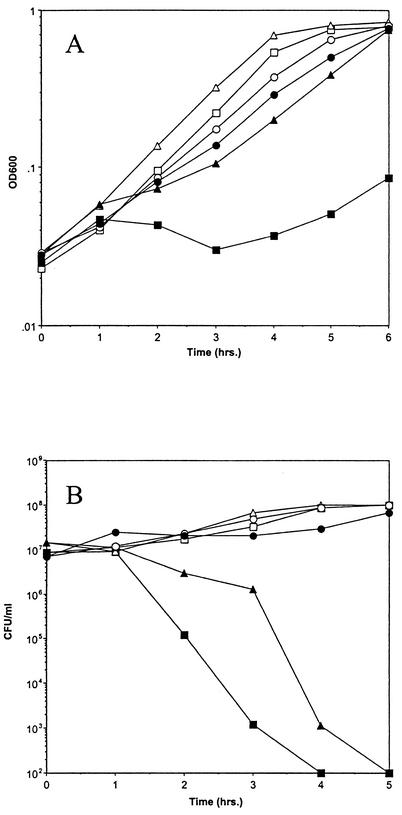
The effect of Fst overproduction on DNA, RNA, and protein synthesis was tested in order to determine what cell functions might be affected. Incorporation of labeled precursors into the acid-insoluble macromolecular fraction was measured over time in the presence and absence of inducing levels of synthetic cAD1 as described in Materials and Methods. As shown in Fig. 1, induction of Fst resulted in simultaneous inhibition of all macromolecular synthesis. Inhibition was apparent within 1 h after induction, concurrent with cessation of cell growth. cAD1 induction of cells containing pAM2005KR, a derivative of pAM2005K deleted for the fst gene (40), had no effect on macromolecular synthesis, proving that this effect was Fst dependent (data not shown). These results suggested that Fst might perturb the cell membrane, thereby affecting membrane potential or some other critical cell function.
FIG. 1.
Inhibition of macromolecular synthesis by Fst induction. OG1X(pAM2005K) cells were cultured in diluted M9 medium in the presence of [3H]thymidine, [14C]uridine, or [35S]methionine as described in Materials and Methods. Samples were taken at the time points indicated and precipitated with trichloroacetic acid, and radioactivity was determined by scintillation counting. Fst production was induced with cAD1 (200 ng/ml) at 1 h Results depicted are representative of multiple experiments. ○, uninduced, [14C]uridine; •, induced, [14C]uridine; □, uninduced, [3H]thymidine; ▪, induced, [3H]thymidine; ▵, uninduced, [35S]methionine; ▴, induced, [35S]methionine.
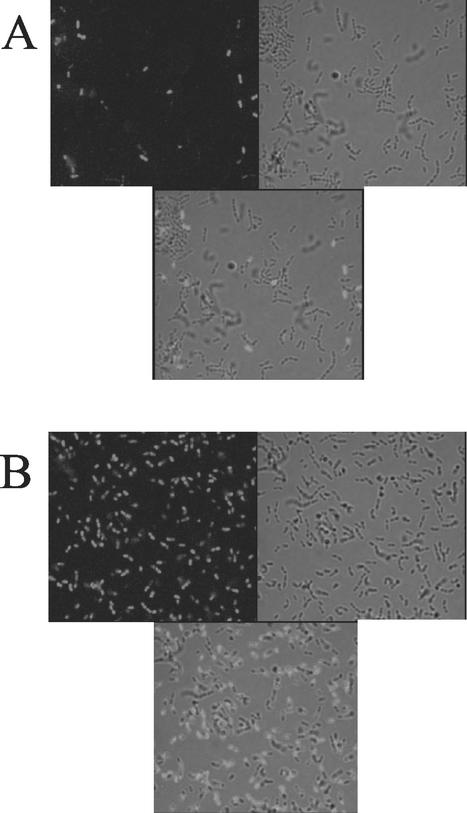
To test the effect of Fst on membrane integrity, cAD1-induced and uninduced OG1X(pAM2005K) cells were stained with Sytox Green. Sytox Green is a DNA stain that cannot pass through the membrane and can only stain cells whose membranes have been disrupted. Results of staining are shown in Fig. 2A (uninduced) and 2B (induced). Although some cells were stained in the uninduced sample, a much larger proportion of cAD1-induced cells were stained. Staining of cAD1-induced OG1X(pAM2005KR) cells was similar to that of uninduced OG1X(pAM2005K) cells, confirming that increased staining was associated with Fst expression (data not shown). Visual examination of three independent fields of a separate staining experiment revealed that no more than 6% of uninduced OG1X(pAM2005K) cells were stained, while between 58% and 67% of induced cells were stained. These results were confirmed by flow cytometry, which showed a shift from a lightly stained population representing ≈80% of events in unexposed cells to a more highly staining population in cells exposed to Fst by cAD1 induction (see Fig. 5).
FIG. 2.
Fluorescence microscopy of Sytox Green-stained cells with and without induction of Fst. Cells were cultured in N2GT medium for 1 h and then induced with either cAD1 (100 ng/ml) or an equal volume of dimethyl sulfoxide for another hour. Cells were harvested, washed, and stained with Sytox Green as described in Materials and Methods. For fluorescence microscopy in A and B, wet mounts were prepared and examined under oil with a 100× objective. For each panel, three views are shown: fluorescence alone (upper left), transmitted light alone (upper right), and an overlay of both (bottom). A, uninduced; B, induced.
FIG. 5.
Flow cytometry analysis of Fst-resistant strains. pAM2005K-containing wild type OG1X and mutant m1 and m7 cells were cultured in N2GT medium for 1 h and then induced with either cAD1 (100 ng/ml) or an equal volume of dimethyl sulfoxide for another hour. Cells were then harvested, washed, and stained with Sytox Green as described in Materials and Methods. Histograms were derived from event data as described in Materials and Methods. Similar results were observed in independent stainings. K−, uninduced OG1X(pAM2005K); K+, induced OG1X(pAM2005K); m1−, uninduced m1(pAM2005K); m1+, induced m1(pAM2005K); m7−, uninduced m7(pAM2005K); m7+, induced m7(pAM2005K)
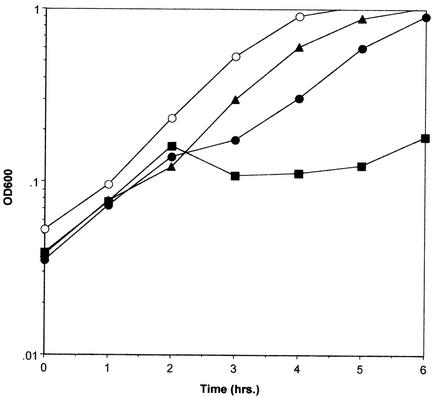
Since Fst and nisin both appear to act at the cell membrane, their combined effect on growth of OG1X(pAM2005K) was determined. Fst was induced first with a submaximal concentration of cAD1, and a subinhibitory dose of nisin was administered 30 min later. As shown in Fig. 3, the level of nisin used had a barely detectable effect on cell growth alone, while cAD1 induction alone under these conditions slowed cell growth marginally. However, addition of nisin to cAD1-induced cells caused some lysis, and cell growth ceased for 3 h. Growth increased after this point due to the accumulation of mutants showing altered response to cAD1 and/or increased resistance to nisin. This effect is indicative of synergy between Fst and nisin. Nisin is the best-studied member of a family of bacteriocins produced by gram-positive bacteria called lantibiotics (see reference 31 for a review). Interestingly, pAD1 encodes its own lantibiotic, cytolysin, that is deleted from pAM2005K. Since cytolysin is not available in pure form, coculture of OG1X(pAM2005K) and a cytolysin producer, FA3333(pCGC), was performed to determine if cytolysin and Fst acted synergistically. Unfortunately, cAD1-induced production of Fst resulted in slow-growing colonies that made interpretation of coculture experiments difficult, and the results of this experiment were ambiguous (data not shown).
FIG. 3.
Combined effects of nisin and Fst on wild-type OG1X cells. OG1X(pAM2005K) cells were cultured in M9 medium. M9 was used in this case because previous experiments showed that the effect of cAD1 induction was reduced in this medium (unpublished results). cAD1 was added at 20 ng/ml at 1 h, and nisin was added at 125 ng/ml at 1.5 h. Cell density was followed by measuring the optical density at 600 nm. Results shown are representative of three independent experiments. ○, untreated control; ▴, nisin-treated cells; •, cAD1-induced cells; ▪, cAD1-induced and nisin-treated cells.
No effect on cell growth was observed when OG1X cells were exposed to concentrations of synthetic Fst up to 200 μg/ml. Addition of nisin to 250 ng/ml did not affect sensitivity to synthetic Fst, indicating that synergism was restricted to internally produced Fst. Increasing the porosity of the cell wall by addition of glycine or subinhibitory levels of lysozyme to the growth medium also had no effect (data not shown).
Effects of Fst on cell morphology.
The only other well-studied antisense RNA-regulated postsegregational killing system is the hok/sok system of the E. coli plasmid R1 (19). Although the Fst and Hok toxins have no amino acid sequence homology, both appear to affect cell membrane integrity. However, the Hok toxin causes leakage of cell contents and the formation of ghost cells. Preliminary examination of cAD1-induced OG1X(pAM2005K) cells by light microscopy showed no evidence of ghost cell formation or any other abnormal cellular morphology. To further examine the effects of Fst on cell morphology, electron microscopy was performed.
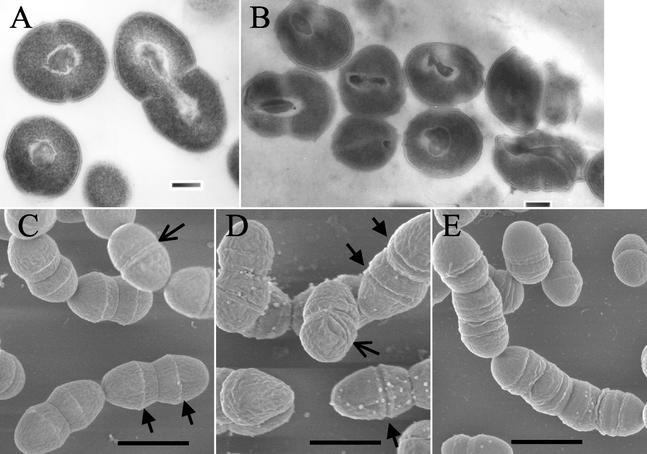
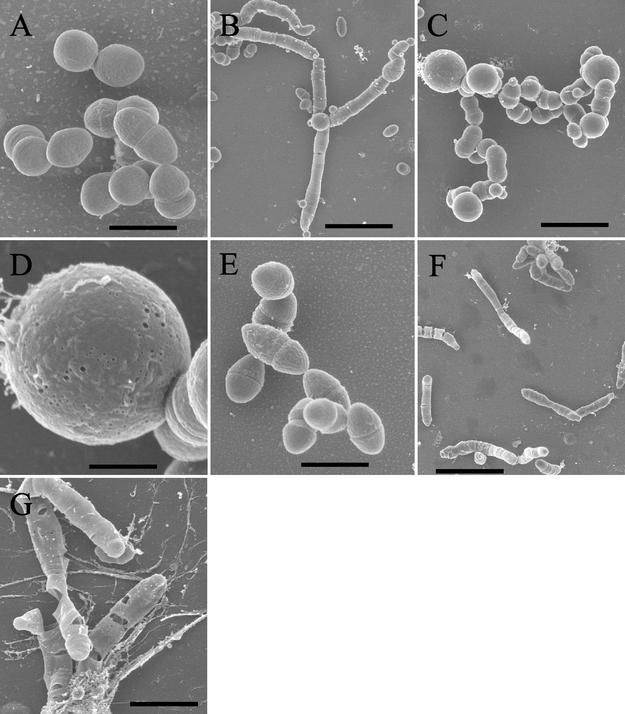
Figures 4A and 4B show representative transmission electron micrographs of uninduced and induced OG1X(pAM2005K), respectively. The uninduced cells showed the typical spherical cell shape of nondividing cells and the classic lancet shape of dividing cells. The electron-dense material at the center of the cell is most likely the nucleoid, which can be seen to be in the process of segregating in the dividing cell. Induced cells showed a variety of ultrastructural abnormalities, including misplaced and inappropriately formed septa and misplaced or, in some cases, absent nuclei. Figures 4C and 4D show representative scanning electron micrographs of uninduced and induced OG1X(pAM2005K), respectively. Again, uninduced cells showed the classic lancet shape and chain formation of cells of the family Streptococcus. In addition, well-formed cell wall bands were observed in most cells, with band splitting occurring at the conclusion of the cell division cycle (25).
FIG. 4.
Effects of Fst induction on cellular morphology. OG1X(pAM2005K) cells were cultured in N2GT and induced after 1 h of growth with 200 ng of synthetic cAD1 per ml for an additional 1 h. Cells were prepared for either transmission electron microscopy or scanning electron microscopy as described in Materials and Methods. A and B show representative transmission electron micrographs of uninduced and induced cells, respectively. Bars in A and B, 200 nm. Induced cells show aberrant cell shape, improper septal placement, and misplaced nucleoids. C and D show representative scanning electron micrographs of uninduced and induced cells, respectively. Arrows at the bottom of C show cell wall bands, and the flared arrow at the top of C indicates a recently split cell wall band as the cell begins the next division cycle. Arrows in D show prematurely split cell wall bands. The flared arrow in D shows an off-center split wall band. E shows filamentation of cells. Bars in C, D, and E, 1, 0.86, and 1.2 μm, respectively.
In induced cells, cell wall bands were rarely seen in dividing cells and were often replaced by troughs, suggesting that the cell wall bands split prematurely. Occasionally, extra division furrows were produced that were off center from the normal split cell wall bands. In some cases aberrant division furrows led to production of filaments with multiple invaginations, suggestive of a failure to complete septation (Fig. 4E). No obvious pores or loss of intracellular material was observed. Therefore, while Fst does not appear to cause lysis or leakage of cell contents, it may affect cell division and chromosomal segregation either directly or indirectly.
Characterization of mutants resistant to Fst.
Spontaneous Fst-resistant mutants were isolated by a two-step, graded exposure to cAD1 as described in detail in Materials and Methods. Growth curves of pAM2005K-containing mutants with and without cAD1 induction revealed that six of the seven mutants, designated m2 to m7, had a similar phenotype. These mutants showed a slightly increased generation time in the absence of cAD1 and very little growth inhibition in the presence of cAD1 compared to wild-type OG1X(pAM2005K) (Table 1). The other mutant, m1, grew as well as the wild type in the absence of cAD1 but showed a cAD1-induced growth inhibition intermediate between that of the wild type and the other six mutants. β-Galactosidase assays were performed to ensure that resistance to pheromone induction was not due to a decreased response to cAD1. β-Galactosidase activities were actually higher in induced mutant cells than in the wild type (Table 1), perhaps because Fst overproduction did not inhibit translation as much in the mutants.
TABLE 1.
Growth rate and cAD1 induction in wild-type and mutant OG1X(pAM2005K) cellsa
| Strain(s) | Avg generation timeb (h) | β-Galactosidase activityc (Miller units) |
|---|---|---|
| Wild type | 0.85 ± 0.09 | 0.55 ± 0.34 |
| Wild type + cAD1 | >4d | 6.07 ± 1.04 |
| m1 | 0.80 ± 0.05 | 2.64 ± 0.93 |
| m1 + cAD1 | 2.20 ± 0.20 | 9.46 ± 1.82 |
| m2 to m7 | 1.08 ± 0.06 | 0.28 ± 0.13 |
| m2 to m7 + cAD1 | 1.43 ± 0.12 | 13.7 ± 1.88 |
Mutants m2, m3, m4, m5, m6, and m7 all showed similar growth rates, so an average growth rate for all strains was determined. β-Galactosidase activity shown was determined only for m7
Defined as the amount of time required for the optical density of the culture to double. Results shown are averages of at least three independent experiments.
β-Galactosidase activity was determined as previously described (41) 1 h after pheromone addition. Results shown are averages of three independent experiments.
Growth of cAD1-induced wild-type OG1X(pAM2005K) essentially ceased approximately 1 h after cAD1 addition, and no doubling of culture optical density was observed over the remaining course of the experiment.
Since it appeared that Fst resistance was partial in all isolated mutants, the membrane integrity of cAD1-induced and uninduced pAM2005K-containing mutant cells was assessed by Sytox Green staining and flow cytometry. As shown in Fig. 5, uninduced cells of both m1 and m7 showed a higher proportion of cells in the highly staining population than the wild-type strain; 22%, 32%, and 36% for the wild-type, m7, and m1 cells, respectively, with a narrow gate on the highly stained population in uninduced wild-type cells. Pheromone induction of mutant cells still resulted in a shift to the highly staining population, but in both mutants the peaks were broader than for the wild type. In fact, the m7 peak was so broad that 18% of events in induced cells fell within a gate drawn around the low-staining population of uninduced wild-type cells, compared to 4% in induced wild-type and 5% of induced m1 cells.
Plasmid-free Fst-resistant mutants are more sensitive to glycine and resistant to nisin and cytolysin.
In the process of preparing plasmid-free mutant strains for electroporation, it was discovered that all of the mutants except m1 were sensitive to glycine. While wild-type OG1X grew well in glycine concentrations up to 7%, the plasmid-free mutants grew poorly in 2% glycine and not at all in higher concentrations. Scanning electron micrographs were prepared to determine the reason for this increased sensitivity to glycine. While mutant cells were consistently somewhat smaller than wild-type cells, no other significant difference was observed when cultures were grown in the absence of glycine (compare Fig. 6A and 6E). Wild-type cells grown in medium containing 2% glycine frequently formed filaments and swelled (Fig. 6B and 6C). The swollen cells showed visible holes, perhaps accounting for the increased ability of glycine-treated cells to take up DNA by electroporation (Fig. 6D). Otherwise, the cells remained structurally intact. In contrast, m7 cells grown in 2% glycine showed filamentation, but the filaments were often fractured in a manner consistent with cell lysis (Fig. 6F and 6G). Glycine is believed to facilitate electroporation of DNA by substituting for alanine in peptidoglycan and reducing the degree of cross-linking (10). Therefore, increased sensitivity to glycine indicates cell envelope changes in the mutants.
FIG. 6.
Effects of glycine on cell morphology of wild-type OG1X and mutant m7. Cultures were grown overnight from a single-colony inoculum in N2GT either without glycine or supplemented with 2% glycine. Cells were harvested and prepared for scanning electron microscopy as described in Materials and Methods. Panel A, wild-type OG1X without glycine; panels B to D, wild-type OG1X with glycine; panel E, mutant m7 without glycine; panels F and G, mutant m7 with glycine. Bars: panels A and E, 1.2 μm; panel B, 4.3 μm; panel C, 2.7 μm; panel D, 0.67 μm; panel F, 5.0 μm; and panel G, 2.0 μm. Panels A and E are at the same scale so that direct size comparisons can be made.
Since the induction of Fst sensitized the cells to nisin, Fst-resistant mutants were tested for resistance to nisin. As shown in Fig. 7A, mutants m1 and m7 were both resistant to nisin in a pattern similar to that observed for resistance to cAD1 induction; m1 was less resistant to nisin than m7. Cross-resistance to the pAD1-encoded cytolysin was also tested by coculture of plasmid-free mutant strains with FA2-2 carrying a cytoloysin clone or an empty vector. Figure 7B shows that while no detectable wild-type OG1X cells survived after 4 h of coculture with a cytolysin producer, m7 mutant cells showed very little loss of viability after 5 h of coculture. Mutant m1 showed an intermediate level of resistance. These results show that the cell envelope changes selected for Fst resistance resulted in cross-resistance to two different lantibiotics.
FIG. 7.
Cross-resistance of Fst-resistant mutants m1 and m7 to the lantibiotics nisin and cytolysin. Panel A, resistance to nisin. Cells were grown in N2GT for 1 h before adding nisin to a concentration of 500 ng/ml. Cell density was followed by measuring the optical density at 600 nm. Resistance was also observed with nisin concentrations of 125 ng/ml, 250 ng/ml, and 1,000 ng/ml and in M9 medium as well as N2GT. Panel B, resistance to cytolysin. Cocultures of OG1X, m1, and m7 with either the cytolysin clone pCGC or vector alone in FA3333 were prepared in N2GT as described in Materials and Methods. Growth on streptomycin-containing agar plates is represented. Similar results were observed in four independent experiments. □, unexposed wild-type OG1X; ▪, nisin- or cytolysin-exposed wild-type OG1X; ▵, unexposed m1; ▴, nisin- or cytolysin-exposed m1; ○, unexposed m7; •, nisin- or cytolysin-exposed m7.
DISCUSSION
The evidence presented in this report indicates that the par-encoded Fst toxin disrupts the integrity of the cell membrane of host cells. Thus, induction of Fst production permeabilized the membrane to Sytox Green, a DNA stain that cannot cross intact membranes. The membrane effects appeared to be linked to an inhibition of macromolecular synthesis, perhaps by disruption of membrane potential or transport of critical nutrients or ions. Unlike the membrane-active Hok toxin of the hok/sok postsegregational killing system, Fst did not result in the formation of ghost cells or leakage of cellular contents. Instead, specific defects in cell division and chromosome segregation were observed. The mechanistic link between the membrane permeability and cytokinetic effects is currently unclear. Fst also sensitized cells to the effects of nisin, a known pore-forming lantibiotic, further indicating that Fst alters the cell membrane.
Synthetic Fst had no effect on E. faecalis growth when added externally to cells in culture, suggesting that the toxin might require activation by intracellular modification or might target features unique to the internal surface of the cytoplasmic membrane. If modification is required, the gene for the enzyme responsible must be chromosomally encoded, since par is capable of postsegregational killing in the absence of any other pAD1-encoded genes (39). Fst does not contain a cysteine residue and so cannot contain the lanthionine ring modification typical of lantibiotics (31).
Fst-resistant mutants had characteristics indicative of changes in the cell envelope, again implicating the cell membrane, or components thereof, as the primary target of Fst function. Mutants demonstrated an increased permeability to Sytox Green and showed an increased sensitivity to glycine, resulting in cell lysis. The fact that mutants still showed increased staining by Sytox Green when exposed to Fst indicates that resistance results from a general change in surface structure rather than loss of a specific Fst target. The mutants were also resistant to two pore-forming lantibiotics, nisin and the broad-spectrum pAD1-encoded cytolysin, perhaps reflecting changes in membrane lipid composition or membrane potential, both of which have been shown to affect nisin function (5, 14).
In conclusion, Fst appears to be a membrane-active peptide used by its host plasmid, pAD1, to ensure its stable inheritance within E. faecalis via a postsegregational killing mechanism. In addition to permeabilizing the cell membrane, Fst inhibits macromolecular synthesis and affects cell division. Whether Fst functions directly to form membrane pores or acts indirectly by interfering with some other essential membrane-localized function has yet to be determined.
Acknowledgments
We acknowledge the technical support of Tara Denke, Melissa Gerhart, Annie Rezac, Suleman Said, Jennifer Kantrovitch, and Richard Duman. We also thank the laboratories of Michael Gilmore and Don Clewell for supplying critical strains and Gerhart Wagner and Kenn Gerdes for helpful discussion.
This work was supported by Public Health Service grant GM55544.
REFERENCES
- 1.Aizenman, E., H. Engelberg-Kulka, and G. Glaser. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine 3′,5′-bispyrophosphate: a model for programmed bacterial cell death. Proc. Natl. Acad. Sci. USA 93:6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.An, F. Y., and D. B. Clewell. 2002. Identification of the cAD1 Sex Pheromone Precursor in Enterococcus faecalis. J. Bacteriol. 184:1880-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernard, P., and M. Couturier. 1992. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J. Mol. Biol. 226:735-745. [DOI] [PubMed] [Google Scholar]
- 4.Booth, M. C., C. P. Bogie, H. G. Sahl, R. J. Siezen, K. L. Hatter, and M. S. Gilmore. 1996. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. Mol. Microbiol. 21:1175-1184. [DOI] [PubMed] [Google Scholar]
- 5.Breukink, E., I. Wiedemann, C. v. Kraaij, O. P. Kuipers, H.-G. Sahl, and B. de Kruijff. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361-2364. [DOI] [PubMed] [Google Scholar]
- 6.Chang, D.-E., D. J. Smalley, and T. Conway. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45:289-306. [DOI] [PubMed] [Google Scholar]
- 7.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. USA 98:14328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clewell, D. B., and G. M. Dunny. 2002. Conjugation and genetic exchange in enterococci, p. 265-300. In M. S. Gilmore (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, D.C.
- 9.Clewell, D. B., P. K. Tomich, M. C. Gawron-Burke, F. A. E., Y. Yagi, and F. Y. An. 1982. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J. Bacteriol. 152:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz-Rodz, A. L., and M. S. Gilmore. 1990. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol. Gen. Genet. 224:152-154. [DOI] [PubMed] [Google Scholar]
- 11.delaHoz, A. B., S. Ayora, I. Sitkiewicz, S. Fernández, R. Pankiewicz, J. C. Alonso, and P. Ceglowski. 2000. Plasmid copy-number control and better-than-random segregation genes of pSM19035 share a common regulator. Proc. Natl. Acad. Sci. USA 97:728-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franch, T., and K. Gerdes. 1996. Programmed cell death in bacteria: translational repression by mRNA end-pairing. Mol. Microbiol. 21:1049-1060. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Garcera, M. J., M. G. L. Elferink, A. J. M. Driessen, and W. N. Konings. 1993. In vitro pore-forming activity of the lantibiotic nisin: role of proton-motive force and lipid composition. J. Biochem. 212:417-422. [DOI] [PubMed] [Google Scholar]
- 15.Gerdes, K. 2000. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182:561-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerdes, K., F. W. Bech, S. T. Jorgensen, A. Lobner-Olesen, P. B. Rasmussen, T. Atlung, L. Boe, O. Karlstrom, S. Molin, and K. von Meyenburg. 1986. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 5:2023-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerdes, K., A. P. Gultyaev, T. Franch, K. Pedersen, and N. D. Mikkelsen. 1997. Antisense RNA-regulated programmed cell death. Annu. Rev. Genet. 31:1-31. [DOI] [PubMed]
- 18.Gerdes, K., A. Nielsen, P. Thorsted, and E. G. Wagner. 1992. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable hok, srnB and pndA effector messenger RNAs. J. Mol. Biol. 226:637-649. [DOI] [PubMed] [Google Scholar]
- 19.Gerdes, K., P. B. Rasmussen, and S. Molin. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 83:3116-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilmore, M. S., P. S. Coburn, S. R. Nallapareddy, and B. E. Murray. 2002. Enterococcal virulence, p. 301-354. In M. S. Gilmore (ed.), The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press, Washington, D.C.
- 21.Greenfield, T. J., E. Ehli, T. Kirshenmann, T. Franch, K. Gerdes, and K. E. Weaver. 2000. The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism. Mol. Microbiol. 37:652-660. [DOI] [PubMed] [Google Scholar]
- 22.Greenfield, T. J., T. Franch, K. Gerdes, and K. E. Weaver. 2001. Antisense RNA regulation of the par postsegregational killing system: structural analysis and mechanism of binding of the antisense RNA, RNAII and its target, RNAI. Mol. Microbiol. 42:527-537. [DOI] [PubMed] [Google Scholar]
- 23.Greenfield, T. J., and K. E. Weaver. 2000. Antisense RNA regulation of the pAD1 par postsegregational killing system requires interaction at the 5′ and 3′ ends of the RNAs. Mol. Microbiol. 37:661-670. [DOI] [PubMed] [Google Scholar]
- 24.Haas, W., B. D. Shepard, and M. S. Gilmore. 2002. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 415:84-87. [DOI] [PubMed] [Google Scholar]
- 25.Higgins, M. L., and G. D. Shockman. 1976. Study of a cycle of cell wall assembly in Streptococcus faecalis by three-dimensional reconstruction of thin sections of cells. J. Bacteriol. 127:1346-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ike, Y., R. A. Craig, B. A. White, Y. Yagi, and D. B. Clewell. 1983. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc. Natl. Acad. Sci. USA 80:5369-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen, R. B., and K. Gerdes. 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 17:205-210. [DOI] [PubMed] [Google Scholar]
- 28.Jiang, Y., J. Pogliano, D. Helinski, and I. Konieczny. 2002. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44:971-979. [DOI] [PubMed] [Google Scholar]
- 29.Kawano, M., T. Oshima, H. Kasai, and H. Mori. 2002. Mol. characterization of long direct repeat (long direct repeats) sequences expressing a stable mRNA encoding for a 35-amino-acid cell killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 45:333-349. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi, I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:3742-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAuliffe, O., R. P. Ross, and C. Hill. 2001. Lantibiotics: structure, biosynthesis and mode of action. FEMS Microbiol. Rev. 25:285-308. [DOI] [PubMed] [Google Scholar]
- 32.Naito, T., K. Kusano, and I. Kobayashi. 1995. Selfish behavior of restriction-modification systems. Science 267:897-899. [DOI] [PubMed] [Google Scholar]
- 33.Pedersen, K., S. K. Christensen, and K. Gerdes. 2002. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 45:501-510. [DOI] [PubMed] [Google Scholar]
- 34.Ruiz-Echevarria, M. J., G. Gimenez-Gallego, R. Sabariegos-Jareno, and R. Diaz-Orejas. 1995. Kid, a small protein of the parD stability system of plasmid R1, is an inhibitor of DNA replication acting at the initiation of DNA synthesis. J. Mol. Biol. 247:568-577. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Thisted, T., N. S. Sorensen, and K. Gerdes. 1995. Mechanism of postsegregational killing: secondary structure analysis of the entire Hok mRNA from plasmid R1 suggests a fold-back structure that prevents translation and antisense RNA binding. J. Mol. Biol. 247:859-873. [DOI] [PubMed] [Google Scholar]
- 37.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 18:4296.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagner, E. G. H., S. Altuvia, and P. Romby. 2002. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 46:361-398. [DOI] [PubMed] [Google Scholar]
- 39.Weaver, K. E. 1995. Enterococcus faecalis plasmid pAD1 replication and maintenance, p. 89-98. In F. Brown (ed.), Genetics of streptococci, enterococci, and lactococci, vol. 85. Karger, Basel, Switzerland. [PubMed]
- 40.Weaver, K. E., and D. B. Clewell. 1989. Construction of Enterococcus faecalis pAD1 miniplasmids: identification of a minimal pheromone response regulatory region and evaluation of a novel pheromone-dependent growth inhibition. Plasmid 22:106-119. [DOI] [PubMed] [Google Scholar]
- 41.Weaver, K. E., and D. B. Clewell. 1988. Regulation of the pAD1 sex pheromone response in Enterococcus faecalis: construction and characterization of lacZ transcriptional fusions in a key control region of the plasmid. J. Bacteriol. 170:4343-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weaver, K. E., K. D. Jensen, A. Colwell, and S. I. Sriram. 1996. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol. Microbiol. 20:53-63. [DOI] [PubMed] [Google Scholar]
- 43.Weaver, K. E., and D. J. Tritle. 1994. Identification and characterization of an Enterococcus faecalis plasmid pAD1-encoded stability determinants which produces two small RNA molecules necessary for its function. Plasmid 32:168-181. [DOI] [PubMed] [Google Scholar]
- 44.Weaver, K. E., K. D. Walz, and M. S. Heine. 1998. Isolation of a derivative of Escherichia coli-Enterococcus faecalis shuttle vector pAM401 temperature sensitive for maintenance in E. faecalis and its use in evaluating the mechanism of pAD1 par-dependent plasmid stabilization. Plasmid 40:225-232. [DOI] [PubMed] [Google Scholar]